ABSTRACT
The Escherichia coli structural maintenance of chromosome (SMC) complex, MukBEF, and topoisomerase IV (TopoIV) interact in vitro through a direct contact between the MukB dimerization hinge and the C-terminal domain of ParC, the catalytic subunit of TopoIV. The interaction stimulates catalysis by TopoIV in vitro. Using live-cell quantitative imaging, we show that MukBEF directs TopoIV to ori, with fluorescent fusions of ParC and ParE both forming cellular foci that colocalize with those formed by MukBEF throughout the cell cycle and in cells unable to initiate DNA replication. Removal of MukBEF leads to loss of fluorescent ParC/ParE foci. In the absence of functional TopoIV, MukBEF forms multiple foci that are distributed uniformly throughout the nucleoid, whereas multiple catenated oris cluster at midcell. Once functional TopoIV is restored, the decatenated oris segregate to positions that are largely coincident with the MukBEF foci, thereby providing support for a mechanism by which MukBEF acts in chromosome segregation by positioning newly replicated and decatenated oris. Additional evidence for such a mechanism comes from the observation that in TopoIV-positive (TopoIV+) cells, newly replicated oris segregate rapidly to the positions of MukBEF foci. Taken together, the data implicate MukBEF as a key component of the DNA segregation process by acting in concert with TopoIV to promote decatenation and positioning of newly replicated oris.
IMPORTANCE
Mechanistic understanding of how newly replicated bacterial chromosomes are segregated prior to cell division is incomplete. In this work, we provide in vivo experimental support for the view that topoisomerase IV (TopoIV), which decatenates newly replicated sister duplexes as a prelude to successful segregation, is directed to the replication origin region of the Escherichia coli chromosome by the SMC (structural maintenance of chromosome) complex, MukBEF. We provide in vivo data that support the demonstration in vitro that the MukB interaction with TopoIV stimulates catalysis by TopoIV. Finally, we show that MukBEF directs the normal positioning of sister origins after their replication and during their segregation. Overall, the data support models in which the coordinate and sequential action of TopoIV and MukBEF plays an important role during bacterial chromosome segregation.
INTRODUCTION
Successful propagation of cells relies on the fidelity of chromosome replication and segregation and the processes that compact and organize the chromosome. In bacteria, much is known about the mechanism of replication and its spatial organization, but far less is known about how the chromosome is organized and segregated (reviewed in references 1 and 2). In extensively studied bacteria, segregation of most genetic loci occurs soon after replication, once the precatenanes that hold newly replicated sister loci together have been removed by the type II topoisomerase, topoisomerase IV (TopoIV) (3–6). TopoIV action on precatenated sisters can be modulated by SeqA, which interacts with newly replicated DNA (7). Structural maintenance of chromosome (SMC) complexes, which are present in most organisms, have been implicated in both bacterial chromosome organization and segregation, although their precise functional role and biochemical action remain unclear (reviewed in references 8 and 9).
The Escherichia coli SMC complex, MukBEF, was identified through a genetic screen, which initially identified a Muk− mutant that generated anucleate cells and displayed temperature-sensitive growth in rich medium (10, 11). MukB is a 170-kDa protein of distinctive SMC architecture that dimerizes through a dimerization hinge located at one end of an ~50-nm long intramolecular coiled-coil and has an ATPase formed by the N- and C-terminal portions of the protein at the other end of the coiled-coil (9, 12, 13). A kleisin-like protein, MukF, bridges the two ATPase heads in a MukB dimer, while a third protein, MukE, binds to MukF. Loss of function of any of these three proteins leads to the same Muk− phenotype. Functional fluorescent fusions of MukB, MukE, or MukF all associate with the replication origin region (ori) of the E. coli chromosome in live-cell imaging assays that observe fluorescent-focus formation (14, 15) in reactions that require ATP binding and hydrolysis by MukBEF (16, 17; our unpublished data). In the absence of functional E. coli MukBEF, oris are relocated from midcell to an old pole in newborn cells, with the whole chromosome undergoing an apparent 90° rotation within a cell (14).
Biochemical experiments in vitro have shown that the MukB hinge interacts with the C-terminal domain of ParC, the catalytic subunit of TopoIV (18–20), leading to the proposal that these proteins collaborate in chromosome disentanglement (20). This interaction stimulates TopoIV-mediated relaxation of negatively supercoiled DNA, but not positively supercoiled DNA in vitro. Because negative supercoils have the same right-handed chirality as replicative precatenanes (see Fig. S1 in the supplemental material), we predict that MukBEF should also stimulate decatenation by TopoIV, the prelude to chromosome segregation, raising the possibility that MukBEF and TopoIV act in sequential steps during chromosome segregation. Nevertheless, an in vitro study failed to demonstrate MukBEF-stimulated decatenation of multiply linked plasmid replicative catenanes, leading to the proposal that the MukBEF stimulation of TopoIV activity is involved in intramolecular chromosomal events rather than in decatenation (21). The failure of MukB to stimulate decatenation in in vitro assays may reflect the fact that the in vitro assay conditions failed to recapitulate in vivo conditions somehow, such as the absence of MukEF proteins or the fact that in vivo proper loading of a complete MukBEF complex onto DNA is required for this stimulation.
The role of TopoIV in decatenation in vivo has been widely documented, as has its ability to relax supercoils in vitro and in vivo (20, 22–25). After TopoIV impairment, DNA replication and reinitiation along with cell growth appear to continue normally, although decatenation of newly replicated loci is blocked as measured by failure to segregate newly replicated sisters (3). Therefore, any action of TopoIV in supercoil removal ahead of a replication fork can be compensated for by the action of other topoisomerases. These observations support the view that the major role of TopoIV in vivo is in decatenation rather than supercoil relaxation (3). Indeed, any MukBEF-stimulated negative supercoil relaxation would compound the in vivo unlinking problem because it would act to increase the overall linkage between duplex strands in the chromosome. Other studies in both prokaryotes and eukaryotes have implicated functional interactions between SMC complexes and type II topoisomerases and have suggested that these are important for decatenation, although it is not always clear whether these interactions also influence the activity of the topoisomerase in regulating supercoiling, which is important for chromosome organization (5, 26–28).
To address whether MukBEF and TopoIV act in a coordinated manner in vivo, we analyzed the functional relationship between MukBEF and TopoIV in live E. coli in which a single round of replication was initiated and completed in the same cell cycle. Functional fusions of the fluorescent protein, mYPet, to the ParC and ParE subunits of TopoIV formed foci that associated specifically with fluorescent foci of functional MukBEF, which frequently localized with the E. coli ori region. The TopoIV foci were dynamic and did not always form discrete spots, probably because of a high dissociation rate of ParC from MukB, making it necessary for us to use a quantitative cumulative distribution method that assessed the distances between the centroid of Gaussian-fitted MukBEF fluorescent foci and the highest pixel intensity exhibited by fluorescent ParC or ParE, which likely marks the cellular site where the complex exhibits the highest residence time. Using this method, we demonstrated that the two subunits of TopoIV colocalize preferentially with MukBEF foci throughout the cell cycle and also in the absence of replication. Specific depletion experiments additionally showed that MukBEF in foci directs TopoIV to the ori region. Additionally, we provide evidence that MukBEF in foci positions ori rather than vice versa. Taken together, the data implicate MukBEF as a key component of the DNA segregation process by acting in concert with TopoIV to promote decatenation and positioning of newly replicated oris.
RESULTS
Topoisomerase IV associates with MukBEF in live E. coli.
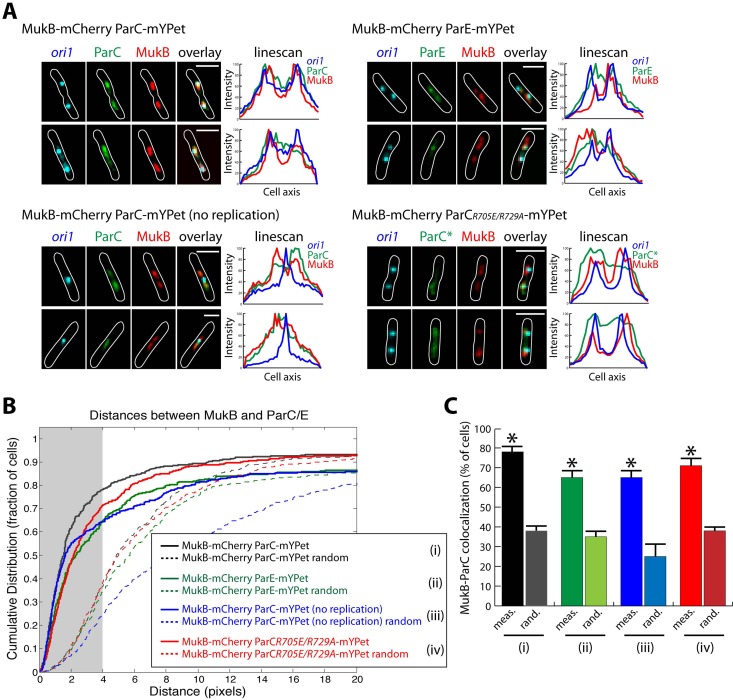
To address whether we could obtain evidence for an interaction of the ParC and ParE subunits of TopoIV with MukBEF in vivo, we replaced the endogenous ParC and ParE genes with functional fluorescent mYPet fusions to ParC and ParE expressed from their endogenous promoters on the E. coli chromosome and analyzed mYPet fluorescence in live cells using wide-field epifluorescence imaging. The cells also expressed MukB-mCherry and had their ori region marked with fluorescent tetracycline repressor (TetR-CFP) bound to an array of tet operators 15 kb counterclockwise (CCW) of oriC (ori1). Cells with the fluorescent fusions showed near-normal flow cytometry profiles and doubling times (see Fig. S2A and S2C in the supplemental material). Fluorescent MukBEF forms foci that are frequently associated with ori, irrespective of whether cells are growing exponentially, or nonreplicating because of a block in replication initiation (Fig. 1A) (14, 15, 17).
FIG 1 .
Association of TopoIV subunits ParC and ParE with MukBEF in vivo. (A) Representative examples of colocalization events between MukB-mCherry and ParC/E-mYPet proteins within E. coli cells. The origin of replication region was identified by the binding of a TetR-CFP protein on a tetO array (ori1) located 15 kb CCW of the replication origin, oriC. The fluorescence profiles of the individual cells show the distribution of fluorescence intensities. Exponential-phase cells were grown in M9-glycerol medium at 30°C prior to imaging. Cells without replication are dnaC(Ts) strains grown at the restrictive temperature for 120 min. Bars, 2 µm. (B) In order to localize the fluorescent foci corresponding to MukB and ParC/E proteins or the ori1 DNA locus within cells, microscopy images were first analyzed using the MicrobeTracker Suite (http://microbetracker.org/) to detect and outline bacterial cells. Cumulative distributions present the distances between the centroids of MukB foci and the brightest ParC/E pixels (Materials and Methods). Colocalization (gray shaded rectangle) is defined as when the MukBEF focus centroid is 4 or less pixels (516 nm) from the brightest ParC/E pixel. (C) The percentages of colocalization between MukBEF and ParC/E were plotted in a histogram. Error bars represent the 95% confidence interval. Asterisks indicate that the measured values (meas.) are statistically significantly different from the random calculated values (rand.) (Materials and Methods).
Although we observed fluorescent ParC-mYPet (ParC fused to mYPet fluorophore) and ParE-mYPet foci in both steady-state and nonreplicating dnaC(Ts) cells (Fig. 1A) (29), they were typically not as well defined as the ori1 and MukBEF foci, and a high background of ParC/ParE outside foci was evident. This is not surprising, given that in vitro analysis has shown a rather weak interaction of ParC with MukB (Kd [dissociation constant] of ~0.5 µM) (18). Initial analysis by simple observation of all three fluorescence channels showed that the ParC-mYPet and ParE-mYPet foci were frequently associated with ori1 and MukBEF, indicative of an in vivo association with MukBEF and/or ori1 (Fig. 1A, images). Analysis of the three fluorescence profiles showed overlapping peaks for MukBEF, ori1, and ParC/E (Fig. 1A, line scans).
In order to establish a quantitative and more objective assessment of the relative localizations of TopoIV and MukBEF, we used automated image analysis that determined the centroid of MukBEF foci by fitting elliptical Gaussian functions. We examined the cumulative distributions of the distances between the brightest ParC/E pixels, which identify the population of ParC/E molecules with the highest residence time and the centroid of the nearest MukBEF focus (Fig. 1B and C; Materials and Methods). For a negative control and to evaluate the level of random coincidence of foci, we performed the same analysis on a simulated random distribution of TopoIV foci within the same cells. The association of TopoIV with MukBEF was now unequivocal: 65% to 78% of the brightest ParC or ParE pixels were located within a distance of 4 pixels (516 nm) from a MukBEF focus, whereas the random distribution yielded 33% and 38%, respectively, for ParE and ParC versus MukBEF. Furthermore, a ParC mutant (ParC with an R-to-E change at position 705 and an R-to-A change at position 729 [ParCR705E/R729A]), which showed an impaired interaction with MukB in vitro (19), retained almost the same level of colocalization with MukBEF, indicative of at least residual binding to MukBEF in vivo (Fig. 1; 72% for mutant ParC compared to 78% for wild-type ParC). Comparable analysis showed similar high levels of colocalization of ParC or ParE with ori1 (see Fig. S2E in the supplemental material; 78% for ParC, 71% for ParE, with reduced colocalization of ParCR705E/R729A with ori1 [61%]), as well as MukBEF with ori1 (Fig. S2D). In our experience, it is not unusual for amino acid substitutions in proteins to lead to loss of in vitro activity, but the proteins retain at least some in vivo activity.
In nonreplicating dnaC(Ts) cells, MukBEF often formed foci close to and on both sides of the single ori1 focus [Fig. 1A, images in the MukB-mCherry ParC-mYPet (no replication) panel; ~66% of single ori1 foci were associated with two such MukBEF foci after 120 min at the restrictive temperature]. In this case, ParC was associated with MukBEF rather than with ori1, demonstrating that the primary association of TopoIV is with MukBEF rather than ori1. The observation that ParE colocalization with MukBEF or ori1 was always lower than that of ParC with MukBEF/ori1, when measured by the cumulative distribution of distances, likely reflects the fact that the ParE association with MukB is via ParC and that there is a significant dissociation rate in vivo between the two TopoIV subunits.
We conclude that the robust TopoIV-MukBEF colocalization demonstrated here likely reflects an interaction of MukBEF present in foci with TopoIV in vivo, thereby recapitulating the characterized in vitro reactions. We are confident that the observed association between ParC/ParE and MukBEF/ori1 is physiologically relevant and not an artifact for the following reasons. ParC-mYPet and ParE-mYPet fluorescent foci form independently at the same cellular positions where they each colocalize with MukBEF-mCherry in cells that retain MukBEF and TopoIV function. ParC-mYPet and ParE-mYPet fluorescent foci form and colocalize with ori1 in the absence of labeled MukBEF (but dependent on the presence of functional MukBEF; see Fig. S2F in the supplemental material). Furthermore, the slightly reduced colocalization of the ParC double point mutant that has impaired interaction with the MukB hinge in vitro likely relates to the physiological relevance of the association between MukBEF and TopoIV. Finally, extensive analysis of a range of MukB and MukE fusions to mYPet, green fluorescent protein (GFP), and mCherry fluorophores has not only failed to show a fluorophore-dependent interaction but demonstrated in vivo localization and function dependent on ATP binding and hydrolysis (16, 17; our unpublished data). Similarly, experiments with the same combinations of fluorophores fused to replisome proteins did not reveal any fluorophore-interaction artifacts (30).
Because of the high background of TopoIV fluorescence, we cannot eliminate the possibility that TopoIV is additionally associated with other regions of the chromosome or with proteins elsewhere; indeed, since many of the MukBEF molecules are not in foci (17), interaction of these molecules with TopoIV could be responsible for at least some of the observed background. Colocalization of ParC foci with the replisome marker, DnaN, was close to random and no higher than that of the association of DnaN with ori1 (data not shown), as expected from our observation that ParC forms foci in dnaC(Ts) cells that are not undergoing replication (Fig. 1A). We are therefore unable to provide evidence that supports the earlier observations that ParC and/or SMC complexes, interact with the bacterial replisome (31–33).
MukBEF within foci positions TopoIV.
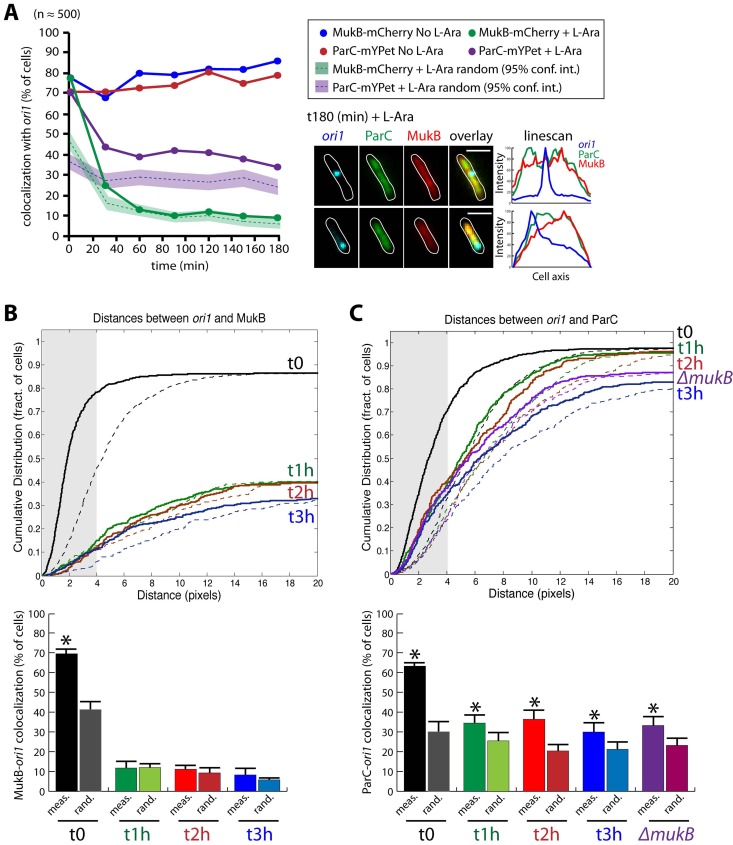
In order to establish whether the observed colocalization between MukBEF and TopoIV is directed by MukBEF, TopoIV, or ori1, we used a MukE degron to deplete functional MukBEF (15). Representative cells in which MukE had been depleted are shown in Fig. 2A (images). The MukB-mYPet signal was now dispersed, whereas the ori1 foci remained intact. All evidence of clear ParC-mYPet focus colocalization with ori1 had also disappeared, with no clear foci elsewhere. Quantitative analysis showed colocalization between MukBEF, or ParC-mYPet, and ori1 had almost completely disappeared after 30 min of MukE depletion (Fig. 2A, left). The cumulative distributions showed that the distance between an ori1 focus and a MukBEF focus or the brightest ParC pixel had become similar to that of the random distribution of localizations after 1 h of MukE depletion and the same as in a ΔmukB strain (Fig. 2B and C; see Fig. S3A in the supplemental material). The loss of ParC foci occurred also after MukE degradation in nonreplicating cells, conditions in which cell growth continued (Fig. S3B). In the absence of functional MukBEF, a low level of ParC localization with ori1 remained (Fig. 2C, bar graph; Fig. S3). This may reflect the fact that action of TopoIV at ori1 is required for timely ori1 segregation in the absence of functional MukBEF.
FIG 2 .
MukB is necessary for TopoIV localization at the origin of replication. (A) Colocalization frequencies (percentages of colocalization determined at a 4-pixel distance) between ori1 versus MukB and ori1 versus ParC were recorded during depletion of the MukE protein. Conditions in the absence or presence of l-arabinose (l-Ara) were plotted. The corresponding random curves were also plotted and show that the measured distances are getting closer to the random positioning during the time course of the experiment. Two representative examples of cells after 180 min of MukE depletion are shown. The values in the 95% confidence interval (95% conf. int.) are shown by purple or green shading. Bars, 2 µm. (B) Cumulative curves of the pairwise distances between ori1 and MukB during the time course of MukE depletion. fract., fraction. (C) Cumulative curves of the pairwise distances between ori1 and ParC during the time course of MukE depletion. In panels B and C, the percentages of colocalization are plotted in the histograms below the graphs (as in Fig. 1C).
TopoIV depletion does not abrogate MukBEF foci.
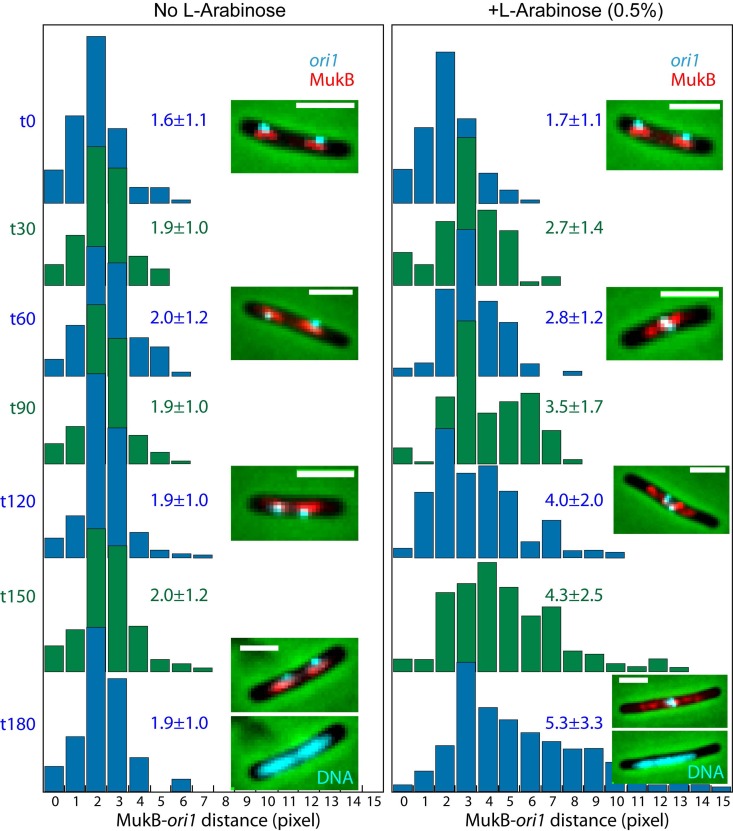
When the complementary experiment was undertaken, in which a degron derivative of ParC was depleted efficiently (see Fig. S4A in the supplemental material), MukBEF foci persisted. By 1 h of depletion, most cells had a single ori1 focus or two closely spaced ori1 foci, consistent with the expected impairment in decatenation of the ori region (Fig. 3, compare left and right panels) (3). In most cells, several MukBEF foci were evident, evenly spaced throughout the nucleoid and placed on both sides of ori1, sometimes with a high background throughout the nucleoid (e.g., see Fig. 3, 120 min with l-Ara). Two to four MukBEF foci were present in most cells, a number similar to the number of ori1s expected in such cells given that replication continues despite the inhibition of chromosome segregation and cell division after TopoIV impairment (3). The apparent regular position of the MukBEF foci within the nucleoid corresponded approximately to where ori1s would have been if their decatenation and segregation had been possible.
FIG 3 .
ParC depletion leads to a dispersion of MukB foci. ParC protein was fused to a degron tag. Upon addition of l-arabinose, ParC was efficiently degraded in less than 1 h (see Fig. S4 in the supplemental material). Images were taken at different time points (from 0 to 180 min) after the addition of l-arabinose, and the pairwise distance between MukBEF foci and ori1 foci was assessed manually using ImageJ software. Means and standard deviations were calculated. Representative examples of cells at each time point are shown. The nucleoid was highlighted by 4′,6′-diamidino-2-phenylindole (DAPI) staining for the examples at t180 (180 min). Bars, 2 µm.
When a similar ParC depletion experiment was repeated in the absence of DNA replication, using a dnaC(Ts) allele at the restrictive temperature, the MukBEF foci persisted, with the majority of cells containing MukBEF foci close to and on either side of the single ori1 focus, as in ParC+ cells (see Fig. S4B in the supplemental material). This confirms that MukBEF foci persist in the absence of TopoIV and that the dispersed multiple MukBEF foci observed in replicating cells are dependent on ongoing DNA replication and/or the consequent accumulation of chromosomal DNA.
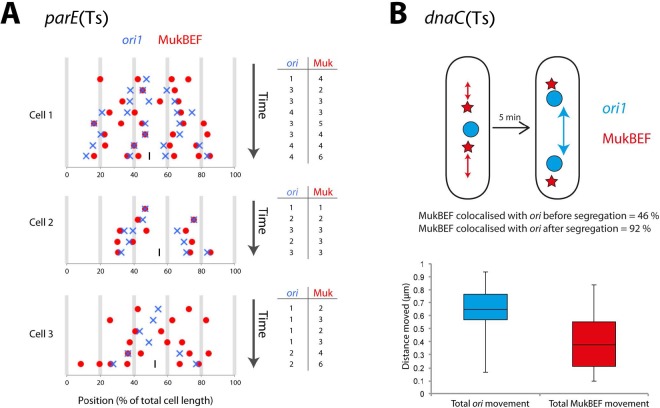
When ParE activity was impaired using a thermosensitive mutation at the restrictive temperature, a similar behavior was observed after 120 min at the restrictive temperature: cells contained a single ori1 focus or closely spaced ori1 foci, with ~4 evenly spaced MukBEF foci throughout the nucleoid (see Fig. S4C in the supplemental material). On return to the permissive temperature, a proportion (~30%) of cells showed a similar distribution of MukBEF foci, but now with oris colocalized. The remaining fraction of cells seemed unable to decatenate and segregate their sister ori1s. To characterize the changes in distribution of MukBEF and ori1 foci during the transition from TopoIV-impaired to functional TopoIV, we undertook time-lapse experiments using parE(Ts) cells in which ori1 and MukBEF were differentially fluorescently labeled (Fig. 4 and Fig. S5). As expected, the temperature shift to 42°C resulted in cells containing either a single ori1 focus close to midcell, or two separated ori1 foci at approximately cell quarter positions. Given that cells had been maintained for just over 1 cell generation at 42°C prior to the shift back to the permissive temperature, we expected each ori1 focus to contain 2 to 4 catenated ori1 regions at shift-down. Again we observed ~30% of cells went on to segregate their ori1 regions and to eventually divide during the 5-h time-lapse experiment at the permissive temperature; in general, these were the shorter cells. In order to simplify the analysis, we focused on cells that started with a single ori1 focus, although the cells starting with two separated ori1 foci showed comparable behavior. We also analyzed only cells that went on to divide into normal looking and growing cells. Typical behavior is shown in the schematic in Fig. 4A, with the primary data for the same three cells shown in Fig. S5. During the 300-min time-lapse experiment, the initial single centrally placed ori1 focus segregated to give 2 to 4 well-separated ori1 foci, with the initial segregation event occurring anywhere between 30 min and 120 min after the shift to the permissive temperature. By the end of the experiment, the segregated ori1 foci were now frequently positioned coincident with or close to a MukBEF focus. We noted that the positions of MukBEF foci before the shift to the permissive temperature were similar to the positions they take up at times immediately prior to cell division. It is tempting to speculate that the MukBEF foci had taken up their final positions before the origins were able to segregate and that this then led to ori positioning; however, this observation is complicated by the fact that the MukBEF foci themselves are mobile during the time-lapse.
FIG 4 .
Regular spacing of ori-independent MukBEF foci after TopoIV impairment and after inhibition of replication initiation. (A) Summary of the time-lapse analysis for 3 representative cells released from a parE(Ts) arrest. Blue × symbols represent origin peaks, and red circles represent MukBEF peaks in the line profiles of intensity along the cells. Time moves down the y axis, and the time points are as follows: 10, 30, 60, 90, 120, 150, 180, and 210 min after release from arrest. For each cell, the analysis is shown up until the point where the cells had clearly divided; septa are marked by a vertical black line on the graph at the last time point. The tables to the right of the three graphs show the number of ori and MukBEF peaks at each time point. The example cells represent 3 cell types observed after parE arrest—cells where the number of Muk foci are equal to (or less than double), double or more than double the number of ori foci—these types are equally represented within the population. (B) Time-lapse following ori1 and MukBEF foci after release from dnaC(Ts) arrest. We measured the total distance moved by ori1 and MukBEF foci along the long axis of the cell during the 5-min interval spanning origin segregation, which is illustrated in the top panel (the blue arrow indicates total distance moved by ori, and the two red arrows combined are the total distance moved by Muk). The distances measured are shown in the box plot below the schematic drawing of the cell; the box indicates the interquartile range with the central line of the box indicating the median value. The whiskers of the plot indicate the minimum and maximum measurements. Colocalization was assessed manually, and it was defined as when the foci centers were less than 2 pixels (258 nm) apart.
MukBEF foci position sister ori regions when the TopoIV-MukBEF association is retained.
In the experiments that exploited TopoIV impairment and in those using TopoIV-positive (TopoIV+) dnaC(Ts) cells, we observed that the association of MukBEF foci with ori1 was reduced, with MukBEF foci taking up regular positions on both sides of ori1. These observations led us to consider that MukBEF focus formation is independent of ori although dependent on the nucleoid, because MukBEF foci always formed on nucleoids rather than in the nucleoid-free space (14). In order to explore the relationship between oris and MukBEF foci, we performed time-lapse experiments with a synchronized TopoIV+ dnaC(Ts) strain released after 105 min at the restrictive temperature, conditions under which we know that the majority of cells have a single unreplicated ori and no associated replisome (29). The cells expressed MukE-mYPet and had their ori region marked with fluorescent Lac repressor (LacI-mCherry) bound to an array of lac operators 15 kb CCW of oriC (ori1). The strain was released into permissive conditions to allow initiation of replication, and ori1 and MukBEF foci were monitored using time-lapse microscopy. We focused on two time points 5 min apart that span the interval in which the replication origin foci segregate (Fig. 4B, top portion). We observed that ori1 is generally colocalized with a MukBEF focus both before and after ori1 segregation; however, there are multiple MukBEF foci at times when there is only a single ori1 focus and not all MukBEF foci are colocalized with ori1. We observed that once the ori1 foci have segregated, the colocalization of MukBEF foci with ori1 foci was increased; before ori1 segregation, 46% of the MukBEF foci colocalized with ori1, whereas after ori1 segregation, 92% of MukBEF foci colocalized with ori1 (colocalization was defined as when the foci centers were less than 2 pixels [258 nm] apart). We also measured the total distances these foci moved along the long axis of the cell during the 5-min interval spanning ori1 segregation. As shown in Fig. 4B, we found that ori1 foci moved a greater total distance (median, 0.65 µm) than the distance moved by the MukBEF foci (median, 0.38 µm). This greater movement of the ori1 foci and the increased localization of MukBEF foci with ori after segregation support the idea that the ori region moves to the position of the MukBEF foci, and therefore, that MukBEF foci play a role in positioning segregated origin regions.
DISCUSSION
We have demonstrated that MukBEF and TopoIV associate in vivo and that this association occurs in the absence of DNA replication and at all stages of the cell cycle. We think it is highly likely that this association reflects the ParC-MukB interaction demonstrated in vitro (18, 19, 21). A key unanswered question regards the functional significance of the in vivo TopoIV-MukBEF association demonstrated here; specifically, does the MukBEF-directed association of TopoIV with ori modulate sister cohesion in the ori region by stimulating decatenation?
Consistent with this hypothesis, earlier work and the work here have demonstrated that in Muk− cells, newly replicated ori1s can exhibit delayed segregation (14) (see Fig. S6 in the supplemental material), suggesting the possibility that the recruitment by MukBEF of TopoIV to the ori region facilitates decatenation of newly replicated sister oris. A quantitative demonstration of this effect was shown when we used a degron to deplete MukE, thereby leading to an abrogation of MukBEF function; the fraction of steady-state cells containing a single ori1 focus increased to 64% from 25% after MukBEF impairment (Fig. S6A). In order to test the hypothesis directly, we analyzed ori1 segregation in the ParCR705E/R729A mutant that is impaired in its interaction with MukBEF in vitro, although any in vivo impairment appears to be modest. Snapshot analysis of steady-state cells showed an increase in the fraction of single ori1 focus cells from 26% to 37% in the mutant compared to the wild type, while time-lapse analysis showed an increase in cohesion time from 18 min to 23 min in the mutant compared to the wild type, when the time of ori1 segregation after replisome appearance was measured (Fig. S6A and S6B). These apparent modest defects in decatenation are consistent with, but do not prove, a model in which the TopoIV-MukBEF interaction directs TopoIV-mediated decatenation to ori. A ParC mutant that is catenation proficient but is totally defective in the in vivo interaction with MukBEF (or the complementary mutant for MukB) would be needed to test the hypothesis directly.
The demonstration in vitro that the relaxation activity of TopoIV on right-handed (negative) supercoiled DNA, but not on left-handed (positive) supercoiled DNA, is stimulated by the interaction with MukBEF (21) suggests that the interaction should enhance decatenation because negative supercoils have the same right-handed chirality as replicative catenanes (see Fig. S1 in the supplemental material). Taken together, the in vitro stimulation of TopoIV activity on a substrate that mimics replicative catenanes and the observation of an in vivo association of MukBEF and TopoIV with ori makes us confident that TopoIV will be directed to ori to remove catenanes from newly replicated sisters. TopoIV may also be associated with the majority of MukBEF molecules that are not present as foci, therefore allowing TopoIV to act globally on the chromosome.
Our observations also lead us to conclude that MukBEF complexes within foci act to position the ori region and to additionally direct a fraction of TopoIV molecules to ori. Ablation of MukBEF leads both to ori mispositioning (14) and to the loss of ParC foci (this paper). Our observation that TopoIV impairment did not prevent MukBEF focus formation makes us confident that it is the MukBEF within foci that positions TopoIV, a conclusion supported by our observation that in dnaC(Ts) cells grown at 37°C, MukBEF foci on either side of ori1 colocalize with ParC rather than ori itself. We do not know what is positioning the MukBEF foci, but the regular patterning is reminiscent of that which occurs when ParAB-parS systems position low-copy plasmids, chemosensory apparatus, or carboxysomes on the nucleoid (1, 34). Since ori positioning and chromosome segregation are disrupted by Muk impairment, the data provide strong support for a model in which the coordinated and sequential action of TopoIV and MukBEF in complexes visualized as foci plays a central role in initiating segregation of newly replicated oris.
An earlier study of TopoIV in E. coli found that the catalytic subunit, ParC, was associated with the replisome, while the other subunit, ParE, was located in nucleoid-free regions of the cell (31). Based on this and other observations, it was proposed that TopoIV activity is regulated temporally, with activity occurring at late stages of the cell cycle when ParC could be released from the replisome and associate with the FtsK translocase (35). We do not know how to reconcile this information with the results obtained in this work and in our earlier work (3), which together show that TopoIV can act at ori through its association with MukBEF. It is difficult to imagine how a single replisome, or FtsK complex, could interact with a substantial proportion of cellular TopoIV. Biochemical characterization has also shown that ParCE complexes are stable during gel filtration and ultracentrifugation (22), and therefore, it seems likely that a substantial proportion of ParC and ParE in cells can form functional topoisomerase molecules. We note that in the cell biology analysis of reference 31, immunocytochemistry was used in fixed rapidly growing cells, a situation where resolution is not at its highest. Finally, the roles of TopoIV in decatenating newly replicated regions of the chromosome and in replicating plasmids, which are positioned in the extranuclear space, have been documented (3, 7, 22, 36). A genetic study of Bacillus subtilis indicated an interaction between the SMC complex and TopoIV (26), while studies of the budding yeast Saccharomyces cerevisiae have implicated functional interactions of both cohesin and condensin with topoisomerase II that are important in chromosome decatenation (27, 28, 37, 38). It therefore seems possible that functional interactions between SMC complexes and topoisomerases are ubiquitous.
MATERIALS AND METHODS
Bacterial strains and growth.
The bacterial strains used in this study are listed in Table 1. The plasmids and oligonucleotides used in this study are shown in Table S1 in the supplemental material. All strains are derivatives of the Escherichia coli K-12 AB1157 strain (39). Fusion of genes with fluorescent or degron tags was performed using the λRed method (40). Fused genes were transferred to generate the final strains through P1 phage transduction (41). For multiple insertions of modified genes, the Kanr gene was removed using site-specific recombination through expression of the Flp recombinase from the pCP20 plasmid (40). All strains were constructed and analyzed several times independently in order to avoid any effect of potential suppressor mutations. dnaC(Ts) cells were grown at 37°C for 120 min to generate a population of cells that could not reinitiate replication, as described previously (15, 29). Depletion of proteins carrying a degron tag necessitates the presence of the SspB protein which is expressed chromosomally under the arabinose-controlled promoter (pAra) in order to control the timing of the depletion (15). tetO and lacO arrays (240 copies) are inserted at 15 kb counterclockwise (CCW) of oriC (ori1).
TABLE 1 .
Bacterial strains used in this studya
| Strain | Genotypeb |
|---|---|
| AU2019 | lacO at ori1 hyg; tetO at ter3 gen; plac-lacI-mCherry at leuB; plac-lacI-tetR-mCerulean at galK; mukE-mYPet dnaC2 thrA::Tn10 |
| ENOX5.130 | mukB::mCherry frt; parC::mYPet kan; plac-tetR::cfp frt; tetO at ori1 gen |
| ENOX5.138 | mukB::mCherry frt; parE::mYPet frt; plac-tetR::cfp kan; tetO at ori1 gen |
| ENOX5.147 | mukB::mCherry frt; parC::mYPet kan; plac-tetR::cfp frt; tetO at ori1 gen; dnaC2(Ts) thrA::Tn10 |
| ENOX5.225 | mukB::mCherry frt; parCR705E/R729A::mYPet kan; plac-tetR::cfp frt; tetO at ori1 gen |
| ENOX5.167 | mukE::degron frt; mukB::mCherry kan; parC::mYPet frt; plac-tetR::cfp frt; tetO at ori1 gen; ΔsspB frt; PAra-sspB frt |
| ENOX5.182 | ΔmukB kan; parC::mYPet frt; plac-tetR::cfp frt; tetO at ori1 gen |
| ENOX5.47 | parC::degron frt; mukB::mCherry frt; plac-tetR::cfp frt; tetO at ori1 gen; ΔsspB frt; PAra-sspB frt |
| ENOX5.41 | parC::degron frt; mukB::mCherry frt; plac-tetR::cfp kan; tetO at ori1 gen; ΔsspB frt; PAra-sspB frt; dnaC2(Ts) thrA::Tn10 |
| ENOX5.56 | parE(Ts); mukB::mCherry frt; plac-tetR::cfp kan; tetO at ori1 gen |
| ENOX5.245 | frt mCherry::dnaN; parC::mYPet kan; plac-tetR::cfp frt; tetO at ori1 gen |
All these strains were constructed in this study.
Abbreviations: kan, kanamycin resistance gene; hyg, hygromycin B resistance gene; gen, gentamicin resistance gene; frt, FLP site-specific recombination site.
Microscopy.
Image captures were performed as described in reference 15. Cells were grown at 30°C in M9 medium supplemented with appropriate amino acids and 0.2% glycerol (M9-gly). For experiments in the absence of replication, strains carrying the dnaC(Ts) allele were shifted 2 h at 37°C (to synchronize cells by allowing completion of existing rounds of replication but preventing any further replication initiation), and the microscope chamber was maintained at 37°C in order to prevent any reinitiation of replication during image acquisition. Induction of SspB protein during depletion experiments was done by addition of 0.5% l-arabinose. For microscopy, cells in exponential phase (A600 ≈ 0.1) were concentrated and laid on a 1% M9-gly agarose pad on a slide. During depletion experiments, the l-arabinose was also maintained in the slides when required.
For the parE(Ts) time-lapse experiments, cells were grown at 30°C in M9-gly to an A600 of ~0.05. The cells were shifted to 42°C for 2 h, and then a sample was spun down and spotted onto a prewarmed 1% M9-gly agarose pad on a slide. The cells were imaged at 30°C for 5 h; the initial images were taken 10 and 30 min after release and then every 30 min up until 300 min.
For the dnaC(Ts) time-lapse experiments (Fig. 4B), cells were grown at 30°C in M9-gly to an A600 of ~0.05. The cells were shifted to 37°C for 105 min (the restrictive temperature for dnaC2). A sample was spotted onto a prewarmed 1% M9-gly agarose pad on a slide. The cells were released to the permissive temperature of 30°C to allow replication initiation and imaged every 5 min for 2 h.
Image analysis.
Images were taken and processed by Metamorph 6.2, and image analysis was done using ImageJ or specific programs run in Matlab.
(i) Semiquantitative analysis of fluorescence distributions.
Fluorescence distributions within cells were plotted as line scans using the Plot Profile command of ImageJ or MicrobeTracker software run in Matlab (42). Maximum intensity values were normalized between 0 and 100% for each channel before plotting. For the parE(Ts) experiments (Fig. 4A), peaks in intensity were defined as peaks when the height of the peak was 5% or more of an increase in intensity above the neighboring points of inflection.
(ii) Quantitative analysis of colocalization.
Cell outlines were first delineated from a phase image using the MicrobeTracker software run in Matlab (see Fig. S7 in the supplemental material). This segmenting analysis created a “mesh” for each cell, within which each pixel is characterized by a specific x,y coordinate (Fig. S7Bi). This step is critical to determine pairwise distances between two independent pixels of different channels within a specific cell (see further steps).
The second step of the analysis was to find the population of fluorescent molecules with the highest residence time within a cell. These populations of molecules assembled as local maxima of fluorescence intensity. As both TetR-CFP (cyan fluorescent protein) bound to the ori1 locus and MukB-mCherry formed well-defined foci, we adapted the automated localization analysis method of Holden et al. (43) that first identified candidate foci above an intensity threshold and subsequently determined their centroids by fitting an elliptical Gaussian function (see Fig. S7Biii and S7Biv in the supplemental material). This kind of analysis was not possible for ParC/E-mYPet signal, as ParC/E did not always assemble discrete foci. In order to localize the population of ParC/E molecules with the highest residence time, we determined the brightest pixel within each cell (Fig. S7Bii). It should be noted that the Gaussian localization analysis for MukBEF and ori1 can identify multiple fluorescent foci within one cell or none at all, but the brightest pixel analysis finds exactly one pixel with the highest intensity for ParC/E.
The third step of the analysis measured the pairwise distances between the brightest ParC/E pixel and the nearest MukBEF or ori1 localization (see Fig. S7C in the supplemental material). To determine the distribution of distances expected from an entirely random localization of ParC/E, we also calculated distances between a pixel randomly positioned within the cell and the nearest MukBEF or ori1 focus (Fig. S7C). For MukBEF-ori1 colocalization analysis with multiple MukBEF foci within each cell, an equal number of random MukBEF localizations was generated per cell, and the smallest pairwise distance was calculated.
In the fourth step, the distances were plotted as cumulative distributions (see Fig. S7D in the supplemental material). A threshold of 4 pixels (516 nm) was chosen to define colocalization. The fraction of cells with colocalizing foci was thus determined from the cumulative distributions at 4-pixel distance with 95% statistical confidence bounds (Fig. S7D). Note that the cumulative distribution curves do not reach 100% of cells even for large distances, because MukBEF or ori1 foci can be absent in some cells; the asymptotic maximum values hence give the fraction of cells that showed MukBEF or ori1 foci. For example, ~90% of wild-type cells displayed MukBEF foci (Fig. 1B), which reduced to ~30% MukBEF foci under MukE degron conditions (Fig. 2B).
(iii) MukB foci distribution during ParC/E depletion.
Because the automated analysis was not sufficiently robust to identify and localize the larger number of closely spaced MukBEF foci during the course of ParC/E depletion, the distances in pixels between centroids of MukB-assembled foci and the ori1 locus were manually measured using ImageJ software and plotted as histograms.
SUPPLEMENTAL MATERIAL
Further details of Materials and Methods. Download
Topology of replicative (pre)catenanes. Duplex DNA has the two strands of the double helix in a right-handed (RH) (+) configuration (bottom panel, right). Precatenanes that interlink the two sister duplexes after replication have the same RH (+) topology (bottom panel). RH (−) supercoils (SCs) have the same RH chirality as (+) precatenanes, whereas the left-handed (LH) (+) SCs that accumulate ahead of a replication fork have the opposite chirality (bottom left panel). Note that one converts an RH (−) SC to an RH (+) replicative catenane by “cutting” the top and bottom of the plectoneme and rejoining (top panel); the sign changes from (−) to (+) because of the relative change in orientation of the red arrows. Hence, conditions that favor the stimulation of RH (−) SC relaxation are expected to stimulate decatenation, since the substrates have the same RH chirality. Download
Cell cycle parameters of fluorescent fusions and the demonstration that TopoIV subunits colocalize with MukBEF foci and ori1. (A) Western blot analysis showing the expression of the full-length versions of the different fusion proteins. (B) Generation times of the strains expressing the different fusion proteins. Cells were grown in LB medium at 30°C. (C) Flow cytometry analysis of cells grown in M9-glycerol medium at 30°C. Exponential-phase cells were analyzed. Strains expressing various versions of ParC/E-fluorescent protein fusions were compared to the AB1157 strain. The table shows the different proportions of 1n, 2n, and >2n content (1n includes chromosomes up to ~50% replicated, while 2n includes chromosomes that are >50% replicated). (D and E) Colocalization events between ori1 versus MukB (D) and ori1 versus ParC/E (E). Cumulative curves and histograms were generated as described in the legend to Fig. 1. In panels D and E, black and gray bars correspond to the MukB-mCherry ParC-mYPet ori1-tetO (TetR-Cfp) strain (i), green bars correspond to the MukB-mCherry ParE-mYPet ori1-tetO (TetR-Cfp) strain (ii), blue bars correspond to the MukB-mCherry ParC-mYPet ori1-tetO (TetR-Cfp) strain in the absence of replication (iii), and red bars correspond to the MukB-mCherry ParCR705E/R729A-mYPet ori1-tetO (TetR-Cfp) strain (iv). (F) Representative examples of cells expressing ParC-mYPet or ParE-mYPet in strains where the origin of replication locus was identified by the binding of a TetR-CFP protein on a tetO array (ori1). Bars, 2 µm. Download
Deletion of MukB or depletion of MukE leads to loss of ParC colocalization with the origin of replication. (A) Representative examples of cells expressing a ParC-mYPet fusion, with ori1 labeled in a ΔmukB strain. Cells were grown at 22°C in M9-glycerol medium. (B) MukE depletion was performed in the absence of replication [dnaC(Ts) allele at 37°C; depletion started after 2 h at 37°C]. Representative examples of cells at different time points after the addition of l-arabinose (0.5%) are shown. The average sizes and standard deviations of 50 independent cells at the different time points are also indicated. Bars, 2 µm. Download
MukB foci persist in E. coli cells upon impairment of topoisomerase IV. (A) Western blot showing the disappearance of ParC protein upon the addition of l-arabinose (0.5%) in cultures of E. coli in LB or M9 medium. (B) The histograms show the distribution of MukB-ori1 distances upon depletion of ParC protein in the absence of replication [dnaC(Ts) allele at 37°C; depletion started after 2 h at 37°C]. (C) Impairment of ParE activity was achieved by using a thermosensitive allele of ParE [parE(Ts)] and shift of growth cultures to 42°C. Return to functional ParE was assayed after shift of the cultures to 30°C. Aliquots were taken at different time points, and the distances (in pixels) between MukB foci and the ori1 locus was assessed manually using ImageJ software. Means and standard deviations were calculated. Representative examples of cells at each time point are represented. Bars, 2 µm. Download
Time-lapse microscopy following ori1 and MukBEF dynamics during the transition from impaired TopoIV to functional TopoIV. Line profile analysis of time-lapse microscopic images for 3 representative cells released from parE(Ts) impairment at 42°C. These are the same 3 cells summarized in Fig. 4. The blue lines represent intensity profiles for ori1, and the red lines represent MukBEF. Cell images are also shown for cell 1. For each cell, the analysis is shown up until the point where the cells had clearly divided; septa are marked by a vertical black line on the graph for the last time point. Download
The TopoIV-MukBEF interaction may direct TopoIV-mediated decatenation to ori. (A) Percentage of cells containing a single ori1 locus in indicated strains. (B) Cumulative distributions describing ori1 locus segregation time after replisome appearance (mCherry-DnaN) in strains expressing ParC-mYPet or ParCR705E/R729A-mYPet fusion. Representative examples of such segregation events are shown. Bars, 2 µm. Download
Analysis of colocalization events. (A) Four different channels were typically imaged during a microscopy experiment: (i) a phase-contrast image, (ii) a fluorescence image in the green channel (GFP filter set) to visualize the mYPet fusion proteins, (iii) a fluorescence image in the blue channel (CFP filter set) to visualize the CFP fusion proteins, and (iv) a fluorescence image in the red channel (mCherry filter set) to visualize the mCherry fusion proteins. (B) Outlines of cells were defined using MicrobeTracker from the phase-contrast image (i), and the generated meshes were used in order to find the brightest pixel by cell in the green channel (ii). Foci assembled by MukB-mCherry (iv) or TetR-CFP (iii) were automatically found using a Gaussian fitting algorithm. (C) Pairwise distances between the closest brightest ParC pixel and the centroids of Gaussian-fitted MukBEF or TetR fluorescent foci were measured (yellow rectangles). Distances between a reference pixel (i.e., from MukB or ori1 focus) and a randomly positioned pixel in the cell were also calculated. (D) The distances were plotted as cumulative distribution curves. Colocalization frequencies were defined as the proportion of cells showing pairwise distances shorter than 4 pixels. Download
Plasmids and oligonucleotides used in this study.
ACKNOWLEDGMENTS
The work was supported by a Wellcome Trust program grant to D.S. (WT083469MA) and by a Wellcome Trust strategic award (091911) supporting advanced microscopy at Micron Oxford (Department of Biochemistry, Dunn School of Pathology, University of Oxford, Oxford, United Kingdom).
We thank Ian Dobbie for his generous support and advice on imaging.
Footnotes
Citation Nicolas E, Upton AL, Uphoff S, Henry O, Badrinarayanan A, Sherratt D. 2014. The SMC complex MukBEF recruits topoisomerase IV to the origin of replication region in live Escherichia coli. mBio 5(1):e01001-13. doi:10.1128/mBio.01001-13.
REFERENCES
- 1. Reyes-Lamothe R, Nicolas E, Sherratt DJ. 2012. Chromosome replication and segregation in bacteria. Annu. Rev. Genet. 46:121–143. 10.1146/annurev-genet-110711-155421 [DOI] [PubMed] [Google Scholar]
- 2. Wang X, Montero Llopis P, Rudner DZ. 2013. Organization and segregation of bacterial chromosomes. Nat. Rev. Genet. 14:191–203. 10.1038/nrg3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang X, Reyes-Lamothe R, Sherratt DJ. 2008. Modulation of Escherichia coli sister chromosome cohesion by topoisomerase IV. Genes Dev. 22:2426–2433. 10.1101/gad.487508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. López V, Martínez-Robles ML, Hernández P, Krimer DB, Schvartzman JB. 2012. Topo IV is the topoisomerase that knots and unknots sister duplexes during DNA replication. Nucleic Acids Res. 40:3563–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sissi C, Palumbo M. 2010. In front of and behind the replication fork: bacterial type IIA topoisomerases. Cell. Mol. Life Sci. 67:2001–2024. 10.1007/s00018-010-0299-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Adams DE, Shekhtman EM, Zechiedrich EL, Schmid MB, Cozzarelli NR. 1992. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell 71:277–288. 10.1016/0092-8674(92)90356-H [DOI] [PubMed] [Google Scholar]
- 7. Joshi MC, Magnan D, Montminy TP, Lies M, Stepankiw N, Bates D. 2013. Regulation of sister chromosome cohesion by the replication fork tracking protein SeqA. PLoS Genet. 9:e1003673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nasmyth K, Haering CH. 2005. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 74:595–648. 10.1146/annurev.biochem.74.082803.133219 [DOI] [PubMed] [Google Scholar]
- 9. Gruber S. 2011. MukBEF on the march: taking over chromosome organization in bacteria? Mol. Microbiol. 81:855–859. 10.1111/j.1365-2958.2011.07764.x [DOI] [PubMed] [Google Scholar]
- 10. Hiraga S, Niki H, Imamura R, Ogura T, Yamanaka K, Feng J, Ezaki B, Jaffé A. 1991. Mutants defective in chromosome partitioning in E. coli. Res. Microbiol. 142:189–194. 10.1016/0923-2508(91)90029-A [DOI] [PubMed] [Google Scholar]
- 11. Niki H, Jaffé A, Imamura R, Ogura T, Hiraga S. 1991. The new gene mukB codes for a 177 KD protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 10:183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Woo JS, Lim JH, Shin HC, Suh MK, Ku B, Lee KH, Joo K, Robinson H, Lee J, Park SY, Ha NC, Oh BH. 2009. Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell 136:85–96. 10.1016/j.cell.2008.10.050 [DOI] [PubMed] [Google Scholar]
- 13. Li Y, Schoeffler AJ, Berger JM, Oakley MG. 2010. The crystal structure of the hinge domain of the Escherichia coli structural maintenance of chromosomes protein MukB. J. Mol. Biol. 395:11–19. 10.1016/j.jmb.2009.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Danilova O, Reyes-Lamothe R, Pinskaya M, Sherratt D, Possoz C. 2007. MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol. Microbiol. 65:1485–1492. 10.1111/j.1365-2958.2007.05881.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Badrinarayanan A, Lesterlin C, Reyes-Lamothe R, Sherratt D. 2012. The Escherichia coli SMC complex, MukBEF, shapes nucleoid organization independently of DNA replication. J. Bacteriol. 194:4669–4676. 10.1128/JB.00957-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nolivos S, Sherratt D. 18 November 2013. The bacterial chromosome: architecture and action of bacterial SMC and SMC-like complexes. FEMS Microbiol. Rev. 10.1111/1574-6976.12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Badrinarayanan A, Reyes-Lamothe R, Uphoff S, Leake MC, Sherratt DJ. 2012. In vivo architecture and action of bacterial structural maintenance of chromosome proteins. Science 338:528–531. 10.1126/science.1227126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Y, Stewart NK, Berger AJ, Vos S, Schoeffler AJ, Berger JM, Chait BT, Oakley MG. 2010. Escherichia coli condensin MukB stimulates topoisomerase IV activity by a direct physical interaction. Proc. Natl. Acad. Sci. USA 107:18832–18837. 10.1073/pnas.1008678107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hayama R, Marians KJ. 2010. Physical and functional interaction between the condensin MukB and the decatenase topoisomerase IV in Escherichia coli. Proc. Natl. Acad. Sci. USA 107:18826–18831. 10.1073/pnas.1008140107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vos SM, Stewart NK, Oakley MG, Berger JM. 2013. Structural basis for the MukB-topoisomerase IV interaction and its functional implications in vivo. EMBO J. 32:2950–2962. 10.1038/emboj.2013.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hayama R, Bahng S, Karasu ME, Marians KJ. 2013. The MukB-ParC interaction affects the intramolecular, not intermolecular, activities of topoisomerase IV. J. Biol. Chem. 288:7653–7661. 10.1074/jbc.M112.418087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peng H, Marians KJ. 1993. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J. Biol. Chem. 268:24481–24490 [PubMed] [Google Scholar]
- 23. Kato J, Nishimura Y, Imamura R, Niki H, Hiraga S, Suzuki H. 1990. New topoisomerase essential for chromosome segregation in E. coli. Cell 63:393–404. 10.1016/0092-8674(90)90172-B [DOI] [PubMed] [Google Scholar]
- 24. Khodursky AB, Peter BJ, Schmid MB, DeRisi J, Botstein D, Brown PO, Cozzarelli NR. 2000. Analysis of topoisomerase function in bacterial replication fork movement: use of DNA microarrays. Proc. Natl. Acad. Sci. U. S. A. 97:9419–9424. 10.1073/pnas.97.17.9419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zechiedrich EL, Cozzarelli NR. 1995. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes Dev. 9:2859–2869. 10.1101/gad.9.22.2859 [DOI] [PubMed] [Google Scholar]
- 26. Tadesse S, Mascarenhas J, Kösters B, Hasilik A, Graumann PL. 2005. Genetic interaction of the SMC complex with topoisomerase IV in Bacillus subtilis. Microbiology (Reading, Engl) 151:3729–3737 [DOI] [PubMed] [Google Scholar]
- 27. Tapia-Alveal C, Outwin EA, Trempolec N, Dziadkowiec D, Murray JM, O’Connell MJ. 2010. SMC complexes and topoisomerase II work together so that sister chromatids can work apart. Cell Cycle 9:2065–2070 [DOI] [PubMed] [Google Scholar]
- 28. Baxter J, Aragón L. 2012. A model for chromosome condensation based on the interplay between condensin and topoisomerase II. Trends Genet. 28:110–117. 10.1016/j.tig.2011.11.004 [DOI] [PubMed] [Google Scholar]
- 29. Withers HL, Bernander R. 1998. Characterization of dnaC2 and dnaC28 mutants by flow cytometry. J. Bacteriol. 180:1624–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ. 2008. Independent positioning and action of Escherichia coli replisomes in live cells. Cell 133:90–102. 10.1016/j.cell.2008.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Espeli O, Levine C, Hassing H, Marians KJ. 2003. Temporal regulation of topoisomerase IV activity in E. coli. Mol. Cell 11:189–201. 10.1016/S1097-2765(03)00013-3 [DOI] [PubMed] [Google Scholar]
- 32. Lindow JC, Kuwano M, Moriya S, Grossman AD. 2002. Subcellular localization of the Bacillus subtilis structural maintenance of chromosomes (SMC) protein. Mol. Microbiol. 46:997–1009. 10.1046/j.1365-2958.2002.03235.x [DOI] [PubMed] [Google Scholar]
- 33. Wang SC, Shapiro L. 2004. The topoisomerase IV ParC subunit colocalizes with the Caulobacter replisome and is required for polar localization of replication origins. Proc. Natl. Acad. Sci. U. S. A. 101:9251–9256. 10.1073/pnas.0402567101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vecchiarelli AG, Han YW, Tan X, Mizuuchi M, Ghirlando R, Biertümpfel C, Funnell BE, Mizuuchi K. 2010. ATP control of dynamic P1 ParA-DNA interactions: a key role for the nucleoid in plasmid partition. Mol. Microbiol. 78:78–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Espeli O, Lee C, Marians KJ. 2003. A physical and functional interaction between Escherichia coli FtsK and topoisomerase IV. J. Biol. Chem. 278:44639–44644. 10.1074/jbc.M308926200 [DOI] [PubMed] [Google Scholar]
- 36. Reyes-Lamothe R, Tran T, Meas D, Lee L, Li AM, Sherratt DJ, Tolmasky ME. 16 October 2013. High-copy bacterial plasmids diffuse in the nucleoid-free space, replicate stochastically and are randomly partitioned at cell division. Nucleic Acids Res. 10.1093/nar/gkt918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Farcas AM, Uluocak P, Helmhart W, Nasmyth K. 2011. Cohesin’s concatenation of sister DNAs maintains their intertwining. Mol. Cell 44:97–107. 10.1016/j.molcel.2011.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Charbin A, Bouchoux C, Uhlmann F. 2014. Condensin aids sister chromatid decatenation by topoisomerase II. Nucleic Acids Res. 42:340–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bachmann BJ. 1972. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev. 36:525–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thomason LC, Costantino N, Court DL. 2007. E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. Chapter 1:Unit 1.17. 10.1002/0471142727.mb0117s79 [DOI] [PubMed] [Google Scholar]
- 42. Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. 2011. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 80:612–627. 10.1111/j.1365-2958.2011.07579.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Holden SJ, Uphoff S, Hohlbein J, Yadin D, Le Reste L, Britton OJ, Kapanidis AN. 2010. Defining the limits of single-molecule FRET resolution in TIRF microscopy. Biophys. J. 99:3102–3111. 10.1016/j.bpj.2010.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Further details of Materials and Methods. Download
Topology of replicative (pre)catenanes. Duplex DNA has the two strands of the double helix in a right-handed (RH) (+) configuration (bottom panel, right). Precatenanes that interlink the two sister duplexes after replication have the same RH (+) topology (bottom panel). RH (−) supercoils (SCs) have the same RH chirality as (+) precatenanes, whereas the left-handed (LH) (+) SCs that accumulate ahead of a replication fork have the opposite chirality (bottom left panel). Note that one converts an RH (−) SC to an RH (+) replicative catenane by “cutting” the top and bottom of the plectoneme and rejoining (top panel); the sign changes from (−) to (+) because of the relative change in orientation of the red arrows. Hence, conditions that favor the stimulation of RH (−) SC relaxation are expected to stimulate decatenation, since the substrates have the same RH chirality. Download
Cell cycle parameters of fluorescent fusions and the demonstration that TopoIV subunits colocalize with MukBEF foci and ori1. (A) Western blot analysis showing the expression of the full-length versions of the different fusion proteins. (B) Generation times of the strains expressing the different fusion proteins. Cells were grown in LB medium at 30°C. (C) Flow cytometry analysis of cells grown in M9-glycerol medium at 30°C. Exponential-phase cells were analyzed. Strains expressing various versions of ParC/E-fluorescent protein fusions were compared to the AB1157 strain. The table shows the different proportions of 1n, 2n, and >2n content (1n includes chromosomes up to ~50% replicated, while 2n includes chromosomes that are >50% replicated). (D and E) Colocalization events between ori1 versus MukB (D) and ori1 versus ParC/E (E). Cumulative curves and histograms were generated as described in the legend to Fig. 1. In panels D and E, black and gray bars correspond to the MukB-mCherry ParC-mYPet ori1-tetO (TetR-Cfp) strain (i), green bars correspond to the MukB-mCherry ParE-mYPet ori1-tetO (TetR-Cfp) strain (ii), blue bars correspond to the MukB-mCherry ParC-mYPet ori1-tetO (TetR-Cfp) strain in the absence of replication (iii), and red bars correspond to the MukB-mCherry ParCR705E/R729A-mYPet ori1-tetO (TetR-Cfp) strain (iv). (F) Representative examples of cells expressing ParC-mYPet or ParE-mYPet in strains where the origin of replication locus was identified by the binding of a TetR-CFP protein on a tetO array (ori1). Bars, 2 µm. Download
Deletion of MukB or depletion of MukE leads to loss of ParC colocalization with the origin of replication. (A) Representative examples of cells expressing a ParC-mYPet fusion, with ori1 labeled in a ΔmukB strain. Cells were grown at 22°C in M9-glycerol medium. (B) MukE depletion was performed in the absence of replication [dnaC(Ts) allele at 37°C; depletion started after 2 h at 37°C]. Representative examples of cells at different time points after the addition of l-arabinose (0.5%) are shown. The average sizes and standard deviations of 50 independent cells at the different time points are also indicated. Bars, 2 µm. Download
MukB foci persist in E. coli cells upon impairment of topoisomerase IV. (A) Western blot showing the disappearance of ParC protein upon the addition of l-arabinose (0.5%) in cultures of E. coli in LB or M9 medium. (B) The histograms show the distribution of MukB-ori1 distances upon depletion of ParC protein in the absence of replication [dnaC(Ts) allele at 37°C; depletion started after 2 h at 37°C]. (C) Impairment of ParE activity was achieved by using a thermosensitive allele of ParE [parE(Ts)] and shift of growth cultures to 42°C. Return to functional ParE was assayed after shift of the cultures to 30°C. Aliquots were taken at different time points, and the distances (in pixels) between MukB foci and the ori1 locus was assessed manually using ImageJ software. Means and standard deviations were calculated. Representative examples of cells at each time point are represented. Bars, 2 µm. Download
Time-lapse microscopy following ori1 and MukBEF dynamics during the transition from impaired TopoIV to functional TopoIV. Line profile analysis of time-lapse microscopic images for 3 representative cells released from parE(Ts) impairment at 42°C. These are the same 3 cells summarized in Fig. 4. The blue lines represent intensity profiles for ori1, and the red lines represent MukBEF. Cell images are also shown for cell 1. For each cell, the analysis is shown up until the point where the cells had clearly divided; septa are marked by a vertical black line on the graph for the last time point. Download
The TopoIV-MukBEF interaction may direct TopoIV-mediated decatenation to ori. (A) Percentage of cells containing a single ori1 locus in indicated strains. (B) Cumulative distributions describing ori1 locus segregation time after replisome appearance (mCherry-DnaN) in strains expressing ParC-mYPet or ParCR705E/R729A-mYPet fusion. Representative examples of such segregation events are shown. Bars, 2 µm. Download
Analysis of colocalization events. (A) Four different channels were typically imaged during a microscopy experiment: (i) a phase-contrast image, (ii) a fluorescence image in the green channel (GFP filter set) to visualize the mYPet fusion proteins, (iii) a fluorescence image in the blue channel (CFP filter set) to visualize the CFP fusion proteins, and (iv) a fluorescence image in the red channel (mCherry filter set) to visualize the mCherry fusion proteins. (B) Outlines of cells were defined using MicrobeTracker from the phase-contrast image (i), and the generated meshes were used in order to find the brightest pixel by cell in the green channel (ii). Foci assembled by MukB-mCherry (iv) or TetR-CFP (iii) were automatically found using a Gaussian fitting algorithm. (C) Pairwise distances between the closest brightest ParC pixel and the centroids of Gaussian-fitted MukBEF or TetR fluorescent foci were measured (yellow rectangles). Distances between a reference pixel (i.e., from MukB or ori1 focus) and a randomly positioned pixel in the cell were also calculated. (D) The distances were plotted as cumulative distribution curves. Colocalization frequencies were defined as the proportion of cells showing pairwise distances shorter than 4 pixels. Download
Plasmids and oligonucleotides used in this study.