ABSTRACT
Type IV pili (Tfp) are expressed by many Gram-negative bacteria to promote aggregation, adhesion, internalization, twitching motility, or natural transformation. Tfp of Neisseria meningitidis, the causative agent of cerebrospinal meningitis, are involved in the colonization of human nasopharynx. After invasion of the bloodstream, Tfp allow adhesion of N. meningitidis to human endothelial cells, which leads to the opening of the blood-brain barrier and meningitis. To achieve firm adhesion, N. meningitidis induces a host cell response that results in elongation of microvilli surrounding the meningococcal colony. Here we study the role of the major pilin subunit PilE during host cell response using human dermal microvascular endothelial cells and the pharynx carcinoma-derived FaDu epithelial cell line. We first show that some PilE variants are unable to induce a host cell response. By engineering PilE mutants, we observed that the PilE C-terminus domain, which contains a disulfide bonded region (D-region), is critical for the host cell response and that hypervariable regions confer different host cell specificities. Moreover, the study of point mutants of the pilin D-region combined with structural modeling of PilE revealed that the D-region contains two independent regions involved in signaling to human dermal microvascular endothelial cells (HDMECs) or FaDu cells. Our results indicate that the diversity of the PilE D-region sequence allows the induction of the host cell response via several receptors. This suggests that Neisseria meningitidis has evolved a powerful tool to adapt easily to many niches by modifying its ability to interact with host cells.
IMPORTANCE
Type IV pili (Tfp) are long appendages expressed by many Gram-negative bacteria, including Neisseria meningitidis, the causative agent of cerebrospinal meningitis. These pili are involved in many aspects of pathogenesis: natural competence, aggregation, adhesion, and twitching motility. More specifically, Neisseria meningitidis, which is devoid of a secretion system to manipulate its host, has evolved its Tfp to signal to brain endothelial cells and open the blood-brain barrier. In this report, we investigate, at the molecular level, the involvement of the major pilin subunit PilE in host cell response. Our results indicate that the PilE C-terminal domain, which contains a disulfide bonded region (D-region), is critical for the host cell response and contains two independent regions involved in host cell signaling.
INTRODUCTION
Neisseria meningitidis is a commensal bacterium of the human nasopharynx that, after bloodstream invasion, is responsible for cerebrospinal meningitis and septicemia. The ability to interact with host cells is essential for meningococcal pathogenesis. Initial binding to human epithelial cells is the first step for rhinopharynx colonization. Interaction with the microvasculature is responsible for the specific aspects of meningococcal pathogenesis—i.e., crossing of the blood-brain barrier, peripheral thrombosis, and purpuric lesions. The ability of N. meningitidis to adhere to human cells relies on several factors, including type IV pili (Tfp) (1), outer membrane proteins, such as Opa and Opc (2–4), and minor adhesins, like NadA, NhhA, or PorB (5–7). In the bloodstream, the polysaccharide capsule, which is essential to bacterial dissemination by inhibiting the bactericidal activity of the complement, prevents the interaction of the outer membrane proteins with their cellular ligands. In the bloodstream, Tfp are believed to be the only factor that allows the initial colonization of the microvasculature by promoting a direct interaction with endothelial cells (1). This was recently confirmed by experiments showing that, in vivo, Tfp are essential (i) to colonize human vessels and (ii) to induce microvasculature lesions and inflammation, both of which are responsible for the clinical symptoms (8, 9). These data clearly demonstrate that, besides adhesion, Tfp induce in vivo an endothelial host cell response that is essential for meningococcal pathogenesis. Data obtained in vitro have demonstrated that the endothelial host cell response following meningococcal Tfp interaction is due to the recruitment and activation of the β2-adrenergic receptor–β-arrestin pathway that triggers the recruitment of host cell components at the site of bacterial adhesion, such as cellular receptor, adhesion molecules, junctional components, proteins of the actin polymerization machinery, and ezrin, which links actin filaments to the cell membrane (10–13). Ezrin is thought to be essential for the accumulation of proteins under colonies (14). The recruitment of these factors leads to the formation of membrane protrusions that enhance cohesion of the meningococcal microcolonies adhering on the apical surface of the host cell and open the paracellular route, allowing invasion of the surrounding tissues (10–13). These data point out the essential role of the Tfp-induced host cell response in meningococcal pathogenesis.
Tfp are long filamentous structures primarily composed of a major pilin subunit PilE. They are shared by many Gram-negative bacteria, such as Pseudomonas aeruginosa, Vibrio cholerae, and enteropathogenic Escherichia coli (EPEC). Pilin subunits consist of an extended hydrophobic N-terminal domain and an α-helical region followed by a globular C-terminal domain containing a disulfide bonded region between the two conserved cysteines (D-region) (for review, see reference 15). Pilin subunits are reversibly assembled into polymeric fibers, and their globular C-terminal domains are exposed on the outer surface of the fiber. Tfp mediate several phenotypes, such as adherence, aggregation, motility, competence, and biofilm formation. In most cases, the C-terminal domain of the structural pilin is responsible for recognition of cellular receptors. For instance, the D-region of the Pseudomonas aeruginosa PAK pilin is involved in adhesion to a biotic surface, through interaction with glycosylated receptor, or an abiotic surface, such as steel (16, 17). The C-terminal domain of V. cholerae pilin is involved in colonization of epithelial cells (18, 19), while the D-region of the bundle-forming pili of EPEC is needed for adhesion to HEp-2 cells (20, 21). N. meningitidis has developed specific strategies to manipulate the host cell. Indeed, meningococcal Tfp were shown to promote remodeling of the host cell plasma membrane (22), while other adhesins of N. meningitidis (such as Opa, Opc, NadA, PorB, and NhhA) are involved in bacterial engulfment inside epithelial cells (for review, see reference 1).
Besides the major pilin PilE, Tfp of N. meningitidis possesses 3 minor pilins, PilV, PilX, and ComP, that are involved in signaling, aggregation, and natural transformation, respectively (11, 13, 23–25). In addition, N. meningitidis has evolved to express a wide diversity of major pilin PilE amino acid sequences. This process, known as antigenic variation (26), involves the recombination of silent pilS cassettes at the pilE locus by resolution of a G4 structure (27). This particular gene and cassette organization favors “parallelized” evolution of pilE (28) and promotes a high rate of variability in the pilE sequence, especially in a “hypervariable” domain containing the surface-exposed D-region (29, 30). Recombination of pilS loci with the pilE locus occurs at a high rate in vivo (31), and N. meningitidis is able to express many different pilin variants during colonization of the same host. To date, variation of PilE primary sequence has only been associated with bacterial aggregation.
The mechanism by which Tfp induce a host cell response is not fully understood. The minor pilin PilV has been proposed as “the” signaling pilin of N. meningitidis in endothelial cells (11). Indeed, a pilV mutated strain is unable to signal to endothelial cells. However, a PilV mutation does not alter the ability of meningococci to signal to epithelial cells (32), thus suggesting that Tfp have another means by which they can promote host cell responses. Here we demonstrate that the major pilin subunit of the Tfp is also a signaling component of N. meningitidis. By engineering various chimeric pilin molecules and performing a structure homology modeling of PilE variants, we show that various hypervariable regions express different host cell specificities carried by two different parts of the D-region. Altogether, these results demonstrate that a regulation of the meningococcal host cell response occurs via antigenic variation.
RESULTS
The major pilin subunit PilE is involved in host cell responses.
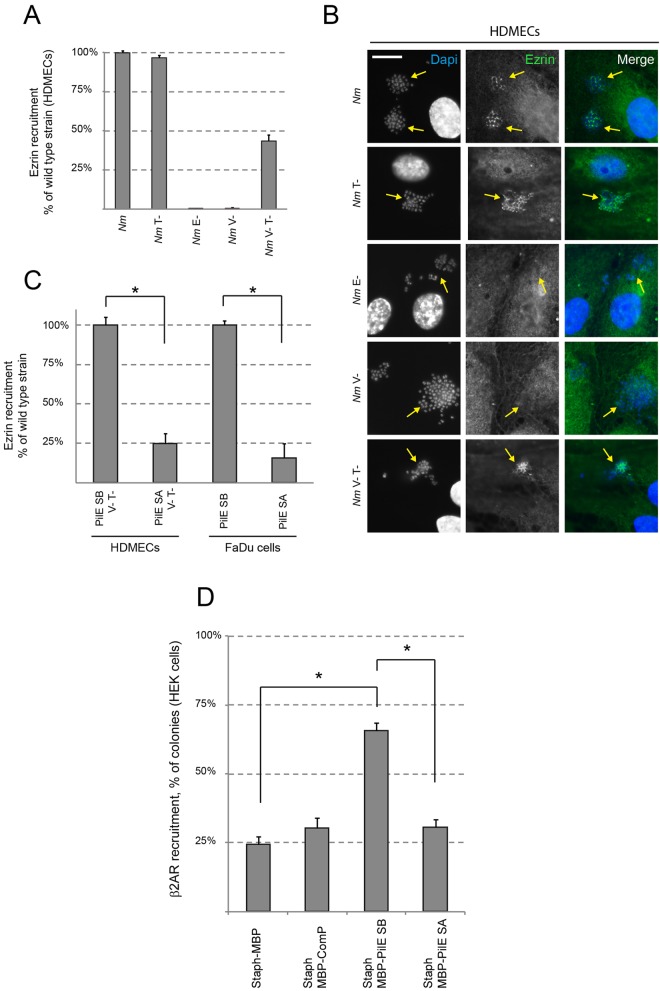
As mentioned above, a pilV-mutated strain is unable to signal to endothelial cells, thus demonstrating the essential role of PilV to induce a host cell response in endothelial cells. However, two sets of data argue that Tfp can induce a host cell response in a PilV-independent manner. (i) It has been clearly shown that PilV is not involved in Tfp signaling in epithelial cells (32), suggesting that Tfp may induce a host cell response by other means. (ii) Considering previous results showing that the loss of phenotypes due to some mutations in the pilin-related machinery could be restored in a pilus retraction-deficient background (PilT−) (33), we tested the ability of a PilT− PilV− strain to signal to endothelial cells. We used ezrin recruitment as the hallmark of meningococcal cell signaling, since ezrin is recruited under the microcolonies after adhesion to both endothelial and epithelial cells. As shown in Fig. 1, a pilT mutation restored, at least partially, the ability of a PilV− strain to induce a response in human dermal microvascular endothelial cells (HDMECs), as determined on the basis of ezrin recruitment (Fig. 1A and B). Altogether, these data clearly demonstrate that Tfp have other means besides PilV to induce a host cell response.
FIG 1 .
The major pilin subunit PilE is involved in endothelial and epithelial cell response. (A and B) HDMECs were infected with strains 2C4.3 and the ΔPilT, ΔPilE, ΔPilV, ΔPilV ΔPilT derivatives (Nm and Nm T-, E-, V-, and V-/T-, respectively). (C) HDMECs and FaDu cells were infected with strain 2C4.3 or the PilV− PilT− derivative, expressing the major pilin subunit PilESB or PilESA (A to C). The host cell response was assessed by monitoring the recruitment of ezrin using an immunofluorescence assay. The ezrin recruitment index was estimated by determining the proportion of colonies that efficiently recruit ezrin at the site of adhesion and expressed as normalized mean values (±standard errors of the mean [SEM]) of three independent experiments in duplicate. *, P < 0.001 (Student’s t test). It is noteworthy that 90% of the bacterial microcolonies of the wild-type (WT) strain 2C4.3 recruit ezrin at the site of adhesion. (B) Ezrin was immunostained (in green), and DNA was stained using DAPI (in blue). Microcolonies are shown by arrows. Scale bars, 10 µm. (D) PilESA is not able to recruit the β2-adrenergic receptor. HEK cells overexpressing the β2-adrenergic receptor tagged with YFP were infected with Staphylococcus aureus coated with anti-MBP antibody and MBP-pilin fusion proteins, as described in Materials and Methods. Receptor recruitment was counted in infected cells and expressed as normalized mean values (±SEM) of three independent experiments in duplicate. *, P < 0.001 (Student’s t test).
Considering that purified PilE molecules have been previously shown to recruit the β2-adrenergic receptor, the Tfp signaling receptor on endothelial cells (13), we hypothesized that the major pilin subunit may be involved in the PilV-independent host cell response and that different pilin variants may have different abilities to signal to cells. Two major pilin variants of strain 8013 have been extensively described—PilESA and PilESB (34). Both of these pilins are known to promote an interaction with epithelial and endothelial cells. However, the major phenotypic difference known is that strains expressing PilESA do not form clumps, unlike strains expressing PilESB (34). These two PilE variants were introduced by allelic exchange in the parental strain as a transcriptional fusion with the kanamycin (Kmr) gene, as previously described (34). Strains expressing these variants were then tested for their ability to induce a host cell response in both epithelial and endothelial cells (FaDu cells and HDMECs, respectively). To address the mechanism of PilV-independent Tfp signaling, infections of endothelial cells were all performed in a PilV− PilT− background, as described in Materials and Methods. As shown in Fig. 1C, the strain expressing the PilESA variant was dramatically impaired in its ability to recruit ezrin but not the strain expressing PilESB. These data demonstrate that the major pilin subunit PilE is the key component involved in the PilV-independent signaling and that only some variants of the major pilin subunit are involved in host cell responses.
Since the signaling receptor has only been identified in endothelial cells, we next aimed at assessing whether the lack of signaling observed in HDMECs with the strain expressing PilESA was due to a lack of interaction of the major pilin subunit with the β2-adrenergic receptor. We subsequently purified the PilESA variant fused to the maltose-binding protein (MBP). Indeed, recombinant PilESB fused to MBP and bound to live staphylococci via anti-MBP antibodies is able to induce the recruitment of the β2-adrenergic receptor overexpressed in HEK-293 cells, as previously shown (13). We subsequently compared the recruitment of the β2-adrenergic receptor by both MBP-PilESB and MBP-PilESA (i.e., the major pilin variants fused to MBP). ComP, another minor pilin involved in DNA uptake, fused to MBP and MBP alone were used as negative controls (Fig. 1D). In agreement with our previous work, MBP-PilESB was able to recruit the endothelial signaling receptor. On the other hand, MBP-PilESA did not recruit the β2-adrenergic receptor (Fig. 1D). These data are consistent with those reported above and strongly suggest that pili expressing the major pilin PilESA variant do not induce a host cell response because of the lack of recruitment of the β2-adrenergic receptor.
The hypervariable D-region of the major pilin subunit is responsible for PilE-mediated host cell responses.
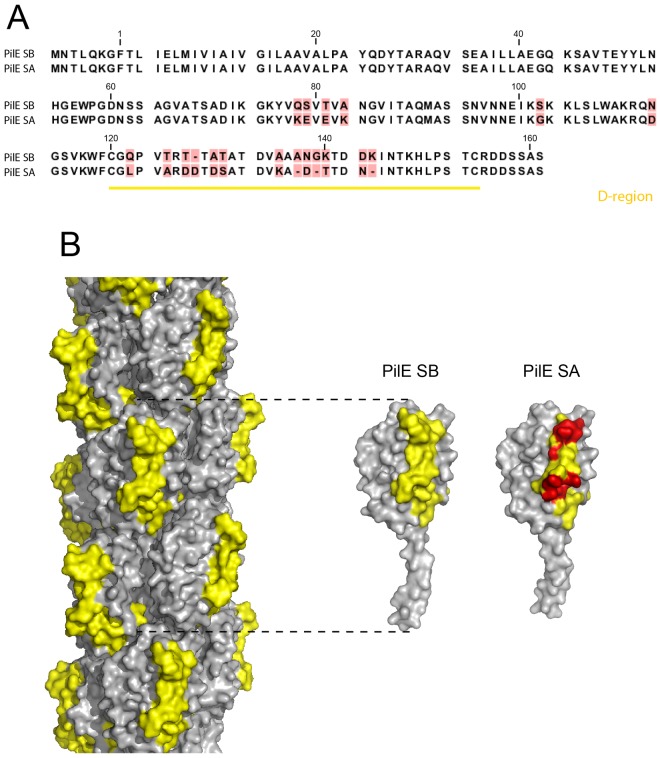
To understand the molecular basis of PilE-dependent signaling, we next compared the sequences of the two pilin variants PilESB and PilESA (Fig. 2A). Out of the 20 differences observed in the peptidic sequence, 13 were in the hypervariable D-region. Modeling of the three-dimensional structures of the PilESB and PilESA variants based on Neisseria gonorrhoeae pilin structure (29) (Fig. 2B; see Fig. S1 in the supplemental material) confirmed that the D-regions are protrusive and that most of the variable residues are exposed at the Tfp surface. We subsequently hypothesized that modifications within the D-region sequence may interfere with the process of signaling to human cells.
FIG 2 .
Structure homology modeling of PilE. (A) ClustalW sequence alignment of the two variants PilESB and PilESA. Unconserved residues between PilESA and PilESB are highlighted in red. The D-region is underlined in yellow. (B) Surface representation of N. meningitidis PilESB filament model and of the PilESB and PilESA subunit models based on the helical reconstruction of the N. gonorrhoeae GC pilus into an electron cryomicroscopy map (PDB code 2HIL). Unconserved residues within the D-region between PilESA and PilESB are highlighted in red.
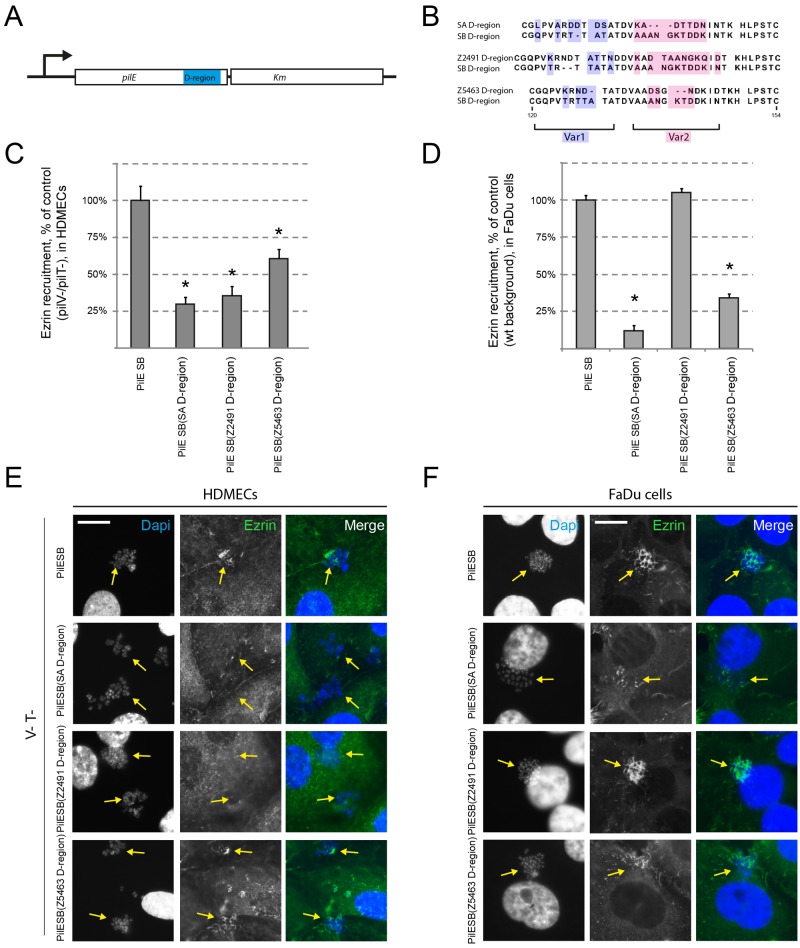
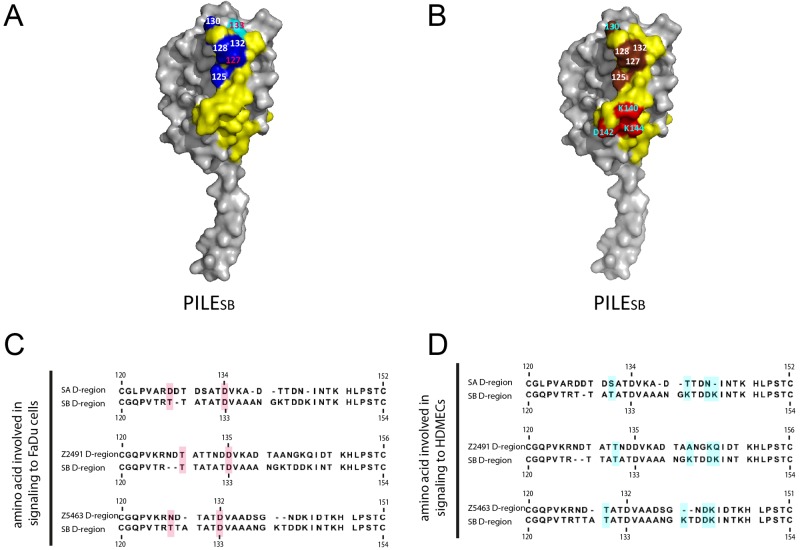
To confirm the role of the D-region in signaling to human cells, we replaced, as described in Materials and Methods, the D-region of the PilESB variant with that of PilESA and with those of two other pilin variants obtained from two serogroup A strains, Z2491 and Z5463. The corresponding alleles were designated PilESB(SA D-region), PilESB(Z2491 D-region), and PilESB(Z5463 D-region), respectively. Alignments of the D-region sequences are shown in Fig. 3B. On the basis of the primary sequence, two regions can be identified among the D-region residues: Var1 from Q122 to T132 and Var2 from A135 to N146. These two regions are linked by two conserved residues (D133 and V134). These pilin variants were then introduced as transcriptional Km fusions into strain 2C4.3, to select for recombination at the pilE locus and not in one of the pilS loci (Fig. 3A). The corresponding strains were tested for PilE expression, competence, aggregation, and adhesion, and all have similar phenotypes (see Table S1).
FIG 3 .
PilESB(D-region) variants are differentially involved in endothelial and epithelial cell responses. (A) Schematic representation of the PilE/Km transcriptional fusions used in this study. (B) ClustalW sequence alignment of the D-region of four different PilE variants: SB, SA, Z2491, and Z5463. Two variable domains (Var1 highlighted in blue and Var2 highlighted in red) were discriminated on the basis of alignment. (C, D, E, and F) HDMECs (C and E) and FaDu cells (D and F) were infected with strains of N. meningitidis expressing PilESB or PilESB(SA D-region), PilESB(Z2491 D-region), and PilESB(Z5463 D-region). HDMECs were infected with the PilV− PilT− derivatives of these strains. (C and D) The ezrin recruitment index was estimated by determining the proportion of colonies that efficiently recruit ezrin at the site of adhesion and expressed as normalized mean values (±SEM) of three independent experiments in duplicate. *, P < 0.002 (Student’s t test). (E and F) Ezrin was immunostained (in green), and DNA was stained using DAPI (in blue). Microcolonies are shown by arrows. Scale bars, 10 µm.
We next assessed the ability of these strains to induce a host cell response in endothelial cells. The strains expressing a PilESB(SA D-region) or PilESB(Z2491 D-region) are defective in Tfp-induced signaling in endothelial cells (Fig. 3C and E). On the other hand, colonies expressing PilESB(Z5463 D-region) are able to recruit ezrin, and the intensity of this recruitment was identical to that of the parental strain expressing PilESB (Fig. 3C and E).
Similar experiments were performed using epithelial cells. Unexpectedly, the strain expressing the pilin PilESB(Z2491 D-region), which is not able to induce recruitment of ezrin in endothelial cells, induced a host cell response identical to that of the PilESB-expressing strain on epithelial cells. On the other hand, colonies expressing PilESB(SA D-region) and PilESB(Z5463 D-region) were unable to recruit ezrin at the site of bacterial adhesion (Fig. 3D and F). Altogether, these data confirm the role of the pilin hypervariable D-region in host cell response and suggest that pilin variants have different ability to induce a host cell response on different cell types.
The hypervariable region of the major pilin subunit encompassed two distinct signaling regions with different specificities.
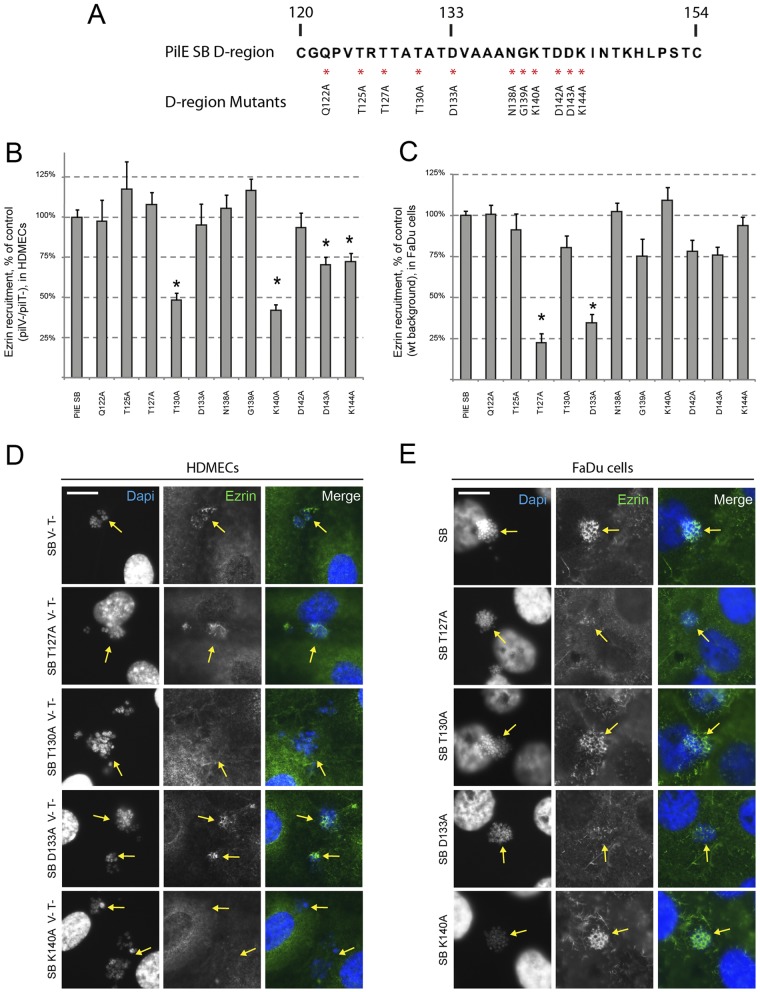
To further characterize the D-region domain of the pilin PilE, point mutations were performed in the sequence of PilESB, and the ability of the corresponding mutated pilin to signal to human cells was tested. We targeted 11 residues of interest and replaced them with alanine using PCR mutagenesis: (i) Q122, T125, T127, and T130 are unconserved residues located in the Var1 region, (ii) D133 is a conserved residue over many pilin variants (30), and (iii) N138, G139, K140, D142, D143, and D144 are unconserved residues located in the Var2 region (Fig. 4A). Each allele was introduced into strain 2C4.3 as a transcriptional fusion with the Kmr gene. All the corresponding strains have similar pilus-related phenotypes (see Table S1). PilV− PilT− derivatives of these strains were engineered to study the host endothelial cells’ response. Four mutants were significantly impaired in ezrin recruitment in HDMECs: the T130A, K140A, D143A, and K144A mutants (Fig. 4B and D). It is noteworthy that these 4 mutants were capable of inducing ezrin recruitment in FaDu cells. However, strains expressing the two T127A and D133A pilin mutants were defective in ezrin recruitment in epithelial cells (Fig. 4C and E). These 6 residues were mapped on the PilESB model (Fig. 5A and B). Residues K140, D143, and K144 are located on the lower part of the D-region. Surprisingly, T130 is not located close to these latter residues. Residues D133 and T127 appear to be located on the top part of the D-region, which is enriched in threonine and aspartic acid (in dark blue and cyan, respectively). This suggests that two different parts of the PilESB D-region are associated with two different signaling specificities.
FIG 4 .
Point mutations reveal residues specifically involved in endothelial or epithelial cell response. (A) Mutated residues of PilESB D-region are indicated by a red asterisk. (B, C, D, and E) HDMECs (B and D) and FaDu cells (C and E) were infected with N. meningitidis strains expressing mutated PilESB. HDMECs were infected with the PilV− PilT− derivatives of these strains. (B and C) The ezrin recruitment index was estimated by determining the proportion of colonies that efficiently recruit ezrin at the site of adhesion and expressed as normalized mean values (±SEM) of three independent experiments performed in duplicate. *P < 0.002 (Student’s t test). (D and E) Ezrin was immunostained (in green), and DNA was stained using DAPI (in blue). Microcolonies are shown by arrows. Scale bars, 10 µm.
FIG 5 .
Point mutations involved in ezrin recruitment are localized on two distinct regions of the D-region. (A and B) Surface representation of PilESB homology models (D-regions are highlighted in yellow). (A) In the top part, threonine and aspartic acid are important for the FaDu cell response. Threonines are highlighted in dark blue and aspartic acids in cyan. The PilESB signaling residues T127 and D133 are in red. (B) The bottom part of the D-region is important for the HDMEC response. The PilESB signaling residues K140, D142, and K144 are highlighted in red, while threonines of the top part are highlighted in brown. Signaling residues are written in blue. (C and D) ClustalW sequence alignment of the D-regions of four different pilin variants: SB, SA, Z2491, and Z5463. Residues involved in the endothelial cell response are highlighted in cyan, and those involved in the epithelial cell response are highlighted in red.
To refine our analysis, we mapped these 6 residues of the PilESB D-region on the D-region sequences of the various PilE variants engineered above (Fig. 5C and D). The two residues (T127 and D133) involved in host cell response on FaDu cells are conserved in the PilESB(Z2491 D-region) sequence. Consistently, pili of this pilin variant are able to promote ezrin recruitment in FaDu cells. We next mapped the four residues T130, K140, D143, and K144, which are involved in host cell response on HDMECs. Residues T130, D143, and K144 are only conserved in PilESB(Z5463 D-region) sequence (Fig. 5D), while residue K140 is only present in the sequence of the PilESB D-region. Consistently, only the strain expressing PilESB(Z5463 D-region) is able to promote ezrin recruitment following adhesion to HDMECs.
To support the importance of these residues in epithelial and endothelial cell signaling, we correlated the percentage of conserved signaling residues with the ezrin recruitment. The results are reported in Fig. S2 in the supplemental material and show that this correlation is statistically significant for both epithelial and endothelial cells (Spearman’s r = 0.8365; r2 = 0.6826). Altogether, these results strongly suggest (i) that the proper localization of a set of threonine and aspartic acid is essential (T127 and D133 in the PilESB model) for the epithelial cell host response and (ii) that a well-organized lysine-aspartic acid-rich motif in the D-region is required for the endothelial cell host response.
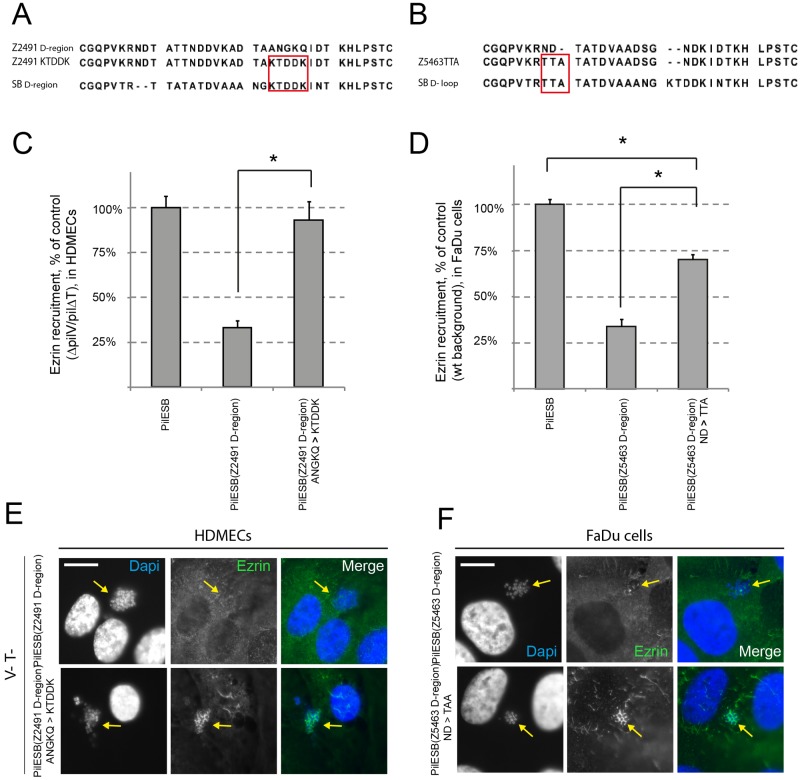
To confirm the above data, chimeric D-regions were engineered. We first modified PilESB(Z2491 D-region), which was unable to induce the endothelial cell response, by replacing A142N143G144K145Q146 with KTDDK (Fig. 6A). This modification of PilESB(Z2491 D-region) should restore a PilESB-like phenotype. The second chimeric pilin was engineered by a modification of N127D128 by TTA in the PilESB(Z5463 D-region) (Fig. 6B). We first confirmed that both mutated strains adhere to cells as PILESB. We then assessed the abilities of these two mutants to recruit ezrin on HDMECs and FaDu cells. A strain expressing the mutated PilESB(Z2491 D-region ANGKQ→KTDDK) is able to recruit ezrin in HDMECs at a level similar to that of a strain expressing PilESB (Fig. 6C and E). A strain expressing the mutated PilESB(Z5463 D-region ND→TTA) partially restored its ability to recruit ezrin on FaDu cells (Fig. 6D and F). Altogether, these results confirm the above hypothesis that the D-region encompasses two signaling regions: one specific for the host epithelial cell response and one specific for the host endothelial cell response.
FIG 6 .
Mutations in PilESB(Z2491 D-region) and PilESB(Z5463 D-region) restore ezrin recruitment. (A and B) Sequence alignment of D-regions from pilin variants SB, Z2491 and Z2491ANGKQ→KTDDK, Z5463, and Z5463ND→TTA. Mutated residues are circled in red. (C to F) HDMECs (C and E) and FaDu cells (D and F) were infected with strains of N. meningitidis expressing PilESB(Z2491 D-region) and PilESB(Z2491 D-region)ANGKQ→KTDDK, PilESB(Z5463 D-region) and PilESB(Z5463 D-region)ND→TTA. HDMECs were infected with the PilV− PilT− derivatives of these strains. (C and D) The ezrin recruitment index was estimated by determining the proportion of colonies that efficiently recruit ezrin at the site of adhesion and expressed as normalized mean values (±SEM) of three independent experiments in duplicate. *, P < 0.002 (Student’s t test). (E and F) Ezrin was immunostained (in green), and DNA was stained using DAPI (in blue). Microcolonies are shown by arrows. Scale bars, 10 µm.
DISCUSSION
Type IV pili are responsible for meningococcal rhinopharyngeal colonization and adhesion of bacterial colonies on the blood vessel wall (8, 9, 11). Regarding endothelial cells, Mikaty et al. have shown that firm adhesion of meningococci requires a PilV-dependent host cell response (11). PilV is required to activate the β2-adrenergic receptor, which promotes the formation of microvilli that surround the colony and protect it from blood flow (13). Here we show that a PilT mutation restored the ability of a PilV-deficient strain to induce signaling in HDMECs, and we demonstrated that in a PilT-deficient strain, PilE induced a host cell response. Interestingly, upon adhesion, the wild-type Tfp switch between a thick conformation and an elongated conformation (35) that is necessary to promote a host cell response (33). We have previously shown that a PilT-deficient strain only expresses Tfp in an elongated conformation (33). Here we hypothesize that a PilT mutation allows the proper presentation of PilE and increases its ability to induce a host cell response.
In this work, we aimed at studying the role of the major structural pilin PilE in inducing host cell membrane reorganization. Here we showed that the pilin variant PilESA is unable to recruit the β2-adrenergic receptor onto endothelial cells and probably the still unknown epithelial signaling receptor. We subsequently demonstrated that the D-region of PilE, which is highly variable, was responsible for the induction of a host cell response. The PilE D-region is also known to be essential for bacterial aggregation and adhesion (34). Here the role of aggregation in Tfp-induced signaling can be questioned. However, statistical analysis revealed that no link exists between aggregation of the strains and variants engineered in this work and their signaling in FaDu cells or HDMECs (data not shown).
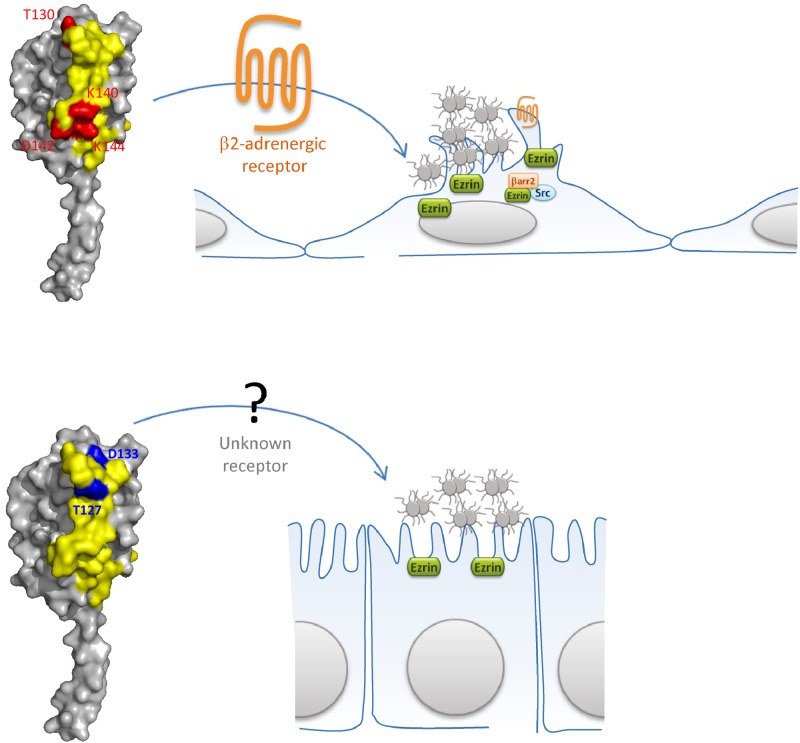
We identified several residues of the PilESB D-region region involved in signaling to different cell types. These residues form two potent host cell response domains exposed at the surface of the fiber—one located in the top part of the D-region and another one located in the lower part (results are summarized in Fig. 7). The former is associated with the epithelial cell host response, whereas the latter is associated with signaling to endothelial cells. These two domains may vary independently by antigenic variation. Indeed, we have observed that a pilin variant of strain Z2491 is unable to induce an endothelial cell response, while it induced an epithelial cell response. These data are consistent with previously reported results demonstrating that the signaling receptor involved in the FaDu cells’ response is different from that of endothelial cells (32). Interestingly, we observed that endothelial cell response required a well-organized lysine- and aspartic acid-enriched domain and a threonine located at the top of the D-region. However, it is reasonable to think that a signaling epitope for endothelial cells may encompass several pilin monomers that may form a binding pocket. This was previously observed for the PilS pilin of Salmonella enterica serovar Typhi (36). Importantly, we and others have already shown that Tfp can transit into two conformations and that adhesion to cells will promote this transition (33, 35, 37). This suggests that one of the two conformations allows the right alignment of pilin monomers, supporting the idea that at least two pilin monomers are involved during interaction with the β2-adrenergic receptors.
FIG 7 .
Schematic representation of PilE-dependent signaling to host cells. In the top panel, the residues T130, K140, D142, and K144 are highlighted in red on the PilESB model. These residues are involved in endothelial cell signaling via the β2-adrenergic receptor. In the lower panel, the residues T127 and D133 are highlighted in blue on the PilESB model. These residues are involved in the epithelial cell response via an unknown receptor. Ezrin polymerization is indicated by the green box, while recruitment of β-arrestin 2 and that of Src is represented as an orange or blue box, respectively.
Moreover, while the nature of the signaling receptor of epithelial cells is still unknown, we hypothesize that there may be more than one possible receptor for epithelial cells; thus, antigenic variation gives to Neisseria meningitidis a large repertoire of possible interactions with host cell receptors. Because Neisseria meningitidis is a commensal of the human nasopharynx, under some circumstances, it may be beneficial for Neisseria meningitidis to express nonsignaling pilins. Consistent with this hypothesis, we observed that the threonine aspartic region is poorly conserved among sequenced PilE variants (30).
This work provides a new insight into the role of PilE during colonization of human tissues. The strategy consisting of a constant renewal of pilE sequences appears to be critical, not only for immune escape but also to broaden the array of targets with which it can interact. Indeed, N. meningitidis has evolved an extraordinary tool to adapt to its host without losing its capacity to spread.
MATERIALS AND METHODS
Bacterial strains and infection conditions.
Strains Z2491 and Z5463 were isolated during the same epidemic in the Gambia in 1983 (38). Z5463PI is a derivative of Z5463 (31). 2C4.3 is a variant of strain 8013, a meningococcal serogroup C strain previously described (34). All derivatives used in this work to test meningococcal host cell interaction were noncapsulated (SiaD−) Opa+ variants of 2C4.3 (2, 12). A noncapsulated Opa+ strain is indeed able to adhere on eukaryotic cells expressing carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) independently of Tfp expression. As previously shown, a piliated noncapsulated Opa+ strain is not affected in its ability to signal to cells via Tfp (Fig. 1) (12). The noncapsulated derivatives were engineered by introduction of a cat resistance cassette into the siaD gene. When needed, PilT− derivatives were engineered by introduction of an ermB resistance cassette into the pilT gene (39). The PilV-deficient derivative was engineered by introduction of an aadA resistance cassette into the pilV gene.
N. meningitidis strains were grown on GCB agar plates (Difco) containing Kellogg’s supplements with appropriate antibiotics in a moist atmosphere containing 5% CO2 at 37°C. The day of infection, a suspension of the bacteria from an overnight culture on a GCB agar plate was adjusted to an optical density at 600 nm (OD600) of 0.05 and incubated for 2 h at 37°C in a prewarmed cell culture medium. Cells were infected with bacteria at a multiplicity of infection (MOI) of 100 for 30 min to allow N. meningitidis adhesion and then washed with cell culture medium and maintained in appropriate fresh medium for 30 min or 2 h.
Cell lines.
Human dermal microvascular endothelial cells (HDMECs) were purchased from Promocell and grown in Promocell endothelial cell growth medium MV supplemented with Promocell endothelial cell growth medium supplement mix and 1% penicillin-streptomycin-amphotericin B (PSA). The pharynx carcinoma-derived FaDu epithelial cells were obtained from the American Type Culture Collection (ATCC HTB-43). FaDu cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (PAA Laboratories) supplemented with 10% fetal bovine serum (FBS) and 1% PSA. Human embryonic kidney 293 (HEK-293) cells were grown in DMEM supplemented with 10% fetal calf serum. HDMECs and FaDu cells were grown in flasks coated with 5 µg/cm2 of rat tail collagen type I (R&D Systems) at 37°C in a humidified incubator in 5% CO2.
Engineering of PilE-Kanr vector.
To introduce a specific pilin variant at the pilE expression locus, a transcriptional fusion of the pilE coding sequence with the aph3′ gene (confers resistance to kanamycin) was engineered as previously described (34). Briefly, the DNA encoding the PilESB variant, the pilE downstream sequence, and the aph3′ coding sequence were amplified from genomic DNA or TOPO 2.1 vector (aph3′ amplification). The pilE gene with its promoter was amplified using the following primers: pilE [EcoRI] FW (CGGAATTCGCCCGCGCACAAGTTTCC) and pilE [BamHI] RV (CGCGGATCCGCGTACCTTAGCTGGCAGATGAAT). The downstream sequence was amplified using pilE 3′ DN [HindIII] FW (CCCAAGCTTGGGCAAGCGGTAAGTGATT TTCCA) and PilE 3′ DN [EcoRI] RV CGGAATTCGGAATTCCATAAAGACCGTCGGGCATCT. The aph3′ coding sequence was amplified by aph3′sp [BamHI] FW CGGGATCCCGGCCGTCTGAAVTGATCAAGAGACAGGATGAGG and aph3′ [HindIII] RV CCCAAGCTTTCAGAAGAACTCGTCAAGAAGGCG. The three fragments were restricted by EcoRI/BamHI, HindIII/EcoRI, and BamHI/HindIII, respectively, and cloned into the EcoRI-restricted pUC19 vector. This construct was then transformed in TOP10 chemically competent Escherichia coli cells (Life Technologies), and vectors from positive white colonies were recovered by Miniprep (Nucleospin II; Macherey-Nagel) before sequencing (GATC Biotech). A sequence-positive clone was conserved and serves as the template for site-directed mutagenesis. The sequence of the pilESA variant fused to the coding sequence of the aph3′ gene was engineered previously (34).
Mutagenesis.
Site-directed mutagenesis was conducted using the GeneTailor site-directed mutagenesis system (Life Technologies) according to the manufacturer’s instructions. The template vector pilESB-Kn, engineered as described above, was methylated and PCR amplified using divergent overlapping primers, one of them containing the mutated site. The PCR product was then transformed in E. coli DH5α-T1R, permitting circularization of the linear PCR product and degradation of the methylated nonmutated template vector. Plasmids from positive clones were recovered by Miniprep (Promega) and sequenced before transformation in N. meningitidis. Three clones from N. meningitidis transformation were isolated, and the pilE locus was sequenced to ensure that the proper mutation was introduced in the sequence.
Exchange of the sequences coding for the D-region domain was performed using PCR mutagenesis and recircularization of the PCR product. Sequences of the PilE D-region of the three strains SA, Z2491, and Z5463 were determined (31, 34). Ten nanograms of template vector pilESB-Kn was amplified using the dedicated primer. Each sequence encoding the D-regions was amplified using the following primers: PilESA D-region Fw (CACCGACGTCAAAGCCGACACCACCGACAACATCAACACCAAGCACCTGCCGTCAACCTGC), PilESA D-region Rv (GCGCTGTCGGTGTCGTCGCGCGCAACCGGCAGTCCGCAGAACCATTTGACCGAACCGTTTTGACG), PilEZ2491 D-region Fw (CTTTGACGTCGTCGTTGGTGGTGGCGGTGTCGTTGCGCTTAACCGGCTGTCCGCAGAACCATTTGACCGAACCGTTTTG), PilEZ2491 D-region Rv (CCGACACCGCCGCCAACGGCAAGCAGATCGACACCAAGCACCTGCCGTCAACCTGCCGCGATGATTCATCTGCCAGCTAAG), and PilEZ5463 D-region Fw (CTGTCGGCGGCGACGTCGGTAGCGGTGTCGTTGCGCTTAACCGGCTGTCCGCAGAACCATTTGACCGAACCGTTTTGACG), and PilEZ5463 D-region Rv (CGGCAACGACAAAATCGACACCAAGCACCTGCCGTCAACCTGCCGCGATGATTCATCTGCCAGCTAAGAG). The PCR products were purified (Promega) and then circularized by ligation using T4 DNA ligase (Fermentas GmbH) and transformed in DH5α. Plasmids from positive clones were recovered by Miniprep (Promega) and sequenced before transformation in N. meningitidis. Three clones from each transformation were kept, and the pilE locus was sequenced.
Immunofluorescence assay.
Infected cells were fixed for 20 min in phosphate-buffered saline (PBS) with 4% PFA and permeabilized for 5 min in PBS containing 0.1% Triton X-100. Cells were then incubated with rabbit polyclonal antiezrin primary antibodies (generously provided by P. Mangeat, CNRS, UMR5539, Montpellier, France) in PBS containing 0.3% bovine serum albumin (BSA). After 3 washes with the same buffer, DAPI (4[prime],6-diamidino-2-phenylindole) was added to Alexa-conjugated secondary antibodies for 1 h at room temperature. After additional washes, coverslips were mounted in Mowiol (Citifluor, Ltd.). Image acquisitions were performed on an inverted microscope equipped with a QIclick digital charge-coupled device (CCD) camera (Qimaging) and Qcapture software (Qimaging).
Quantitative analysis of the bacterial host cell interactions and statistical analysis.
The Tfp-mediated signaling to host cells was estimated by determining the proportion of colonies that efficiently recruit ezrin in a “‘honeycomb shape’” just under the colonies. Ezrin is considered the marker of the meningococcal host cell response. At least 50 colonies were scrutinized per coverslip. Each experiment was performed in duplicate and repeated at least 3 times. Data were examined for significance using Student’s t test (GraphPad Prism 5 software). Results were then expressed as normalized values to ezrin recruitment using the control strain. Correlation Spearman’s tests were performed using GraphPad Prism 5 software. A correlation is considered statistically positive when r is >0.5 and P is <0.05 (confidence interval). In that case the goodness of fit is given (r2). A goodness of fit equal to 1 means that 100% of value X is due to value Y.
Expression and purification of recombinant MBP-pilin proteins.
Recombinant MBP-pilin fusion proteins were produced as described before (13, 24). Briefly, fragments of pilESB, pilESA, and comP lacking the region coding for the first 28 amino acid residues were amplified by PCR and subcloned in pMAL-p2X vector (New England Biolabs). The resulting plasmids, pMAL-pilESB, pMAL-pilESA, and pMAL-comP, contain in frame the malE gene, which encodes the maltose-binding protein (MBP), followed by the factor Xa protease recognition site and the truncated pilin coding regions. These plasmids were transformed in the PAP5198 E. coli strain (generously provided by Olivera Francetic, Institut Pasteur, Paris, France), deleted for the periplasmic enzymes: OmpT, Prt, and DegP. The fusion proteins were purified on amylose resin (New England Biolabs).
Coating of Staphylococcus aureus cells with MBP-pilin fusion proteins and infection.
S. aureus ATCC 25923 cells expressing specific receptors for the Fc domain of IgG immunoglobulins were used for this experiment, as described before (13). Briefly, about 108 bacteria were incubated with the anti-MBP rabbit polyclonal antibody for 20 min at room temperature. Bacteria were then centrifuged at 4,000 rpm for 5 min, washed with warm LB 3 times, and incubated with 2.5 µg of each recombinant fusion protein for 20 min at room temperature. After several washes using DMEM supplemented with 10% fetal calf serum, bacteria were incubated for 30 min with HEK293 cells previously transfected with a plasmid encoding the yellow fluorescent protein (YFP)-tagged human β2-adrenergic receptor and the myc-tagged β-arrestin 2 (40). Cells were then fixed using 4% PFA in PBS and analyzed by immunofluorescence assay for β2-adrenergic receptor–YFP recruitment underneath S. aureus colonies.
Detection of the pilin PilE by immunoblotting.
Whole-cell lysates were washed with ice-cold PBS and lysed in modified radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 25 mM HEPES, 2 mM EDTA, 1% [wt/vol] SDS) buffer containing a protease inhibitor cocktail (Fermentas GmbH). Equal amounts of whole-cell lysates were then boiled and analyzed by SDS-PAGE. After transfer on nitrocellulose (Optitran BA-S 83; Millipore), blots were probed with anti-PilE (SM1) (41) antibody or an anti Rmp4 and a secondary antimouse antibody coupled to horseradish peroxidase (HRP) (Cell signaling). Immunoblots were revealed by enhanced chemiluminescence (ECLplus and Hyperfilm ECL; Thermo Scientific and GE Healthcare Life Sciences, respectively). Expression of PilE was normalized to that of Rmp4, an inner membrane protein used as a loading control.
Aggregation assay.
Aggregation of each strain was assessed in a 24-well plate in DMEM with 10% FBS. Briefly, on the day of infection, a suspension of bacteria from an overnight culture on a GCB agar plate was adjusted to an OD600 of 0.05 and incubated for 1 h at 37°C in a 24-well plate in prewarmed DMEM–10% FBS under agitation. Then the aggregates were mechanically disrupted by being pipetted up and down 10 times. Bacteria were incubated for 1 more hour without agitation, and images of aggregates were acquired on an inverted microscope.
Adhesion assay.
Adhesion of each capsulated strain was assessed (i) on HDMECs under shear stress to recapitulate the in vivo condition and (ii) on FaDu cells under the static condition.
HDMECs were grown on disposable flow chambers (15μ-Slide VI; IBIDI) coated with 5 µg/cm2 of rat tail collagen type I. A total of 107 capsulated PilV− PilT− bacteria were allowed to adhere for 20 min under a shear stress of 0.2 dyne/cm2, and then the chambers were washed several times and fixed in PBS–4% PFA. Bacteria were stained using DAPI, and then 10 images per chamber were acquired on an inverted microscope. Individual bacteria were counted using ImageJ.
FaDu cells were grown in a 24-well plate. A total of 2.106 capsulated bacteria were allowed to adhere for 2 h under static conditions, and then the cells were washed six times with phosphate-buffered saline (PBS) to remove nonadherent bacteria. Cells were incubated for 10 min in PBS–0.2% saponin, harvested, and vortexed in order to dissociate the bacteria. Bacteria were enumerated by plating dilutions on GCB agar. The number of CFU was then compared to that of the control strain of the same experiment. CFU determination was performed at least twice in duplicate.
Homology modeling of the pilin PilE.
Structural models of the different N. meningitidis PilE variants were generated thanks to the high sequence identity (78 to 82%) with the known X-ray structure of N. gonorrhoeae major pilin GC (PDB code 2HI2). The program Modeler from the server ModBase (42) was used to generate the different homology models of the PilE variants, and each of the models shows a reliable ModPipe protein quality score (MPQS): 1.84 < MPQS <1.89. (The MPQS is explained in more detail below.) The same symmetrical operation parameters as the reconstructed GC pilus filament (PDB code 2HIL) were applied to generate the PilE filament model (29). Pictures were generated using the molecular graphics system PyMOL (Schrödinger, LLC). Note that structural modeling and comparison of the C-terminal domain of the pilin variants are not hampered by the apparent flexibility of the N-terminal α-helix domain, presumably induced by the hinge at Pro 22.
The ModPipe protein quality score (MPQS) is a composite score comprising sequence identity to the template, coverage, and the three individual scores E value, z-Dope, and GA341. An MPQS of >1.1 is considered reliable (http://modbase.compbio.ucsf.edu/modbase-cgi/index.cgi).
SUPPLEMENTAL MATERIAL
Structure homology modeling of PilESB and PilESA based on the helical reconstruction of N. gonorrhoeae GC pilus into electron cryomicroscopy map (PDB code 2HI2). The homology models of PilESB and PilESA monomers are indicated with the D-region residues in yellow and the unconserved residues throughout the protein in red. Download
Statistical analysis by Spearman’s correlation test between ezrin recruitment index and the percentage of conserved signaling residues. Download
Phenotypes of each strain.
ACKNOWLEDGMENTS
We are grateful to Alain Charbit, Emmanuelle Bille, and Guillain Mikaty for helpful discussion. We thank Anne Cavau for technical support. We thank Anne Jamet for technical advice on flow experiments.
This work was supported by ANR grant ANR-11-JSV3-0002 and the grant program EMERGENCE from La Mairie de Paris.
Footnotes
Citation Miller F, Phan G, Brissac T, Bouchiat C, Lioux G, Nassif X, Coureuil M. 2014. The hypervariable region of meningococcal major pilin PilE controls the host cell response via antigenic variation. mBio 5(1):e01024-13. doi:10.1128/mBio.01024-13.
REFERENCES
- 1. Carbonnelle E, Hill DJ, Morand P, Griffiths NJ, Bourdoulous S, Murillo I, Nassif X, Virji M. 2009. Meningococcal interactions with the host. Vaccine 27(Suppl 2):B78–B89. 10.1016/j.vaccine.2009.04.069 [DOI] [PubMed] [Google Scholar]
- 2. Virji M, Makepeace K, Ferguson DJ, Achtman M, Moxon ER. 1993. Meningococcal Opa and Opc proteins: their role in colonization and invasion of human epithelial and endothelial cells. Mol. Microbiol. 10:499–510. 10.1111/j.1365-2958.1993.tb00922.x [DOI] [PubMed] [Google Scholar]
- 3. de Vries FP, van der Ende A, van Putten JP, Dankert J. 1996. Invasion of primary nasopharyngeal epithelial cells by Neisseria meningitidis is controlled by phase variation of multiple surface antigens. Infect. Immun. 64:2998–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Virji M, Evans D, Hadfield A, Grunert F, Teixeira AM, Watt SM. 1999. Critical determinants of host receptor targeting by Neisseria meningitidis and Neisseria gonorrhoeae: identification of Opa adhesiotopes on the N-domain of CD66 molecules. Mol. Microbiol. 34:538–551. 10.1046/j.1365-2958.1999.01620.x [DOI] [PubMed] [Google Scholar]
- 5. Serruto D, Adu-Bobie J, Scarselli M, Veggi D, Pizza M, Rappuoli R, Aricò B. 2003. Neisseria meningitidis App, a new adhesin with autocatalytic serine protease activity. Mol. Microbiol. 48:323–334. 10.1046/j.1365-2958.2003.03420.x [DOI] [PubMed] [Google Scholar]
- 6. Capecchi B, Adu-Bobie J, Di Marcello F, Ciucchi L, Masignani V, Taddei A, Rappuoli R, Pizza M, Aricò B. 2005. Neisseria meningitidis NadA is a new invasin which promotes bacterial adhesion to and penetration into human epithelial cells. Mol. Microbiol. 55:687–698 [DOI] [PubMed] [Google Scholar]
- 7. Rechner C, Kühlewein C, Müller A, Schild H, Rudel T. 2007. Host glycoprotein Gp96 and scavenger receptor SREC interact with PorB of disseminating Neisseria gonorrhoeae in an epithelial invasion pathway. Cell Host Microbe 2:393–403. 10.1016/j.chom.2007.11.002 [DOI] [PubMed] [Google Scholar]
- 8. Join-Lambert O, Lecuyer H, Miller F, Lelievre L, Jamet A, Furio L, Schmitt A, Pelissier P, Fraitag S, Coureuil M, Nassif X. 2013. Meningococcal interaction to microvasculature triggers the tissular lesions of Purpura fulminans. J. Infect. Dis. 208:1590–1597 [DOI] [PubMed] [Google Scholar]
- 9. Melican K, Michea Veloso P, Martin T, Bruneval P, Dumenil G. 2013. Adhesion of Neisseria meningitidis to dermal vessels leads to local vascular damage and purpura in a humanized mouse model. PLoS Pathog. 9:e1003139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Merz AJ, Enns CA, So M. 1999. Type IV pili of pathogenic Neisseriae elicit cortical plaque formation in epithelial cells. Mol. Microbiol. 32:1316–1332. 10.1046/j.1365-2958.1999.01459.x [DOI] [PubMed] [Google Scholar]
- 11. Mikaty G, Soyer M, Mairey E, Henry N, Dyer D, Forest KT, Morand P, Guadagnini S, Prévost MC, Nassif X, Duménil G. 2009. Extracellular bacterial pathogen induces host cell surface reorganization to resist shear stress. PLoS Pathog. 5:e1000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coureuil M, Mikaty G, Miller F, Lécuyer H, Bernard C, Bourdoulous S, Duménil G, Mège RM, Weksler BB, Romero IA, Couraud PO, Nassif X. 2009. Meningococcal type IV pili recruit the polarity complex to cross the brain endothelium. Science 325:83–87. 10.1126/science.1173196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coureuil M, Lécuyer H, Scott MG, Boularan C, Enslen H, Soyer M, Mikaty G, Bourdoulous S, Nassif X, Marullo S. 2010. Meningococcus hijacks a β2-adrenoceptor/beta-arrestin pathway to cross brain microvasculature endothelium. Cell 143:1149–1160. 10.1016/j.cell.2010.11.035 [DOI] [PubMed] [Google Scholar]
- 14. Eugène E, Hoffmann I, Pujol C, Couraud PO, Bourdoulous S, Nassif X. 2002. Microvilli-like structures are associated with the internalization of virulent capsulated Neisseria meningitidis into vascular endothelial cells. J. Cell Sci. 115:1231–1241 [DOI] [PubMed] [Google Scholar]
- 15. Giltner CL, Nguyen Y, Burrows LL. 2012. Type IV pilin proteins: versatile molecular modules. Microbiol. Mol. Biol. Rev. 76:740–772. 10.1128/MMBR.00035-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sheth HB, Lee KK, Wong WY, Srivastava G, Hindsgaul O, Hodges RS, Paranchych W, Irvin RT. 1994. The pili of Pseudomonas aeruginosa strains PAK and PAO bind specifically to the carbohydrate sequence beta GalNAc(1–4)beta Gal found in glycosphingolipids asialo-GM1 and asialo-GM2. Mol. Microbiol. 11:715–723. 10.1111/j.1365-2958.1994.tb00349.x [DOI] [PubMed] [Google Scholar]
- 17. Giltner CL, van Schaik EJ, Audette GF, Kao D, Hodges RS, Hassett DJ, Irvin RT. 2006. The Pseudomonas aeruginosa type IV pilin receptor binding domain functions as an adhesin for both biotic and abiotic surfaces. Mol. Microbiol. 59:1083–1096. 10.1111/j.1365-2958.2005.05002.x [DOI] [PubMed] [Google Scholar]
- 18. Kirn TJ, Lafferty MJ, Sandoe CM, Taylor RK. 2000. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol. Microbiol. 35:896–910. 10.1046/j.1365-2958.2000.01764.x [DOI] [PubMed] [Google Scholar]
- 19. Krebs SJ, Taylor RK. 2011. Protection and attachment of Vibrio cholerae mediated by the toxin-coregulated pilus in the infant mouse model. J. Bacteriol. 193:5260–5270. 10.1128/JB.00378-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baldini MM, Kaper JB, Levine MM, Candy DC, Moon HW. 1983. Plasmid-mediated adhesion in enteropathogenic Escherichia coli. J. Pediatr. Gastroenterol. Nutr. 2:534–538. 10.1097/00005176-198302030-00023 [DOI] [PubMed] [Google Scholar]
- 21. Humphries RM, Donnenberg MS, Strecker J, Kitova E, Klassen JS, Cui L, Griener TP, Mulvey GL, Armstrong GD. 2009. From alpha to beta: identification of amino acids required for the N-acetyllactosamine-specific lectin-like activity of bundlin. Mol. Microbiol. 72:859–868. 10.1111/j.1365-2958.2009.06679.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Merz AJ, So M. 1997. Attachment of piliated, Opa- and Opc- gonococci and meningococci to epithelial cells elicits cortical actin rearrangements and clustering of tyrosine-phosphorylated proteins. Infect. Immun. 65:4341–4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Winther-Larsen HC, Hegge FT, Wolfgang M, Hayes SF, van Putten JP, Koomey M. 2001. Neisseria gonorrhoeae PilV, a type IV pilus-associated protein essential to human epithelial cell adherence. Proc. Natl. Acad. Sci. U. S. A. 98:15276–15281. 10.1073/pnas.261574998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hélaine S, Carbonnelle E, Prouvensier L, Beretti JL, Nassif X, Pelicic V. 2005. PilX, a pilus-associated protein essential for bacterial aggregation, is a key to pilus-facilitated attachment of Neisseria meningitidis to human cells. Mol. Microbiol. 55:65–77 [DOI] [PubMed] [Google Scholar]
- 25. Wolfgang M, van Putten JP, Hayes SF, Koomey M. 1999. The comP locus of Neisseria gonorrhoeae encodes a type IV prepilin that is dispensable for pilus biogenesis but essential for natural transformation. Mol. Microbiol. 31:1345–1357. 10.1046/j.1365-2958.1999.01269.x [DOI] [PubMed] [Google Scholar]
- 26. Gibbs CP, Reimann BY, Schultz E, Kaufmann A, Haas R, Meyer TF. 1989. Reassortment of pilin genes in Neisseria gonorrhoeae occurs by two distinct mechanisms. Nature 338:651–652. 10.1038/338651a0 [DOI] [PubMed] [Google Scholar]
- 27. Cahoon LA, Seifert HS. 2009. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science 325:764–767. 10.1126/science.1175653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Andrews TD, Gojobori T. 2004. Strong positive selection and recombination drive the antigenic variation of the PilE protein of the human pathogen Neisseria meningitidis. Genetics 166:25–32. 10.1534/genetics.166.1.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. 2006. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell 23:651–662. 10.1016/j.molcel.2006.07.004 [DOI] [PubMed] [Google Scholar]
- 30. Cehovin A, Winterbotham M, Lucidarme J, Borrow R, Tang CM, Exley RM, Pelicic V. 2010. Sequence conservation of pilus subunits in Neisseria meningitidis. Vaccine 28:4817–4826. 10.1016/j.vaccine.2010.04.065 [DOI] [PubMed] [Google Scholar]
- 31. Omer H, Rose G, Jolley KA, Frapy E, Zahar JR, Maiden MC, Bentley SD, Tinsley CR, Nassif X, Bille E. 2011. Genotypic and phenotypic modifications of Neisseria meningitidis after an accidental human passage. PLoS One 6:e17145. 10.1371/journal.pone.0017145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lécuyer H, Nassif X, Coureuil M. 2012. Two strikingly different signaling pathways are induced by meningococcal type IV pili on endothelial and epithelial cells. Infect. Immun. 80:175–186. 10.1128/IAI.05837-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brissac T, Mikaty G, Duménil G, Coureuil M, Nassif X. 2012. The meningococcal minor pilin PilX is responsible for type IV pilus conformational changes associated with signaling to endothelial cells. Infect. Immun. 80:3297–3306. 10.1128/IAI.00369-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nassif X, Lowy J, Stenberg P, O’Gaora P, Ganji A, So M. 1993. Antigenic variation of pilin regulates adhesion of Neisseria meningitidis to human epithelial cells. Mol. Microbiol. 8:719–725. 10.1111/j.1365-2958.1993.tb01615.x [DOI] [PubMed] [Google Scholar]
- 35. Biais N, Higashi DL, Brujic J, So M, Sheetz MP. 2010. Force-dependent polymorphism in type IV pili reveals hidden epitopes. Proc. Natl. Acad. Sci. U. S. A. 107:11358–11363. 10.1073/pnas.0911328107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Balakrishna AM, Saxena AM, Mok HY, Swaminathan K. 2009. Structural basis of typhoid: Salmonella typhi type IVb pilin (PilS) and cystic fibrosis transmembrane conductance regulator interaction. Proteins 77:253–261. 10.1002/prot.22500 [DOI] [PubMed] [Google Scholar]
- 37. Baker JL, Biais N, Tama F. 2013. Steered molecular dynamics simulations of a type IV pilus probe initial stages of a force-induced conformational transition. PLoS Comput. Biol. 9:e1003032. 10.1371/journal.pcbi.1003032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Achtman M, Neibert M, Crowe BA, Strittmatter W, Kusecek B, Weyse E, Walsh MJ, Slawig B, Morelli G, Moll A. 1988. Purification and characterization of eight class 5 outer membrane protein variants from a clone of Neisseria meningitidis serogroup A. J. Exp. Med. 168:507–525. 10.1084/jem.168.2.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pujol C, Eugène E, Marceau M, Nassif X. 1999. The meningococcal PilT protein is required for induction of intimate attachment to epithelial cells following pilus-mediated adhesion. Proc. Natl. Acad. Sci. U. S. A. 96:4017–4022. 10.1073/pnas.96.7.4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Angers S, Salahpour A, Joly E, Hilairet S, Chelsky D, Dennis M, Bouvier M. 2000. Detection of beta 2-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET). Proc. Natl. Acad. Sci. U. S. A. 97:3684–3689. 10.1073/pnas.97.7.3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Virji M, Heckels JE, Watt PJ. 1983. Monoclonal antibodies to gonococcal pili: studies on antigenic determinants on pili from variants of strain P9. J. Gen. Microbiol. 129:1965–1973 [DOI] [PubMed] [Google Scholar]
- 42. Pieper U, Webb BM, Barkan DT, Schneidman-Duhovny D, Schlessinger A, Braberg H, Yang Z, Meng EC, Pettersen EF, Huang CC, Datta RS, Sampathkumar P, Madhusudhan MS, Sjölander K, Ferrin TE, Burley SK, Sali A. 2011. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 39:D465–D474. 10.1093/nar/gkq1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structure homology modeling of PilESB and PilESA based on the helical reconstruction of N. gonorrhoeae GC pilus into electron cryomicroscopy map (PDB code 2HI2). The homology models of PilESB and PilESA monomers are indicated with the D-region residues in yellow and the unconserved residues throughout the protein in red. Download
Statistical analysis by Spearman’s correlation test between ezrin recruitment index and the percentage of conserved signaling residues. Download
Phenotypes of each strain.