ABSTRACT
To establish a replicative niche during its infectious cycle between the intestinal lumen and tissue, the enteric pathogen Salmonella enterica serovar Typhimurium requires numerous virulence genes, including genes for two type III secretion systems (T3SS) and their cognate effectors. To better understand the host-pathogen relationship, including early infection dynamics and induction kinetics of the bacterial virulence program in the context of a natural host, we monitored the subcellular localization and temporal expression of T3SS-1 and T3SS-2 using fluorescent single-cell reporters in a bovine, ligated ileal loop model of infection. We observed that the majority of bacteria at 2 h postinfection are flagellated, express T3SS-1 but not T3SS-2, and are associated with the epithelium or with extruding enterocytes. In epithelial cells, S. Typhimurium cells were surrounded by intact vacuolar membranes or present within membrane-compromised vacuoles that typically contained numerous vesicular structures. By 8 h postinfection, T3SS-2-expressing bacteria were detected in the lamina propria and in the underlying mucosa, while T3SS-1-expressing bacteria were in the lumen. Our work identifies for the first time the temporal and spatial regulation of T3SS-1 and -2 expression during an enteric infection in a natural host and provides further support for the concept of cytosolic S. Typhimurium in extruding epithelium as a mechanism for reseeding the lumen.
IMPORTANCE
The pathogenic bacterium Salmonella enterica serovar Typhimurium invades and persists within host cells using distinct sets of virulence genes. Genes from Salmonella pathogenicity island 1 (SPI-1) are used to initiate contact and facilitate uptake into nonphagocytic host cells, while genes within SPI-2 allow the pathogen to colonize host cells. While many studies have identified bacterial virulence determinants in animal models of infection, very few have focused on virulence gene expression at the single-cell level during an in vivo infection. To better understand when and where bacterial virulence factors are expressed during an acute enteric infection of a natural host, we infected bovine jejunal-ileal loops with S. Typhimurium cells harboring fluorescent transcriptional reporters for SPI-1 and -2 (PinvF and PssaG, respectively). After a prescribed time of infection, tissue and luminal fluid were collected and analyzed by microscopy. During early infection (≤2 h), bacteria within both intact and compromised membrane-bound vacuoles were observed within the epithelium, with the majority expressing SPI-1. As the infection progressed, S. Typhimurium displayed differential expression of the SPI-1 and SPI-2 regulons, with the majority of tissue-associated bacteria expressing SPI-2 and the majority of lumen-associated bacteria expressing SPI-1. This underscores the finding that Salmonella virulence gene expression changes as the pathogen transitions from one anatomical location to the next.
INTRODUCTION
The intestinal mucosa is located at an important crossroads of dynamic interactions between the intestinal microbiota, vital absorptive cells, transient as well as resident immune cells, and pathogenic organisms. Intestinal villi extend into the luminal milieu and provide a selective barrier against luminal contents, remove injured or aged epithelial cells via controlled sloughing or extrusion, educate naive immune cells to intestinal symbiotic bacteria, and monitor the local environment for pathogenic threats (1, 2). Appropriate immunologic and cellular responses to the autochthonous intestinal microbial populations, as well as general luminal conditions, are important for the health of the organism.
Disruption of the autochthonous population plays an important role in the establishment and propagation of infection for several pathogens of the alimentary tract, of which Salmonella enterica serovar Typhimurium has received significant attention (3–7). This member of the Enterobacteriaceae family is a food-borne pathogen that elicits clinically and pathologically similar disease outcomes in humans and cattle (8–10). Animal models for this localized gastroenteric infection include neonatal bovines and streptomycin-treated mice (11, 12). In the bovine model, bacterial invasion of intestinal tissue occurs as early as 15 min after exposure and typically affects phagocytic and nonphagocytic cells (13). Ileal Peyer’s patch phagocytes, likely tissue-associated dendritic cells and M cells, capture and deliver invading S. Typhimurium cells to the local mesenteric lymph node to educate and recruit T cells for return to the site of infection (14, 15). The initial hours of acute Salmonella infection in humans and cattle are similarly characterized by polymorphonuclear cell (PMN) infiltration into the lamina propria and then PMN efflux and transit through the intestinal epithelium into the lumen, luminal fluid accumulation, epithelial cell shedding, and villus blunting (16, 17). Similar features of mucosal damage have also been described for S. Typhimurium infection of rabbits and rhesus monkeys (18, 19).
S. Typhimurium employs two type III secretion systems (T3SS) to mediate their interactions with host cells (20, 21). T3SS-1 and T3SS-2 are encoded in Salmonella pathogenicity islands 1 and 2 (SPI-1 and SPI-2, respectively). Genetic deletion of SPI-1 or SPI-2 can abrogate the virulence and ability of Salmonella to invade, colonize, or replicate within host cells (10, 11, 22, 23). The SPI-1 and SPI-2 regulons are induced under different environmental conditions. Expression of the SPI-1 regulon is controlled by numerous proteins, including invF and hilA, and induced extracellularly (24), consistent with its role in invasion of nonphagocytic host cells, such as enterocytes. Proteins encoded by genes within the SPI-1 regulon include structural components of the T3SS-1 apparatus and several type III effectors that modulate macropinocytosis at the plasma membrane, trafficking of the nascent Salmonella-containing vacuole (SCV), and intracellular replication (25). Following invasion into nonphagocytic cells, S. Typhimurium down-regulates SPI-1 and induces the SPI-2 regulon (26). SPI-2-encoded T3SS-2 translocates effector proteins that are required for maturation and maintenance of the SCV (25). Although the SCV has been considered the predominant site of intracellular replication for Salmonella, recent studies have identified a distinct population of S. Typhimurium cells that hyper-replicate in the cytosol of epithelial cells (27, 28). In contrast to the SPI-2-induced vacuolar population, these cytosolic bacteria are induced for SPI-1 and flagellated. In a polarized epithelial cell model, these “invasion-primed” intracellular bacteria are released into the extracellular milieu when the host cell is extruded from the monolayer. In murine gall bladders, extruding epithelial cells were shown to also contain cytosolic invasion-primed S. Typhimurium cells (27). In addition, enterocytes containing large numbers of S. Typhimurium cells have been observed within rabbit ileal mucosa (18) and chick ileocecal mucosa (29). It has been proposed that this bacterial population may play an important role in cell-to-cell transmission and/or dissemination in vivo (27).
Although it is clear that SPI-1 and, to a lesser extent, SPI-2 are required for the induction of pathological changes during acute enteric infection (10, 30), the timing and location of bacterial gene expression in vivo have received little attention and are poorly understood. Here we have addressed this question using the well-established neonatal bovine ileal loop model. Calves were infected with S. Typhimurium harboring transcriptional fusions to representative genes from SPI-1 (invF) or SPI-2 (ssaG) for various times before tissue was harvested for transmission electron and confocal microscopy. We report the presence of both vacuole membrane-bound and -compromised bacteria within the epithelium and dissemination of invasion-primed S. Typhimurium by epithelial extrusion, particularly early during infection. In addition, our single-cell expression analysis revealed a distinct temporal and spatial segregation of SPI-1- and SPI-2-positive bacteria during intestinal colonization.
RESULTS AND DISCUSSION
S. Typhimurium interactions with the epithelium during early infection.
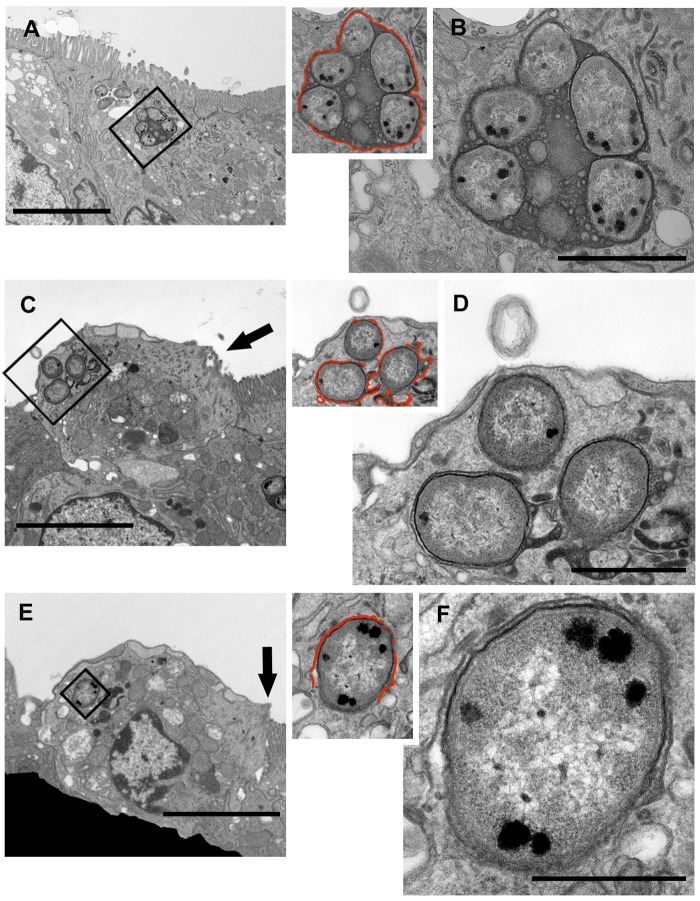
To observe interactions between S. Typhimurium and the host epithelium during early infection, bovine ileal loops were harvested at 2 h postinoculation (p.i.) and processed for transmission electron microscopy (TEM). Analysis of infected tissue revealed numerous examples of S. Typhimurium cells colonizing enterocytes, goblet cells, and other cells within the epithelium (Fig. 1; see also Fig. S1 in the supplemental material). In most instances, infected host cells remained part of the epithelium; however, examples of epithelial cells with remnant microvilli, seemingly undergoing extrusion and containing S. Typhimurium cells, were also observed (Fig. 1C and E, arrows). We further noted that while many intracellular bacteria were enclosed by a vacuolar membrane (Fig. 1B and S2A and B), others were not (Fig. 1C to F and S2C to E). Instead, the integrity of the vacuole around these bacteria appeared to be compromised (defined as >25% of the bacterial surface not being associated with a vacuolar membrane), affording access to the host cytosol. Approximately 13% of S. Typhimurium cells were observed in such compromised vacuoles (n = 173 bacteria). By electron microscopy, we were unable to identify a bovine epithelial cell laden with cytosolic S. Typhimurium cells, as has been described for chick ileocecal mucosa (31) and polarized human intestinal epithelial monolayers (27). However, our TEM analysis does support previous observations of the presence of S. Typhimurium in distinct intracellular compartments in epithelial cells and cellular extrusion acting as a mechanism for bacterial dissemination into the lumen (18, 27). TEM analysis of infected tissue samples from 8 h p.i. revealed S. Typhimurium bacteria located in the lamina propria within intact vacuoles, compromised vacuoles, or vacuoles apparently free of any discernible host membrane (Fig. S3).
FIG 1 .
In enterocytes, S. Typhimurium cells are surrounded by intact and compromised vacuolar membranes. Representative transmission electron micrograph of infected epithelial cells at 2 h p.i. The vacuolar membrane is highlighted with a red line in the inset. (A and B) Series of images demonstrating S. Typhimurium cells enclosed within an intact membrane (C to F). Infected epithelial cells likely undergoing cellular extrusion. At least one bacterium in each cell has a compromised vacuolar membrane (indicated by discontinuous red lines in the insets). (C and E) Arrows indicate remnant microvilli. Bars, 5 µm (A, C, and E), 1 µm (B, D, and G), and 0.5 µm (F).
Further TEM analysis of intracellular S. Typhimurium within intact and compromised vacuoles from 2-h-p.i. samples revealed the presence of numerous vesicular bodies within the SCV lumen (Fig. S1). These vesicles ranged in size from <50 nm to >200 nm. Intracellular and extracellular Gram-negative bacteria are known to secrete spherical vesicles, called outer membrane vesicles (OMV), that are 50 to 250 nm in diameter and often have an electron-dense luminal content by electron microscopy, consistent with what we report here (32). Budding or recently formed OMV originating from the pathogen were found associated with all intracellular S. Typhimurium cells analyzed (Fig. S1, arrowheads), complementing the findings from a previous in vivo study of a human Salmonella isolate in chicken ileum (33). OMV were typically found free within the SCV lumen (Fig. S1J) or adjacent to or apparently spanning the SCV membrane (Fig. S1K, arrowhead, and S2C, arrow). Larger, more-electron-lucent membrane structures were also noted within the SCV (Fig. S1J and K and S2E, chevrons), sometimes apparently fusing with or blebbing from the vacuolar membrane (indicated in Fig. S2E, chevron). It is unclear if these larger vesicles originate from the pathogen or the host.
SPI-1 and -2 expression during acute infection.
To further our understanding of bacterial virulence gene expression during acute infection in vivo, we used S. Typhimurium harboring reporter plasmids for either the SPI-1 or the SPI-2 regulon in the bovine ileal loop model. The plasmids encode a destabilized green fluorescent protein (GFP) variant, GFP[LVA] (half-time, ~40 min), that is under the control of gene promoters from either SPI-1 (PinvF) or SPI-2 (PssaG). InvF is a transcriptional activator of SPI-1 (34), whereas SsaG is a structural component of T3SS-2 (35). In broth cultures, wild-type bacteria harboring pMPMA3∆Plac-PinvF-GFP[LVA] exhibited green fluorescence when grown under SPI-1-inducing conditions (Luria-Bertani [LB]–Miller broth, 3.5 h) but not under SPI-2-inducing conditions (low phosphate magnesium [LPM], pH 5.8, 14 h) (Fig. S4A). Fluorescence was not observed under SPI-1-inducing conditions upon deletion of hilA, a master regulator of the SPI-1 regulon (36). Conversely, bacteria harboring pMPMA3∆Plac-PssaG-GFP[LVA] were fluorescent only when grown under SPI-2-inducing conditions, and this was dependent on ssrB (Fig. S4), part of a two-component regulatory system that is absolutely required for expression of the SPI-2 regulon (37). Upon examination of infected ileal loop tissues, we found that the intrinsic fluorescence of GFP[LVA] was often too weak to detect by confocal microscopy. To circumvent this, we used rabbit polyclonal anti-GFP antibodies to amplify the fluorescence signal associated with individual bacteria. Mammalian tissue culture invasion assays were used to assess the expression kinetics and frequencies of GFP-positive bacteria and to determine that intrinsic fluorescence and antibody amplification of the GFP[LVA] signal were comparable to those of methods of reporter activity quantification (Fig. S4B). Furthermore, at both 2 h and 8 h p.i. in the in vivo model, luminal fluid accumulation and bacterial burdens in tissue, mucus, and fluid samples from loops infected with S. Typhimurium harboring the GFP reporter plasmids were consistent with those of wild type (WT)-infected loops (Fig. S5). Additionally, plasmids were retained throughout the duration of an 8-h infection (Fig. S5). Collectively, these experiments validated the use of pMPMA3∆Plac-PinvF-GFP[LVA] and pMPMA3∆Plac-PssaG-GFP[LVA] as accurate transcriptional reporters of the SPI-1 and SPI-2 regulons, respectively, in vitro.
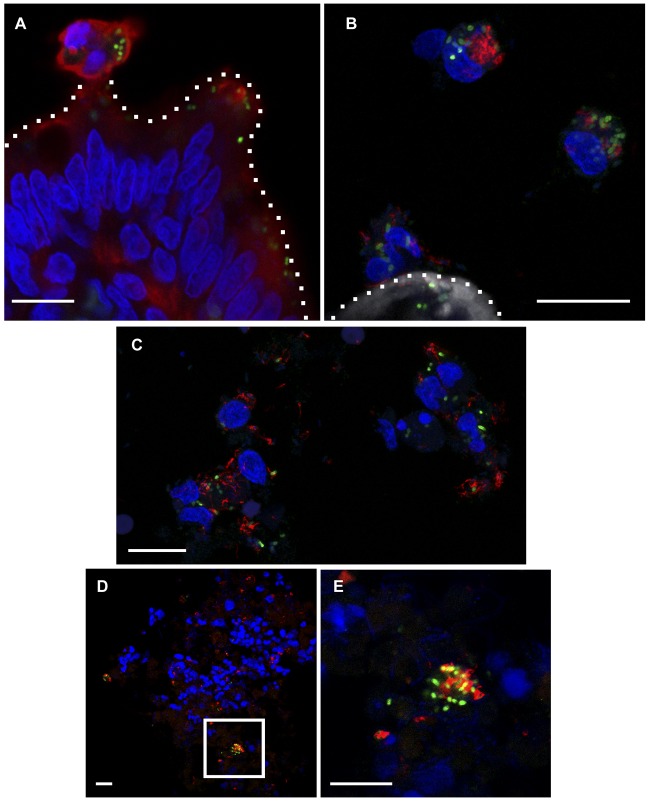
Confocal microscopy analysis of villus tips and cells immediately adjacent to tissue revealed numerous instances of extruding or sloughed cells positive for staining by cytokeratin 8, an intermediate filament protein found in epithelial cells containing PinvF-positive S. Typhimurium cells at 2 h (Fig. 2A). In bovine cells found adjacent to villus tips, many of the bacteria also immunostained for flagellin (FliC) (Fig. 2B). Salmonella has previously been associated with epithelial cell extrusion in polarized monolayers, rabbit ileal loops, and mouse gall bladders (18, 27). Such extrusion has been postulated as a dissemination mechanism, since bacteria within these dying cells are induced for SPI-1 and flagellated (18). Our observations provide further evidence for the presence SPI-1-induced, flagellated S. Typhimurium bacteria associating with host cells sloughing from villus tips in vivo. Infected cells adjacent to the villus often contained multiple regions positively stained for DNA, suggesting either nuclear degradation of a single cell or multiple cells clumping together (Fig. 2A and B). Extruding epithelial cells or near-tissue PinvF-positive bacteria were rarely observed at 8 h p.i., likely due to the significant villus blunting and immune responses seen at this later time point.
FIG 2 .
PinvF-positive, flagellated S. Typhimurium bacteria are associated with host cells proximal to villus tips and in the lumen. Ligated jejunal-ileal loops were infected with S. Typhimurium cells harboring PinvF-GFP[LVA] for 2 h. (A) Confocal image showing an extruded epithelial cell (cytokeratin 8 positive) containing numerous PinvF-GFP-positive bacteria. Green, anti-GFP; red, anti-cytokeratin 8; blue, DNA. (B) Confocal image showing several bovine cells adjacent to a villus tip containing a significant burden of flagellin-positive and PinvF-GFP positive Salmonella cells. Green, anti-GFP; red, anti-FliC; shades of gray, phalloidin for actin; blue, DNA. For A and B, dotted lines indicate the apical surface of the villus tip. (C) Confocal image of cells adjacent to villus tips revealing flagellin-positive and PinvF-GFP-positive bacteria intimately associated with bovine cells sloughed from the tissue. Green, anti-GFP; red, anti-FliC; blue, DNA. (D) Confocal image of luminal fluid content showing PinvF-GFP-positive, flagellated Salmonella bacteria found in clusters within a heterogeneous milieu of eukaryotic and prokaryotic cells. Green, anti-GFP; red, anti-FliC; blue, DNA. (E) Enlarged inset from panel D. Bars, 10 µm.
Immunostaining of the luminal fluid samples for GFP and FliC and fluorescent staining of DNA with DAPI (4′,6-diamidino-2-phenylindole) revealed a heterogeneous population of PinvF-positive and -negative S. Typhimurium cells, other bacterial species, and host cells (Fig. 2D and E). Interestingly, PinvF-positive S. Typhimurium appeared to aggregate in this environment (Fig. 2E) and also immunostained for flagellin (Fig. 2B and C). Stecher et al. used a reporter plasmid expressing GFPmut2 under the control of the fliC promoter in a streptomycin-treated mouse model of colitis and showed there a gradation of fliC expression by S. Typhimurium in the gut at 1 day p.i. (38). Approximately half of the bacteria in the cecal lumen were GFP positive, whereas almost all were GFP positive (92%) when in close proximity to the cecal epithelium. In an independent study of flagellin expression during systemic salmonellosis, Cummings et al. used a fliC::gfp mut3 reporter to show that ~60% of S. Typhimurium cells in the Peyer’s patches transcribed fliC at 7 days p.i. but that none in the mesenteric lymph nodes or spleen were GFP positive (39). Our finding that luminal S. Typhimurium cells are flagellated supports the concept of an anatomical restriction of FliC expression to the gut.
We did not detect any PssaG-positive bacteria in luminal fluid samples at either 2 h or 8 h p.i. (Fig. S6). This is in contrast to the findings of a recent report that identified a very small population (1.34% of the total) of SPI-2-induced S. Typhimurium cells in the lumen of the cecum during a mouse model of colitis. Approximately two-thirds of these bacteria were extracellular; the remaining cells were within luminal CD18+ neutrophils (40). Using recombinase-based in vivo expression technology (RIVET), another group has also reported that SPI-2 is expressed prior to bacterial penetration of the epithelial layer in a murine model of salmonellosis (41). The considerable differences in pathology and host response between the bovine and mouse models of salmonellosis, in addition to infection parameters and detection methods, may account for the discrepancy between these observations.
Virulence gene expression within gut tissue.
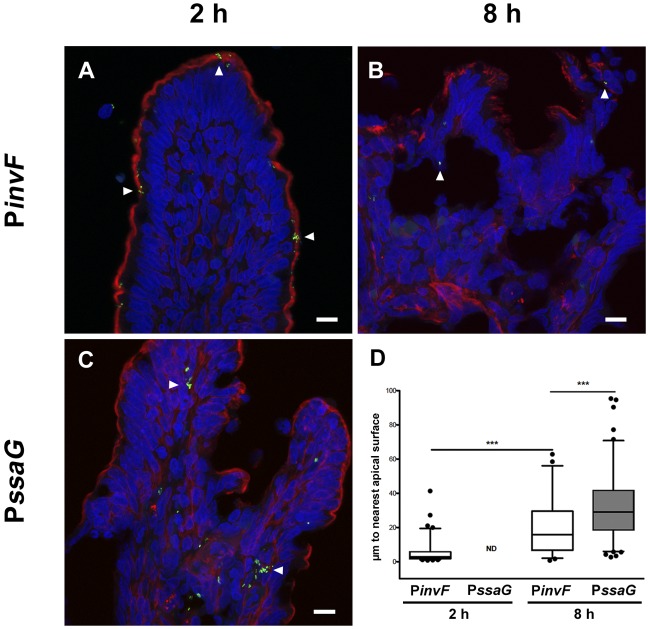
During enteric infections, many different cell types are targets for S. Typhimurium, including enterocytes, goblet cells, macrophages, and dendritic cells (42, 43). However, it is unclear when these particular host cell-bacterium interactions occur during the course of acute infection. To monitor the physical location of PinvF- and PssaG-induced S. Typhimurium cells, GFP-positive bacteria were quantified in two ways by confocal microscopy: (i) by determining the distance of tissue-associated bacteria from the nearest apical surface, denoted by phalloidin staining (Fig. 3), and (ii) by categorizing the subtissue localization of GFP-positive bacteria (Fig. 4).
FIG 3 .
Temporal and spatial distribution of PinvF- and PssaG-positive S. Typhimurium cells. Ligated jejunal-ileal loops were infected with S. Typhimurium cells harboring either PinvF-GFP[LVA] or PssaG-GFP[LVA]. (A to C) Representative confocal images of tissue stained for GFP (green), phalloidin (for actin, red), and DNA (blue), with GFP-positive bacteria indicated by white arrowheads. (A) PinvF-GFP[LVA] infection at 2 h p.i.; (B) PinvF-GFP[LVA] infection at 8 h p.i.; (C) PssaG-GFP[LVA] infection at 8 h p.i.; (D) box-and-whisker plot of tissue-associated PinvF- or PssaG-GFP[LVA]-positive S. Typhimurium cells and their distance from the nearest apical surface (assessed from phalloidin staining). Whiskers represent the 5-to-95% confidence interval. • symbols represent outlier data points. ***, P < 0.001. ND, not determined. Numbers of GFP-positive bacteria assessed for distance to the luminal surface were as follows: 82 bacteria at 2 h for PinvF-GFP[LVA]-infected cells, 50 bacteria at 8 h for PinvF-GFP[LVA]-infected cells, and 199 bacteria at 8 h for PssaG-GFP[LVA]-infected cells (values are from samples from at least three animals).
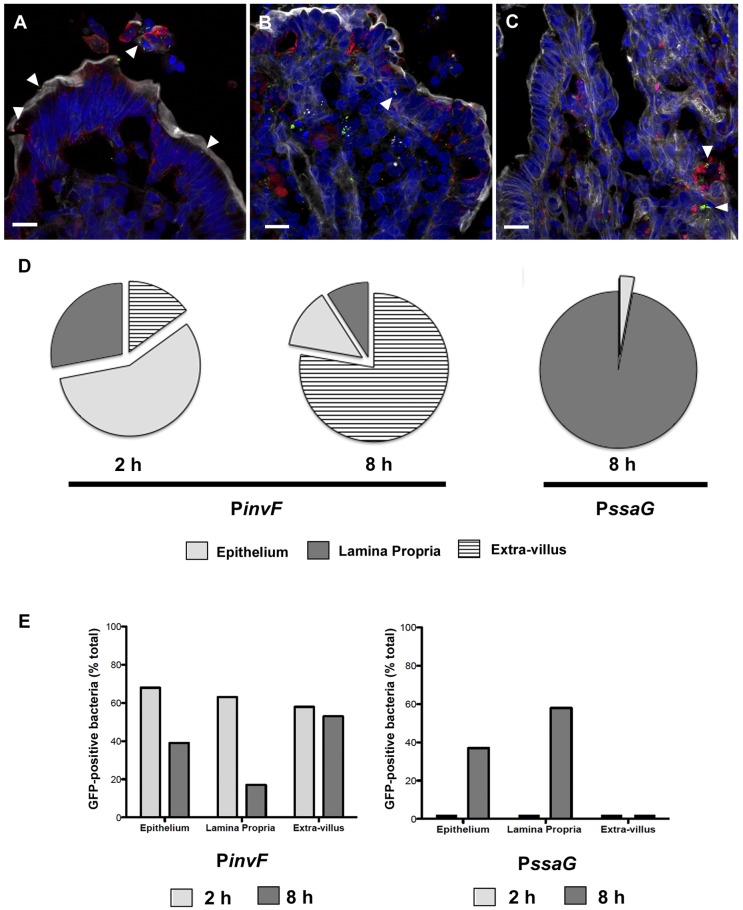
FIG 4 .
PinvF- and PssaG-positive S. Typhimurium bacteria predominantly associate with different host cell types. Ligated jejunal-ileal loops were infected with S. Typhimurium cells harboring either PinvF-GFP[LVA] or PssaG-GFP[LVA] and processed for confocal microscopy. (A) Representative image of PinvF-GFP[LVA] infection at 2 h p.i. (B and C) PssaG-GFP[LVA] infection at 8 h p.i. (A and B) Green, anti-GFP; red, anti-cytokeratin 8; shades of gray, phalloidin for actin; blue, DNA. (C) Green, anti-GFP; red, anti-HLA-DRα; shades of gray, phalloidin for actin; blue, DNA. Bars = 10 µm. Arrowheads indicate GFP-positive S. Typhimurium bacteria within epithelial cells (A and B) or phagocytes (C). (D) The locations of PinvF- and PssaG-positive bacteria were scored as in the epithelium (light gray), extravillus (hatched), or lamina propria (dark gray). Percentages of bacteria in the extravillus/epithelium/lamina propria were as follows: 15/57/28% for PinvF-GFP[LVA]-infected specimens at 2 h (n = 439), 77/13/9% for PinvF-GFP[LVA]-infected specimens at 8 h (n = 298), and 0/3/97% for PssaG-GFP[LVA]-infected specimens at 8 h (n = 308). (E) Proportions of total bacteria positive for either PinvF- or PssaG-GFP in the epithelium, lamina propria, or extravillous space at 2 h (light gray) or 8 h p.i. (dark gray). Percentages of PinvF-GFP-positive bacteria at 2 h/8 h were as follows: 68/39% in the epithelium, 63/17% in the lamina propria, and 58/53% in the extravillous space. Percentages of PssaG-GFP-positive bacteria at 2 h/8 h were as follows: 0/37% in the epithelium, 0/58% in the lamina propria, and 0/0% in the extravillous space.
At 2 h p.i., PinvF-positive bacteria were largely associated with the epithelium (57%) (Fig. 4A and D) and typically situated <10 µm from the nearest apical surface (Fig. 3A and D). The remaining PinvF-positive bacteria were distributed between the extravillous milieu (15%) and the lamina propria (28%), implying that the SPI-1 regulon is expressed after the initial interaction of bacteria with enterocytes. PinvF-positive bacteria were still detected at 8 h p.i., (Fig. 3B), although at a reduced frequency compared to that at 2 h p.i., indicating that at a relatively late stage in acute enteritis, SPI-1 is down-regulated but continues to be expressed. The vast majority of PinvF-positive bacteria were outside the villus (77%) at 8 h p.i., typically within sloughed-off cells in the mucosal layer immediately adjacent to the tissue (Fig. 4D). Of the tissue-associated PinvF-positive bacteria at 8 h p.i., there was more variation in their distances from the apical surface than at 2 h p.i. Some were within 10 µm of the apical surface, but others were found much deeper within the tissue (Fig. 3D). In concordance, tissue-associated PinvF-positive bacteria were similarly distributed between the epithelium and lamina propria at 8 h p.i. (Fig. 4D).
To better understand the prevalence of SPI-1 gene expression within the subtissue compartments over time, we determined the proportion of GFP-positive S. Typhimurium cells in the epithelium, lamina propria, and extravillous space. In the epithelium and lamina propria, we observed a marked decrease in the proportion of PinvF-GFP[LVA] expression from 2 h p.i. to 8 h p.i.: 68% (n = 298) to 39% (n = 85) in the epithelium and 63% (n = 171) to 17% (n = 162) in the lamina propria (Fig. 4E). A previous report has observed a similar decrease in expression of another gene within SPI-1, sicA, in the murine intestine after oral infection (44). In contrast, the proportion of PinvF-GFP[LVA]-positive bacteria remained constant from 2 h to 8 h in the extravillous space, with 58% (n = 93) positive at the early time point and 53% (n = 389) positive later in the infection (Fig. 4E). Historically, a compelling role for SPI-1 in the colonization of nonphagocytic cells, such as epithelial cells, has been well established (45–47). Much less is known about this pathogenicity island and phagocytic cells, but SPI-1 does contribute to cell death in macrophages and dendritic cells and to nitric oxide production in macrophages (48, 49). Our tissue localization data indicate that both enterocytes and cells within the lamina propria are colonized by SPI-1-induced S. Typhimurium during acute enteritis.
For the SPI-2 regulon, no tissue-associated PssaG-positive bacteria were observed at 2 h (Fig. S6B and see above). In contrast, PssaG-positive bacteria were prevalent at 8 h p.i. and strongly associated with subepithelial tissue (97%, n = 308), especially cells positive for HLA-DRα (Fig. 4C), which is a cell surface receptor found on dendritic cells, B cells, and monocytes/macrophages. These immune-originating cell types are often found near the central lacteal. This preferential localization of PssaG-positive S. Typhimurium cells to the lamina propria was also reflected in the distance of the bacteria from the apical surface (Fig. 3C and D); GFP-positive bacteria were significantly further from the apical surface than PinvF-positive S. Typhimurium cells at the same time point. Despite this strong association with the lamina propria, in some instances, PssaG-positive bacteria were found within cytokeratin 8-positive epithelial cells (Fig. 4B). Proportionally, 58% (n = 518) of bacteria in the lamina propria were PssaG positive at 8 h p.i., while only 37% (n = 27) were positive in the epithelium (Fig. 4E). Intracellular induction of SPI-2 is known to occur in both nonphagocytic and phagocytic cells (50, 51), in agreement with the tissue localization that we report here.
In this work, we have utilized a natural-host model of infection, i.e., bovine, ligated jejunal-ileal loops, to better understand host-pathogen interactions during acute enteritis. One striking observation was that of extruding or sloughing enterocytes harboring S. Typhimurium during early stages of infection. Bacteria within these extruded cells expressed SPI-1 and flagella. We have previously reported a similar phenotype in polarized epithelial monolayers (27). Epithelial cell turnover is a normal process in healthy, uninfected tissue that must be exquisitely regulated; an imbalance can lead to states of acute or chronic pathological disturbances in the gut. Interestingly, gastrointestinal infections are often associated with altered rates of epithelial cell extrusion in the gut. For example, after oral inoculation of calves, enteropathogenic Escherichia coli (EPEC) induces enterocyte exfoliation in the terminal rectum (52). Likewise, extensive epithelial shedding into the lumen of the small intestine is observed upon Vibrio parahaemolyticus infection of infant rabbits (53). Perhaps increased epithelial cell shedding is a common host defense mechanism against enteric pathogens.
In order to establish a successful infection, pathogens must precisely regulate virulence gene expression, both temporally and spatially. By documenting bacterial gene expression at the single-cell level in vivo, we have demonstrated for the first time a distinct segregation of SPI-1- and SPI-2-induced S. Typhimurium cells over a time course of acute intestinal infection. PinvF-positive bacteria were found predominantly within enterocytes early during infection but were mostly limited to the extravillous space as the infection progressed, likely due to immunological responses (i.e., PMNs, professional phagocytes, etc.) and pathological responses (i.e., villus blunting, epithelial sloughing) by the host. Extruded epithelial cells contained SPI-1-induced, flagellated bacteria, suggestive of reseeding of the lumen with invasive bacteria (26). In contrast, SPI-2-induced bacteria were almost exclusively found within the lamina propria and only at 8 h p.i. Notably, less than half of the tissue-associated Salmonella cells were induced for either SPI-1 or SPI-2 at 2 h or 8 h p.i., although it is likely that at earlier time points, a larger proportion of the bacterial population is induced for SPI-1, given the central role that this pathogenicity island plays in enterocyte entry and colonization (45, 54). Variable gene expression in genetically identical populations of Salmonella is well recognized and described for broth culture conditions (55–57) and infection of tissue culture cells (26–28, 58) but has only lately received attention in vivo (59, 60). Heterogeneity is most certainly dictated by both host and bacterial factors (61), and the complex nature of the gut suggests a high likelihood of population heterogeneity during colonization. For example, movement from the anaerobic lumen of the intestine to the zone of relative oxygenation at the epithelial surface is considered a trigger for T3SS activation on the surface of Shigella flexneri, the causative agent of dysentery (62). Heterogeneity in virulence gene expression in vivo has also recently been reported for Vibrio cholerae, the etiological agent of cholera. During infection of rabbit ileal loops, only bacteria that are closest to the epithelial surface, not those in the luminal fluid, express tcpA, a structural subunit of the toxin-coregulated pilus (63). The existence of these subpopulations of bacteria in vivo underscores the importance of single-cell studies for analyzing gene expression. Future studies directed at deciphering the distinct microenvironments within the gut and how bacteria respond to each of these environments is key to our understanding of the complexities of pathogen-host interactions during gastroenteritis.
MATERIALS AND METHODS
Bacterial strains and culture.
Salmonella enterica serotype Typhimurium derivative IR715 was transformed by electroporation with the plasmid pMPMA3ΔPlac, containing a destabilized version of green fluorescent protein (GFP[LVA]) under the control of either the invF or the ssaG promoter (26, 64). A promoterless GFP[LVA] plasmid (EMPTY-GFP) served as a vector control (26). Due to the reduced stability of GFP[LVA] (~40 min in S. Typhimurium), these plasmids are valid reporters for transient gene expression (26). Bacterial cultures were grown either in shaking Luria-Bertani (LB) broth or on LB agar plates containing nalidixic acid (50 mg/liter) with carbenicillin (100 mg/liter) when appropriate. Bacterial inocula for the ligated ileal loop surgeries were prepared as described previously (22). Briefly, IR715, pMPMA3ΔPlac-PinvF-GFP[LVA] (PinvF), and pMPMA3ΔPlac-PssaG-GFP[LVA] (PssaG) were grown in LB broth with appropriate antibiotics for 14 h at 37°C at 220 rpm in a shaking incubator (model 24; New Brunswick Scientific). Cultures were then diluted 1:100 in LB broth containing carbenicillin (100 mg/liter), where appropriate, and incubated as described above for 4 h. Bacteria in the exponential phase of growth were quantified using a Genesys 10S visible-light spectrophotometer (Thermo Scientific) and diluted to 109 CFU in 3 ml of LB broth. Bacterial densities were confirmed by plating on LB agar plates with appropriate antibiotics.
Animals and surgeries on bovine, ligated jejunal-ileal loops.
Surgeries on ligated jejunal-ileal loops were performed as described previously (22, 65). Brangus calves 4 weeks of age and 45 to 55 kg were used in accordance with the Texas A&M University International Animal Care and Use Committee (IACUC) animal use policies and approved under Animal Use Protocol 2011-077. Calves were obtained from the Texas A&M University Veterinary Medical Park and received colostrum prior to isolation. Animals were fed antibiotic-free milk replacer twice daily and water ad libitum. Prior to surgery, calves were twice tested for Salmonella spp. in fecal excretions. Rectal swabs were collected immediately after isolation and again 1 week prior to surgery. Swabs were placed in tetrathionate broth (BBL) overnight and subsequently streaked onto XLT-4 agar plates (BBL). All calves were negative for Salmonella species colonies on XLT-4 plates after 48 h of incubation. Loops from seven calves were utilized for this study, with at least three independent loops for each bacterial strain.
For the surgical procedure, calves were fasted for 12 h prior to surgery. Anesthesia was induced with propofol (Abbot Laboratories), followed by intubation and maintenance with isoflurane (Isoflo; Abbot Laboratories) for the duration of the experiment. After laparotomy, the distal jejunum and ileum were externalized and 20 to 30 loops, each ~6 cm in length, were formed with a 1-cm spacer loop in between. Bacterial cultures of 3 ml containing 1 × 109 total CFU were prepared as described above and loaded into a 5-ml syringe with a 26-gauge needle and kept on ice until inoculation into the loop via intraluminal injection. Cultures were inoculated into separate loops as WT (S. Typhimurium IR715), LB (LB broth), PinvF (IR715 harboring pMPMA3ΔPlac-PinvF-GFP[LVA]), PssaG (IR715 pMPMA3ΔPlac-PssaG-GFP[LVA]), or Empty (IR715 pMPMA3ΔPlac-null-GFP[LVA]). Following inoculation, the loops were returned to the body cavity (the surgical incision was temporarily secured) and maintained at approximately 37°C. Infections were allowed to continue for 2 h or 8 h before excision and processing for bacteriology and confocal and electron microscopy.
Pathology, bacteriology, and plasmid retention.
To assess the level of tissue-associated S. Typhimurium cells, two 6-mm biopsy punches (0.1 g) from the Peyer’s patch portion of the loop (antimesenteric side of the intestinal mucosa) were collected from each loop. Extracellular bacteria were removed by washing the samples three times in sterile phosphate-buffered saline (PBS), followed by incubation for 1 h in 10 µg/ml gentamicin in PBS. The biopsy specimens were then homogenized and diluted in PBS (10-fold) before being plated on selective LB agar plates. For samples containing WT S. Typhimurium, LB agar plates contained 50 mg/liter nalidixic acid (LB-NAL). PinvF or PssaG S. Typhimurium samples were plated on LB-NAL plates, and LB-NAL was supplemented with 100 mg/liter carbenicillin (LB-NAL/CARB). LB control samples were plated on LB-NAL plates. Plates were incubated overnight at 37°C, and colonies were then counted. Data are reported as log numbers of CFU per mg of tissue. To quantify S. Typhimurium cells in the mucus, ileal loops were opened and tissue surfaces were scraped to collect mucus. Scrapings were placed in a preweighed container, reweighed, serially diluted, and spread on LB-NAL or LB-NAL/CARB selective plates, and colonies were counted the next day. Data are reported as log numbers of CFU per mg of mucus. For quantification of S. Typhimurium cells in the luminal fluid, luminal contents were collected from intact loops and quantified for weight and volume, 10-fold serially diluted in sterile PBS, and plated on either LB-NAL or LB-NAL/CARB plates as described above; the plates were incubated overnight at 37°C and colonies counted the next day. Data are reported as log numbers of CFU per volume of fluid.
Tissue and fluid fixation for microscopy.
For each loop, 6-mm biopsy specimens were taken from the Peyer’s patch tissue and placed into 10% buffered formalin for confocal microscopy or 2.5% glutaraldehyde, 2.5% formaldehyde in 0.1 M sodium cacodylate buffer for transmission electron microscopy (TEM) for 24 h. Samples for confocal microscopy were then floated into 20% sucrose with 0.05% sodium azide and stored at 4°C until use. Samples for TEM were placed into 0.1 M sodium cacodylate buffer and kept at 4°C until processed further.
Fluid samples were removed from loops as described above, and 100 µl of a sample was placed in 500 µl of 10% buffered formalin overnight. Following fixation, samples were centrifuged at 10,000 × g for 10 min, the supernatant was removed, and the pellet was gently resuspended in 20% sucrose with 0.05% sodium azide. Samples were stored at 4°C until processed further.
Confocal microscopy.
Once infused with 20% sucrose, tissue samples were enrobed with optimum-cutting-temperature compound (Sakura Finetek USA, Inc.) and snap-frozen in liquid nitrogen. Frozen samples were sectioned at 10 µm on an OTF/AS 5000 Cryostat (Vibratome). For immunostaining, sections were rehydrated with PBS for 5 min, followed by permeabilization and blocking with 2% normal donkey serum, 1% bovine serum albumin (BSA), 0.1% Triton X-100, and 0.05% Tween 20 in PBS for 45 min at room temperature (RT). Primary antibodies were diluted in 1% BSA, 0.1% Triton X-100, and 0.05% Tween 20 in PBS overnight at 4°C, followed by 3 washes for 5 min each in 0.05% Tween 20 in PBS (PBST). Secondary antibodies were diluted in 0.1% Triton X-100 in 0.05% Tween 20 in PBS and incubated for 1 h at RT. Sections were washed 3 times for 5 min each in PBST, coated in SlowFade gold antifade reagent with 4',6-diamidino-2-phenylindole (DAPI; Life Technologies), and covered with a coverslip. Samples were cured overnight at RT. Slides were viewed at appropriate fluorescent wavelengths on a Carl Zeiss LSM 510 META NLO Multiphoton confocal microscope (Texas A&M University) or an LSM 710 confocal laser scanning microscope (Rocky Mountain Laboratories, National Institutes of Health). Images were processed and rendered with ImageJ (W. S. Rasband, National Institutes of Health, Bethesda, MD) and assembled using Adobe Photoshop CS3 or Elements 9 (ACD Systems).
Antibodies and reagents.
Primary antibodies for indirect immunofluorescence staining were mouse monoclonal anti-FliC (1:100; BioLegend), rabbit polyclonal anti-Salmonella O-antigen group B factors 1, 4, 5, and 12 (1:200; Difco), rabbit polyclonal anti-GFP (1:1,000; Life Technologies), mouse monoclonal anti-major histocompatibility complex class II DR alpha DA6.147 (HLA-DRα) (1:100; Santa Cruz), and mouse monoclonal anti-human cytokeratin 8 CK3-3E4 (1:200; Miltenyi Biotech). Fluorescent secondary antibodies used included Alexa Fluor 488-, 568-, or 647-conjugated secondary antibodies (1:1,000; Life Technologies) and DyLight 488-conjugated secondary antibodies (1:500; KPL). Alexa Fluor 647 phalloidin was used at a 1:50 dilution (Life Technologies).
Tissue localization of SPI-1- and SPI-2-induced bacteria.
Tissue sections from 2-h or 8-h infections with PinvF or PssaG strains from multiple calves were immunostained with anti-cytokeratin 8 and anti-GFP antibodies. The locations of GFP-positive bacteria were designated (i) “extravillus” if bacteria were in the lumen, (ii) “epithelium” if they were in cytokeratin-positive cells, or (iii) “lamina propria” if bacteria were in the tissue beneath the epithelium. At least 20 villi were scored for each time point and infection. Distances of tissue-associated PinvF- or PssaG-positive bacteria to the nearest apical surface were quantified with the measurement tool in ImageJ and analyzed with Prism 5 (GraphPad Software Inc.). Data were displayed in box-and-whisker plots, with the whiskers representing the 5th-to-95th percentiles. Data were analyzed statistically by a one-way analysis of variance followed by Tukey’s multiple-comparison test, with a P of <0.01 considered significant.
Electron microscopy.
Tissue samples fixed for TEM were postfixed for 1.3 h in 1% OsO4 reduced with 0.35% K4[Fe(CN)6] and buffered with 0.1 M sodium cacodylate. The samples were dehydrated in an ascending ethanol gradient and embedded in epoxy resin. Thin sections (60 to 90 nm) were prepared with a Leica EM UC6 ultramicrotome and poststained with uranyl acetate and lead citrate. The sections were viewed and imaged with a Morgagni 268 transmission electron microscope (FEI). Images were cropped, and exposure was optimized and sharpened in Photoshop Elements 9 (Adobe).
Analysis of GFP reporter expression.
Bacteria were grown under SPI-1- or SPI-2 inducing conditions as follows. The SL1344 wild type (66), ∆hilA mutant (67), or ssrB::kan mutant (68) harboring pMPMA3∆Plac-PinvF-GFP[LVA] or pMPMA3∆Plac-PssaG-GFP[LVA] was grown overnight with shaking in LB-Miller broth at 37°C and then subcultured 1:33 in LB-Miller broth for 3.5 h with shaking at 37°C to induce SPI-1. For SPI-2 induction, bacteria were grown in LB-Miller broth with shaking for 4 to 5 h at 37°C. Bacteria (0.5 ml) were centrifuged, washed once in low-phosphate, low-magnesium (LPM) medium, pH 5.8 (69), and resuspended in an equal volume of LPM medium. Bacteria were then diluted 1:50 in LPM medium and grown for 14 h with shaking at 37°C.
Cultures were centrifuged at 8,000 × g for 2 min, and bacterial pellets were washed once in PBS and then fixed in 1% paraformaldehyde (PFA) for 10 min at room temperature. Bacteria were washed in PBS and then stained with mouse monoclonal anti-S. Typhimurium group B lipopolysaccharide (LPS; 1:200 dilution, clone 1E6; Meridian Life Science), followed by Alexa Fluor 568-conjugated goat anti-mouse IgG (1:400 dilution). After being washed once in PBS, bacteria were mounted on glass slides using Mowiol 4-88 (Calbiochem) and viewed on a Leica DM4000 upright fluorescence microscope.
HeLa epithelial cells (ATCC CCL-2) and RAW264.7 macrophage-like cells (ATCC TIB-71) were obtained from the American Type Culture Collection and used within 15 passages of receipt. HeLa cells were grown in Eagle’s minimum essential medium (EMEM; Corning Cellgro) containing 10% heat-inactivated fetal calf serum (FCS; Invitrogen). RAW264.7 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Corning Cellgro) containing 10% heat-inactivated FCS. Cells were seeded on acid-washed glass coverslips in 24-well plates at 6 × 104 cells/well (HeLa) or 2 × 105 cells/well (RAW264.7). SPI-1-induced bacteria were grown to late log phase as described above, centrifuged at 8,000 × g for 2 min, and then resuspended in Hanks’ buffered saline solution (HBSS; Corning Cellgro). Bacteria were added to epithelial and macrophage-like cells at multiplicities of infection of 50 and 10, respectively, for 10 min. Noninternalized bacteria were removed by three washes with HBSS and cells incubated in growth medium until 30 min p.i. Then, growth medium containing 50 µg/ml gentamicin was added for 1 h, followed by growth medium containing 10 µg/ml gentamicin for the remainder of the experiment. Infected monolayers were fixed in 2.5% PFA for 10 min at 37°C and then permeabilized and blocked in 10% normal goat serum–0.2% Triton X-100–PBS for 20 min. Primary antibodies were rabbit anti-GFP (1:1,000 dilution; Molecular Probes) and mouse monoclonal anti-S. Typhimurium group B LPS (1:2,000 dilution, clone 1E6; Meridian Life Science). Secondary antibodies were Alexa Fluor 488-conjugated goat anti-rabbit IgG and Alexa Fluor 568-conjugated goat anti-mouse IgG (1:800 dilution; Molecular Probes). Coverslips were mounted on glass slides in Mowiol, and the percentages of GFP-positive bacteria were scored by fluorescence microscopy.
SUPPLEMENTAL MATERIAL
S. Typhimurium invasion of bovine jejunal-ileal tissue at 2 h p.i. (A to D) Electron micrographs of a goblet cell containing several S. Typhimurium bacteria within the epithelium of an intestinal villus; (B) higher magnification of the S. Typhimurium bacteria revealing a vacuolar membrane surrounding the organisms and numerous vesicles within the SCV; (C and D) high magnification of outer membrane vesicles or blebs from S. Typhimurium (arrowheads); (E to G) series of electron micrographs of an epithelial cell containing several S. Typhimurium bacteria; (G) high magnification of S. Typhimurium bacteria with numerous outer membrane vesicles (arrowheads); (H to K) series of electron micrographs of a sloughing epithelium with numerous intracellular S. Typhimurium bacteria; (J) high magnification of an SCV containing abundant, large electron-lucent vesicular structures (chevron); (K) high magnification of an SCV with numerous OMV on the surfaces of S. Typhimurium cells (arrowhead) and electron-lucent vesicular structures (chevron). Bars, 10 µm (A, E, and H) and 1 µm (B and F). Download
Identification of intact or compromised vacuolar membrane. Electron micrographs of S. Typhimurium bacteria within the epithelium at 2 h p.i. Red lines shown in insets illustrate continuous or interrupted vacuolar membranes surrounding bacteria. (A and B) Examples of S. Typhimurium cells within intact SCVs that contain numerous vesicular structures in the lumen; (C to E) examples of compromised vacuolar membranes surrounding S. Typhimurium cells; (E) membrane blebbing/fusion of a large electron-lucent vesicle with an SCV membrane is indicated by the chevron. Bars, 1 µm (A to C) and 0.5 µm (D and E). Download
S. Typhimurium within the lamina propria at 8 h p.i. (A and C) Electron micrographs of intracellular S. Typhimurium bacteria contained within intact or compromised vacuoles of cells residing in the lamina propria; (B) higher magnification of S. Typhimurium cells from panel A, showing organisms in an intact vacuole, with one organism undergoing degradation; (D) higher magnification of image in panel C showing Salmonella cells associated with compromised host vacuoles or without any apparent host membrane structure. The host vacuolar membrane is highlighted in red in the middle panels. Bars, 5 µm (A and C) or 1 µm (B and D). Download
Characterization of destabilized GFP reporters. (A) Expression of invF and ssaG under different in vitro growth conditions. Wild-type, ∆hilA, and ssrB::kan S. Typhimurium strains harboring the pMPMA3∆Plac-PinvF-GFP[LVA] or pMPMA3-PssaG-GFP[LVA] plasmid were grown in LB-Miller broth for 3.5 h (SPI-1-inducing conditions) or low phosphate, low magnesium (LPM) broth, pH 5.8, for 14 h (SPI-2-inducing conditions). Bacteria were immunostained with mouse anti-S. Typhimurium group B LPS antibodies. Representative images are shown for each bacterial strain (GFP, green channel; LPS, red channel). (B) Intracellular expression of invF and ssaG in mammalian cells. HeLa epithelial or RAW264.7 macrophage-like cells were infected over a time course with wild-type or ssrB::kan S. Typhimurium cells harboring the pMPMA3∆Plac-PinvF-GFP[LVA] or pMPMA3-PssaG-GFP[LVA] plasmid. Mammalian cells and bacteria were permeabilized with Triton X-100, and then bacteria were immunostained with mouse anti-S. Typhimurium group B LPS antibodies. Where indicated, GFP[LVA] was detected by rabbit polyclonal anti-GFP amplification. The percentage of GFP-positive bacteria was scored by fluorescence microscopy. Values are means ± standard deviations (SD) from three independent experiments. Filled circles, wild-type bacteria (intrinsic GFP fluorescence); open circles, wild-type bacteria (anti-GFP detection); filled triangles, ssrB::kan bacteria (intrinsic GFP fluorescence). The asterisk indicates significantly different data, per Student’s t test (P < 0.05). Download
Assessment of pathology and bacterial burden in ligated jejunal-ileal loops. (A) To assess luminal fluid accumulation, a hallmark of S. Typhimurium infection, fluid from loops infected with WT bacteria or WT bacteria harboring pMPMA3∆Plac-PinvF-GFP[LVA], pMPMA3∆Plac-PssaG-GFP[LVA], or pMPMA3∆Plac-EMPTY-GFP[LVA] was collected and weighed after 2 h or 8 h of infection. Loops inoculated with LB medium only (uninfected control) were also measured. Data are reported as a ratio of the weight of the fluid collected (grams) to the length of the loop (centimeters). Levels of fluid accumulation in loops infected with S. Typhimurium, regardless of plasmid or GFP expression, were not significantly different. Control loops contained little or no fluid, resulting in a low fluid-to-length ratio. Results are reported as means ± SD. (B) To assess the tissue, mucus, and fluid association of WT bacteria harboring the pMPMA3ΔPlac plasmid (solid bars) compared to those of WT bacteria (striped bars), infected samples were collected after 2 h or 8 h of infection, quantified, homogenized (if necessary), serially diluted, and plated on LB agar plates. Equivalent bacterial numbers were found for all cases. To assess for any loss of the pMPMA3ΔPlac plasmid after 8 h of infection, bacteria were plated as described above on LB agar plates containing nalidixic acid (Nal) (unshaded) or nalidixic acid and carbenicillin (Nal/Carb) (shaded). No loss of plasmid, as assessed by carbenicillin resistance, was observed at either infection time point in the tissue, fluid, or mucus samples. Data are reported as means ± SD of the log numbers of CFU per mg tissue, g/cm fluid, or mg mucus, respectively. Download
Fluid and tissue immunofluorescence micrographs of S. Typhimurium cells show heterogeneous populations of virulence gene expression. (A and B) Luminal fluid (A) or tissue (B) collected from jejunal-ileal loops inoculated with S. Typhimurium PssaG-GFP[LVA] for 2 h were immunostained with anti-GFP (green) and anti-Salmonella group B O antigen (red). DNA is shown in blue. Note the lack of GFP-positive bacteria in both samples. (C to F) Fluid and tissue collected from 8-h loops infected with S. Typhimurium PssaG-GFP[LVA] were immunostained with anti-GFP (green) and anti-Salmonella group B O antigen (red). DNA is shown in blue. GFP-positive bacteria were not detected in the fluid sample (C) but were prevalent in tissue (D). (E and F) Increased magnifications of tissues at different z axis locations. Bars, 10 µm. Download
ACKNOWLEDGMENTS
This work was funded by grants to A.J.B. and L.G.A. from the National Institute of Allergy and Infectious Diseases (A144170 and AI076246). L.A.K. is supported by startup funds from the Paul G. Allen School of Global Animal Health. O.S.-M. is supported by the Intramural Research Program of the National Institutes of Allergy and Infectious Diseases. Confocal and electron microscopy performed at the Image Analysis Laboratory, College of Veterinary Medicine, Texas A&M University, were funded by an NIH NCRR Shared Instrumentation grant (1S10RR22532-01). R.C.L. was partially supported by a College of Veterinary Medicine, Texas A&M University, Postdoctoral Trainee Research award. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Clay Ashley and Destiny Taylor at the Veterinary Medical Park, College of Veterinary Medicine, Texas A&M University, for their assistance during surgery. We thank Robert Alaniz for helpful conversations and critical review of the manuscript.
Footnotes
Citation Laughlin RC, Knodler LA, Barhoumi R, Payne HR, Wu J, Gomez G, Pugh R, Lawhon SD, Bäumler AJ, Steele-Mortimer O, Adams LG. 2014. Spatial segregation of virulence gene expression during acute enteric infection with Salmonella enterica serovar Typhimurium. mBio 5(1):e00946-13. doi:10.1128/mBio.00946-13.
REFERENCES
- 1. Ashida H, Ogawa M, Kim M, Mimuro H, Sasakawa C. 2012. Bacteria and host interactions in the gut epithelial barrier. Nat. Chem. Biol. 8:36–45. 10.1038/nchembio.741 [DOI] [PubMed] [Google Scholar]
- 2. Madara JL. 1990. Maintenance of the macromolecular barrier at cell extrusion sites in intestinal epithelium: physiological rearrangement of tight junctions. J. Membr. Biol. 116:177–184. 10.1007/BF01868675 [DOI] [PubMed] [Google Scholar]
- 3. Raffatellu M, Santos RL, Chessa D, Wilson RP, Winter SE, Rossetti CA, Lawhon SD, Chu H, Lau T, Bevins CL, Adams LG, Bäumler AJ. 2007. The capsule encoding the viaB locus reduces interleukin-17 expression and mucosal innate responses in the bovine intestinal mucosa during infection with Salmonella enterica serotype Typhi. Infect. Immun. 75:4342–4350. 10.1128/IAI.01571-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kaiser P, Hardt WD. 2011. Salmonella typhimurium diarrhea: switching the mucosal epithelium from homeostasis to defense. Curr. Opin. Immunol. 23:456–463. 10.1016/j.coi.2011.06.004 [DOI] [PubMed] [Google Scholar]
- 5. Littman DR, Pamer EG. 2011. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe 10:311–323. 10.1016/j.chom.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt WD. 2007. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5:2177–2189. 10.1371/journal.pbio.0050244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Bäumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. 10.1038/nature09415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang S, Kingsley RA, Santos RL, Andrews-Polymenis H, Raffatellu M, Figueiredo J, Nunes J, Tsolis RM, Adams LG, Bäumler AJ. 2003. Molecular pathogenesis of Salmonella enterica serotype Typhimurium-induced diarrhea. Infect. Immun. 71:1–12. 10.1128/IAI.71.1.1-12.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Santos RL, Tsolis RM, Bäumler AJ, Smith R, III, Adams LG. 2001. Salmonella enterica serovar Typhimurium induces cell death in bovine monocyte-derived macrophages by early sipB-dependent and delayed sipB-independent mechanisms. Infect. Immun. 69:2293–2301. 10.1128/IAI.69.4.2293-2301.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang S, Santos RL, Tsolis RM, Stender S, Hardt W-D, Bäumler AJ, Adams LG. 2002. The Salmonella enterica serotype Typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves. Infect. Immun. 70:3843–3855. 10.1128/IAI.70.7.3843-3855.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang S, Adams LG, Nunes J, Khare S, Tsolis RM, Bäumler AJ. 2003. Secreted effector proteins of Salmonella enterica serotype Typhimurium elicit host-specific chemokine profiles in animal models of typhoid fever and enterocolitis. Infect. Immun. 71:4795–4803. 10.1128/IAI.71.8.4795-4803.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Rüssmann H, Hardt WD. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71:2839–2858. 10.1128/IAI.71.5.2839-2858.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nunes JS, Lawhon SD, Rossetti CA, Khare S, Figueiredo JF, Gull T, Burghardt RC, Bäumler AJ, Tsolis RM, Andrews-Polymenis HL, Adams LG. 2010. Morphologic and cytokine profile characterization of Salmonella enterica serovar Typhimurium infection in calves with bovine leukocyte adhesion deficiency. Vet. Pathol. 47:322–333. 10.1177/0300985809358037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chan SS, Mastroeni P, McConnell I, Blacklaws BA. 2008. Salmonella infection of afferent lymph dendritic cells. J. Leukoc. Biol. 83:272–279. 10.1189/jlb.0607401 [DOI] [PubMed] [Google Scholar]
- 15. Voedisch S, Koenecke C, David S, Herbrand H, Förster R, Rhen M, Pabst O. 2009. Mesenteric lymph nodes confine dendritic cell-mediated dissemination of Salmonella enterica serovar Typhimurium and limit systemic disease in mice. Infect. Immun. 77:3170–3180. 10.1128/IAI.00272-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Santos RL, Tsolis RM, Bäumler AJ, Adams LG. 2003. Pathogenesis of Salmonella-induced enteritis. Braz. J. Med. Biol. Res. 36:3–12. 10.1590/S0100-879X2003000100002 [DOI] [PubMed] [Google Scholar]
- 17. Darwin KH, Miller VL. 1999. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 12:405–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wallis TS, Starkey WG, Stephen J, Haddon SJ, Osborne MP, Candy DC. 1986. The nature and role of mucosal damage in relation to Salmonella typhimurium-induced fluid secretion in the rabbit ileum. J. Med. Microbiol. 22:39–49. 10.1099/00222615-22-1-39 [DOI] [PubMed] [Google Scholar]
- 19. Kent TH, Formal SB, Labrec EH. 1966. Salmonella gastroenteritis in rhesus monkeys. Arch. Pathol. 82:272–279 [PubMed] [Google Scholar]
- 20. Thiennimitr P, Winter SE, Bäumler AJ. 2012. Salmonella, the host and its microbiota. Curr. Opin. Microbiol. 15:108–114. 10.1016/j.mib.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Santos RL, Raffatellu M, Bevins CL, Adams LG, Tükel C, Tsolis RM, Bäumler AJ. 2009. Life in the inflamed intestine, Salmonella style. Trends Microbiol. 17:498–506. 10.1016/j.tim.2009.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lawhon SD, Khare S, Rossetti CA, Everts RE, Galindo CL, Luciano SA, Figueiredo JF, Nunes JE, Gull T, Davidson GS, Drake KL, Garner HR, Lewin HA, Bäumler AJ, Adams LG. 2011. Role of SPI-1 secreted effectors in acute bovine response to Salmonella enterica serovar Typhimurium: a systems biology analysis approach. PLoS One 6:e26869. 10.1371/journal.pone.0026869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Müller AJ, Kaiser P, Dittmar KE, Weber TC, Haueter S, Endt K, Songhet P, Zellweger C, Kremer M, Fehling HJ, Hardt WD. 2012. Salmonella gut invasion involves TTSS-2-dependent epithelial traversal, basolateral exit, and uptake by epithelium-sampling lamina propria phagocytes. Cell Host Microbe 11:19–32. 10.1016/j.chom.2011.11.013 [DOI] [PubMed] [Google Scholar]
- 24. Ellermeier JR, Slauch JM. 2007. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 10:24–29. 10.1016/j.mib.2006.12.002 [DOI] [PubMed] [Google Scholar]
- 25. Moest TP, Méresse S. 2013. Salmonella T3SSs: successful mission of the secret(ion) agents. Curr. Opin. Microbiol. 16:38–44. 10.1016/j.mib.2012.11.006 [DOI] [PubMed] [Google Scholar]
- 26. Ibarra JA, Knodler LA, Sturdevant DE, Virtaneva K, Carmody AB, Fischer ER, Porcella SF, Steele-Mortimer O. 2010. Induction of Salmonella pathogenicity island 1 under different growth conditions can affect Salmonella-host cell interactions in vitro. Microbiology 156:1120–1133. 10.1099/mic.0.032896-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Knodler LA, Vallance BA, Celli J, Winfree S, Hansen B, Montero M, Steele-Mortimer O. 2010. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc. Natl. Acad. Sci. U. S. A. 107:17733–17738. 10.1073/pnas.1006098107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Malik-Kale P, Winfree S, Steele-Mortimer O. 2012. The bimodal lifestyle of intracellular Salmonella in epithelial cells: replication in the cytosol obscures defects in vacuolar replication. PLoS One 7:e38732. 10.1371/journal.pone.0038732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Popiel I, Turnbull PC. 1985. Passage of Salmonella enteritidis and Salmonella thompson through chick ileocecal musoca. Infect. Immun. 47(3):786–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tobar JA, Carreño LJ, Bueno SM, González PA, Mora JE, Quezada SA, Kalergis AM. 2006. Virulent Salmonella enterica serovar Typhimurium evades adaptive immunity by preventing dendritic cells from activating T cells. Infect. Immun. 74:6438–6448. 10.1128/IAI.00063-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Popiel I, Turnbull PC. 1985. Passage of salmonella enteritidis and Salmonella Thompson through chick ileocecal mucosa. Infect. Immun. 47:786–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuehn MJ, Kesty NC. 2005. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 19:2645–2655. 10.1101/gad.1299905 [DOI] [PubMed] [Google Scholar]
- 33. Yashroy RC. 2007. Mechanism of infection of a human isolate Salmonella (3,10:r:– ) in chicken ileum: ultrastructural study. Indian J. Med. Res. 126:558–566 [PubMed] [Google Scholar]
- 34. Darwin KH, Miller VL. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181:4949–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Valdivia RH, Falkow S. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007–2011. 10.1126/science.277.5334.2007 [DOI] [PubMed] [Google Scholar]
- 36. Bajaj V, Hwang C, Lee CA. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18:715–727. 10.1111/j.1365-2958.1995.mmi_18040715.x [DOI] [PubMed] [Google Scholar]
- 37. Walthers D, Carroll RK, Navarre WW, Libby SJ, Fang FC, Kenney LJ. 2007. The response regulator SsrB activates expression of diverse Salmonella pathogenicity island 2 promoters and counters silencing by the nucleoid-associated protein H-NS. Mol. Microbiol. 65:477–493. 10.1111/j.1365-2958.2007.05800.x [DOI] [PubMed] [Google Scholar]
- 38. Stecher B, Barthel M, Schlumberger MC, Haberli L, Rabsch W, Kremer M, Hardt WD. 2008. Motility allows S. typhimurium to benefit from the mucosal defence. Cell. Microbiol. 10:1166–1180. 10.1111/j.1462-5822.2008.01118.x [DOI] [PubMed] [Google Scholar]
- 39. Cummings LA, Wilkerson WD, Bergsbaken T, Cookson BT. 2006. In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Mol. Microbiol. 61:795–809. 10.1111/j.1365-2958.2006.05271.x [DOI] [PubMed] [Google Scholar]
- 40. Loetscher Y, Wieser A, Lengefeld J, Kaiser P, Schubert S, Heikenwalder M, Hardt WD, Stecher B. 2012. Salmonella transiently reside in luminal neutrophils in the inflamed gut. PLoS One 7:e34812. 10.1371/journal.pone.0034812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brown NF, Vallance BA, Coombes BK, Valdez Y, Coburn BA, Finlay BB. 2005. Salmonella pathogenicity island 2 is expressed prior to penetrating the intestine. PLoS Pathog. 1:e32. 10.1371/journal.ppat.0010032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Finlay BB, Brumell JH. 2000. Salmonella interactions with host cells: in vitro to in vivo. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:623–631. 10.1098/rstb.2000.0603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Geddes K, Cruz F, Heffron F. 2007. Analysis of cells targeted by Salmonella type III secretion in vivo. PLoS Pathog. 3:e196. 10.1371/journal.ppat.0030196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bumann D. 2002. Examination of Salmonella gene expression in an infected mammalian host using the green fluorescent protein and two-colour flow cytometry. Mol. Microbiol. 431269–1283. 10.1046/j.1365-2958.2002.02821.x [DOI] [PubMed] [Google Scholar]
- 45. Galán JE, Curtiss R., III 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. U. S. A. 86:6383–6387. 10.1073/pnas.86.16.6383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Steele-Mortimer O, Brumell JH, Knodler LA, Méresse S, Lopez A, Finlay BB. 2002. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell. Microbiol. 4:43–54. 10.1046/j.1462-5822.2002.00170.x [DOI] [PubMed] [Google Scholar]
- 47. Bueno SM, Wozniak A, Leiva ED, Riquelme SA, Carreño LJ, Hardt WD, Riedel CA, Kalergis AM. 2010. Salmonella pathogenicity island 1 differentially modulates bacterial entry to dendritic and non-phagocytic cells. Immunology 130:273–287. 10.1111/j.1365-2567.2009.03233.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hernandez LD, Pypaert M, Flavell RA, Galán JE. 2003. A salmonella protein causes macrophage cell death by inducing autophagy. J. Cell Biol. 163:1123–1131. 10.1083/jcb.200309161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Drecktrah D, Knodler LA, Galbraith K, Steele-Mortimer O. 2005. The Salmonella SPI1 effector SopB stimulates nitric oxide production long after invasion. Cell. Microbiol. 7:105–113. 10.1111/j.1462-5822.2004.00436.x [DOI] [PubMed] [Google Scholar]
- 50. Beuzón CR, Méresse S, Unsworth KE, Ruíz-Albert J, Garvis S, Waterman SR, Ryder TA, Boucrot E, Holden DW. 2000. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 19:3235–3249. 10.1093/emboj/19.13.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cirillo DM, Valdivia RH, Monack DM, Falkow S. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175–188. 10.1046/j.1365-2958.1998.01048.x [DOI] [PubMed] [Google Scholar]
- 52. Nart P, Naylor SW, Huntley JF, McKendrick IJ, Gally DL, Low JC. 2008. Responses of cattle to gastrointestinal colonization by Escherichia coli O157:H7. Infect. Immun. 76:5366–5372. 10.1128/IAI.01223-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ritchie JM, Rui H, Zhou X, Iida T, Kodoma T, Ito S, Davis BM, Bronson RT, Waldor MK. 2012. Inflammation and disintegration of intestinal villi in an experimental model for Vibrio parahaemolyticus-induced diarrhea. PLoS Pathog. 8:e1002593. 10.1371/journal.ppat.1002593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tsolis RM, Adams LG, Hantman MJ, Scherer CA, Kimbrough T, Kingsley RA, Ficht TA, Miller SI, Bäumler AJ. 2000. SspA is required for lethal Salmonella enterica serovar Typhimurium infections in calves but is not essential for diarrhea. Infect. Immun. 68:3158–3163. 10.1128/IAI.68.6.3158-3163.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Clark L, Perrett CA, Malt L, Harward C, Humphrey S, Jepson KA, Martinez-Argudo I, Carney LJ, La Ragione RM, Humphrey TJ, Jepson MA. 2011. Differences in Salmonella enterica serovar Typhimurium strain invasiveness are associated with heterogeneity in SPI-1 gene expression. Microbiology 157:2072–2083. 10.1099/mic.0.048496-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Clark CG, Kruk TM, Bryden L, Hirvi Y, Ahmed R, Rodgers FG. 2003. Subtyping of Salmonella enterica serotype Enteritidis strains by manual and automated PstI-SphI ribotyping. J. Clin. Microbiol. 41:27–33. 10.1128/JCM.41.1.27-33.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sturm A, Heinemann M, Arnoldini M, Benecke A, Ackermann M, Benz M, Dormann J, Hardt WD. 2011. The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type III secretion system 1. PLoS Pathog. 7:e1002143. 10.1371/journal.ppat.1002143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Helaine S, Thompson JA, Watson KG, Liu M, Boyle C, Holden DW. 2010. Dynamics of intracellular bacterial replication at the single cell level. Proc. Natl. Acad. Sci. U. S. A. 107:3746–3751. 10.1073/pnas.1000041107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ackermann M, Stecher B, Freed NE, Songhet P, Hardt WD, Doebeli M. 2008. Self-destructive cooperation mediated by phenotypic noise. Nature 454:987–990. 10.1038/nature07067 [DOI] [PubMed] [Google Scholar]
- 60. Diard M, Garcia V, Maier L, Remus-Emsermann MN, Regoes RR, Ackermann M, Hardt WD. 2013. Stabilization of cooperative virulence by the expression of an avirulent phenotype. Nature 494:353–356. 10.1038/nature11913 [DOI] [PubMed] [Google Scholar]
- 61. Helaine S, Holden DW. 2013. Heterogeneity of intracellular replication of bacterial pathogens. Curr. Opin. Microbiol. 16:184–191. 10.1016/j.mib.2012.12.004 [DOI] [PubMed] [Google Scholar]
- 62. Marteyn B, West NP, Browning DF, Cole JA, Shaw JG, Palm F, Mounier J, Prévost MC, Sansonetti P, Tang CM. 2010. Modulation of Shigella virulence in response to available oxygen in vivo. Nature 465:355–358. 10.1038/nature08970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nielsen AT, Dolganov NA, Rasmussen T, Otto G, Miller MC, Felt SA, Torreilles S, Schoolnik GK. 2010. A bistable switch and anatomical site control Vibrio cholerae virulence gene expression in the intestine. PLoS Pathog. 6:e1001102. 10.1371/journal.ppat.1001102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Andersen JB, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, Molin S. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Santos RL, Zhang S, Tsolis RM, Bäumler AJ, Adams LG. 2002. Morphologic and molecular characterization of Salmonella typhimurium infection in neonatal calves. Vet. Pathol. 39:200–215. 10.1354/vp.39-2-200 [DOI] [PubMed] [Google Scholar]
- 66. Hoiseth SK, Stocker BA. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239. 10.1038/291238a0 [DOI] [PubMed] [Google Scholar]
- 67. Main-Hester KL, Colpitts KM, Thomas GA, Fang FC, Libby SJ. 2008. Coordinate regulation of Salmonella pathogenicity island 1 (SPI1) and SPI4 in Salmonella enterica serovar Typhimurium. Infect. Immun. 76:1024–1035. 10.1128/IAI.01224-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Worley MJ, Ching KH, Heffron F. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol. Microbiol. 36:749–761. 10.1046/j.1365-2958.2000.01902.x [DOI] [PubMed] [Google Scholar]
- 69. Coombes BK, Brown NF, Valdez Y, Brumell JH, Finlay BB. 2004. Expression and secretion of Salmonella pathogenicity island-2 virulence genes in response to acidification exhibit differential requirements of a functional type III secretion apparatus and SsaL. J. Biol. Chem. 279:49804–49815. 10.1074/jbc.M404299200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S. Typhimurium invasion of bovine jejunal-ileal tissue at 2 h p.i. (A to D) Electron micrographs of a goblet cell containing several S. Typhimurium bacteria within the epithelium of an intestinal villus; (B) higher magnification of the S. Typhimurium bacteria revealing a vacuolar membrane surrounding the organisms and numerous vesicles within the SCV; (C and D) high magnification of outer membrane vesicles or blebs from S. Typhimurium (arrowheads); (E to G) series of electron micrographs of an epithelial cell containing several S. Typhimurium bacteria; (G) high magnification of S. Typhimurium bacteria with numerous outer membrane vesicles (arrowheads); (H to K) series of electron micrographs of a sloughing epithelium with numerous intracellular S. Typhimurium bacteria; (J) high magnification of an SCV containing abundant, large electron-lucent vesicular structures (chevron); (K) high magnification of an SCV with numerous OMV on the surfaces of S. Typhimurium cells (arrowhead) and electron-lucent vesicular structures (chevron). Bars, 10 µm (A, E, and H) and 1 µm (B and F). Download
Identification of intact or compromised vacuolar membrane. Electron micrographs of S. Typhimurium bacteria within the epithelium at 2 h p.i. Red lines shown in insets illustrate continuous or interrupted vacuolar membranes surrounding bacteria. (A and B) Examples of S. Typhimurium cells within intact SCVs that contain numerous vesicular structures in the lumen; (C to E) examples of compromised vacuolar membranes surrounding S. Typhimurium cells; (E) membrane blebbing/fusion of a large electron-lucent vesicle with an SCV membrane is indicated by the chevron. Bars, 1 µm (A to C) and 0.5 µm (D and E). Download
S. Typhimurium within the lamina propria at 8 h p.i. (A and C) Electron micrographs of intracellular S. Typhimurium bacteria contained within intact or compromised vacuoles of cells residing in the lamina propria; (B) higher magnification of S. Typhimurium cells from panel A, showing organisms in an intact vacuole, with one organism undergoing degradation; (D) higher magnification of image in panel C showing Salmonella cells associated with compromised host vacuoles or without any apparent host membrane structure. The host vacuolar membrane is highlighted in red in the middle panels. Bars, 5 µm (A and C) or 1 µm (B and D). Download
Characterization of destabilized GFP reporters. (A) Expression of invF and ssaG under different in vitro growth conditions. Wild-type, ∆hilA, and ssrB::kan S. Typhimurium strains harboring the pMPMA3∆Plac-PinvF-GFP[LVA] or pMPMA3-PssaG-GFP[LVA] plasmid were grown in LB-Miller broth for 3.5 h (SPI-1-inducing conditions) or low phosphate, low magnesium (LPM) broth, pH 5.8, for 14 h (SPI-2-inducing conditions). Bacteria were immunostained with mouse anti-S. Typhimurium group B LPS antibodies. Representative images are shown for each bacterial strain (GFP, green channel; LPS, red channel). (B) Intracellular expression of invF and ssaG in mammalian cells. HeLa epithelial or RAW264.7 macrophage-like cells were infected over a time course with wild-type or ssrB::kan S. Typhimurium cells harboring the pMPMA3∆Plac-PinvF-GFP[LVA] or pMPMA3-PssaG-GFP[LVA] plasmid. Mammalian cells and bacteria were permeabilized with Triton X-100, and then bacteria were immunostained with mouse anti-S. Typhimurium group B LPS antibodies. Where indicated, GFP[LVA] was detected by rabbit polyclonal anti-GFP amplification. The percentage of GFP-positive bacteria was scored by fluorescence microscopy. Values are means ± standard deviations (SD) from three independent experiments. Filled circles, wild-type bacteria (intrinsic GFP fluorescence); open circles, wild-type bacteria (anti-GFP detection); filled triangles, ssrB::kan bacteria (intrinsic GFP fluorescence). The asterisk indicates significantly different data, per Student’s t test (P < 0.05). Download
Assessment of pathology and bacterial burden in ligated jejunal-ileal loops. (A) To assess luminal fluid accumulation, a hallmark of S. Typhimurium infection, fluid from loops infected with WT bacteria or WT bacteria harboring pMPMA3∆Plac-PinvF-GFP[LVA], pMPMA3∆Plac-PssaG-GFP[LVA], or pMPMA3∆Plac-EMPTY-GFP[LVA] was collected and weighed after 2 h or 8 h of infection. Loops inoculated with LB medium only (uninfected control) were also measured. Data are reported as a ratio of the weight of the fluid collected (grams) to the length of the loop (centimeters). Levels of fluid accumulation in loops infected with S. Typhimurium, regardless of plasmid or GFP expression, were not significantly different. Control loops contained little or no fluid, resulting in a low fluid-to-length ratio. Results are reported as means ± SD. (B) To assess the tissue, mucus, and fluid association of WT bacteria harboring the pMPMA3ΔPlac plasmid (solid bars) compared to those of WT bacteria (striped bars), infected samples were collected after 2 h or 8 h of infection, quantified, homogenized (if necessary), serially diluted, and plated on LB agar plates. Equivalent bacterial numbers were found for all cases. To assess for any loss of the pMPMA3ΔPlac plasmid after 8 h of infection, bacteria were plated as described above on LB agar plates containing nalidixic acid (Nal) (unshaded) or nalidixic acid and carbenicillin (Nal/Carb) (shaded). No loss of plasmid, as assessed by carbenicillin resistance, was observed at either infection time point in the tissue, fluid, or mucus samples. Data are reported as means ± SD of the log numbers of CFU per mg tissue, g/cm fluid, or mg mucus, respectively. Download
Fluid and tissue immunofluorescence micrographs of S. Typhimurium cells show heterogeneous populations of virulence gene expression. (A and B) Luminal fluid (A) or tissue (B) collected from jejunal-ileal loops inoculated with S. Typhimurium PssaG-GFP[LVA] for 2 h were immunostained with anti-GFP (green) and anti-Salmonella group B O antigen (red). DNA is shown in blue. Note the lack of GFP-positive bacteria in both samples. (C to F) Fluid and tissue collected from 8-h loops infected with S. Typhimurium PssaG-GFP[LVA] were immunostained with anti-GFP (green) and anti-Salmonella group B O antigen (red). DNA is shown in blue. GFP-positive bacteria were not detected in the fluid sample (C) but were prevalent in tissue (D). (E and F) Increased magnifications of tissues at different z axis locations. Bars, 10 µm. Download