Summary
Mitochondria have long been implicated in the pathogenesis of Parkinson’s disease (PD). Mutations in the mitochondrial kinase PINK1 that reduce kinase activity are associated with mitochondrial defects and result in an autosomal recessive form of early onset PD. Therapeutic approaches for enhancing the activity of PINK1 have not been considered since no allosteric regulatory sites for PINK1 are known. Here we show that an alternative strategy, a neo-substrate approach involving the ATP analog kinetin triphosphate (KTP), can be used to increase the activity of both PD related mutant PINK1G309D and PINK1wt. Moreover, we show that application of the KTP precursor kinetin to cells results in biologically significant increases in PINK1 activity, manifest as higher levels of Parkin recruitment to depolarized mitochondria, reduced mitochondrial motility in axons, and lower levels of apoptosis. Discovery of neo-substrates for kinases could provide a heretofore-unappreciated modality for regulating kinase activity.
Introduction
Parkinson’s disease (PD) is characterized by the loss of dopaminergic (DA) neurons in the substantia nigra, a region in the midbrain that is critical for motor control (Lang and Lozano, 1998). Mitochondrial dysfunction has been closely linked to PD via several mechanisms (Nunnari and Suomalainen, 2012; Rugarli and Langer, 2012), including mutations in the mitochondria-specific kinase PTEN Induced Kinase 1 (PINK1) (Valente et al., 2004) and mitochondria-associated E3 ubiquitin ligase Parkin (Kitada et al., 1998). PINK1 plays an important role in repairing mitochondrial dysfunction by responding to damage at the level of individual mitochondria. In healthy mitochondria, PINK1 is rapidly degraded by the protease ParL (Meissner et al., 2011); but in the presence of inner membrane depolarization, PINK1 is stabilized on the outer membrane, where it recruits and activates Parkin (Narendra et al., 2010), blocks mitochondrial fusion and trafficking (Clark et al., 2006; Deng et al., 2008; Wang et al., 2011), and ultimately triggers mitochondrial autophagy (Geisler et al., 2010; Narendra et al., 2008; Youle and Narendra, 2011). The PINK1 pathway has also been linked to the induction of mitochondrial biogenesis and the reduction of mitochondria-induced apoptosis in neurons, the latter phenotype due at least in part to the effect of PINK1 on mitochondrial motility, a neuron specific phenotype (Deng et al., 2005; Petit et al., 2005; Pridgeon et al., 2007; Shin et al., 2011; Wang et al., 2011).
Individuals homozygous for PINK1 loss-of-function mutations can develop a form of early onset PD that results from highly selective DA neuronal loss and, in at least one clinical case, shares the Lewy-body pathology of sporadic PD (Gautier et al., 2008; Geisler et al., 2010; Haque et al., 2008; Henchcliffe and Beal, 2008; Petit et al., 2005; Samaranch et al., 2010). Recent work has shown that of 17 clinically relevant PINK1 mutations, those mutants that affect catalytic activity but do not affect cleavage or subcellular localization have the most dramatic effect on neuron viability, further supporting a role for PINK1 activity in the prevention of neurodegeneration (Song et al., 2013). One of the most common of the catalytic mutants, PINK1G309D, shows a ~70% decrease in kinase activity and abrogates the neuroprotective effect of PINK1 (Petit et al., 2005; Pridgeon et al., 2007). However, lower PINKG309D catalytic activity can be rescued by overexpression of PINK1wt, and increasing PINK1 activity by PINK1wt overexpression has been shown to reduce staurosporine- and oxidative-stress-induced apoptosis in multiple cell lines, suggesting that enhanced PINK1 activity could be an effective therapeutic strategy for PD (Arena et al., 2013; Deng et al., 2005; Kondapalli et al., 2012; Petit et al., 2005; Pridgeon et al., 2007).
Recognizing the therapeutic potential of PINK1/Parkin pathway activation, we began investigating mechanisms for the pharmacological activation of PINK1. Small molecule activation of kinases is typically accomplished by binding allosteric regulatory sites: for example, natural products such as phorbol esters bind to the lipid binding domain of PKCs and recruit the kinase to the membrane (Castagna et al., 1982; Nishizuka, 1984); separately, the AMP activated protein kinase (AMPK) is activated by binding of AMP to an allosteric site (Ferrer et al., 1985; Hardie et al., 2012). However, PINK1 contains no known small molecule binding sites. Another potential strategy might involve manipulation of protein interaction sites or the active site, given that synthetic ligands have been identified which bind to the protein docking sites on the kinase PDK1 (Hindie et al., 2009; Wei et al., 2010), and, separately, that Src activity can be controlled by chemical complementation of an active site catalytic residue, allowing ATP to be accepted only when imidazole was provided to mutant Src (Ferrando et al., 2012; Qiao et al., 2006). However these approaches were not applicable to PINK1 as no structural data for PINK1 is available.
We next turned our attention to sites within PINK1 known to alter its function or stability. Four PINK1 disease associated mutations, including G309D, occur in an unusual insertion in the canonical kinase fold. PINK1, as well as several orthologs, share 3 such large (>15 AA) insertions in the N-terminal kinase domain (Figure S1A) (Cardona et al., 2011; Mills et al., 2008) that provides the majority of contacts to the adenine ring of ATP. Inserts in the active site of several enzymes have been shown to alter substrate specificity. In one example, the deubiquitinase UCH-L5 can hydrolyze larger ubiquitin chains only when a >14 AA loop is present in the active site (Zhou et al., 2012). In another example, protein engineering of alkyl guanine DNA alkyltransferase through insertion of a loop into the active site allows for recognition of an enlarged O6-modified guanine substrate not accepted by the enzyme without the loop insertion (Heinis et al., 2006). In light of these findings, the three insertions in PINK1’s adenine binding N-terminal subdomain led us to believe that PINK1 might also exhibit altered substrate specificity.
In considering the possibility that nucleotides other than ATP could be substrates for kinases, we noticed that CK2 can utilize multiple substrates as phospho-donors: GTP as well as ATP, though its activity with GTP is much lower (Niefind et al., 1999). Though it is uncommon for eukaryotic protein kinases to accept alternative substrates in the ATP binding site, kinases engineered with a single mutation to the gatekeeper residue often tolerate ATP analogs with substitutions at the N6 position (Liu et al., 1998; Shah et al., 1997). Importantly, no wildtype kinase we had previously studied had shown the ability to accept N6 modified ATP analogs (Figure S1B).
We discovered that, unlike any kinase we have studied, PINK1 accepts the neo-substrate N6 furfuryl ATP (kinetin triphosphate or KTP) with higher catalytic efficiency than its endogenous substrate, ATP. We also found that the metabolic precursor of this neo-substrate (kinetin) can be taken up by cells and converted to the nucleotide triphosphate form, which leads to accelerated Parkin recruitment to depolarized mitochondria, diminished mitochondrial motility in axons, and suppression of apoptosis in human derived neural cells, all in a PINK1 dependent manner.
Results
PINK1 accepts N6 modified ATP analog kinetin triphosphate (KTP)
We expressed both PINK1wt and PINK1G309D GST tagged kinase domain (156–496PINK1) in E. coli (Figure S2A) and performed kinase assays with a series of neo-substrate analogs. As expected, PINK1G309D displayed reduced activity with ATP; interestingly, however, incubation with N6 furfuryl ATP (kinetin triphosphate or KTP) (Figure 1A) led to increased levels of transphosphorylation of the mitochondrial chaperone hTRAP1 (residues 60–704) (Figure 1B) and autophosphorylation with both PINK1G309D and PINK1wt (Figures 1C,D). Using a phosphopeptide capture and release strategy (Blethrow et al., 2008; Hertz et al., 2010), we were able to identify the T257 autophosphorylation site (Kondapalli et al., 2012) using KTP as the phospho-donor for PINK1 (Figure 1E), which showed that this neo-substrate is capable of supporting bona fide PINK1-dependent substrate phosphorylation.
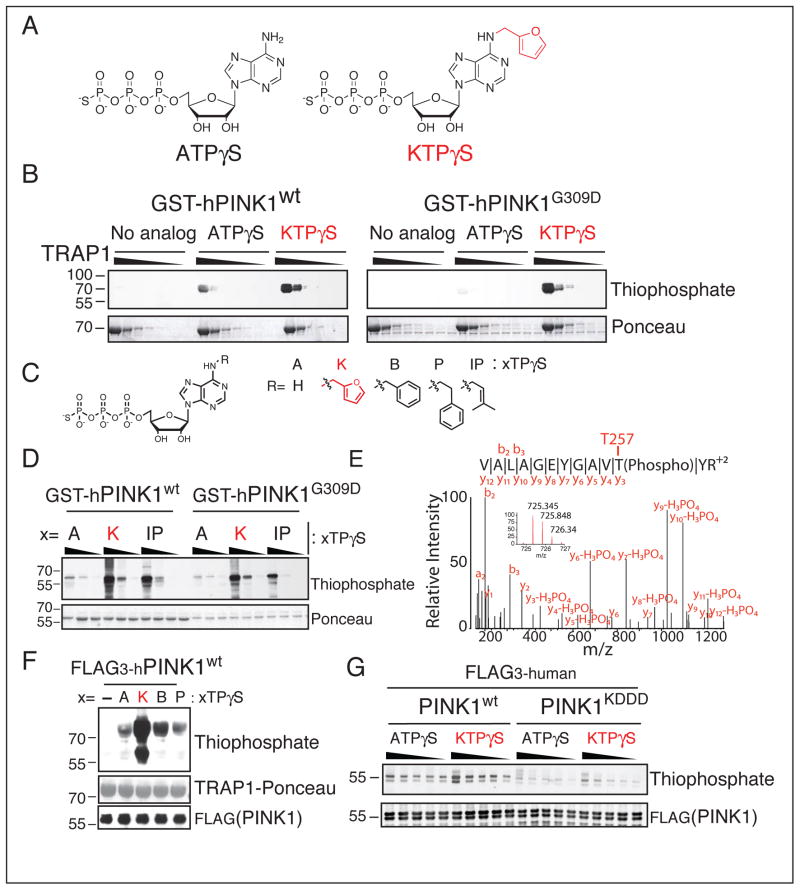
Figure 1. Neo-substrate Kinetin Triphosphate (KTP) amplifies PINK1 kinase activity in-vitro.
(A) Chemical structure of kinase substrate adenosine triphosphate with gamma thiophosphate (ATPγS) and neo-substrate kinetin triphosphate gamma thiophosphate (KTPγS).
(B–D) PINK1 kinase assay (B), with substrate 60–704TRAP1 (1 mg/ml decreasing by 1/3) and 500 μM indicated nucleotide (C) or (D), PINK1 alone (4.3 μM) 100, 200, 400 μM nucleotide analyzed by immunoblotting for thiophospho labeled TRAP1 and PINK1.
(E) PINK1 autophosphorylation site identified by specific peptide capture and LCMSMS found only with PINK1wt and KTPγS indicating this nucleotide is utilized as a bona fide substrate
(F) SF21 produced PINK1 was incubated with 60–704TRAP1 and the indicated nucleotide (structure shown in panel C) and analyzed as in (B–D)
(G) PINK1 kinase assay with indicated nucleotide (250 to 1250 μM in increments of 250 μM) analyzed as in (B–D) reveals much reduced phosphorylation activity with PINK1KDDD (lanes 11–20), increased autophosphorylation was seen with neo-substrate KTPγS over endogenous substrate ATPγS.
PINK1 is intrinsically highly unstable, and especially so when produced in bacteria (Beilina et al., 2005); therefore, in order to confirm the PINK1-dependency of the observed kinase activity, we took several steps to optimize PINK1 expression. We constructed several FLAG3 tagged truncation variants of PINK1 and induced expression using baculovirus infected SF21 insect cells (Figure S2B). C-terminally tagged 112–581PINK1FLAG3 expressed the most soluble protein. However, the amount was far below what we required for biochemical characterization (Figure S2C). Hypothesizing that PINK1 might require interaction with other proteins to fold properly, we co-expressed proteins known to associate with PINK1 such as DJ-1, PARKIN, and TRAP1. Co-expression of full length TRAP1 dramatically increased the stability of PINK1 (Figure S2C). This finding enabled us to express larger amounts of properly folded PINK1wt, PINK1G309D and a kinase dead PINK1kddd (residues 112–581 with K219A, D362A, and D384A (Figure S1A)). In line with our initial observations, SF21-produced-PINK1 activity is also enhanced using KTP but not other nucleotides (Figure 1F). We confirmed that PINK1kddd has severely compromised activity (Figure 1G and S2D), and were able to show that PINK1wt could autophosphorylate with a 3.9 ±1.3 fold higher (p=0.02;t-test) Vmax and a higher KM (27.9 ± 4.9 μM vs. 74.6 ± 13.2 μM) for the neo-substrate KTP versus ATP (Figures 1G and S2E,F). As the previous assays utilized ATP with a γ-thiophosphate as a tracer, we wanted to confirm the activity using an orthogonal tracer, γ-32P ATP to visualize PINK1 activity. Therefore, we generated KTP with a γ-32P labeled phosphate, and were able to see that using this orthogonally labeled version of KTP PINK1wt transphosphorylation of 60–704TRAP1 is increased relative to γ-32P ATP (Figure S2G).
Since KTP would have to compete with millimolar intracellular ATP concentrations in order to function, we performed a competition assay with γ-thiophosphate labeled KTP versus ATP. We found that the KTPγS signal persisted even when ATP was present at 4 fold greater concentration than KTPγS (2 mM vs. 0.5 mM) (Figure S2H) suggesting that PINK1 can be activated by KTP in the presence of cellular ATP.
KTP is produced in human cells upon treatment with KTP precursor kinetin
Our results showing in vitro increases in PINK1 activity using KTP led us to investigate the ability to achieve enhanced activity of PINK1 in cells. One major challenge to activating PINK1 in cells is that ATP analogs like KTP are not membrane permeable; however, previous work has shown that certain cytokinins can be taken up by human cells and converted to the nucleotide triphosphate form (Ishii et al., 2003). Additionally, recent work in cells expressing hypomorphic mutant CDK2 alleles showed that the activity of CDK2 could be increased in cells by providing nucleotide analog precursors that can be converted to the nucleotide triphosphate form and are able to fit into the hypomorphic CDK2 active site (Merrick et al., 2011).
The first step in bioconversion is ribosylation of the cytokinin to a 5′-monophosphate form, which can be mediated by adenine phospho ribosyl transferase (APRT)(Kornberg et al., 1955; Lieberman et al., 1955b) (Figure 2A). Following established protocols (Parkin et al., 1984), we incubated either adenine, kinetin or negative control N9 methyl-kinetin (9MK) with 5′-phosphoribosyl pyrophosphate (PRPP) and APRT and assayed the reaction by LCMS (Figure 2B). Adenine converted rapidly to AMP, achieving near-complete conversion (84%) after 10 minutes of reaction time; kinetin’s conversion to KMP (Figure 2B,C) was markedly slower, requiring 150 minutes for half to be converted. The control compound 9MK is not converted to KMP even after 16 hours of incubation (Figure 2B) whereas kinetin was completely converted to KMP in this time frame. This experiment demonstrated that the ribosylation of kinetin is biosynthetically possible using the enzymatic route for AMP production.
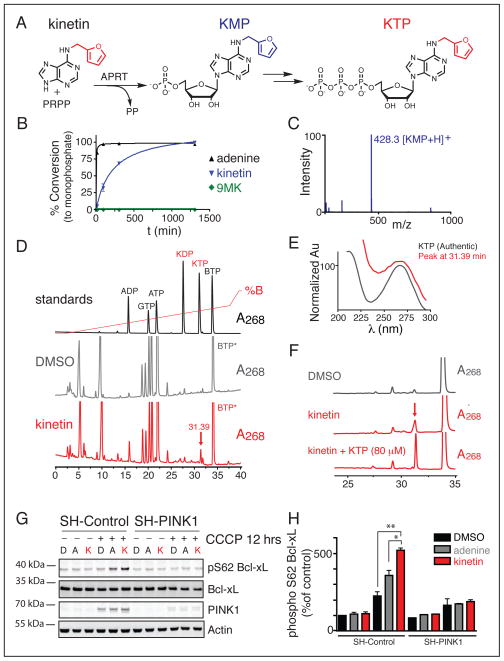
Figure 2. Neo-substrate precursor kinetin leads to KTP in human cells and activates PINK1 dependent phosphorylation of Bcl-xL.
(A) Schematic of the ribosylation reaction of kinetin to KMP via APRT and PRPP followed by cellular conversion to KTP.
(B) LCMS analysis of production of either AMP (adenine) or KMP (kinetin and 9MK) by LCMS analysis reveals that kinetin can undergo APRT mediated ribosylation to KMP, but N9 methyl-kinetin cannot.
(C) LCMS analysis of kinetin reaction; major peak represents KMP+H peak at 428.3 m/z.
(D) HPLC analysis of standards (ADP, GTP, ATP, KDP, KTP, BTP) or cellular lysate of DMSO or kinetin treated HeLa cells with 250 μM BTP* addition reveals a novel peak present in kinetin treated cells at 31.39 minutes.
(E) Absorbance spectrum of peak at 31.39 minutes in kinetin treated cells compared to absorbance spectrum of KTP in standard.
(F) Zoom of HPLC analysis of DMSO or kinetin treated cell lysate with BTP addition, or kinetin cell lysate with BTP addition and KTP addition reveals an increase in the peak that co-elutes with KTP (186%) where BTP decreases to 76% of original area suggesting the peak is KTP.
(G) Immunoblot analysis with the indicated antibodies for phosphorylation on serine 62 of Bcl-xL reveals an increase following kinetin treatment.
(H) Quantitative analysis of data shown in G where a significant increase in the phosphorylation of Bcl-xL on serine 62 with pre-treatment with kinetin versus DMSO or adenine (p=0.01, p=0.04;t-test) in SH-Control expressing cells not in SH-PINK1 cells. (P values shown are the result of two-tailed student’s t-test, *P<0.05 **P≤0.01) (all values shown are mean ± sem)
Our next step was to ask whether KMP could be converted to the nucleotide triphosphate form (KTP) required for it to serve as a neo-substrate for PINK1. Work by many labs has demonstrated that ribosyl nucleotide analogs can be phosphorylated to nucleotide triphosphate forms by endogenous enzymes (Krishnan et al., 2002; Lieberman et al., 1955a; Ray et al., 2004). To confirm the presence of intracellular KTP following incubation with kinetin, we adapted an ion-pairing HPLC analysis method on a reverse phase C18 column (Figure 2D) according to established methods (Vela et al., 2007). After treatment with kinetin or DMSO, cells were lysed and analyzed for the presence of peaks eluting at the retention time of KTP. An internal standard of BTP (denoted by * in Figures 2D,F, S3A,B) was added following lysis. Analysis of the kinetin-treated cells revealed a peak that co-elutes (offset by a consistent amount (Figure S3A)) with synthetic KTP, a result not seen in any of our controls (Figure 2D and Figure S3A,B). The UV absorbance maximum measured with a diode array detector is the same as KTP (Figure 2E)(absorbance peak at 268 nm) and significantly different than that of either ATP or GTP (Figure S3C) both of which have significantly different retention times. In a separate HPLC analytical run an aliquot of authentic KTP (to 80 μM) was added to authenticate the putative KTP peak (Figure 2F). The peak grew to 186% of its original area, while the peak of the standard BTP decreased to 76% of its original area, suggesting that these were the same substance. From these data we calculate an ATP concentration of 1950 ± 421 μM and KTP concentration of 68 ± 13 μM (3 biological replicates Table 1), is produced upon incubation with kinetin.
Table 1.
| Concentration | |
|---|---|
| ATP | 1950 ± 421 μM |
| KTP | 68 ± 13.3 μM |
Kinetin increases phosphorylation of PINK1 substrate anti apoptotic protein Bcl-xL
Bcl-xL is a member of the Bcl-2 protein family that plays a key regulatory role in mitochondrial-induced apoptosis (Adams and Cory, 1998; Gross et al., 1999). PINK1 phosphorylates Bcl-xL at serine 62 in response to mitochondrial depolarization blocking cleavage to a pro-apoptotic form (Arena et al., 2013). We therefore measured PINK1-dependent phosphorylation of Bcl-xL in human SH-SY5Y cells following CCCP-induced depolarization. We analyzed DMSO, 50 μM adenine or 50 μM kinetin treated SH-SY5Y cells at 12 hours post 50 μM CCCP addition (Figure 2G) and observed a significant (p=0.01, p=0.04;t-test) (Figure 2G,H) increase in phospho Bcl-xL (S62) only in kinetin treated cells compared to DMSO or adenine, where PINK1 expression has not been silenced by shRNA. These results confirm that CCCP mediated depolarization will induce PINK1 dependent phosphorylation on S62, and suggest that kinetin stimulates this activity in a PINK1 dependent manner.
Kinetin accelerates Parkin recruitment to depolarized mitochondria in a PINK1 dependent manner
Parkin recruitment to depolarized mitochondria is PINK1 dependent; (Narendra et al., 2010) therefore, we postulated that enhancement of PINK1 activity might accelerate this process. We treated cells with either kinetin or adenine (Figure 3A) and measured Parkin localization following CCCP mediated mitochondria depolarization. HeLa cells, which have low levels of endogenous PINK1 and PARKIN, were transfected with PINK1wt or PINK1G309D, mCherryParkin, and mitochondrial-targeted GFP (mitoGFP). After 48 hours of incubation with 25 μM adenine, kinetin or equivalent DMSO (Figure 3B), we imaged every five minutes following CCCP-mediated depolarization of mitochondria (Figures 3C,D and S4A–F) and calculated the percentage of GFP labeled mitochondria with mCherryParkin associated (Figures 3E,F and Table 2).
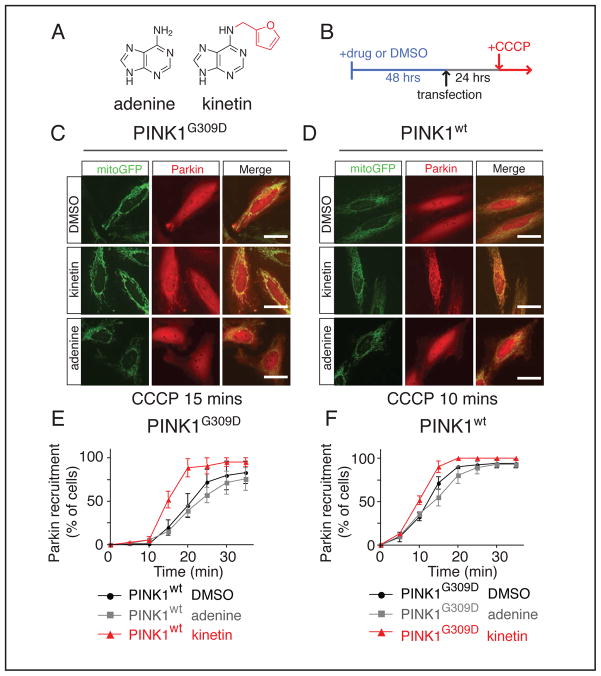
Figure 3. PINK1 neo-substrate kinetin accelerates PINK1 dependent Parkin recruitment in cells.
(A) Chemical structure of adenine or kinase neo-substrate precursor kinetin.
(B) Schematic depicting HeLa cell drug treatment.
(C–D) HeLa cells treated with indicated drug, co-transfected with mitoGFP, mCherryParkin, and indicated PINK1 construct imaged at either 10 or 15 minutes after 5 μM CCCP addition.
(E–F) Kinetin treated cells reached R50 significantly faster than with adenine or DMSO (all data shown is mean ± sem) by two-way ANOVA analysis kinetin has an effect in both cases when compared to adenine (wt; F=25.41 p<0.0001, G309D; F=31.89, p<0.0001) and DMSO (wt; F=21.94 p<0.0001, G309D; F=12.79, p<0.0011), no significant difference for DMSO adenine in either case (at least 150 cells/experiment n=3 experiments-all values are mean ± sem).
Table 2.
50% Recruitment Times (R50 in min) mCherry Parkin to depolarized mitochondria.
| DMSO | adenine | kinetin | |
|---|---|---|---|
| PINK1wt | 14 ± 1 min | 15 ± 1 min | 10 ± 2 min |
| PINK1G309D | 20 ± 2 min | 23 ± 2 min | 15 ± 2 min |
In line with previous reports (Narendra et al., 2010), transfection of PINK1G309D slowed the 50% recruitment (R50) time of mCherryParkin to depolarized mitochondria (23±2min vs. 15±1min R50) (Figure 3E and Table 2). The addition of kinetin, but not adenine, decreased the R50 for Parkin in PINK1G309D cells from 23±2 to 15±2min, and also decreased the R50 for PINK1wt cells from 15±1 to 10±2min (Figure 3E,F and Table 2). Using an image analysis algorithm (Figure S4G,H) that quantified the time dependent change in co-localization, we found that PINK1wt expressing cells achieved a maximum change in co-localization of 0.112 with DMSO or adenine and 0.13 with kinetin treatment (Figure S4I). PINK1G309D expressing cells treated with DMSO or adenine achieved delta co-localization of 0.076, but upon addition of kinetin returned to near- PINK1wt levels (0.124) (Figure S4J). These results suggested significant rescue of PINK1G309D activity using kinetin. Two-way ANOVA analysis revealed that kinetin has an effect in both cases (wt; F=24.10 p<0.0001, G309D; F=54.14, p<0.0001). Additionally, in line with in vitro results in which benzyl triphosphate (BTP) did not activate PINK1 as robustly as KTP (Figure 1F), benzyl adenine is in general less active than kinetin in cells, although it also demonstrated some acceleration of Parkin recruitment (Data not shown). However, benzyl adenine has been shown to be cytotoxic in other assays (Ishii et al., 2002), therefore, despite PINK1 activation potential of benzyl adenine we decided to focus on KTP to use as a neo-substrate to amplify PINK1 activity.
To test the PINK1-dependency of our findings, we assayed PINK1 activity by using phospho-specific antibodies raised against the PINK1-specific S65 phosphosite on Parkin (Kondapalli et al., 2012). We observed a small but reproducible increase in the phosphorylation level of Parkin following CCCP treatment in a PINK1-dependent manner (Figure S5A,B). In a finding that supported our co-localization results, we also found that the addition of neo-substrate kinetin (p=0.03; t-test), but not adenine (p=0.30; t-test), to PINK1G309D mutant expressing cells increased the phosphorylation levels of Parkin (Figure S5C,D). The addition of an adenosine kinase inhibitor (AKI) blocking the conversion of kinetin to KTP prevented this effect (p=0.31;t-test) (Figure S5D).
Kinetin blocks mitochondrial motility in axons in a PINK1 dependent manner
Increasing PINK1 activity markedly decreases the mobility of axonal mitochondria and this is thought to be the first step in the sequestration and removal of damaged mitochondria (Wang et al., 2011). To determine if PINK1 activation by kinetin also decreases mitochondrial motility, we examined the mobility of axonal mitochondria in rat hippocampal neurons co-transfected with mitoGFP to identify mitochondria and N-terminal mCherry-tagged Synaptophysin to identify axons (Hua et al., 2011; Nakamura et al., 2011). Cells were pre-treated for 48 hours with 50 μM kinetin, adenine, 9-methyl-kinetin (9MK shown in Figure 4A) or equivalent DMSO, and mitochondrial motility was imaged live. Kymographs were generated (Figure 4B–E) using standard techniques. Kinetin markedly inhibited the percentage of moving mitochondria when compared to DMSO (p=0.0005;t-test)(Figure 4D,F). In contrast, kinetin analog 9-methyl-kinetin (9MK) (Figure 4A,E,F), which cannot be converted to a nucleotide triphosphate form, did not affect mitochondrial motility when compared to DMSO (p=0.86;t-test). Kinetin also produced a small decrease in the velocity of mitochondria that remain in motion (p=0.03;t-test)(Figure 4G).
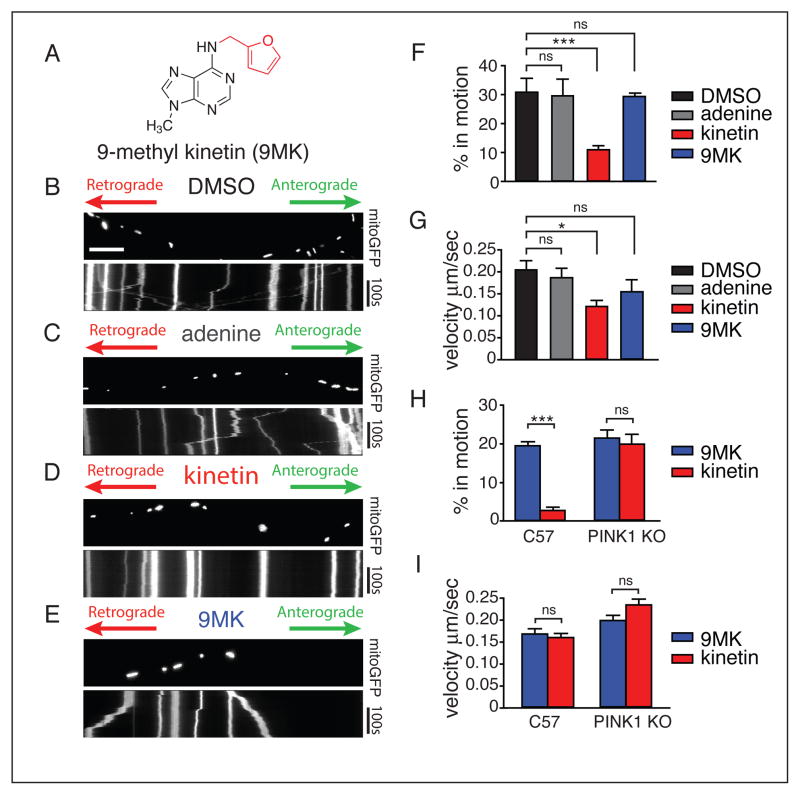
Figure 4. Kinetin halts axonal mitochondrial motility in a PINK1 dependent manner.
(A) Chemical structure of negative control non-metabolizable kinetin analog 9-methyl-kinetin
(B–E) Kymograph for analysis of mitochondrial movement in representative PINK1wt expressing rat derived hippocampal axons transfected with mitoGFP. Scale bar represents 10 μm.
(F) The percentage of time each mitochondrion was in motion was determined and averaged. Kinetin significantly blocks mitochondrial motility whereas 9MK has no effect (DMSO-kinetin, P=0.0005; DMSO-9MK, P=0.86)
(G) Kinetin induces a small decrease in velocity (DMSO-kinetin, P=0.03; DMSO-9MK, P=0.24).
(H) Kymograph for analysis of mitochondrial movement in C57BL/6 shows a response to kinetin (kinetin 9MK, P<0.0001), whereas in PINK1 knockout derived hippocampal axons kinetin has no effect (kinetin 9MK, P=0.64).
(I) Both kinetin and 9MK have no effect on mitochondrial velocity of moving mitochondrion (C57BL/6 kinetin-9MK, P=0.64; PINK1 KO, P=0.074) (all values are mean ± sem, analysis was two tailed students t-test)
See also Figure S5
To confirm that kinetin decreases mitochondrial motility through effects on PINK1, we also performed experiments in hippocampal neurons derived from wild-type (C57BL/6) and PINK1 KO mice (Xiong et al., 2009) after treatment with DMSO, kinetin or 9MK. Consistent with the results in rat neurons, kinetin also significantly decreased mitochondrial motility in control neurons (p<0.0001;t-test) when compared with either 9MK or DMSO (Figure 4H and S5E). However, kinetin had no effect on the motility of mitochondria in PINK1 KO neurons (Figure 4H) (p=0.64;t-test), and, unlike rat derived neurons, kinetin had no effect on the velocity of mitochondria that remain in motion (Figure 4I). These data suggest that kinetin can block mitochondrial motility in a PINK1 dependent manner and that the metabolism of kinetin to KTP is a necessary precursor for that effect.
Kinetin decreases apoptosis induced by oxidative stress in human derived neuronal cells in a PINK1 dependent manner
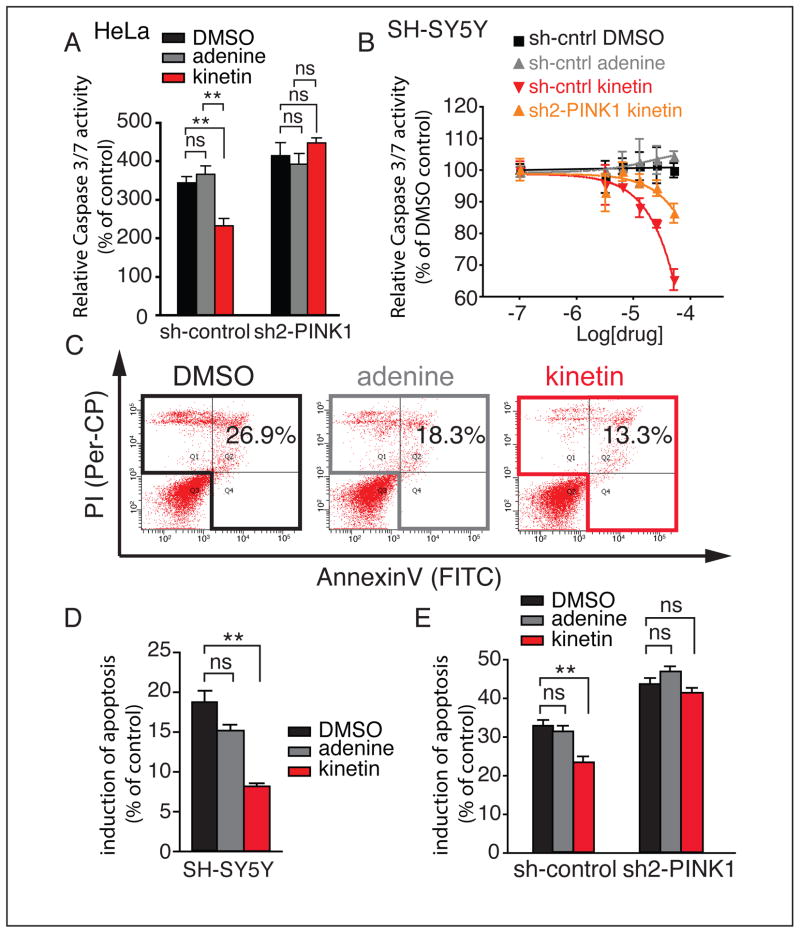
Previous studies have shown that PINK1 expression can block apoptosis in response to proteosomal stress induced by the proteosome inhibitor MG132 (Klinkenberg et al., 2010; Wang et al., 2007). Caspase 3/7 activity is an early marker for apoptosis therefore, to assess the ability of kinetin to amplify PINK1 activity to block apoptosis, we measured caspase 3/7 activity using a fluorogenic caspase 3/7 peptide cleavage assay in HeLa cells. We treated HeLa cells transfected with Parkin with DMSO or 25 μM adenine or kinetin for 48 hours, followed by 1 μM MG132 for 12 hours. We found that kinetin significantly (p=0.005, p=0.004;t-test) reduced Caspase 3/7 cleavage versus DMSO or adenine pre-treatment, and that knockdown of PINK1 abrogated this effect (Figure 5A and S6A).
Figure 5. Kinetin inhibits oxidative stress induced apoptosis in human cells in a PINK1 dependent manner.
(A) Caspase 3/7 cleavage activity assay following pre-treatment with DMSO, adenine or kinetin for 48 hours followed by MG132 treatment for 12 hours compared to no MG132 treated cells reveals reduced Caspase 3/7 activity (p=0.005, p=0.004;t-test) only in sh-control expressing cells not in cells expressing shRNA against PINK1.
(B) Caspase 3/7 cleavage activity of SH-SY5Y cells pre-treated with indicated concentration of adenine or kinetin for 96 hours followed by MG132 treatment for 12 hours in all conditions. Two-way ANOVA analysis revealed that kinetin has an effect when compared to DMSO or adenine (Figure 5B) (DMSO; F=34.95 p<0.0001;adenine; F=38.37 p<0.0001) but no statistically significant effect (DMSO; F=3.552 p=0.084;adenine; F=1.7 p=0.215) in cells expressing an shRNA against PINK1
(C) SH-SY5Y cells pre-treated as above, were stained with FITC conjugated Annexin V and propidium iodide and analyzed by FACS.
(D) Quantification of (C) shows kinetin treated cells have significantly lower induction of apoptosis (DMSO-kinetin, P=0.0023) but adenine had no effect (DMSO-adenine, P=0.09).
(E) Indicated SH-SY5Y cell lines were treated as is (A). Kinetin treated cells had significantly lower induction of apoptosis only when PINK1 was present (normalized to DMSO control untreated cells)(sh-control DMSO-kinetin, P=0.008) (sh-PINK1 DMSO-kinetin, P=0.23)(sh-Control DMSO-adenine, P=0.48; sh-PINK1 DMSO-adenine, P=0.23) (all values are mean ± sem, analysis was Wilcoxon t-test)
See also Figure S6
We then tested whether kinetin would be tolerated by cultured DA neurons. Previous work has shown KTP precursor kinetin to be extremely well tolerated in both mouse models and in human clinical testing (Axelrod et al., 2011; Shetty et al., 2011). To confirm these results, we treated DA neurons with 50 μM kinetin or adenine, and measured cell density after 10 days. Kinetin and adenine have no effect on cell density, indicating that neither promotes apoptosis of cultured DA neurons (Figure S6B).
We next utilized patient-derived neuroblastoma SH-SY5Y cells, which are also known to exhibit decreased apoptosis upon overexpression of PINK1 (Deng et al., 2005; Klinkenberg et al., 2010). We performed a dose-response assay in which SH-SY5Y cells were treated with increasing concentrations of kinetin, adenine or DMSO for 96 hours, 1 μM MG132 for 16 hours, followed by analysis for Caspase 3/7 cleavage activity. As in HeLa cells kinetin pre-treatment significantly decreased Caspase 3/7 cleavage in SH-SY5Y cells (Figure 5B). Two-way ANOVA analysis revealed that kinetin has an effect when compared to DMSO or adenine (Figure 5B) (DMSO; F=34.95 p<0.0001;adenine; F=38.37 p<0.0001) only in cells where we did not silence PINK1 expression via stable shRNA expression (DMSO; F=3.552 p=0.084;adenine; F=1.7 p=0.215) (Figure 5B and S6C,D) despite a small visible effect, probably due to incomplete knockdown of PINK1 (Figure S6C)). These experiments suggest that the kinetin-induced reduction in Caspase 3/7 cleavage activity is PINK1 dependent.
To assay later stages of apoptosis in SH-SY5Y cells, we utilized an independent FACS based method to measure cellular apoptosis. In addition to proteosomal stress, PINK1 over-expression is known to block apoptosis induced by H2O2 treatment (Deng et al., 2005; Gautier et al., 2008; Petit et al., 2005; Pridgeon et al., 2007). SH-SY5Y cells were treated with 50 μM kinetin, adenine or DMSO for 96 hours, followed by 400 μM H2O2 treatment for 24 hours. Using a cytometry-based FACS assay utilizing annexin V and propidium iodide staining, we determined the percentage of apoptotic cells after treatment with DMSO, adenine or kinetin (Figure 5C). We observed a significant decrease in the total amount of apoptotic cells following kinetin treatment (Figure 5D) (DMSO vs. kinetin p=0.0023;Wilcoxon T test), but no significant change with adenine (DMSO vs. adenine, p=0.09;Wilcoxon T test). Additionally, we observed a significant drop in apoptosis in shRNA control lentivirus infected cells (DMSO vs. kinetin p=0.008;Wilcoxon T test), but no kinetin effect with infection of a lentivirus expressing PINK1-silencing shRNA (Figures 5E and S6C) (DMSO vs. kinetin, p=0.23;Wilcoxon T test). These results demonstrate a PINK1-dependent anti-apoptotic effect, and suggest that kinetin can activate PINK1 to block oxidative stress induced apoptosis of human neural cells.
Discussion
Our investigation of a neo-substrate approach to modulating PINK1 activity has yielded three significant findings: 1) that the ATP analog kinetin triphosphate (KTP) can be used by both PINK1wt and PINK1G309D; 2) that KTP enhances both PINK1wt and PINK1G309D activity in vitro and in cells, and in the case of the latter version, returns it to near-wt catalytic efficiency; and 3) that KTP precursor kinetin can be applied to neurons to enhance PINK1 activity in several biologically relevant paradigms.
The finding that kinetin (or its nucleotide triphosphate form, KTP) can restore PINK1G309D catalytic activity to near-wt levels in-vitro and in cells, in light of the fact that mutations in PINK1 produce PD in humans, raises the possibility that kinetin may be used to treat patients who have mutant PINK1. Further, because kinetin has already been shown to be well tolerated in human trials (for familial dysautonomia, an unrelated splicing disorder), and since previous work in mice has shown that kinetin can cross the blood-brain barrier and achieve pharmacologically significant concentrations (Axelrod et al., 2011; Shetty et al., 2011), these results have the potential for near-term clinical relevance.
Although mutations in PINK1 are a rare cause of PD, the development of an effective disease modifying therapy for any neurodegenerative disease would be a tremendous advance, and could provide important therapeutic insights into disease modifying strategies for other types of PD. In fact, the finding that increasing PINK1 activity beyond endogenous levels can protect against a variety of apoptotic stressors (Klinkenberg et al., 2010; Petit et al., 2005; Pridgeon et al., 2007), and that kinetin can also reproduce this protection by enhancing endogenous PINK1 function, raises the possibility that enhancing PINK1 activity may also have therapeutic potential for idiopathic PD.
Current kinase targeted drugs are striking for the single modality of regulating kinase function—inhibition. However, a wide range of kinase dysregulation in disease is caused by a lack of kinase activity: desensitization of insulin receptor kinase in diabetes (Kulkarni et al., 1999); inactivation of the death associated protein kinase (DAPK) in cancer (Kissil et al., 1997); inactivation of the LKB1 tumor suppressor kinase in cancer (Gao et al., 2011); and decreased PINK1 activity in early-onset Parkinson’s Disease. Although many examples of inactive kinases causing disease have been uncovered, there have been no therapeutic approaches for enhancing kinase activity using alternative substrates. Our insights into the potential for manipulating kinase-dependent cellular processes via a specifically targeted neo-substrate may presage the ability to treat other diseases resulting from kinase misregulation with a novel class of neo-substrate kinase activators.
Experimental Procedures
Detailed methods for dopamine neuron cultures, PINK1 shRNA lentivirus production and apoptosis assays are found in Extended Experimental Procedures.
Western blot analysis
Western blots analysis was carried out as described (Ultanir et al., 2012) with the indicated antibodies. See Supplementary Information for details.
Expression, purification and enzymatic characterization of PINK1
H. sapiens PINK1 kinase domain (PINK1, residues 156–496 all plasmids obtained from Addgene) with an N-terminal GST tag was expressed using a pGEX vector using standard techniques. H. sapiens PINK1 kinase domain with c-terminal extension (PINK1, residues 112–581) with a C-terminal FLAG3 tag was co-expressed with full length H.sapiens TRAP1 in the baculovirus/Sf21 insect cell system. Following lysis, PINK1112–581 kinase was purified using magnetic M2 FLAG affinity resin (Sigma) with the kinase reaction performed on beads after no more than 2 hours following lysis. The reaction was performed using 50 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 3 mM MnCl2, 0.5 mM DTT and 1 mg/ml substrate if not otherwise indicated. After reaction at room temperature with rotation, kinase reaction was quenched with 50 mM EDTA and reacted with 1.5 mM p-nitrobenzylmesylate (PNBM) and identified by immunoblot with anti thiophosphate ester antibody (Epitomics) (Allen et al., 2007).
Identification of PINK1 autophosphorylation site by LC/MS/MS
Protocol was carried out as described (Ultanir et al., 2012). See Supplementary Information for details.
Enzymatic production of KMP in vitro
Reaction with APRT, PRPP and either adenine, kinetin or 9MK were carried out as described (Parkin et al., 1984). See Supplementary Information for details.
HPLC analysis for KTP production in cells
HeLa cells were incubated with the indicated drug for 96 hours and nucleotides were extracted and analyzed as described (Ray et al., 2004; Vela et al., 2007). See Supplementary Information for details.
Parkin mitochondrial translocation assay
HeLa cells were grown in DMEM supplemented with 10% FBS. Log phase cells were plated in 24 well plates with glass coverslips (Mattek) pretreated with fibronectin. Cells were pretreated with 25 μM of the indicated compound, followed by transfection with MitoGFP, mCherryParkin, and full length PINK1FLAG3 in a 1:4:2 ratio using Fugene 6 (Promega). Fields of cells were selected by expression of MitoGFP (6 fields/well-3 wells/condition) and imaged at five-minute intervals following depolarization with 5 μM CCCP. Quantification was performed according to published protocols (Narendra et al., 2010) and by implementing a Matlab based script (See Supplementary Information for details and for script).
Mitochondrial motility assay
All experiments were carried out according to IACUC guidelines and UCSF IACUC approved all experiments before execution. Primary hippocampal cultures were prepared from early postnatal (P0–P1) rat or mouse (C57BL6 or Pink1 −/−) pups, co-transfected by electroporation (Amaxa) with mitochondrial targeted GFP (mitoGFP) to visualize mitochondria, and mCherry fused to synaptophysin (mCherrySynaptophysin) to visualize axons. Cells were pre-treated for 48 hours with 50 μM kinetin, adenine, 9-methyl-kinetin or equivalent DMSO at day 9 and imaged live in Tyrode’s medium (in mM: 127 NaCl, 10 HEPES-NaOH, pH 7.4, 30 glucose, 2.5 KCl, 2 CaCl2, 2 MgCl2) with a 60x water immersion objective on a Nikon Ti-E inverted microscope. Images were captured every 2 s for a total of 200s, and kymographs were generated from each live-imaging movie with Metamorph software (version 7.7.3.0). Mitochondria were considered moving if they travelled more than 0.67 μm during the 200 s imaging.
Supplementary Material
Highlights.
PINK1 activity is amplified by neo-substrate kinetin triphosphate (KTP)
KTP amplifies activity of PINK1wt and Parkinson’s disease related mutant PINK1G309D
Kinetin blocks mitochondrial motility in a PINK1 dependent manner
Kinetin inhibits apoptosis of human neurons in a PINK1 dependent manner
Acknowledgments
We thank Laura Lavery for providing recombinantly expressed TRAP1, Michael Lopez for synthesis of 9-methyl-kinetin, Xiaoxi Zhuang (U of Chicago) for C57BL/6 background PINK1 −/− mice and Valerie Ohman for excellent administrative assistance. We thank Zachary A. Knight, Robert H. Edwards and Daniel de Roulet Jr. for critical reading of the manuscript. N.T.H. was supported by a MJFF Rapid Response Award and a Genentech predoctoral grant. K.M.S. was supported by RO1 EB001987, and early work was supported by the Michael J. Fox Foundation. K.N. and A.B. were supported by a Burroughs-Wellcome Medical Scientist Fund Career Award and grant awards 1KO8NS062954-01A1 and P30NS069496 from the National Institute of Neurological Disorders and Stroke. Mass spectrometry was made possible by NIH grants NCRR RR015804 and NCRR RR001614. Imaging data were collected at the Nikon Imaging Center at QB3/UCSF.
Footnotes
Supplemental Data including Extended Experimental Procedures, six figures, the MATLAB co-localization script, and Supplemental References and can be found with this article online at www.cell.com
References
- Adams JM, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- Allen JJ, Li M, Brinkworth CS, Paulson JL, Wang D, Hubner A, Chou WH, Davis RJ, Burlingame AL, Messing RO, et al. A semisynthetic epitope for kinase substrates. Nat Methods. 2007;4:511–516. doi: 10.1038/nmeth1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arena G, Gelmetti V, Torosantucci L, Vignone D, Lamorte G, De Rosa P, Cilia E, Jonas EA, Valente EM. PINK1 protects against cell death induced by mitochondrial depolarization, by phosphorylating Bcl-xL and impairing its pro-apoptotic cleavage. Cell Death Differ. 2013 doi: 10.1038/cdd.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod FB, Liebes L, Gold-Von Simson G, Mendoza S, Mull J, Leyne M, Norcliffe-Kaufmann L, Kaufmann H, Slaugenhaupt SA. Kinetin improves IKBKAP mRNA splicing in patients with familial dysautonomia. Pediatr Res. 2011;70:480–483. doi: 10.1203/PDR.0b013e31822e1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilina A, Van Der Brug M, Ahmad R, Kesavapany S, Miller DW, Petsko GA, Cookson MR. Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc Natl Acad Sci U S A. 2005;102:5703–5708. doi: 10.1073/pnas.0500617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blethrow JD, Glavy JS, Morgan DO, Shokat KM. Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc Natl Acad Sci U S A. 2008;105:1442–1447. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona F, Sanchez-Mut JV, Dopazo H, Perez-Tur J. Phylogenetic and in silico structural analysis of the Parkinson disease-related kinase PINK1. Hum Mutat. 2011;32:369–378. doi: 10.1002/humu.21444. [DOI] [PubMed] [Google Scholar]
- Castagna M, Takai Y, Kaibuchi K, Sano K, Kikkawa U, Nishizuka Y. Direct activation of calcium-activated, phospholipid-dependent protein kinase by tumor-promoting phorbol esters. J Biol Chem. 1982;257:7847–7851. [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Deng H, Dodson MW, Huang H, Guo M. The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci U S A. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Jankovic J, Guo Y, Xie W, Le W. Small interfering RNA targeting the PINK1 induces apoptosis in dopaminergic cells SH-SY5Y. Biochem Biophys Res Commun. 2005;337:1133–1138. doi: 10.1016/j.bbrc.2005.09.178. [DOI] [PubMed] [Google Scholar]
- Ferrando IM, Chaerkady R, Zhong J, Molina H, Jacob HK, Herbst-Robinson K, Dancy BM, Katju V, Bose R, Zhang J, et al. Identification of targets of c-Src tyrosine kinase by chemical complementation and phosphoproteomics. Mol Cell Proteomics. 2012;11:355–369. doi: 10.1074/mcp.M111.015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer A, Caelles C, Massot N, Hegardt FG. Activation of rat liver cytosolic 3-hydroxy-3-methylglutaryl coenzyme A reductase kinase by adenosine 5′-monophosphate. Biochem Biophys Res Commun. 1985;132:497–504. doi: 10.1016/0006-291x(85)91161-1. [DOI] [PubMed] [Google Scholar]
- Gao Y, Ge G, Ji H. LKB1 in lung cancerigenesis: a serine/threonine kinase as tumor suppressor. Protein Cell. 2011;2:99–107. doi: 10.1007/s13238-011-1021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier CA, Kitada T, Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci U S A. 2008;105:11364–11369. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Treis A, Skujat D, Weber SS, Fiesel FC, Kahle PJ, Springer W. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy. 2010;6:871–878. doi: 10.4161/auto.6.7.13286. [DOI] [PubMed] [Google Scholar]
- Gross A, McDonnell JM, Korsmeyer SJ. BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 1999;13:1899–1911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- Haque ME, Thomas KJ, D’Souza C, Callaghan S, Kitada T, Slack RS, Fraser P, Cookson MR, Tandon A, Park DS. Cytoplasmic Pink1 activity protects neurons from dopaminergic neurotoxin MPTP. Proc Natl Acad Sci U S A. 2008;105:1716–1721. doi: 10.1073/pnas.0705363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinis C, Schmitt S, Kindermann M, Godin G, Johnsson K. Evolving the substrate specificity of O6-alkylguanine-DNA alkyltransferase through loop insertion for applications in molecular imaging. ACS Chem Biol. 2006;1:575–584. doi: 10.1021/cb6003146. [DOI] [PubMed] [Google Scholar]
- Henchcliffe C, Beal MF. Mitochondrial biology and oxidative stress in Parkinson disease pathogenesis. Nat Clin Pract Neurol. 2008;4:600–609. doi: 10.1038/ncpneuro0924. [DOI] [PubMed] [Google Scholar]
- Hertz NT, Wang BT, Allen JJ, Zhang C, Dar AC, Burlingame AL, Shokat KM. Chemical Genetic Approach for Kinase-Substrate Mapping by Covalent Capture of Thiophosphopeptides and Analysis by Mass Spectrometry. Current Protocols in Chemical Biology. 2010;2:15–36. doi: 10.1002/9780470559277.ch090201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindie V, Stroba A, Zhang H, Lopez-Garcia LA, Idrissova L, Zeuzem S, Hirschberg D, Schaeffer F, Jorgensen TJ, Engel M, et al. Structure and allosteric effects of low-molecular-weight activators on the protein kinase PDK1. Nat Chem Biol. 2009;5:758–764. doi: 10.1038/nchembio.208. [DOI] [PubMed] [Google Scholar]
- Hua Z, Leal-Ortiz S, Foss SM, Waites CL, Garner CC, Voglmaier SM, Edwards RH. v-SNARE composition distinguishes synaptic vesicle pools. Neuron. 2011;71:474–487. doi: 10.1016/j.neuron.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y, Hori Y, Sakai S, Honma Y. Control of differentiation and apoptosis of human myeloid leukemia cells by cytokinins and cytokinin nucleosides, plant redifferentiation-inducing hormones. Cell Growth Differ. 2002;13:19–26. [PubMed] [Google Scholar]
- Ishii Y, Sakai S, Honma Y. Cytokinin-induced differentiation of human myeloid leukemia HL-60 cells is associated with the formation of nucleotides, but not with incorporation into DNA or RNA. Biochim Biophys Acta. 2003;1643:11–24. doi: 10.1016/j.bbamcr.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Kissil JL, Feinstein E, Cohen O, Jones PA, Tsai YC, Knowles MA, Eydmann ME, Kimchi A. DAP-kinase loss of expression in various carcinoma and B-cell lymphoma cell lines: possible implications for role as tumor suppressor gene. Oncogene. 1997;15:403–407. doi: 10.1038/sj.onc.1201172. [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Klinkenberg M, Thurow N, Gispert S, Ricciardi F, Eich F, Prehn JH, Auburger G, Kogel D. Enhanced vulnerability of PARK6 patient skin fibroblasts to apoptosis induced by proteasomal stress. Neuroscience. 2010;166:422–434. doi: 10.1016/j.neuroscience.2009.12.068. [DOI] [PubMed] [Google Scholar]
- Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, Macartney TJ, Deak M, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Lieberman I, Simms ES. Enzymatic synthesis of purine nucleotides. J Biol Chem. 1955;215:417–427. [PubMed] [Google Scholar]
- Krishnan P, Fu Q, Lam W, Liou JY, Dutschman G, Cheng YC. Phosphorylation of pyrimidine deoxynucleoside analog diphosphates: selective phosphorylation of L-nucleoside analog diphosphates by 3-phosphoglycerate kinase. J Biol Chem. 2002;277:5453–5459. doi: 10.1074/jbc.M109025200. [DOI] [PubMed] [Google Scholar]
- Kulkarni RN, Bruning JC, Winnay JN, Postic C, Magnuson MA, Kahn CR. Tissue-specific knockout of the insulin receptor in pancreatic beta cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell. 1999;96:329–339. doi: 10.1016/s0092-8674(00)80546-2. [DOI] [PubMed] [Google Scholar]
- Lang AE, Lozano AM. Parkinson’s disease. First of two parts. N Engl J Med. 1998;339:1044–1053. doi: 10.1056/NEJM199810083391506. [DOI] [PubMed] [Google Scholar]
- Lieberman I, Kornberg A, Simms ES. Enzymatic synthesis of nucleoside diphosphates and triphosphates. J Biol Chem. 1955a;215:429–440. [PubMed] [Google Scholar]
- Lieberman I, Kornberg A, Simms ES. Enzymatic synthesis of pyrimidine nucleotides; orotidine-5′-phosphate and uridine-5′-phosphate. J Biol Chem. 1955b;215:403–451. [PubMed] [Google Scholar]
- Liu Y, Shah K, Yang F, Witucki L, Shokat KM. A molecular gate which controls unnatural ATP analogue recognition by the tyrosine kinase v-Src. Bioorg Med Chem. 1998;6:1219–1226. doi: 10.1016/s0968-0896(98)00099-6. [DOI] [PubMed] [Google Scholar]
- Meissner C, Lorenz H, Weihofen A, Selkoe DJ, Lemberg MK. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J Neurochem. 2011;117:856–867. doi: 10.1111/j.1471-4159.2011.07253.x. [DOI] [PubMed] [Google Scholar]
- Merrick KA, Wohlbold L, Zhang C, Allen JJ, Horiuchi D, Huskey NE, Goga A, Shokat KM, Fisher RP. Switching Cdk2 on or off with small molecules to reveal requirements in human cell proliferation. Mol Cell. 2011;42:624–636. doi: 10.1016/j.molcel.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RD, Sim CH, Mok SS, Mulhern TD, Culvenor JG, Cheng HC. Biochemical aspects of the neuroprotective mechanism of PTEN-induced kinase-1 (PINK1) J Neurochem. 2008;105:18–33. doi: 10.1111/j.1471-4159.2008.05249.x. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Nemani VM, Azarbal F, Skibinski G, Levy JM, Egami K, Munishkina L, Zhang J, Gardner B, Wakabayashi J, et al. Direct membrane association drives mitochondrial fission by the Parkinson disease-associated protein alpha-synuclein. J Biol Chem. 2011;286:20710–20726. doi: 10.1074/jbc.M110.213538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niefind K, Putter M, Guerra B, Issinger OG, Schomburg D. GTP plus water mimic ATP in the active site of protein kinase CK2. Nat Struct Biol. 1999;6:1100–1103. doi: 10.1038/70033. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y. The role of protein kinase C in cell surface signal transduction and tumour promotion. Nature. 1984;308:693–698. doi: 10.1038/308693a0. [DOI] [PubMed] [Google Scholar]
- Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin DW, Leung HB, Schramm VL. Synthesis of nucleotides with specific radiolabels in ribose. Primary 14C and secondary 3H kinetic isotope effects on acid-catalyzed glycosidic bond hydrolysis of AMP, dAMP, and inosine. J Biol Chem. 1984;259:9411–9417. [PubMed] [Google Scholar]
- Petit A, Kawarai T, Paitel E, Sanjo N, Maj M, Scheid M, Chen F, Gu Y, Hasegawa H, Salehi-Rad S, et al. Wild-type PINK1 prevents basal and induced neuronal apoptosis, a protective effect abrogated by Parkinson disease-related mutations. J Biol Chem. 2005;280:34025–34032. doi: 10.1074/jbc.M505143200. [DOI] [PubMed] [Google Scholar]
- Pridgeon JW, Olzmann JA, Chin LS, Li L. PINK1 Protects against Oxidative Stress by Phosphorylating Mitochondrial Chaperone TRAP1. PLoS Biol. 2007;5:e172. doi: 10.1371/journal.pbio.0050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Molina H, Pandey A, Zhang J, Cole PA. Chemical rescue of a mutant enzyme in living cells. Science. 2006;311:1293–1297. doi: 10.1126/science.1122224. [DOI] [PubMed] [Google Scholar]
- Ray AS, Vela JE, Olson L, Fridland A. Effective metabolism and long intracellular half life of the anti-hepatitis B agent adefovir in hepatic cells. Biochem Pharmacol. 2004;68:1825–1831. doi: 10.1016/j.bcp.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Rugarli EI, Langer T. Mitochondrial quality control: a matter of life and death for neurons. EMBO J. 2012;31:1336–1349. doi: 10.1038/emboj.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaranch L, Lorenzo-Betancor O, Arbelo JM, Ferrer I, Lorenzo E, Irigoyen J, Pastor MA, Marrero C, Isla C, Herrera-Henriquez J, et al. PINK1-linked parkinsonism is associated with Lewy body pathology. Brain. 2010;133:1128–1142. doi: 10.1093/brain/awq051. [DOI] [PubMed] [Google Scholar]
- Shah K, Liu Y, Deirmengian C, Shokat KM. Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proc Natl Acad Sci U S A. 1997;94:3565–3570. doi: 10.1073/pnas.94.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty RS, Gallagher CS, Chen YT, Hims MM, Mull J, Leyne M, Pickel J, Kwok D, Slaugenhaupt SA. Specific correction of a splice defect in brain by nutritional supplementation. Hum Mol Genet. 2011;20:4093–4101. doi: 10.1093/hmg/ddr333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, Troconso JC, Dawson VL, Dawson TM. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson’s disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Jang S, Park J, Bang S, Choi S, Kwon KY, Zhuang X, Kim E, Chung J. Characterization of phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1) mutations associated with Parkinson’s disease in mammalian cells and Drosophila. J Biol Chem. 2013 doi: 10.1074/jbc.M112.430801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ultanir SK, Hertz NT, Li G, Ge WP, Burlingame AL, Pleasure SJ, Shokat KM, Jan LY, Jan YN. Chemical genetic identification of NDR1/2 kinase substrates AAK1 and Rabin8 Uncovers their roles in dendrite arborization and spine development. Neuron. 2012;73:1127–1142. doi: 10.1016/j.neuron.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Vela JE, Olson LY, Huang A, Fridland A, Ray AS. Simultaneous quantitation of the nucleotide analog adefovir, its phosphorylated anabolites and 2′-deoxyadenosine triphosphate by ion-pairing LC/MS/MS. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;848:335–343. doi: 10.1016/j.jchromb.2006.10.063. [DOI] [PubMed] [Google Scholar]
- Wang HL, Chou AH, Yeh TH, Li AH, Chen YL, Kuo YL, Tsai SR, Yu ST. PINK1 mutants associated with recessive Parkinson’s disease are defective in inhibiting mitochondrial release of cytochrome c. Neurobiol Dis. 2007;28:216–226. doi: 10.1016/j.nbd.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Gao X, Warne R, Hao X, Bussiere D, Gu XJ, Uno T, Liu Y. Design and synthesis of benzoazepin-2-one analogs as allosteric binders targeting the PIF pocket of PDK1. Bioorg Med Chem Lett. 2010;20:3897–3902. doi: 10.1016/j.bmcl.2010.05.019. [DOI] [PubMed] [Google Scholar]
- Xiong H, Wang D, Chen L, Choo YS, Ma H, Tang C, Xia K, Jiang W, Ronai Z, Zhuang X, et al. Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J Clin Invest. 2009;119:650–660. doi: 10.1172/JCI37617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZR, Zhang YH, Liu S, Song AX, Hu HY. Length of the active-site crossover loop defines the substrate specificity of ubiquitin C-terminal hydrolases for ubiquitin chains. Biochem J. 2012;441:143–149. doi: 10.1042/BJ20110699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.