Abstract
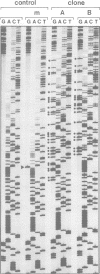
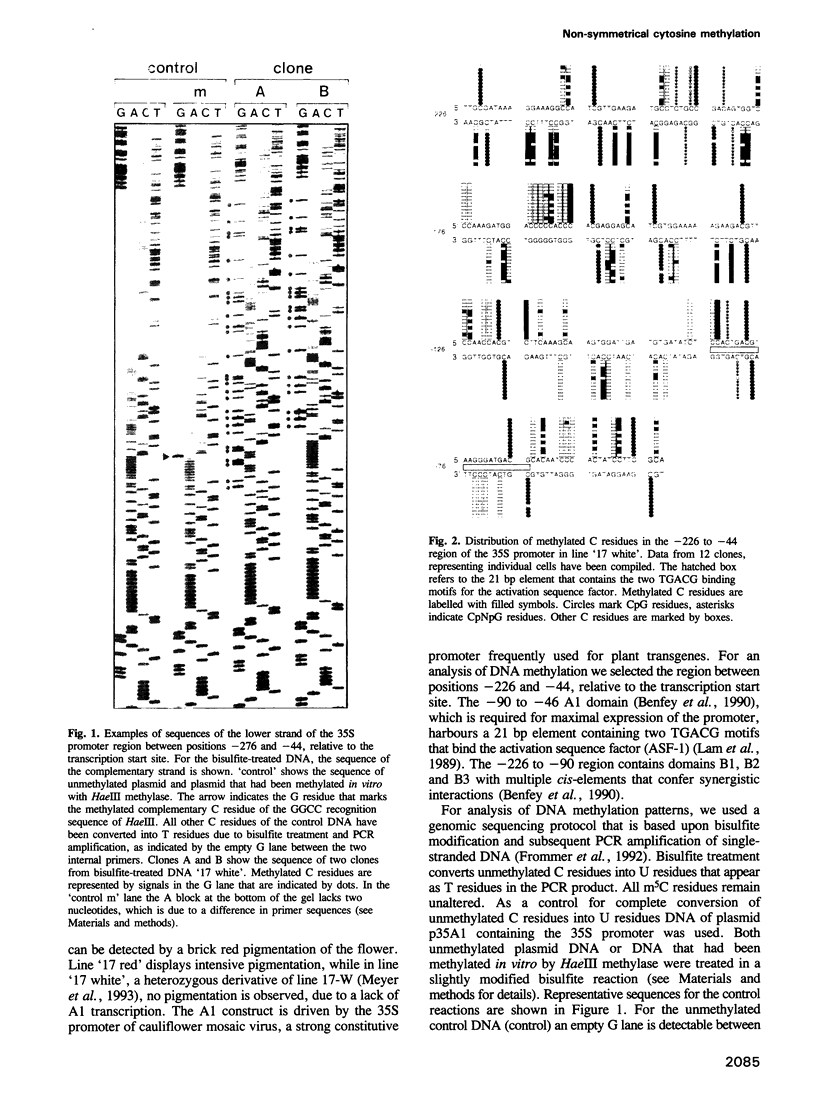
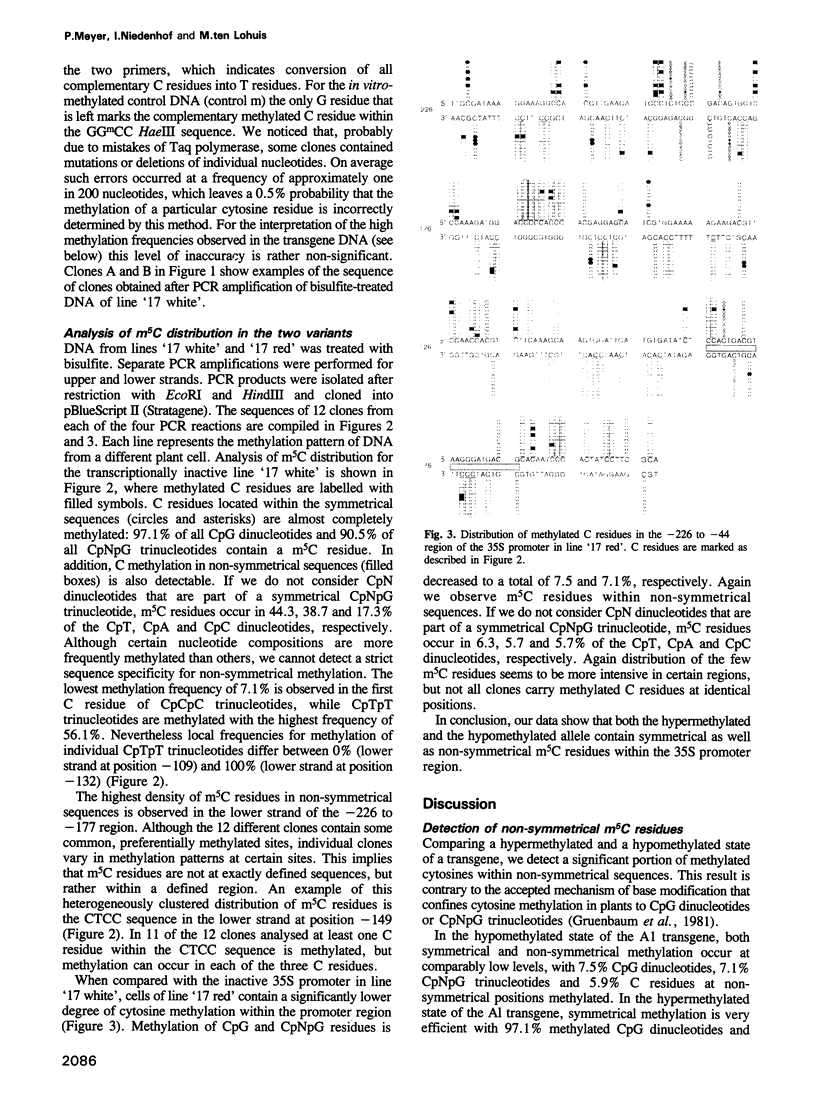
A considerable proportion of cytosine residues in plants are methylated at carbon 5. According to a well-accepted rule, cytosine methylation is confined to symmetrical sequences such as CpG and CpNpG, which provide the signal for faithful transmission of symmetrical methylation patterns by maintenance methylase. Using a genomic sequencing technique, we have analysed cytosine methylation patterns within a hypermethylated and a hypomethylated state of a transgene in Petunia hybrida. Examination of a part of the transgene promoter revealed that in both states m5C residues located within non-symmetrical sequences could be detected. Non-symmetrical C residues in the two states were methylated at frequencies of 5.9 and 31.9%, respectively. Methylation appeared to be distributed heterogeneously, but some DNA regions were more intensively methylated than others. Our results show that at least in a transgene, a heterogeneous methylation pattern, which does not depend on symmetry of target sequences, can be established and conserved.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benfey P. N., Ren L., Chua N. H. Combinatorial and synergistic properties of CaMV 35S enhancer subdomains. EMBO J. 1990 Jun;9(6):1685–1696. doi: 10.1002/j.1460-2075.1990.tb08292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Deumling B. Sequence arrangement of a highly methylated satellite DNA of a plant, Scilla: A tandemly repeated inverted repeat. Proc Natl Acad Sci U S A. 1981 Jan;78(1):338–342. doi: 10.1073/pnas.78.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- Finnegan E. J., Dennis E. S. Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res. 1993 May 25;21(10):2383–2388. doi: 10.1093/nar/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss H. M., Roberts C. J., Claeys K. M., Selker E. U. Abnormal chromosome behavior in Neurospora mutants defective in DNA methylation. Science. 1993 Dec 10;262(5140):1737–1741. doi: 10.1126/science.7505062. [DOI] [PubMed] [Google Scholar]
- Frommer M., McDonald L. E., Millar D. S., Collis C. M., Watt F., Grigg G. W., Molloy P. L., Paul C. L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982 Feb 18;295(5850):620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Naveh-Many T., Cedar H., Razin A. Sequence specificity of methylation in higher plant DNA. Nature. 1981 Aug 27;292(5826):860–862. doi: 10.1038/292860a0. [DOI] [PubMed] [Google Scholar]
- Holliday R., Pugh J. E. DNA modification mechanisms and gene activity during development. Science. 1975 Jan 24;187(4173):226–232. [PubMed] [Google Scholar]
- Holliday R. The inheritance of epigenetic defects. Science. 1987 Oct 9;238(4824):163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- Kane S. E., Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol Cell Biol. 1985 Sep;5(9):2298–2306. doi: 10.1128/mcb.5.9.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E., Benfey P. N., Gilmartin P. M., Fang R. X., Chua N. H. Site-specific mutations alter in vitro factor binding and change promoter expression pattern in transgenic plants. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7890–7894. doi: 10.1073/pnas.86.20.7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E., Beard C., Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993 Nov 25;366(6453):362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Li E., Bestor T. H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992 Jun 12;69(6):915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Matzke M. A., Primig M., Trnovsky J., Matzke A. J. Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. EMBO J. 1989 Mar;8(3):643–649. doi: 10.1002/j.1460-2075.1989.tb03421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P., Heidmann I., Forkmann G., Saedler H. A new petunia flower colour generated by transformation of a mutant with a maize gene. Nature. 1987 Dec 17;330(6149):677–678. doi: 10.1038/330677a0. [DOI] [PubMed] [Google Scholar]
- Meyer P., Heidmann I., Niedenhof I. Differences in DNA-methylation are associated with a paramutation phenomenon in transgenic petunia. Plant J. 1993 Jul;4(1):89–100. doi: 10.1046/j.1365-313x.1993.04010089.x. [DOI] [PubMed] [Google Scholar]
- Pröls F., Meyer P. The methylation patterns of chromosomal integration regions influence gene activity of transferred DNA in Petunia hybrida. Plant J. 1992 Jul;2(4):465–475. doi: 10.1046/j.1365-313x.1992.t01-20-00999.x. [DOI] [PubMed] [Google Scholar]
- Rhounim L., Rossignol J. L., Faugeron G. Epimutation of repeated genes in Ascobolus immersus. EMBO J. 1992 Dec;11(12):4451–4457. doi: 10.1002/j.1460-2075.1992.tb05546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14(1):9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Russell P. J., Rodland K. D., Rachlin E. M., McCloskey J. A. Differential DNA methylation during the vegetative life cycle of Neurospora crassa. J Bacteriol. 1987 Jun;169(6):2902–2905. doi: 10.1128/jb.169.6.2902-2905.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K., Okada N. 5-Methylcytidylic modification of in vitro transcript from the rat identifier sequence; evidence that the transcript forms a tRNA-like structure. Nucleic Acids Res. 1985 Oct 25;13(20):7195–7206. doi: 10.1093/nar/13.20.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U., Fritz D. Y., Singer M. J. Dense nonsymmetrical DNA methylation resulting from repeat-induced point mutation in Neurospora. Science. 1993 Dec 10;262(5140):1724–1728. doi: 10.1126/science.8259516. [DOI] [PubMed] [Google Scholar]
- Selker E. U. Premeiotic instability of repeated sequences in Neurospora crassa. Annu Rev Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- Simon D., Stuhlmann H., Jähner D., Wagner H., Werner E., Jaenisch R. Retrovirus genomes methylated by mammalian but not bacterial methylase are non-infectious. Nature. 1983 Jul 21;304(5923):275–277. doi: 10.1038/304275a0. [DOI] [PubMed] [Google Scholar]
- Vongs A., Kakutani T., Martienssen R. A., Richards E. J. Arabidopsis thaliana DNA methylation mutants. Science. 1993 Jun 25;260(5116):1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Wang R. Y., Gehrke C. W., Ehrlich M. Comparison of bisulfite modification of 5-methyldeoxycytidine and deoxycytidine residues. Nucleic Acids Res. 1980 Oct 24;8(20):4777–4790. doi: 10.1093/nar/8.20.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]