Abstract
Neurobiologists address neural structure, development and function at the level of “macrocircuits” (how are different brain compartments interconnected, what overall pattern of activity do they produce), and at the level of “microcircuits” (how does connectivity and physiology of individual neurons and their processes within a compartment determine the functional output of this compartment). Work in our lab aims at reconstructing the developing Drosophila brain at both levels. Macrocircuits can be approached conveniently by reconstructing the pattern of brain lineages, which form groups of neurons whose projections form cohesive fascicles interconnecting the compartments of the larval and adult brain. The reconstruction of microcircuits requires serial section electron microscopy, due to the small size of terminal neuronal processes and their synaptic contacts. Because of the amount of labor that traditionally comes with this approach, very little is known about microcircuitry in brains across the animal kingdom. Many of the problems of serial EM reconstruction is now solvable with digital image recording and specialized software for both image acquisition and post-processing. In this paper we introduce our efforts to reconstruct the small Drosophila larval brain, and discuss our results in light of the published data on neuropile ultrastructure in other animal taxa.
Keywords: Drosophila, brain, lineage, connectivity, elctron microscopy
Introduction
Studies of nervous system architecture and function typically approach the brain at two different levels of resolution: the levels of macrocircuitry and microcircuitry. The macrocircuitry-oriented approach asks questions like: what functions can be attributed to brain compartments such as the mammalian primary visual cortex or lateral geniculate nucleus; what is the connectivity between these and other brain compartments. By contrast, the study of microcircuitry zooms in on neurons, their dendrites, axons and synapses. Thus, the way in which a given neuron is tuned to a specific input stimulus, or the pattern of activity triggered in this neuron when providing a specific input, depends on the distribution of excitatory and inhibitory synapses that connect the neuron with its neighbors (Douglas and Martin, 1998; Silberberg et al., 2002; Silberberg, 2008; Toledo-Rodriguez et al., 2005). The analysis of microcircuits is of great importance. All acts of fine motor control, memory formation and cognition can only be understood if the microcircuitry within the brain compartments dealing with these functions is known. Likewise, the insight into psychiatric disease mechanisms and their pharmacological treatment requires brain microcircuitry to be known. For example, recent findings suggest that diseases like schizophrenia can be understood in terms of abnormalities in the microcircuitry of the prefrontal cortex (Winterer and Weinberger, 2004; Rolls et al., 2008).
This useful conceptualization of the nervous system as being comprised of interconnected, structurally defined compartments, constituting microcircuits integrated into macro-circuits, also applies to the brain of invertebrates, such as the fruit fly Drosophila. The nervous system of Drosophila (and insects in general) is formed by a relatively small number of genetically and structurally defined modules, the neural lineages. The ventral nerve cord and subesophageal ganglion (containing the circuits controlling locomotion, flight, and feeding) are built of approximately 80 bilaterally symmetric pairs of lineages; the central brain, a mostly sensory and associative center, is formed by 100 paired lineages (Goodman and Doe, 1993; Younossi-Hartenstein et al., 1996; Urbach and Technau, 2003; Truman et al., 2004). Each lineage is derived from an asymmetrically dividing stem cell, called neuroblast, that is born in the early embryo (Hartenstein et al., 2008a, b; Fig.1A). Neuroblasts and the lineages they produce represent genetic modules (“units of gene expression”). The expression pattern of more than forty transcription factors in specific embryonic neuroblasts has been described (Urbach and Technau, 2003). A given transcription factor becomes active in one, or a small number of, neuroblasts; a particular neuroblast thereby acquires a “genetic address”, consisting of a specific set of transcription factors active in this cell. It is thought that this genetic address will essentially be involved in shaping the morphology and function of the lineage of neurons produced by the neuroblast.
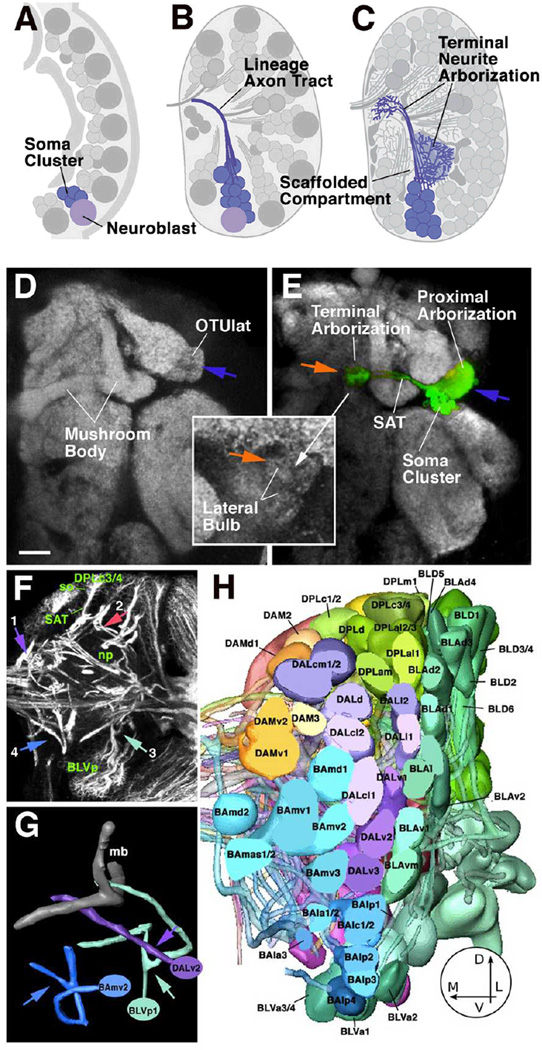
Fig. 1.
Developmental and structural characteristics of wild-type lineages. A-C: Lineages as units of gene expression, projection, and connectivity. Stereotyped population of neuroblasts generates neurons in the embryo and larva (A). Neurons belonging to one lineage form a cohesive cluster and project their axons in one fascicle (B). Terminal branches of neurons of one lineage arborize in specific neuropile compartments (C). D, E: Z-projection of adult brain hemisphere labeled with anti-Bruchpilot (Nc82; Kittel et al., 2006) to visualize neuropile compartments (white). In E, one lineage, DALcl1, is labeled by expression of GFP. Note dense proximal arborization restricted to lateral domain of optic tubercle (OTUlat; one of the optic foci); distal arborization is restricted to the lateral bulb, one of the input regions of the central complex. F: Z-projection of 10 successive 1µm confocal cross sections at level of central neuropile. Secondary lineages, their axon tracts (secondary axon tracts; SATs) and neuropile fascicles formed by convergence of SATs are labeled with anti-Neurotactin antibody (white). Clusters of somata (so) belonging to lineages are located in the cortex; axon tracts project centripetally into the neuropile (np). Arrows point at lineages representing the types of SAT trajectories observed: SAT is unbranched and enters the neuropile in a straight course (1; DPMm lineage) or after a sharp turn at the cortex-neuropile boundary (2; DPLc3/4). (3) SAT bifurcates into two branches at cortex-neuropile boundary (BLVp1/2); (4) distal part of SAT bifurcates in neuropile (BAmv2). G: Digital models of three representative lineage tracts illustrating typical branching behavior of SATs [DALv2: straight unbranched entry into neuropile; BLVp1: bifurcation at point of entry into neuropile (arrowhead); BAmv2: bifurcation in distal leg of SAT]. H: 3D digital models of all clusters of neuronal somata representing all lineages of one brain hemisphere; anterior view. The polar region of the cortex was removed for clearer view of lineages. (panels F-H modified from from Fung et al., 2009).
Bar: 20µm
Lineages also form structural modules. Thus, neurons that belong to one lineage remain together throughout development, forming compact clusters of cells (Fig.1B). More importantly, axons emitted by neurons of one lineage also form a coherent fascicle, the primary and secondary lineage axon tracts. This means that neurons of one lineage share their principal trajectory; they form a “unit of projection” (Fig.1C–G). Lineages thereby represent the most appropriate structural/developmental units of brain macro-circuitry. Based on their characteristic location and axon tract we have generated an atlas of all lineages of the central brain for the larval stage (Pereanu and Hartenstein, 2006; Fig.1H). Attempts are currently under way to link the larval lineage map with the adult stage, when each lineage has completed its terminal arborization (Pereanu et al., 2009).
We will in the following briefly summarize our recent findings pertaining to the structure and development of neural lineages of the Drosophila brain. We will then outline our approach to reconstruct microcircuitry in the fly brain using computer-aided serial electron microscopy, an approach that is guided by the lineage-centered macro-architectural map of the fly brain. Finally, we will describe first results shedding light on Drosophila microcircuitry, and discuss these data in the context of brain evolution.
Lineage based analysis of Drosophila brain structure and development
Using clonal marking techniques and specific Gal4 driver lines (Brand and Perrimon, 1993), we have analyzed representative Drosophila brain lineages at all stages of development (Larsen et al., 2009). Most lineages have a number of important characteristics in common; our focus was on these generic lineage features. The early neurons of a lineage generated during embryogenesis (primary neurons; 15–20 neurons per neuroblast for the large majority of lineages) stay together as a coherent cluster (Fig.2A, B). Likewise, axons of each lineage form a coherent bundle (primary axon tract; PAT) that follows a stereotyped pathway in the neuropile (Younossi-Hartenstein et al., 2006). Apoptotic cell death removes an average of 30–40% of neurons from the primary lineages around the time of hatching. Secondary neurons generated later, during the larval period, also form clusters that stays together all the way to the adult stage, and that form cohesive axon bundles (secondary axon tract; SAT). SATs project into the neuropile compartment visited by the corresponding primary axon tract (Fig.2C, F). SATs develop into the long fiber fascicles that interconnect the different compartments of the adult brain (Fig.2D, E). These fascicles can be easily recognized by labeling brains with global markers. For example, labeling with synaptic markers like the anti-DNcad or anti-Brp antibody, fascicles (which only contain long fibers and lack synapses) stand out as signal negative channels (Fig.2G, H).
Fig. 2.
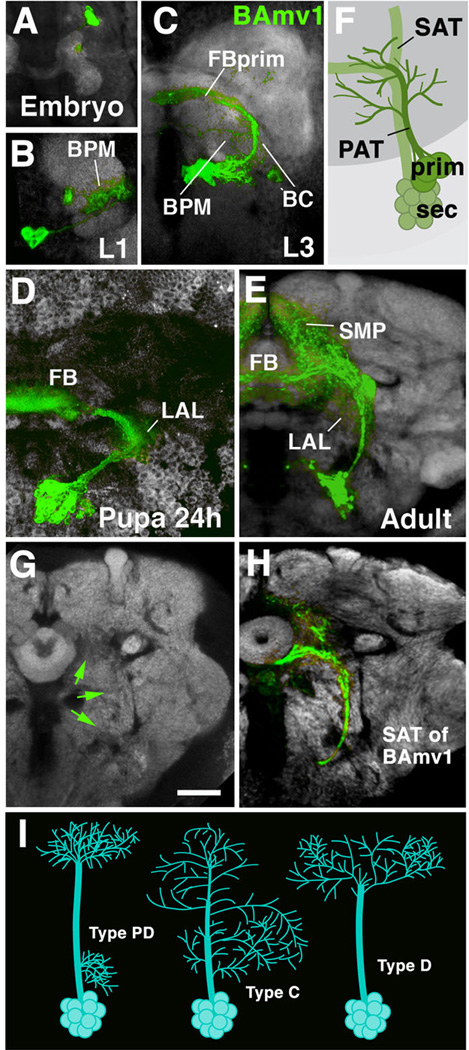
Morphogenesis of a brain lineage from embryo to adult. A-E: Z-projections of confocal sections of one brain hemisphere in which BAmv1 lineage is labeled by GFP driven by the line per-Gal4 (Kaneko and Hall, 2000). BAmv1 has a conspicuous crescent-shaped tract, projecting first posteriorly, then dorsolaterally, and finally dorso-medially towards the primordium of the fan-shaped body (FBprim), which forms part of the CPM compartment of the larval brain. Arborizations of primary neurons occur in BC, BPM and FBprim compartments (B, C). Secondary axons follow the same trajectory and branch in the lateral accessory lbe (LAL), fan-shaped body (FB), and superior medial protocerebrum (SMP; D, E). F: Cartoon illustrating that secondary axon tract (SAT) typically fasciculates with, or at least grows close to, primary axon tract (PAT) of the corresponding lineage. G, H: Secondary axon tracts develop into long fiber bundles of adult brain. G shows frontal confocal section of adult brain hemisphere labeled with anti-Bruchpilot (Nc82; neuropile; white). In H, the secondary neurons of the BAmv1 lineage are labeled by GFP (driven by per-Gal4). Note that the coherent secondary axon tract of BAmv1 now forms a long fiber bundle which is visible as a Nc82-negative (i.e., synapse-free) “tunnel”, indicated by green arrows in G.[SF1] I: schematic representation of different types of lineages encountered in Drosophila brain (PD: separate proximal and distal arborization; C: continuous arborization; D: distal arborization).
Bar: 20µm (all photographic panels at same scale)
In regard to overall arbor geometry, we distinguish between three types of lineages (Larsen et al., 2009; Fig.2I). Type PD (“Proximal-Distal”) lineages are characterized by distinct, spatially separate proximal (close to soma) and distal arborizations. Type C (“Continuous”) lineages have arborizations distributed more or less evenly along the entire length of their tract. Type D lineages (“Distal”) lack proximal arborizations; their fiber tract extends for a considerable distance into the neuropile before branching into a more or less complex distal arbor.
Arborizations of lineages, in particular those of type PD, are restricted to distinct neuropile compartments; we propose that compartments are “scaffolded” by individual lineages, or small groups thereof. Primary lineages set up the compartment map already in the embryo. Compartments then grow during the larval period simply by an increase in arbor volume of primary neurons. Arbors of secondary neurons form within the larval compartments, resulting in smaller compartment subdivisions and additional, adult specific compartments. We proposed that each compartment has its own “scaffolding lineage” (or set of scaffolding lineages) which would be defined in the following manner: (i) during development, the outgrowth of neurites from a lineage S generates the compartment S’. If S is deleted, S’ also does not form (this has been shown for the calyx, the compartment scaffolded by the four MB lineages (Ito et al., 1997), and for compartments of the central complex (Lovick and Hartenstein, unpublished). (ii)The arborization of lineage S forms a dense matrix of terminal fibers on which synapses of S neurons themselves, as well as extrinsic neurons that enter compartment S’ from the outside, are made. Again, the calyx provides an example in this case: electron microscopic investigations have shown that the majority of the postsynaptic terminal neurites belong to neurons of the MB lineages (Yasuyama et al., 2002). Our lab and others have found numerous other lineages whose proximal (or sometimes distal, axonal) terminal arborizations, in the adult brain, are highly focused in a small compartment. As example in point Fig.1D, E shows the labeled lineage DALcl1 with dense terminal arborizations in a small subcompartment, the lateral optic tubercle (OTUlat), and terminal arborizations confined even more narrowly to the lateral bulb, an input region of the central complex.
In summary, our data support the idea that lineages form the developmental/ anatomical substrate of Drosophila brain macro-circuitry: compartments and the long axon tracts interconnecting them can be assigned to specific (sets of) lineages. The reconstruction and digital representation of all lineages, including their axonal and dendritic arborization and interconnecting axon fascicle, will add up to a complete map of the fly brain macro-circuitry.
Analysis of Microcircuitry
The anatomical reconstruction of microcircuitry requires to document, at the level of single synapses and neuronal processes, how neurons in a given (small) volume of the brain are interconnected. Due to their small size, which lies in the range of 0.1–0.5µm, fine neurites and their synaptic contacts can be conclusively shown only electron microscopically. However, the acquisition of complete series of EM sections and their photographic documentation and analysis requires a considerable effort, and therefore studies of microcircuitry have traditionally been restricted to small parts of neurons or neuropile compartments in insects or other invertebrates (e.g., Watson and Burrows, 1983; Meinertzhagen and O’Neil, 1991; Yasuyama et al., 2002). The relatively complete reconstruction of the miniature C. elegans central nervous system, containing less than 400, unbranched neurons, represents a notable exception (White et al., 1986). The problem of image acquisition and reconstruction is now solvable with digital image recording and specialized software for image acquisition and post-processing, and we and other groups have begun to generate stacks of digitized images from serial EM sections. These stacks can be segmented and analyzed in their entirety, which makes it possible to reconstruct the way a neuron (or neurite) is connected to its immediate neighbors.
It should be pointed out that even with the digital technology available today, digital serial EM-based analysis is still feasible only for relatively small objects in the <mm range. That means that for a large vertebrate brain, only small “aliquots” of brain tissue can be processed. But this maybe sufficient, if one assumes (and evidence for this assumption is accumulating; see work reviewed in Kozloski et al., 2001; Silberberg et al., 2002) that large brain areas like the neocortex are essentially built in a stereotypical manner; that means that the microcircuitry characteristic of one cortical area is highly similar to that in a different area, so that many of the principles of microcircuitry that emerge from the analysis of one tissue “aliquot” can be generalized to other cortical domains. In our view, this concept does not argue against location specific properties: it merely suggests that it is practical to first undertake a comprehensive analysis of stereotyped aspects of microcircuitry; this then will make it easier to get at the additional, site specific properties.
Insect brains offer the advantage of a much smaller size. The early first instar larval (L1) brain of Drosophila, formed by approximately 1500 differentiated and functional nerve cells (Larsen et al., 2009), has a diameter of approximately 50µm. The neuropile that forms the center of a brain hemisphere, measures less than 30µm. Given these specifications, we are currently working with approximately 500 sections of 60nm thickness; a more complete series of sections that includes both brain hemispheres and the ventral nerve cord, is under way. We aimed at a resolution of approximately 2–3nm per pixel; synaptic contacts and fine processes (diameter of about 100nm), or even synaptic vesicles (20–30nm), can be clearly resolved at this resolution. The size of the digitized image of one section contains approximately 15,000×15,000 pixels. We have packed the entire post-processing pipeline in our own custom software TrakEM2, which is based on ImageJ, an NIH-sponsored image processing platform (www.ini.uzh.ch/~acardona/trakem2. html). The software allows us to stitch individual photographs covering one section into one seamless montage, to register montages of each section, and to navigate the resulting stack of sections efficiently (Saalfeld et al., 2009; Cardona et al., 2009).
Microcircuitry data analysis and reconstruction: Merging EM and LM data
When browsing EM sections, the staggering amount of data collected becomes very difficult to process without an a priori “low resolution knowledge” of the architecture of the object, i.e., the L1 Drosophila brain. The navigation of TEM images must be guided by known, labeled and registered confocal stacks which provide cues regarding the position of major “macroarchitectural” landmarks like axon bundles or neuropile compartment boundaries, and thus provide the necessary context for the analysis of microarchitectural components of the neuropile. In other words: If the user wants to zoom in on the pattern of connectivity in a small volume of neuropile, he/she needs to know what compartment this volume is located in, or what are the lineages and major tracts that connect the compartment to the rest of the brain. The TrakEM2 pipeline is designed to embed the analysis of the EM stack at the microcircuit level into the light microscopically derived macroarchitectural framework.
We have defined a set of lineages, compartments and major axon tracts that can be recognized at all developmental stages and the adult, and which serve as macro-architectural landmarks. We could identify in our preliminary L1 EM stack most of these landmarks, including compartments, neuropile fascicles, and individual lineage tracts (Fig.3A). Thus, the lineage associated primary axon tracts of the L1 brain contain 6–20 tightly bundled, straight axons that are visible in the EM stack if one knows where to look for them (Fig.3B). Moreover, PATs of multiple lineages converge to form major axonal “thoroughfares” in the neuropile, such as the antenno-cerebral tract, peduncle, or longitudinal central tract. These tracts are all associated with high concentrations of glial processes, which further facilitates their recognition in the EM stack. Glial densities also define many of the compartment boundaries (Younossi-Hartenstein et al., 2003). After identifying and segmenting the macroarchitectural landmarks in the EM stack they become the objects of an intrinsic macro-model. Thus, with the help of the drawing tool, label fields are created on each section; the label fields stacked along the z-axis form a given object for which a surface is generated and which can be displayed as an interactive digital 3D model.
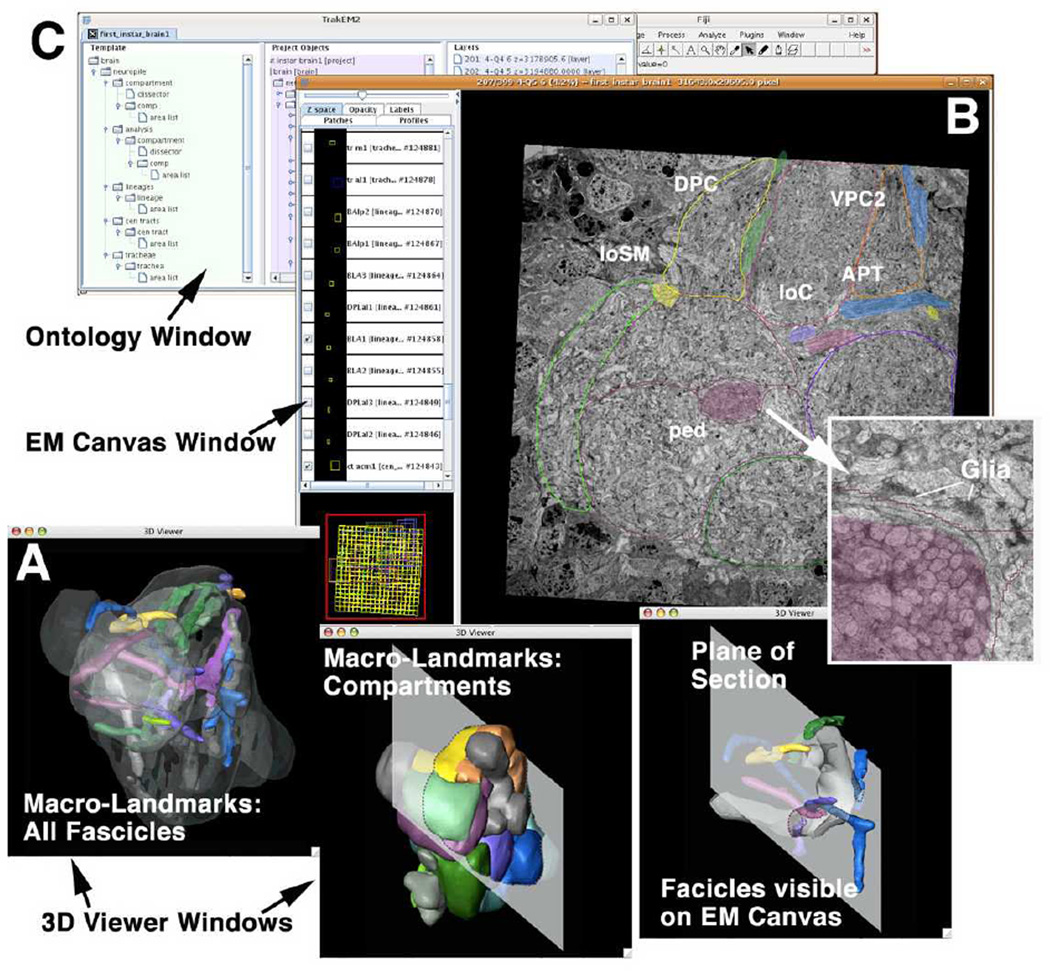
Fig. 3.
The graphical user interface of TrakEM2. The EM canvas presents the EM stack. The user can scroll and navigate through all sections in the “Google Map” style. Identified structures (e.g., compartments and lineage tracts) can be segmented. The ontology window displays all segmented objects as a hierarchically organized list. The interactive 3D viewer windows show selected objects segmented from the EM stack.
Figure 3 shows the current graphic user interface of TrakEM2. It is composed of three simultaneously active windows: the ontology window (Fig.3C); the EM (raw data) canvas (Fig.3B); the 3D viewer windows (Fig.3A). All objects of the model are displayed as a hierarchical list in the ontology window; here, individual objects can be activated or inactivated, and properties of the digital rendering of each object, such as color or transparency, can be changed. Activating an object in the ontology window will lead to its appearance in the EM canvas window, as well as in the 3D viewer. In both EM canvas and 3D viewer windows, one can further interact with the object in manifold ways. As a result, TrakEM2 allows us to focus on any part of the neuropile in the EM canvas, and at the same time look up in the intrinsic macro-model (displayed in the 3D viewer) where we are, and what lineages or tracts are close by.
Quantitative analysis of connectivity in “micro-volumes”
Given that the problems of sectioning, EM imaging and data handling of complete L1 brains are in principle solved, our ultimate goal is to reconstruct all neurons and their connections in their entirety. However, even a complete stack without any gaps will be difficult to register “perfectly” in toto, given the current technology. “Perfectly” would imply that, given the thinness of terminal processes, any pixel of the 15,000×15,000pixel montage has to be less than 25 pixel (=about 100nm) removed from its “true” x/y position. Considering the fact that sectioning, heating (electron beam) and handling add up to deformations of all sections (independent of each other), it will be understandable that it is currently impossible to register section montages with that accuracy. The strategy is therefore to break down the overall EM stack volume into smaller “micro-volumes”, in the range of 5×5x5µm, and reconstruct these. TrakEM2 allows us to efficiently “cut out” micro-volumes from the total stack and re-register them automatically, as well as manually, at an accuracy level high enough for segmenting even fine processes. Multiple micro-volumes can be sampled from any desired position, guided by the Macro-landmarks. It should be noted that, even irrespective of the technical factors that suggest the micro-volume approach, this approach seems also conceptually well justified. Thus, as pointed out for the vertebrate brain above, it stands to reason that the Drosophila brain is structured in a stereotypical way: the biological “algorithm” underlying the neuronal wiring in a given micro-volume may well resemble that of a neighboring volume. It seems appropriate to sample and compare micro-volumes from different compartments, formed by different lineages and reached by different types of sensory input, before eventually reconstruct the entire neuropile in toto. Fundamental parameters of microcircuitry can be gained from the micro-volumes:
what types of neurites (diameter, shape, branching behavior) do exist?
what is the density/directionality of neurites (of different types) within a micro-volume
what is the density of branchpoints; how is branching of different neurites coordinated?
what is the density and distribution of input/output synapses, relative to neurite type, neurite directionality, branching behavior?
In the following section we will summarize our recent findings (Cardona et al., 2009) that start providing answers to these questions.
Drosophila brain microcircuitry: neurite profiles and distribution of synapses
The brain neuropile contains neurites connected by synapses, as well as glia. Recognizable by their lamellar shape and high electron density, glial processes form more or less continuous sheaths around the neuropile surface, as well as around many compartments and fascicles. Within neuropile compartments, fine glial processes are interspersed with terminal neurites and synapses. Neurites come in several different classes that relate in a systematic way to pre-and postsynaptic contact sites (Fig.4). We distinguish between axiform neurites, varicose neurites, globular neurites and dendritiform neurites (Fig.4A–C). These generic classes of neurites can be found in all compartments of the brain. Axiform neurites (axis = axle) are straight, unbranched processes of even diameter, ranging between 0.2 and 0.4µm. These processes typically form bundles. The primary axon tract (PAT) emitted by the neurons belonging to one lineage consist of axiform neurites. Within the neuropile, thick bundles of axiform neurites (created by the coalescence of multiple PATs) form the long fiber tracts, such the antenno-protocerebral tract. Varicose neurites (varix = dilated vein) make up most of the volume of the neuropile. They represent branched processes that vary in diameter and change direction. Along their length, thin segments (0.2–0.4µm) alternate with swellings (“varicosities”) that measure 0.5–1.5µm. Globular neurites (globus = round body) contain one (terminal) or several large diameter (1.5–3µm) round or irregularly shaped segments. These formations structurally and functionally resemble closely the endplates, or “boutons”, of motor axons. Dendritiform neurites (dendron = tree) are thin, highly branched processes. They change direction frequently. Short, terminal branches of these “trees” are in the range of 0.1µm.
Fig. 4.
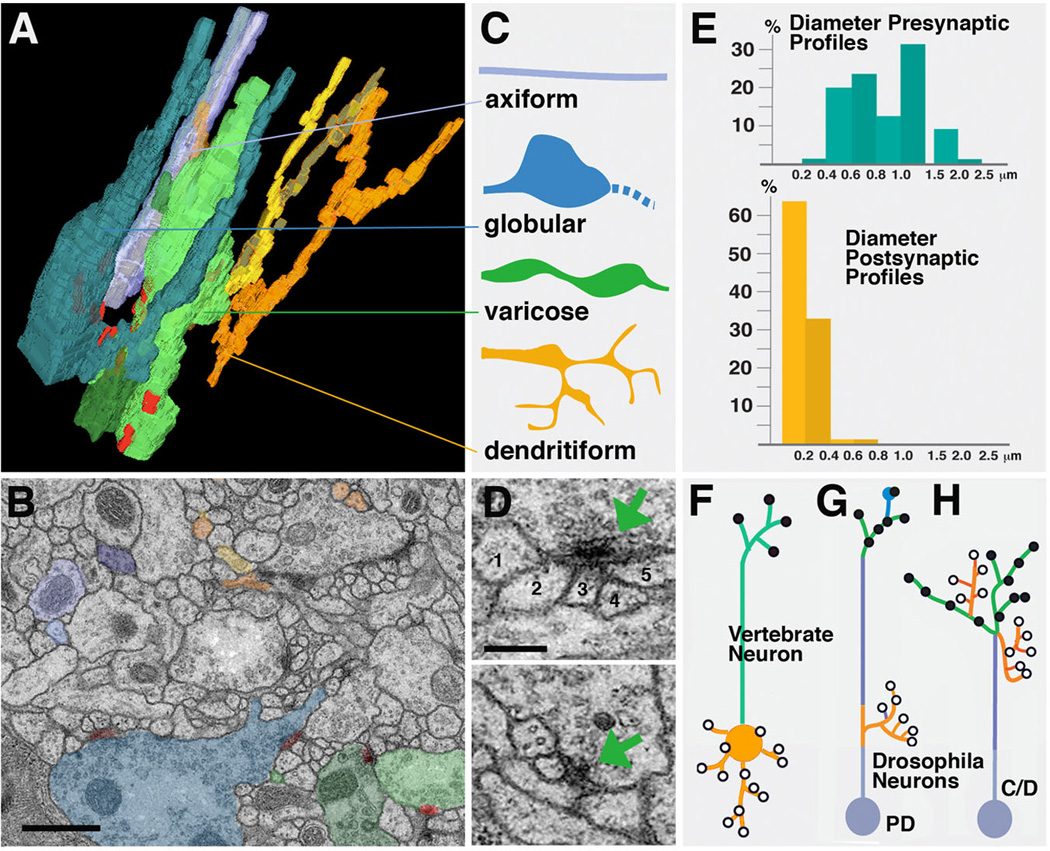
Drosophila neuropile ultrastructure. A-C: Types of neurite profiles. A shows 3D digital model of several short neurites segmented from one micro-volume. B: representative EM section of microvolume in which profiles of neurites modeled in A are shaded in the corresponding colors. C: Schematic depiction of types of neurites. Axiform neurites (light blue) are straight, unbranched processes of intermediate (0.2–0.4µm) diameter. Globular neurites and varicose neurites (dark blue and green) have alternating segments of intermediate (0.2–0.4µm) and large diameter (0.5–1.5µm for varicose; 1–3µm for globular neurites). Dendritiform neurites (yellow, brown, orange) are highly branched and thin (< 0.2µm). D: Section of two typical polyadic synapses. Green arrow points at presynaptic specialization, consisting of the T-bar and synaptic vesicles. Presynapses are contacted by multiple, thin branches of dendritiform processes. Numbers 1–5 in upper panel denote profiles of thin, postsynaptic profiles (dendritiform neurites). #2–5 have clear, direct contact to presynaptic zone; #1 is located right adjacent to the presynaptic site, and represents a case that may or may not represent a postsynaptic neurite of this synapse. E: Correlation between frequency of presynaptic and postsynaptic sites and neurite diameter. Presynaptic sites (blue; top) are predominantly found on large diameter profiles, which correspond to thick segments of varicose and globular neurites. These neurites represent terminal axonal branches. Postsynaptic profiles (yellow, bottom) almost exclusively belong to thin dendritiform neurites; they represent terminal dendritic branches. F-H: Distribution of axonal and dendritic branches. F: Polarized vertebrate neuron with dendrite/soma compartment carrying postsynaptic sites, and axon compartment with presynaptic sites. G: Drosophila type PD neuron which comes close to the vertebrate pattern, with postsynaptic dendritiform processes concentrated proximally and presynaptic varicose/globular processes distally. H: In Drosophila type C and D neurons, terminal axons and dendrites are intermingled.
Bars: 0.5µm (B); 0.2µm (D)
Synapses are defined by the characteristic presynaptic site, an electron dense patch of membrane bordered by the T-bar, a cytoplasmic specialization involved in tethering and docking of synaptic vesicles (Feeney et al., 1998; Kittel et al., 2006; Prokop and Meinertzhagen, 2006; Fig.4D). Synaptic vesicles can be observed at or near presynaptic sites. Presynaptic contact sites are relatively uniform in size, ranging from 0.1–0.3µm, and are very predominantly found on large diameter segments of neurites, i.e., the swellings of varicose neurites and globular neurites (Fig.4E); these neurites then constitute the output, or axonal, branches within the neuropile. Postsynaptic sites are less conspicuous membrane densities lacking T-bars or synaptic vesicles; they are found almost exclusively on thin dendritiform neurites and, occasionally, thin side branches of varicose neurites. The discrepancy in diameter between pre- and postsynaptic neurites naturally creates the characteristic polyadic synapse: the relatively large size of the presynaptic element creates a “platform” that is in direct contact with multiple, thin postsynaptic elements (Fig.4D). It should be noted that for most synapses, it is difficult to infer exactly how many postsynaptic partners exist. Thus, postsynaptic membrane specializations are very subtle, and one can only base the assertion that a given profile represents a postsynaptic partner on whether or not its membrane is in direct contact with the presynaptic membrane (density). This, in numerous cases, is ambiguous (e.g., Fig.4D).
In vertebrates, neurons show three different compartments, soma, dendrite, and axon (Fig.4F). Dendrites arise from the soma, and are in terms of structure (e.g., microtubule cytoskeleton) and function similar to the soma, and different from the axon (Peters et al., 1976; Baas and Yu, 1996). This cell biological distinction between dendrite/soma and axon goes hand in hand with a functional distinction: dendrites and soma carry mostly postsynaptic membrane specializations; axons are specialized to conduct axon potentials, and carry presynaptic sites at their terminals. The situation is different in insect central neurons. Here, the soma emits a single neurite, which in the neuropile forms numerous branches that are dendritic (i.e., postsynaptic), axonal (i.e., presynaptic), or, frequently, mixed (both post and presynaptic sites intermingled). For this reason, one can strictly speaking not refer to axons or dendrites, but just to neurites, when referring to neuronal processes in the Drosophila CNS. There clearly exist neurons (those that are part of PD lineages; see Fig.1I; Fig.4G) which resemble to some extent the typical vertebrate neuron, in the sense that they have postsynaptic (dendritic) branches proximally, close to the soma; a long, unbranched “axon” projects away from these dendrites and ends in multiple axonal terminal branches carrying presynaptic sites. The antennal projection neurons with dendrites in the antennal lobe (primary olfactory center; Stocker, 1994; Lai et al., 2008) and axons in the calyx of the mushroom body are an example in case. But we estimate that the majority of brain lineages are of type C or D; how are pre- and postsynaptic sites arranged in neurons belonging to these lineages?
Axiform neurites in any of the micro-volumes reconstructed are essentially devoid of synapses. They form the long axon fascicles interconnecting compartments (“macrocircuitry”). These processes then would most closely correspond to the long axons of vertebrates. The swellings of varicose and globular neurites contain almost all presynaptic sites; these then are the terminal axonal branches. The fine processes of dendritiform branches carry postsynaptic sites. Note that, at least at the level of several microns (which for the 30µm L1 brain is a lot!), neurites are very predominantly either dendritic or varicose/globular (axonal). This does not mean that a given neuron could not have both types of branches close to each other; but at the level of a given branch, input and output is separated (Fig.4H). This rule is only violated by the occasional thin ‘side projections” that occur on or near varicosities, and that carry postsynaptic sites contacting nearby presynaptic neurites (Cardona et al., 2009; see Fig.6D).
Fig. 6.
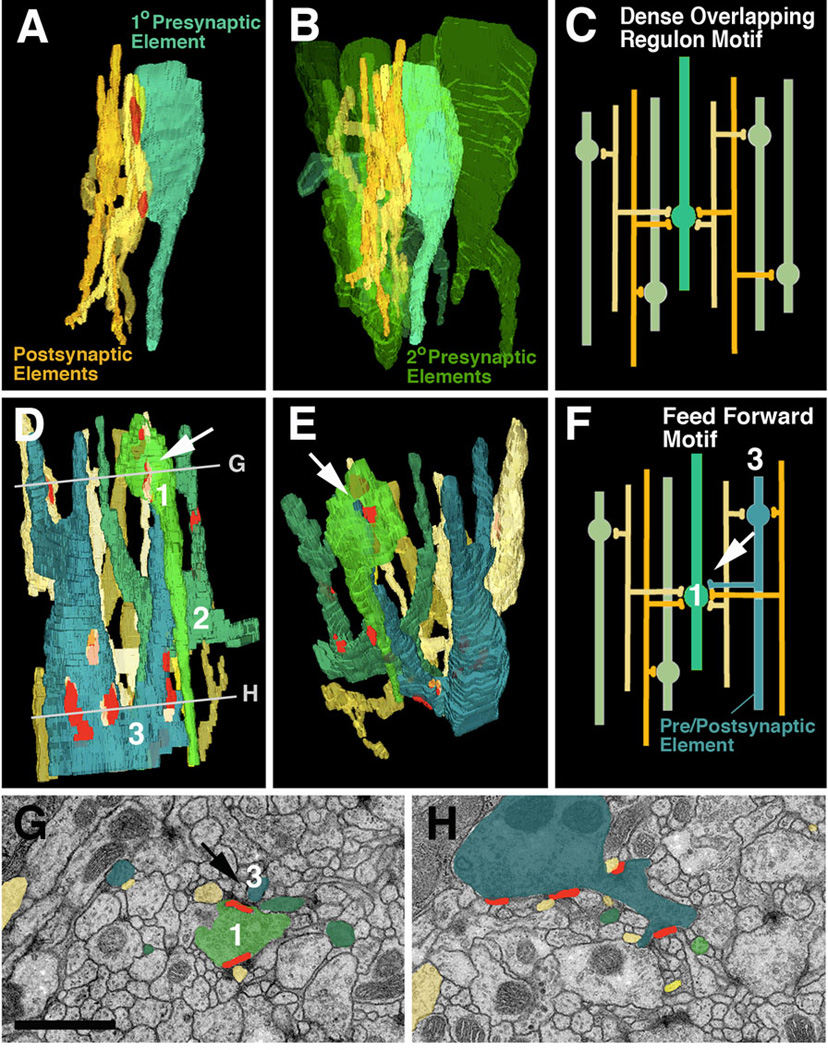
Network motifs that are most frequently encountered in micro-volumes. A-C: Dense overlapping regulon motif. A: Segment of one “primary” presynaptic element (turquoise; varicose neurite) which contacts five postsynaptic elements (dendritiform neurites) at two synapses. B: Same configuration of pre- and postsynaptic elements as in A; several other “secondary” presynaptic elements (green) which form synapses with the same dendrites as the primary axon are shown. C: schematic representation of this network motif. D-H: Feed forward motif. D, E, F: 3D digital models and schematic modelof segments of three varicose neurites which form predominantly presynaptic contacts (terminal axons). The blue element has a thin branch that is postsynaptic to the light-green element (white arrow in D-F). Shown are also the dendritiform postsynaptic neurites that are postsynaptic to both green and blue varicose neurites. G, H: EM sections at levels shown by lines in panel D. G represents top level and shows the synapse between pre-/postsynaptic element (blue) and presynaptic element (green; arrow).
Bar: 1µm
Microcircuitry: Density, branching and directionality of neurites
The above characterization of neurite ultrastructure applies to all regions sampled so far (6 micro-volumes: calyx, peduncle periphery, spur, CPL compartment, larval optic neuropile, ventral nerve cord). But there are significant quantitative differences. We will summarize findings from three microvolumes, the calyx (input region of mushroom body), spur (output region of mushroom body), and dorsolateral ventral nerve cord.
Afferent axons of the calyx are the antennal projection neurons receiving olfactory input in the antennal lobe (Stocker, 1994; Ramaekers et al., 2005). We can identify in the calyx micro-volume (Fig.5A) the largely parallel array of varicose neurites, carrying presynaptic sites, as being derived from the incoming fiber bundle (antenno-protocerebral tract) carrying the antennal projection neurons. In overall volume, varicose/globular afferents make up approximately 16% of the neuropile. At a medium diameter of 0.38µm (reducing the shape of these neurites to smooth cylinders) this corresponds to a density of 4.2 varicose neurites terminating in or passing through a volume of 1µm3. Counting the number of branch points (for varicose neurites) in the microvolume yields a density of branches at 1 branch every 4µm. Presynaptic sites are entirely restricted to the varicose/globular swellings of neurites. We find a density of about 4 presynaptic sites per 1µm3. Forming bundles between the varicose (and globular) presynaptic profiles are the fine terminal fibers of denritiform neurites, mostly derived from mushroom body neurons (Yasuyama et al., 2002). Synapses are mostly dyadic-tetradic, with 2–4 fine fibers “winding” around swellings of varicose/globular neurites and participating in several synapses. Connectivity is highly local: individual varicose/globular neurites interact with multiple dendrites in their immediate vicinity.
Fig. 5.
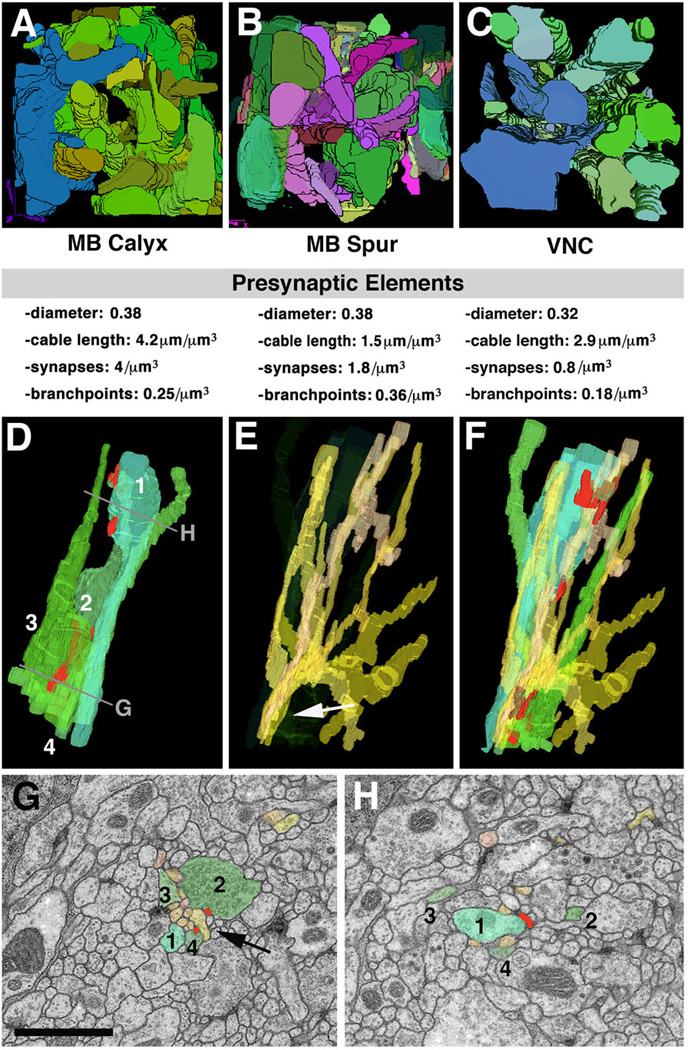
Structural network properties in Drosophila brain neuropile. Panels of the top row (A-C) show 3D digital models of presynaptic varicose/globular neurites from three different micro-volumes. A, B: micro-volumes from calyx (A; input region) and spur (B; output region) of mushroom body. In both micro-volumes, branches of terminal axonal neurites follow all directions. C: micro-volume from dorso-lateral neuropile of ventral nerve cord. All neurites are oriented predominantly along longitudinal axis. Shown below each panel are some core parameters of neurite profiles seen in micro-volumes. Note that average diameter of presynaptic elements (varicose and globular neurites) are very similar between the different regions. Density of presynaptic sites and branch points is significantly lower in VNC compared to mushroom body. D-H: Typical trajectories of presynaptic and postsynaptic terminal branches. D shows 3D digital models of four neighboring presynaptic neurites (1–4). Neurite 1 has a varicosity near top of panel (arrow); varicosities of the other neurites are more basally. E: bundle of dendritiform neurites extending in vicinity of terminal axons shown in D. F: terminal axons and dendrites shown together. G, H: EM sections close to top and bottom of VNC microvolume (levels of section shown in D). Profiles corresponding to the elements shown in models D-F are shaded in corresponding colors. As shown here, groups of dendritiform neurites (typically ranging between 6 and 10) form tight bundles in between adjacent preterminal axons (arrow in E, G). After forming synaptic contacts, dendritiform neurites typically splay apart (E) to then regroup with other dendrites in different configurations.
Bar: 1µm
Neuropile ultrastructure appears different in the spur, an output region of the mushroom body (Fahrbach, 2006), or the ventral nerve cord. Interestingly, the spur is characterized by the absence of intrinsic glia. Varicose neurites are aligned in bundles that travel along all three cardinal axes (Fig.5B). Thin terminal filaments of dendritiform neurites are much less frequent than in the calyx (and most other compartments). The lateral connectivity is much more pronounced than in the calyx: if one visualizes all neurites connected to a given synapse, they reach throughout the entire microvolume. Intriguing are also the synaptic geometries: in most cases, two presynaptic sites and 2–3 postsynaptic sites are clustered together (not shown). We speculate that both widefield connectivity and multi-input synapses are elements of a microcircuitry required for the specialized function (associative learning) of the mushroom body.
In the dorsolateral domain of the ventral nerve cord, all neurites are predominantly arranged parallel to the longitudinal axis (Fig.5C, D). Axonal cable length is 2.9µm per µm3 (axiform plus varicose/globular neurites). The branching density is low, with 0.18 branches per 1µm3. The high quality of the VNC volume allowed us to reconstruct the pattern of thin dendritiform neurites with great confidence. These neurites have a much higher branch density than terminal axons (Fig.5E, F). As a result, overall dendritic cable length exceeds that of axonal elements by a factor of almost 10. Dendritiform processes show an interesting convergence-divergence pattern (Fig.5E–H). At a given level, dendritiform processes are not scattered evenly across the section, but form several bundles of 5–10 processes each; one such bundle is highlighted by arrows in Fig.5E, G. Bundles typically run between the loosely packed terminal, varicose axons, with which they form synapses. However, dendrites of a given bundle stay together only for a short interval (<1µm); subsequently, they diverge and redistribute (compare Fig.5G and H), coming together with other dendritform processes in new combinations. This behavior is in contrast to that of axons: axiform neurites (long axons) form tight bundles where neighborhood relationships between neurites is maintained over many µm; terminal varicose/globular axons are more loosely packed, but like axiform processes run largely parallel to each other and maintain their position relative to each other (Fig.5D, G, H).
Connectivity and network motifs in the VNC microvolume
As a result of the high branching density of dendritiform processes, as well as the fact that each presynaptic site contacted multiple postsynaptic profiles, neurites within a microvolume are highly interconnected (as will be shown in the next section, this feature distinguishes the Drosophila neuropile from vertebrate brain neuropile). The most common type of network is shown in Fig.6A–C. In the example shown here, one presynaptic element forms two adjacent presynaptic sites on one varicosity. Contacting these sites are six postsynaptic dendritiform processes. Quite frequently, as in thiis example, a given dendrite contacts its presynaptic partner multiple times at adjacent synapses. Each dendrite forms multiple branches that connect to other presynaptic partners; in the average, a dendrite received input from 2.1 axons within the 85µm3 VNC microvolume. Fig.6B illustrates the presynaptic element, its associated six postsynaptic elements, and five additional presynaptic elements that are in contact with the same postsynaptic elements. This type of connectivity has been defined as dense overlapping regulon motif (Reigl et al., 2004; Alon, 2007): a given input element diverges onto multiple targets, and at the same time, each target element receives input from multiple presynaptic elements (Fig.6C). The large majority of presynaptic neurites and postsynaptic neurites within the VNC were engaged in dense overlapping regulon motifs.
Less frequently we encountered a type of connection labeled as feed forward motif. An example is shown in Fig.6D–H: Among the postsynaptic partners of one of the varicose neurite (1, light green) are thin branches of two other varicose neurites (2, dark green; 3, blue). In addition, axon A forms input onto several dendrites, which at the same time receive input from B and C. In other words: an afferent neurite is at the same time presynaptic to one neurite and postsynaptic to another, neighboring neurite; both elements are presynaptic to a common dendritic element. It will be informative to investigate how such network motifs are distributed throughout the brain, and are correlated to different sensory modalities or types of output.
Neuropile architecture in mammalian cortex and fly brain: a first comparison
Quantitative statements about the architecture and connectivity of the neuropile of vertebrate brains, in particular mammalian cortex were mainly based on statistical analyses of light microscopic preparations (e.g., Golgi stained prepartions) and representative EM sections (e.g., Braitenberg and Schüz, 1998). Recently, the first reconstruction of a microvolume of mammalian neuropile has become available (Chklovskii et al., in preparation). What can one learn from the comparison of these data with our data on Drosophila larval brain?
Fig.7 presents EM sections of mouse neocortex (A) and Drosophila brain (L1 larva: B; adult: C). Plotting the frequency of profiles with different diameters shows quite similar distributions in mouse and fly (Fig.7D). In particular, the average diameter of axon shafts and synapse bearing varicosities, as well as presynaptic sites themselves, is quite conserved. The most conspicuous difference between the two species lies in the size of dendrites. In mouse cortex, dendrite shafts are large, measuring in average almost one micrometer (Braitenberg and Schüz, 1998; Fig.7D, E). Many dendrites are considerably thicker (e.g., examples shown in Fig.7A). Dendrites of pyramidal cells (and some other classes of neurons) bear spines, club-shaped processes of 1–2µm length and an average diameter of approximately 0.4µm. Synapses are found on dendritic shafts and spines; synapses are typically monadic, which naturally follows from the fact that the presynaptic element is of equal size, or even smaller, than the postsynaptic element (arrow in Fig.7A). The comparison between mouse and fly in regard to axonal and dendritic cable length per volume unit is also informative (Fig.7E). In mouse, axonal cable adds up to approximately 4mm per 1000µm3; the length of dendrites is 0.5mm. Axonal cable length in Drosophila lies in the same range; dendritic cable length, on the other hand is ten times higher, which is a reflection of the fact that dendritic processes are very thin and highly branched. Interestingly, judging from the inspection of EM photographes (e.g., Peters et al., 1976) processes in the 0.1µm range seem to be quite numerous also in mammalian brain, but the nature of these thin neurites is unclear.
Fig. 7.
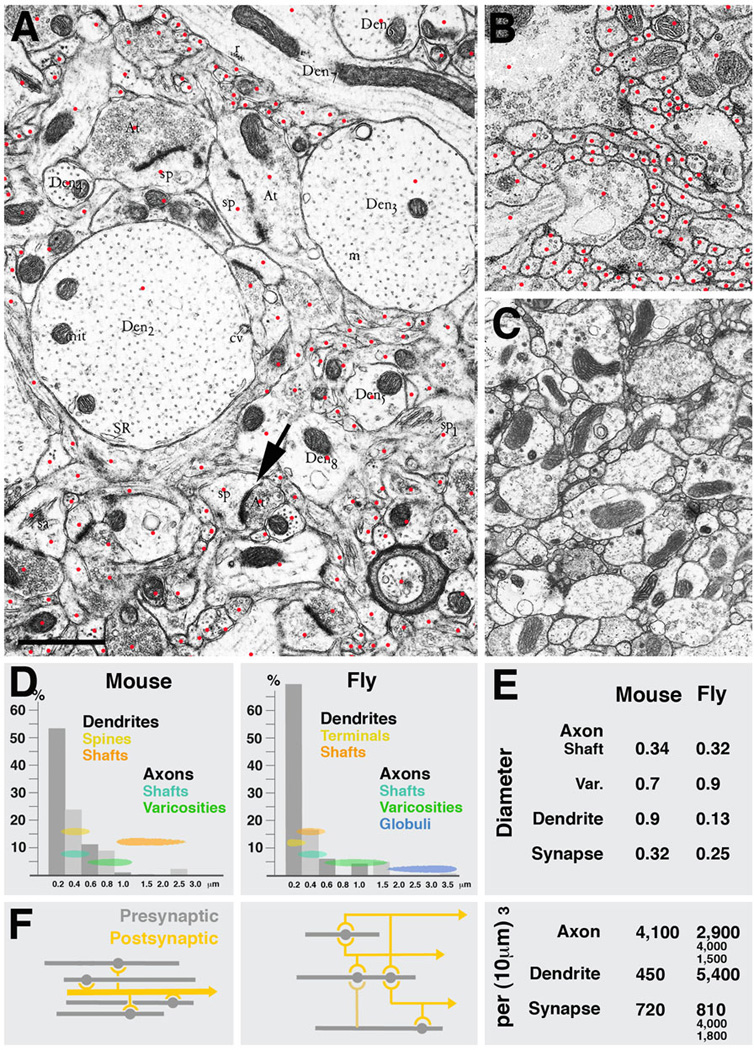
Parameters of microcircuitry in mammalian neocortex and Drosophila brain. A-C: Representative EM sections of mouse neocortex (A), Drosophila larval brain (B) and Drosophila adult brain (C) shown at the same scale. Red dots in A and B indicate profiles of individual sectioned neurites. D: Frequency distribution of neurites with different diameters in mammalian cortex and Drosophila brain. Indicated are also the range of diameters that correspond to different neuropile elements (yellow/orange: dendrites; green/blue: axons). E: Comparison of several core parameters in mouse and Drosophila neuropile. In both systems, terminal axons are varicose neurites which form presynaptic sites on their varicosities. Diameters of varicosities are in the range of 0.5 to 1.5µm (average in mouse: 0.7µm; in Drosophila: 0.9µm); the thin segments of terminal axons have an average diameter of approximately 0.33µm. The diameter of synapses (presynaptic sites) is also quite similar in both systems (0.32µm in mammalian cortex, 0.25µm in fly brain. Also the overall cable length of terminal axons per volume unit is comparable: 4,100µm per 1000µm3 in mouse, and between 1500 and 4,000µm in different micro-volumes of fly brain. The major difference between mouse and fly neuropile lies in the size and branching density of dendrites. In Drosophila, dendrites are very thin (average diameter: 0.13µm) and densely branched; in mammalian brain, dendrites are thick (average diameter: 0.9µm; see even thicker examples of dendrites in panel A) and branches are much further apart. This is also reflected in the dendritic cable length which is 450mm per 1000µm3 in mouse and more than 10fold higher in Drosophila. Lower branch density as well as the absence of polyadic synapses in mouse cortex neuropile also results in a considerably less dense connectivity, schematically shown in F. Shown for Drosophila is the dense overlapping regulon motif, in which the large majority of neurite segments encountered in any micro-volume of 100µm3 or more is engaged. In a mammalian cortical micro-volume of that size, dendrite segments are unbranched; the only type of connectivity is convergence, whereby multiple terminal axons converge on a dendritic segment that happens to be within their range.
Bar: 1µm
As described in the previous section, the overall synaptic density per volume unit seems to be quite different in different parts of the fly brain, ranging from approximately one per µm3 (VNC) to more than four per µm3 (input region of mushroom body). In mammalian cortex, synaptic density is towards the lower end of this range, with 0.72/µm3 (Braitenberg and Schüz, 1998). Approximately the same value was extracted from the micro volume of rat hippocampus (Chklovskii et al., in preparation). Here, one axonal varicosity typically contained a single synapse; in Drosophila, most varicosities had between two and four synaptic sites. The higher density of synapses in Drosophila is accompanied by a higher number of branch points. In the rat hippocampus micro-volume, measuring 8×8×8µm, very vew axons or dendrites (out of hundreds) had any branches, not counting dendritic spines. Similarly, statistical analysis of Golgi preparations yielded typical distances of 10µm and higher between branch points (Braitenberg and Schüz, 1998). By contrast, in Drosophila, terminal axons had one branch point every 4µm (mushroom body calyx), 2.8µm (mushroom body, spur) or 7.5µm (VNC), respectively. Dendritic branch density is even higher; in the VNC microvolume, dendritiform neurites had approximately twice as many branchpoints as varicose/globular neurites.
The fact that the spacing of branches and synapses is significantly higher in the Drosophila brain compared to mammalian brain is reflected in the presence of a much higher connectedness of neurites in the former. As described in the previous section, the majority of axons engaged in networks containing both dendrites and other axons within a microvolume of less than 100µm3. This is not the case in mammalian microvolumes of comparable, or even larger, size (Chklovskii et al., in preparation). Here, the only type of connectivity is represented by a convergence of axonal segments onto isolated dendritic segments (Fig.7H). However, as opposed to the fly brain, very few axonal segments will form input to more than one dendrite, so that network motifs like the dense overlapping regulon motif or feed forward motif, do not emerge. What this simply means is that the modules of mammalian brain that house microcircuits are considerably larger than in Drosophila. In vertebrates, neurons reach much higher numbers than in the Drosophila brain. Furthermore, dendrites are larger in diameter, and polyadic synapses are largely absent. That has the result that connectivity occupies more space: in Drosophila, one presynaptic neurite reaches up to six postsynaptic elements in a single synapse. In mammalian brain, for the same purpose, six individual synapses, spaced apart by intervals of several µm, would have to be formed. One may speculate that, evolutionarily, each neuron grew in axonal and dendritic length, to accommodate the higher number of synapses that had to form to connect a given neuron to a certain fraction of other neurons. As a result of this numerical increase in cell number, cable length, and synapse number, branches of dendrites and axons are spaced much further apart, such that in a volume of 8×8×8µm almost no branchpoints (and consecutively no networks) occur.
A glimpse at the phylogeny of neuropile architecture
Systematic studies of neuropile architecture and connectivity from serial EM sections have not been carried out for other animals, with the exception of the CNS of the nematode C. elegans (White et al., 1986). On the other hand, there is a rich literature documenting neurites and synapses in representative EM sections. These neuropile elements exist in all metazoans. Measurements of diameters of neurites and synapses, taken from representative photographs published in the literature (mollusk: Nagy and Elekes, 2000; annelid: Riehl and Schlue, 1998; vertebrate: Peters et al., 1976; nematode: White et al., 1986; acoela: our material) yielded figures similar to those for Drosophila. Presynaptic sites measured 0.2–0.35um in diameter. Neurite numbers, as in Drosophila, showed a peak in the range below 0.2µm diameter (Fig.8). In addition, a substantial number of neurites were in the range of 0.4–0.8µm. Presynaptic sites are typically located on profiles with a diameter of 0.4–0.8µm; these presynaptic sites contact a variable number of thin, postsynaptic dendrites. Large profiles serving as postsynaptic sites (as in vertebrates) are reported rarely. This finding, which should be substantiated with further systematic serial reconstruction of microvolumes, suggests that the pattern of neurites shown here for Drosophila, with thin, highly branched dendrites which engage with medium-sized, varicose axons through polyadic synapses may be primitive. The appearance of “gigantic” dendrites, accompanied by a significant increase in the number of neurons and overall cable length, appears to be an innovation of the vertebrate clade.
Fig. 8.
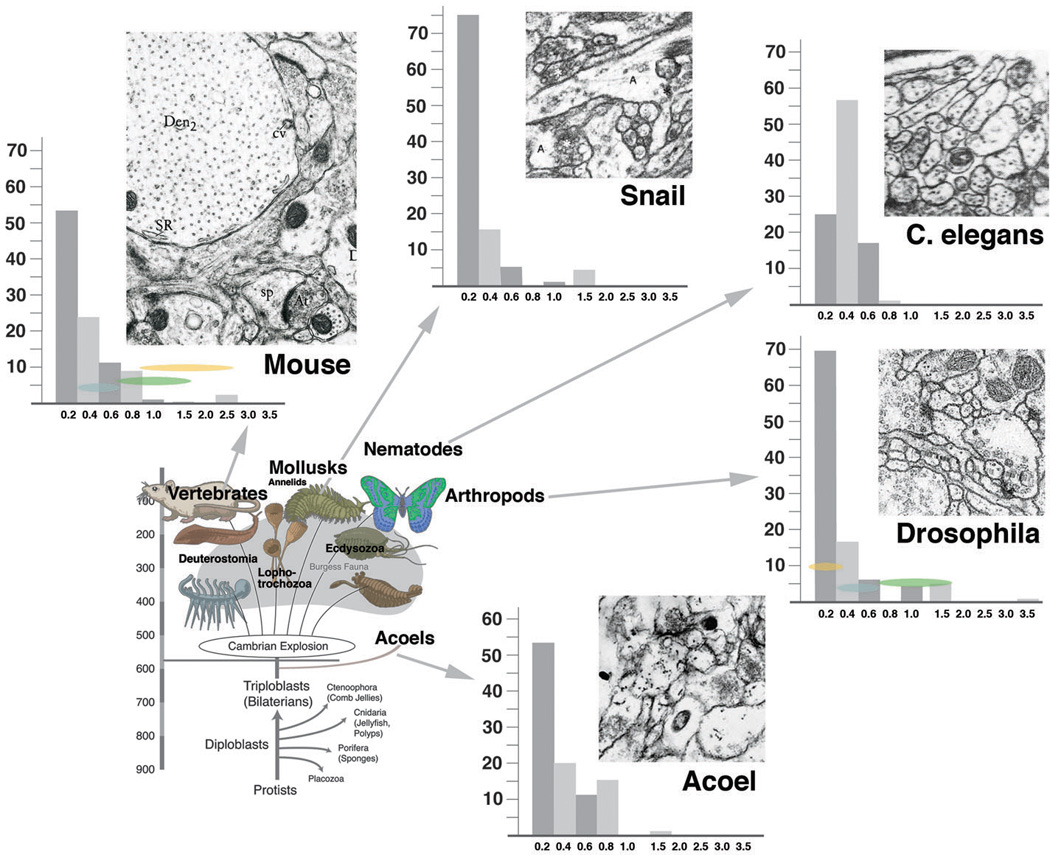
Frequency distribution of neurites with different diameters across multiple phyla. Bottom left: highly schematic phylogenetic diagram depicting basal metazoa and major groups of bilaterians. Top and left: representative EM sections of neuropile of five phyla [chordata: mouse (from Peters et al., 1976); mollusca: pond snail (Nagy and Elekes, 2000); nematoda: C. elegans (White et al., 1986); arthropoda: Drosophila (our material); acoela: Neochildia (our material); all shown at same scale]. Next to EM sections are histograms depicting size distribution of neurite profiles. In all phyla except nematoda, thin profiles (< 0.2µm) represent majority of neurites, followed by intermediate profiles (0.4–0.6µm) and large profiles (> 0.6µm). For details, see text.
C. elegans with its miniturerized CNS, consisting of less than 400 neurons, presents an interesting case with respect to neuropile architecture. Thin profiles below 0.2µm are almost entirely absent; the large majority of neurites have a diameter of 0.3–0.6µm. This anomaly among invertebrate taxa is accompanied by the fact that neurons of C. elegans are almost all unbranched (White et al., 1986; Fig.8 bottom right). Most somata project a single neurite which carries clusters of intermingled presynaptic and postsynaptic sites. In other words, thin terminal dendritiform branches as in other taxa do not exist in the worm.
Outlook
We propose that the three dimensional reconstruction of neuropile architecture and connectivity will provide an important tool for the study of brain function and development. The technology we present here is well adapted to handle small volumes of neuropile, but a number of technical improvements are under way that will bring larger volumes (in the size range of adult fly brains) into the range of feasibility. The improvements are directed at three major bottlenecks of the procedure: better registration, automatic segmentation and automatic feature extraction. Programs are in the trial phase that allow for automatic recognition of profile boundaries and subsequent segmentation (Chklosvskii et al., in preparation). A step in microvolume analysis that is currently as labour intensive as segmentation (which takes 5–10 working days per volume), is extracting relevant information from the segmented objects. For example, we currently input manually what synapse belongs to what neurite, and manually generate lists that encapsulate the connectivity pattern. We are working on adding features to the EM canvas window that fully automate these tasks. Thus, after segmentation of neurite profiles, synapses can be assigned automatically to the underlying 3D reconstructed neuronal arborizations that host them. With not much extra effort than merely segmentation, a complete wiring diagram of the micro volume can be built. The next step would be to generate dynamic models, where lengths and diameters of neurites, intersynapse distances, and other parameters are taken into account to predict the temporal component of activity flow. We anticipate that these models of small volumes will provide suitable material for computational neuroscientists to build more accurate and heuristically valuable large scale models of brain function.
Acknowledgements
We thanks many friends and collaborators, notably Dmitrii Chklovskii, Johannes Schindelin, James Truman, and Wayne Pereanu for helpful discussions and comments. Parts of this work were supported by NIH Grant R01 NS054814 to VH.
References
- Alon U. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Baas PW, Yu W. A composite model for establishing the microtubule arrays of the neuron. Mol. Neurobiol. 1996;12:145–161. doi: 10.1007/BF02740651. [DOI] [PubMed] [Google Scholar]
- Braitenberg V, Schüz A. Cortex: Statistics and Geometry of Neuronal Connectivity. 2nd. Springer: Berlin Heidelberg; 1998. [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Cardona A, Saalfeld S, Preibisch S, Schindelin J, Tomancak P, Cheng A, Potter C, Carragher B, Hartenstein V. Micro- and macroarchitectural analysis of fly brain by computer-assisted serial section electron microscopy. 2009 doi: 10.1371/journal.pbio.1000502. in preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chklovskii D, et al. Can proximity predict connectivity? Analysis of 3D organization of hippocampal neuropil at ultrastructural resolution. in prep. [Google Scholar]
- Douglas R, Martin KAC. Neocortex. In: Shepherd GM, editor. The synaptic organization of the brain. Oxford University Press; 1998. pp. 459–511. [Google Scholar]
- Fahrbach SE. Structure of the mushroom bodies of the insect brain. Annu. Rev. Entomol. 2006;51:209–232. doi: 10.1146/annurev.ento.51.110104.150954. [DOI] [PubMed] [Google Scholar]
- Feeney CJ, Karunanithi S, Pearce J, Govind CK, Atwood HL. Motor nerve terminals on abdominal muscles in larval flesh flies, Sarcophaga bullata: comparisons with Drosophila. J. Comp. Neurol. 1998;402:197–209. [PubMed] [Google Scholar]
- Fung S, Wang F, Spindler S, Hartenstein V. Drosophila E-cadherin and its binding partner Armadillo/ β-catenin are required for axonal pathway choices in the developing larval brain. Dev. Biol. 2009;332:371–382. doi: 10.1016/j.ydbio.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CS, Doe CQ. Embryonic development of the Drosophila central nervous system. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 1131–1206. [Google Scholar]
- Hartenstein V, Spindler S, Pereanu W, Fung S. The development of the Drosophila larval brain. Adv. Exp. Med. Biol. 2008a;628:1–31. doi: 10.1007/978-0-387-78261-4_1. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Cardona A, Pereanu W, Younossi-Hartenstein A. Modeling the developing Drosophila brain: Rationale, Technique and Application. BioScience. 2008b;58:823–836. [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J. Comp. Neurol. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Kittel RJ, Wichmann C, Rasse TM, Fouquet W, Schmidt M, Schmid A, Wagh DA, Pawlu C, Kellner RR, Willig KI, Hell SW, Buchner E, Heckmann M, Sigrist SJ. Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science. 2006;312:1051–1054. doi: 10.1126/science.1126308. [DOI] [PubMed] [Google Scholar]
- Kozloski J, Hamzei-Sichani F, Yuste R. Stereotyped position of local synaptic targets in neocortex. Science. 2001;293:868–872. doi: 10.1126/science.293.5531.868. [DOI] [PubMed] [Google Scholar]
- Lai SL, Awasaki T, Ito K, Lee T. Clonal analysis of Drosophila antennal lobe neurons: diverse neuronal architectures in the lateral neuroblast lineage. Development. 2008;135:2883–2893. doi: 10.1242/dev.024380. [DOI] [PubMed] [Google Scholar]
- Larsen C, Shy D, Spindler S, Fung S, Younossi-Hartenstein A, Hartenstein V. Patterns of growth, axonal extension and axonal arborization of neuronal lineages in the developing Drosophila brain. Dev. Biol. 2009 doi: 10.1016/j.ydbio.2009.06.015. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinertzhagen IA, O’Neil SD. Synaptic organization of columnar elements in the lamina of the wild type in Drosophila melanogaster. J. Comp. Neurol. 1991;305:232–263. doi: 10.1002/cne.903050206. [DOI] [PubMed] [Google Scholar]
- Nagy T, Elekes K. Embryogenesis of the central nervous system of the pond snail Lymnea stagnalis L. An ultrastructural study. J. Neurocyt. 2000;29:43–60. doi: 10.1023/a:1007112130414. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Hartenstein V. Neural lineages of the Drosophila brain: A 3D digital atlas of the pattern of lineage location and projection at the late larval stage. J. Neurosci. 2006;26:5534–5553. doi: 10.1523/JNEUROSCI.4708-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W, Jennett A, Younossi-Hartenstein A, Hartenstein V. A development-based compartmentalization of the Drosophilacentral brain. 2009 doi: 10.1002/cne.22376. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Palay S, Webster HE. The fine structure of the nervous system. Philadelphia London Toronto: WB SaundersCcompany; 1976. [Google Scholar]
- Prokop A, Meinertzhagen IA. Development and structure of synaptic contacts in Drosophila. Semin. Cell Dev.Biol. 2006;17:20–30. doi: 10.1016/j.semcdb.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Ramaekers A, Magnenat E, Marin EC, Gendre N, Jefferis GS, Luo L, Stocker RF. Glomerular maps without cellular redundancy at successive levels of the Drosophila larval olfactory circuit. Curr. Biol. 2005;15:982–992. doi: 10.1016/j.cub.2005.04.032. [DOI] [PubMed] [Google Scholar]
- Riehl B, Schlue WR. Morphological organization of neuropile glial cells in the central nervous system of the medicinal leech (Hirudo medicinalis) Tissue Cell. 1998;30:177–186. doi: 10.1016/s0040-8166(98)80066-9. [DOI] [PubMed] [Google Scholar]
- Reigl M, Alon U, Chklovskii DB. Search for computational modules in the C. elegans brain. BMC Biol. 2004;2:2–25. doi: 10.1186/1741-7007-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls ET, Loh M, Deco G, Winterer G. Computational models of schizophrenia and dopamine modulation in the prefrontal cortex. Nat. Rev. Neurosci. 2008;9:696–709. doi: 10.1038/nrn2462. [DOI] [PubMed] [Google Scholar]
- Saalfeld S, Cardona A, Hartenstein V, Tomancák P. CATMAID: Collaborative Annotation Toolkit for Massive Amounts of Image Data. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp266. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberberg G. Polysynaptic subcircuits in the neocortex: spatial and temporal diversity. Curr. Opin. Neurobiol. 2008;18:332–337. doi: 10.1016/j.conb.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Silberberg G, Gupta A, Markram H. Stereotypy in neocortical microcircuits. Trends Neurosci. 2002;25:227–230. doi: 10.1016/s0166-2236(02)02151-3. [DOI] [PubMed] [Google Scholar]
- Stocker RF. The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res. 1994;275:3–26. doi: 10.1007/BF00305372. [DOI] [PubMed] [Google Scholar]
- Toledo-Rodriguez M, El Manira A, Wallén P, Svirskis G, Hounsgaard J. Cellular signalling properties in microcircuits. Trends Neurosci. 2005;28:534–540. doi: 10.1016/j.tins.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Truman JW, Schuppe H, Shepherd D, Williams DW. Developmental architecture of adult-specific lineages in the ventral CNS of Drosophila. Development. 2004;131:5167–5184. doi: 10.1242/dev.01371. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development. 2003;130:3621–3637. doi: 10.1242/dev.00533. [DOI] [PubMed] [Google Scholar]
- Watson AH, Burrows M. The morphology, ultrastructure, and distribution of synapses on an intersegmental interneuron of the locust. J. Comp. Neurol. 1983;214:154–169. doi: 10.1002/cne.902140205. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Phil. Trans. R. Soc. Lond. B. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Winterer G, Weinberger DR. Genes, dopamine and cortical signal-to-noise ratio in schizophrenia. Trends Neurosci. 2004;27:683–690. doi: 10.1016/j.tins.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Yasuyama K, Meinertzhagen IA, Schürmann FW. Synaptic organization of the mushroom body calyx in Drosophila melanogaster. J. Comp. Neurol. 2002;445:211–226. doi: 10.1002/cne.10155. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Nassif C, Green P, Hartenstein V. Early neurogenesis of the Drosophila brain. J. Comp. Neurol. 1996;370:313–329. doi: 10.1002/(SICI)1096-9861(19960701)370:3<313::AID-CNE3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Salvaterra P, Hartenstein V. Early development of the Drosophila brain IVLarval neuropile compartments defined by glial septa. J. Comp. Neurol. 2003;455:435–450. doi: 10.1002/cne.10483. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Shy D, Hartenstein V. The embryonic formation of the Drosophila brain neuropile. J. Comp. Neur. 2006;497:981–998. doi: 10.1002/cne.20884. [DOI] [PubMed] [Google Scholar]