Abstract
Recent advances in technology have led to a dramatic increase in the number of available transcription factor ChIP-seq and ChIP-chip data sets. Understanding the motif content of these data sets is an important step in understanding the underlying mechanisms of regulation. Here we provide a systematic motif analysis for 427 human ChIP-seq data sets using motifs curated from the literature and also discovered de novo using five established motif discovery tools. We use a systematic pipeline for calculating motif enrichment in each data set, providing a principled way for choosing between motif variants found in the literature and for flagging potentially problematic data sets. Our analysis confirms the known specificity of 41 of the 56 analyzed factor groups and reveals motifs of potential cofactors. We also use cell type-specific binding to find factors active in specific conditions. The resource we provide is accessible both for browsing a small number of factors and for performing large-scale systematic analyses. We provide motif matrices, instances and enrichments in each of the ENCODE data sets. The motifs discovered here have been used in parallel studies to validate the specificity of antibodies, understand cooperativity between data sets and measure the variation of motif binding across individuals and species.
INTRODUCTION
Chromatin immunoprecipitation (ChIP) (1) followed by hybridization to an array (ChIP-chip) (2,3) or sequencing (ChIP-seq) (4) enables the genome-wide identification of the binding locations of transcription factors (TFs) present in a given condition and cell type or tissue. As these technologies have matured, their use has become increasingly widespread. The resolution of these experimental techniques can be as low as 300 bp for ChIP-chip (5) and 50 bp for ChIP-seq (6), depending on the experimental design (e.g. fragment size, paired-end sequencing) and algorithmic processing of the raw data.
The use of these technologies on a variety of factors across many cell types has increasingly highlighted the complex nature of TF activity, often violating the simple model of a factor binding to its recognition pattern (motif) in isolation: binding has been shown to be dynamic across cell types, requiring the coordinated binding of cofactors or specific configurations of the underlying chromatin. Moreover, TF binding frequently occurs in the absence of any discernible motif instance (7,8) or to ‘hot-spots’ where several factors are simultaneously found (9). Understanding this complex binding necessitates identifying the underlying sequence features responsible. To address this need, we have performed a systematic, motif-centric analysis of hundreds of TF binding experiments made available as part of the human ENCODE project (8,10). As part of this, we provide a collection of motifs for each assayed factor, both taken from the literature and through de novo discovery, and also an annotation of motif instances genome-wide, which may be used to pinpoint the specific regulatory bases in regions bound by TFs.
We found that no single algorithm or database comprehensively assays the motifs relevant to the binding diversity surveyed by ENCODE. Therefore, our approach was to collect motifs from several literature sources (11–16) and supplement them with motifs discovered de novo on the data sets themselves using five established tools (17–21). Although this general approach of using multiple motif discovery tools is popular [e.g. (22–24)], its application to this number of data sets is unprecedented and permits the identification of TFs that are likely to be interacting or participating in common pathways.
This work is accompanied by a web interface for browsing the discovered and literature motifs along with their enrichments (Figure 1; http://compbio.mit.edu/encode-motifs). In addition to the browsing interface, we provide several data files including all motif matrices and their matches to the genome, as well as software to compute enrichments and perform unified motif discovery with the five tools we use. Together, these permit both analyses of individual factors (e.g. to identify cooperating TFs) in addition to systematic analysis (e.g. to examine differences between TFs). Moreover, the breadth of data sets available enables systematic comparisons and analyses that are not possible when only one or a few factors are studied in isolation.
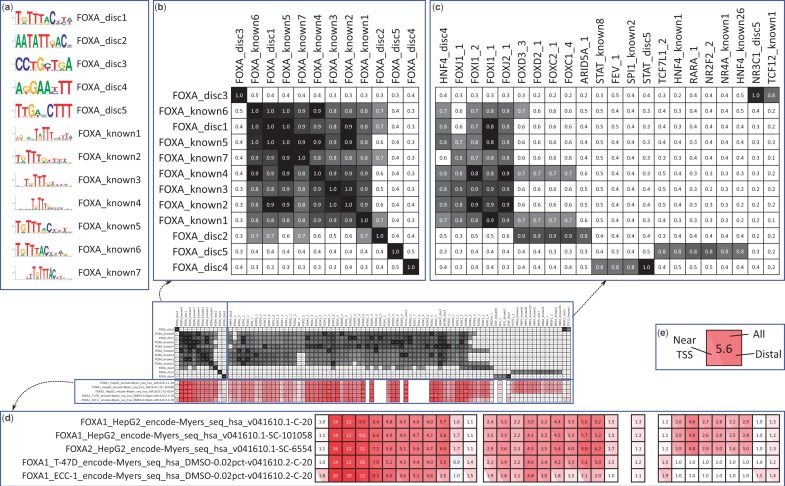
Figure 1.
Pipeline output for FOXA factor group, an example that highlights different aspects of the resource (in the interest of space, only selected columns are shown in the enlargements). (a) The known and discovered motifs for the factor group, drawn with WebLogo (25). Because the original orientation is arbitrary, motifs are flipped as necessary to increase the similarity of the displayed orientations. (b and c) Similarity between the motifs for this factor group and the motifs: (b) for this factor group, (c) for all other motifs that match at least one motif for this factor group with similarity 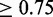 . Motif similarities are shown in black/white scale with gray starting at 0.65. (d) Enrichments of motifs in the data sets for this factor group. Data sets are named indicating the factor, cell type/stage, lab code, followed by values to differentiate data sets and make them unique. Red is used for enrichment, blue is used for depletion (which is rare in this study). Enrichments are not shown for motifs when control motifs could not be generated or there was not sufficient information content to match at a
. Motif similarities are shown in black/white scale with gray starting at 0.65. (d) Enrichments of motifs in the data sets for this factor group. Data sets are named indicating the factor, cell type/stage, lab code, followed by values to differentiate data sets and make them unique. Red is used for enrichment, blue is used for depletion (which is rare in this study). Enrichments are not shown for motifs when control motifs could not be generated or there was not sufficient information content to match at a 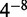 P-value. (e) Magnified enrichment heatmap value. The three triangles represent the background regions used for enrichment; the top triangle is all regions, the left triangle is those within 2 kb of an annotated TSS and the right triangle is those outside the 2 kb window (the regions used in the left and right triangles partition that of the top triangle). The number shown is the enrichment in the inclusive background. Here we see an apparent contradiction: a higher enrichment for the union than the parts. This occurs because the higher counts permit a smaller confidence interval around the ratios used to compute the enrichment. Heatmaps are ordered using hierarchical clustering followed by optimal leaf ordering (26). The Supplementary Results include an analysis of the robustness of the motif similarity metric and the enrichment values.
P-value. (e) Magnified enrichment heatmap value. The three triangles represent the background regions used for enrichment; the top triangle is all regions, the left triangle is those within 2 kb of an annotated TSS and the right triangle is those outside the 2 kb window (the regions used in the left and right triangles partition that of the top triangle). The number shown is the enrichment in the inclusive background. Here we see an apparent contradiction: a higher enrichment for the union than the parts. This occurs because the higher counts permit a smaller confidence interval around the ratios used to compute the enrichment. Heatmaps are ordered using hierarchical clustering followed by optimal leaf ordering (26). The Supplementary Results include an analysis of the robustness of the motif similarity metric and the enrichment values.
Later in the text, we describe the details of how the resource was generated and conduct an initial analysis to provide examples of its usage and to highlight potentially interesting results.
MATERIALS AND METHODS
Our goals were to produce a resource that (i) contains a comprehensive collection of relevant motifs for each factor; (ii) avoids repetitive, weakly enriched motifs that do not contribute to the in vivo specificity of the factor or its partners; and (iii) excludes variants of the same motif, particularly among the discovered motifs. With this in mind, we conducted motif discovery separately on each data set using five motif discovery tools and manually placed all its data sets into ‘factor groups’ on the basis of known motifs and homology (Figure 2). Known motifs from the literature and the top 10 most enriched discovered motifs (excluding duplicates) were collected for each factor group (see Supplementary Methods) and named as TF_known# for known motifs and TF_disc# for discovered motifs, where TF denotes the factor group (e.g. FOXA, CTCF, etc.). Known motifs were ordered arbitrarily, whereas the discovered motifs were ordered in descending order of the enrichment value that was used for their selection.
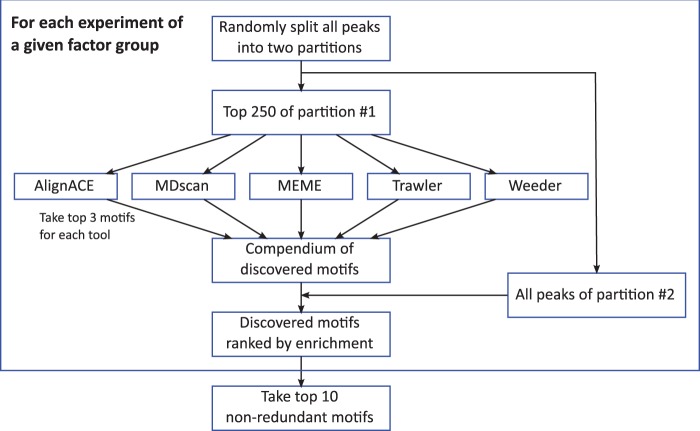
Figure 2.
Outline of motif discovery pipeline. Input regions for each data set are randomly partitioned into two groups. The top 250 regions of one of the partitions are scanned for motifs using five de novo motif discovery tools. These motifs are evaluated using the peaks from the other partitioned and pooled across data sets for a factor group to produce the final list of discovered motifs for each factor group.
The 427 ENCODE experiments analyzed correspond to 123 TFs, which we place into 84 factor groups (Figure 3a). We failed to discover an enriched motif for only 12 of the 84 factor groups, of which 9 lack DNA binding domains (BRF, CTBP2, HDAC8, KAT2A, NELFE, SUPT20H, SUZ12, WRNIP1 and XRCC4) as identified by UniProt (27), and 6 have all their data sets flagged as unreliable based on various quality metrics [BRF, KAT2A, NELFE, NR4A, SUPT20H and ZZZ3; see (A. Kundaje, L.Y. Jung, P.V. Kharchenko, B. Wold, A. Sidow, S. Batzoglou and P.J. Park, in preparation)]. Of these factor groups, only NR4A has a previously identified known motif.
Figure 3.
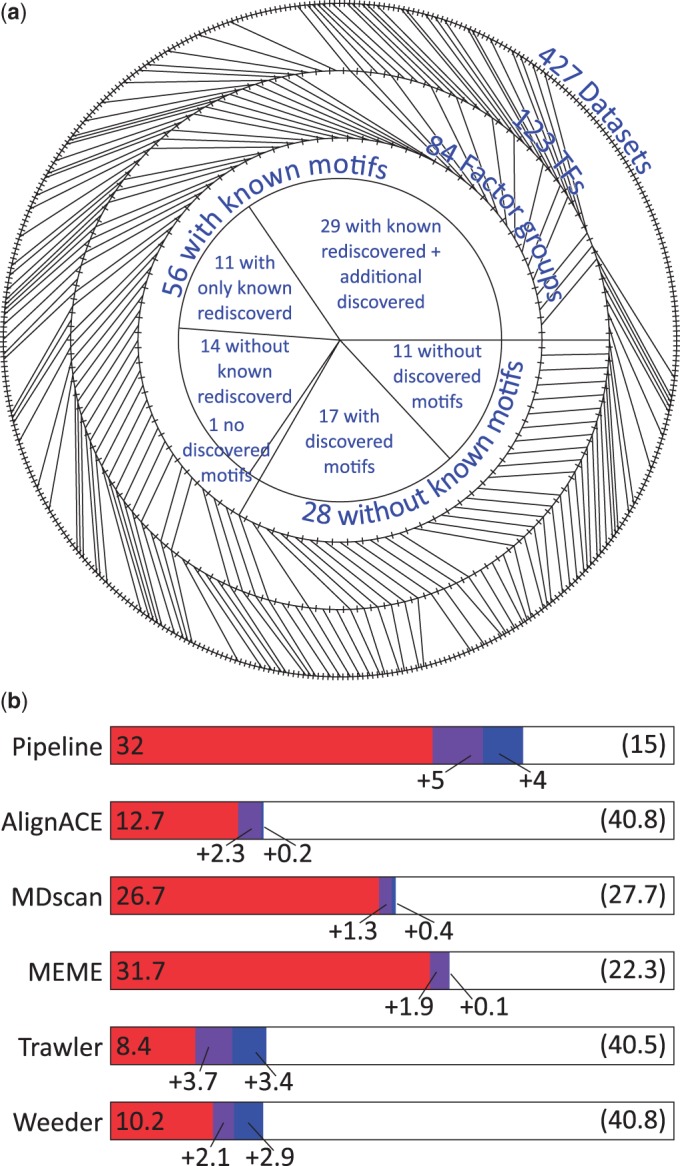
(a) Summary of input data used. The outside ring indicates the experimental data sets (one tick for each of 427), which are separated into 123 transcription factors (second ring). The TFs are further grouped into 84 factor groups (third ring). We are able to find a matching discovered motif for 41 of the 56 factor groups with a known motif; 29 of these 41 factor groups have additional discovered motifs that may be associated with cofactors. For all but 1 of the 15 factor groups where the known motif is not recovered we still find enriched discovered motifs. We also discovered enriched motifs for 17 of the 28 factor groups without a known motif. (b) Recovery of known motifs by each of the discovery tools. Performance of discovery in terms of number of factor groups for which the known motif was recovered. A motif is considered a match if it matches any of the known motifs for a factor group (see Supplementary Methods for details on how matches are computed). The number of additional factors that have a match is shown with each additional motif (only three motifs are taken from each individual method, whereas we have up to 10 for the pipeline). The number of factor groups with no motif match is shown in parenthesis. When multiple data sets exist for a factor group, the fraction that matches is used in computing its contribution for computing the performance of the individual tools.
We exclude from the discussion below motifs that we consider unlikely to be relevant to our analysis, while maintaining them as part of the overall resource where they may be useful. These include 46 discovered motifs that are either low-complexity (e.g. dinucleotide repeats) or consistently have weak enrichment (<2) and do not match known motifs (Supplementary Table S1). These are likely a consequence of slight biases in the discovery pipeline, or are due to real, but relatively weak, specificity for the factor. We also exclude an additional 36 motifs that have a weak similarity to the known motif for the factor but for which a better matching and enriched motif is also found (Supplementary Table S2). These are most frequently seen for longer motifs that can be broken up into recognizable, but globally dissimilar, patterns that are not captured by our automatic exclusion criteria (see Supplementary Methods). Together, these represent 28% of the 293 discovered motifs.
RESULTS
Using motif similarity metrics, we are able to link the discovered motifs directly to the TFs that recognize them through their known motifs. Here we use these inferred relationships between TFs to make specific biological insights, illustrating the types of analyses that our resource enables. In the interest of clarity, most descriptions of TFs will be omitted, but may be found along with further references at RefSeq (28) and Entrez (29).
Recovery of known specificity for TFs
Most of the known literature motifs we collect are derived from biochemical in vitro assays. Thus, they provide a largely independent, although somewhat imperfect way to evaluate the performance of our discovered motifs. Recovery of known motifs varies significantly by method, but taking the most enriched motif (our pipeline) is competitive with the best single method (Figure 3b). Overall, our pipeline found a motif matching a previously characterized literature motif for 41 of the 56 factor groups with a known motif.
One of the most striking observations of this analysis is how frequently other distinct motifs were also found. For 29 of these 41 factor groups other motifs are found, even after manually excluding redundant or repetitive motifs, and for 9 factor groups one or more of these discovered motifs is ranked higher than the motif matching a known motif (see Supplementary Table S3). In the next section, we will analyze the additional motifs we found for these factors, which in many cases identify factors known to interact, either cooperatively or competitively.
For the remaining 15 of 56 factor groups with a known motif (e.g. HSF, NANOG, PBX3, SREBP and TAL1) the known motif is not found at all, including NR4A where no enriched motif is discovered. Frequently this is because the known motif itself is not enriched and may not accurately capture the specificity of the factor in vivo. For example, the ‘known’ EP300 motif from Transfac was likely built on a specific bound region of EP300 and would not accurately capture its binding in all cell types where it interacts with a variety of factors and has no DNA binding domain of its own (we avoided removing such motifs to prevent bias in the database). Likewise, we do not discover a motif that matches the known ZBTB33 specificity, and moreover the known motif itself is not enriched at all in the bound regions.
Although some known motifs were of apparently low quality, we largely found our database of known motifs to be relatively comprehensive and had difficulty finding matches to novel motifs outside it. An exception is ZNF263_disc1, which does not match a motif in our database, but does roughly match the specificity for ZNF263 indicated in (30) despite only having weak enrichment (1.8-fold).
Although the motifs that match each other (either known or discovered) generally have similar enrichments, in some cases we find substantially higher enrichment for some motif variants over others (Figure 4 and Supplementary Table S3). For example, NFE2_disc1 matches the known NFE2 motif, but has a 76-fold maximal enrichment across NFE2 data sets, compared with 56-fold enrichment for the most enriched known NFE2 motif. Different known motifs for the same factor often show a broad range in enrichment: MEF2 has six motifs described in Transfac, with an enrichment differential of as much as 4-fold consistently across data sets. This enrichment analysis provides a systematic way to choose among variants of a motif.
Figure 4.

Comparison of known versus discovered motifs (selected where discovered better enriched than known; all factor groups with a discovered motif matching a known motif in Supplementary Table S3). Displayed is the known and discovered motif with the maximum enrichment across all data sets for a factor group. Only the discovered motifs that match a known motif for a factor group are considered. The maximum enrichment is indicated for each factor and, in parenthesis, the ‘raw’ enrichment for the same data set without the use of the shuffle motifs for correction.
We also saw varying enrichment of the known motif, depending on the specific data set for a factor group. For example, CTCF_known2 is enriched in CTCF data sets in a range from 30- to 78-fold on identically processed data. This may be a result of varying quality of the samples across data sets or may be a consequence of true biological differences.
Identifying the sequence specificity for factors that were previously uncharacterized is of particular interest. In all, 17 factor groups had no known motif but now have discovered enriched motifs (BCL, BDP1, CCNT2, CHD2, CTCFL, HDAC2, HMGN3, RAD21, SETDB1, SIRT6, SMARC, SMC3, SP2, SIN3A, THAP1, TRIM28 and ZNF263). These discovered motifs may represent the direct or indirect (e.g. through cofactors) DNA binding specificity.
Shared motifs suggest interacting relationships
We find that most factors have motifs for other factors enriched in their binding sites (summarized in Supplementary Table S4). This may occur due to (i) cooperative binding of the two factors to the same locations; (ii) interfering binding between factors where one binds near the other to prevent binding; (iii) some similarity in motif specificity; (iv) the two factors functioning on a similar set of genes (e.g. ones specific to one tissue), without directly interacting; or (v) the factors binding to similar genomic regions (e.g. near genes). Our analysis does not directly rule out any of these possibilities; however, (iii) is generally verifiable using our motif similarity metrics and (v) can be examined by inspecting only the TSS-proximal enrichment.
The motif most enriched in multiple data sets was the TPA DNA response element (TRE; TGA[C/G]TCA), which is recognized by the AP1 TF when it is formed by FOS/JUN dimers (31) and other factors including MAF and NFE2. The enrichment of the TRE in a data set is often stronger than that of even the known in vitro sequence specificity and may arise from a number of phenomena, including (i) a cooperatively interaction with AP1, (ii) competition with AP1 for the same binding sites, leading to a potentially repressive role for the TF or (iii) reuse of binding sites due to, for example, accessibility of chromatin. We find a motif matching the TRE motif for 20 factor groups (AP1_disc3, AP2_disc1, BATF_disc1, BCL_disc2, CTCF_disc8-9, EP300_disc1, GATA_disc2, HMGN3_disc1, IRF_disc2, MAF_disc1, MEF2_disc3, MYC_disc3, NFE2_disc1, NR3C1_disc2, PRDM1_disc2, RXRA_disc3, SMARC_disc1, STAT_disc2, TCF7L2_disc1 and TRIM28_disc1).
We found that the enrichment of the TRE to be particularly notable for a few factors. GATA and AP1 have known cooperative binding (32). TFs in the SMARC factor group are members of the SWI/SNF chromatin remodeling complex (33), which is necessary for proper regulation by FOS/JUN dimers (34); and TCF7L2_disc1, which matches the TRE, is more enriched than the known TCF7L2 motif (TCF7L2_disc2) in only the TCF7L2 colorectal cancer cell line HCF-116 data set, consistent with the known interaction of JUN and TCF7L2 during intestinal cancer development (35).
AP1 also binds to the cAMP response element (CRE; TGACGTCA) when the dimer is formed by ATF3/JUN (31) and this is the motif we find as AP1_disc1. However, AP1_disc3 (which matches the TRE) is the most enriched motif in FOS data sets. Interestingly, ATF3_disc1 is not the CRE, but rather the E-box (see later in text). We do, however, find a variant of the CRE (with additional specificity) as ATF3_disc2. The most enriched discovered motif for E2F, E2F_disc1 also matches the CRE and is highly enriched in all data sets.
MYC is a critical regulator, which recognizes the E-box sequence. To aid in comparisons, we include MAX, which forms complexes with MYC, and USF1/2, which also recognizes the E-box sequence, in the MYC factor group. We find multiple motifs enriched in MYC binding sites, highlighting the multifunctional role MYC and the other E-box recognizing proteins play. We found a version of the E-box with additional specificity (MYC_disc1) that was highly enriched in USF1/2 bound regions (max 98-fold for USF2 versus <9-fold enrichment for MYC/MAX). This motif was more enriched than the known E-box motifs, including known USF motifs, in many USF data sets. We find a second, less specific E-box motif (MYC_disc2), which shows more even enrichment across factors. We also find discovered motifs of other factors matching the E-box, including SIN3A_disc2 (discussed later in text), NFE2_disc2-3 and SIRT6_disc1. It is notable that although SIRT6 is a chromatin-associated protein without a known DNA binding domain (36), the only discovered motif matches the E-box (with 16-fold enrichment in SIRT6 bound regions), suggesting that MYC or another E-box recognizing factor may play an important, but indirect, chromatin-related role.
Motif enrichment is able to identify both positive and negative interactions for the same factor. For example, SIN3A, a corepressor known to interact with a number of proteins, has discovered motifs matching REST (SIN3A_disc1 and more weakly disc3–4) and MYC (SIN3A_disc2). These are consistent with SIN3A’s known involvement in repression by REST (37) and SIN3A being a known antagonist for MYC (38).
Morever, MYC_disc4 matches RFX5 and is enriched particularly for MAX-bound regions in H1-hESC and GM12878, and MYC_disc5 matches the CEBPB known motif and is enriched in MYC regions bound in unstimulated K562 cells. MXI1, which was not included in the MYC factor group although it does interact with MAX to bind to MYC-MAX sites (39), has MXI1_disc1 that matches RFX5 in both the K562 and HeLa-S3 cell lines.
We analyzed six IRF family data sets: IRF1 binding in K562 cells stimulated by IFNa (viral innate response) or IFNg (viral, bacterial and tumor control); IRF3 in HepG2, GM12878 and HeLa-S3; and IRF4 in GM12878. The most strongly enriched motif (IRF_disc1, matching NFY) is highly enriched (>20-fold) for all three IRF3 data sets and IRF1 in K562 under IFNg stimulation. This suggests that binding of IRF to NFY sites occurs only under specific conditions and by only some IRF members and potentially expands on the previously documented interaction of NFY and IRF2 at a single promoter (40). IRF_disc4, which matches SP1, is enriched in the same cell types, albeit at much lower levels. IRF_disc3, which matches the known IRF consensus, shows weak-to-no enrichment in these data sets, but shows an enrichment of 8.8-fold for IRF1 bound regions in K562 cells under IFNa stimulation and 3.1-fold enrichment for IRF4 bound regions in GM12878. IRF_disc2, which matches the TRE, is enriched primarily in GM12878 regions bound by IRF4. The known SPI1 motif matches IRF_disc5, and reciprocally SPI1_disc2 matches the IRF motif, consistent with the importance of SPI1 in hematopoietic development (41).
Beyond the discovered motif for IRF, several other discovered motifs (AP1_disc2, CEBP_disc2, E2F_disc4, PBX3_disc1, RFX5_disc2 and SP1_disc1-2) match the known NFY specificity (CCAAT). These discovered motifs are consistent with several known interactions of NFY. RFX5 promotes the cooperative binding between RFX and NFY (42), CEBPB and NFY interact in at least one promoter (43) and SP1 and NFY are known to interact (44). E2F_disc4 has particularly high enrichment in E2F4 data sets, consistent with the cooperative role E2F4 and NFY play in cell cycle regulation (45).
STAT factors are involved in regulating number of growth-related functions. We analyze STAT1, STAT2 and STAT3 here in the context of GM12878, HeLa-S3, MCF10A-Er-Src and K562 cells. We find relatively consistent enrichment of the STAT full site (TTCCNGGAA), which STAT_disc1 matches, while finding weak enrichment for just the half-site (TTCC). We also find motifs involved in other proliferative functions including STAT_disc2, which is particularly enriched in STAT3 data sets and matches the TRE, consistent with STAT3 being one of the many interaction partners for AP1 (46). STAT_disc3 matches the IRF consensus and has enrichment that is particularly high in STAT1 and STAT2 data sets stimulated by IFNa, highlighting the cooperativity of STAT factors and IRF in immune functions. STAT_disc4 is a match to the CEBPB motif and is found enriched in STAT3 data sets, consistent with the known cooperative role for these two factors (47).
TFs with ETS domains are highly conserved and involved in several cellular processes [reviewed in (48)]. A number of TFs have discovered motifs that match the ETS consensus, including EGR1_disc2, GATA_disc3, MEF2_disc2, NRF1_disc2, NR2C2_disc1 and PAX5_disc4. These discovered motifs are supported by known interactions between GATA and ETS in sea squirts (49), MEF2 and the ETS factor PEA3 (50) and NR2C2 with the ETS factor ELK4 (51). Moreover, PAX5 and ETS factors have shared roles in the development of B-cells (52,53). Looking at the discovered ETS motifs, we find that ETS_disc8 matches the known motif for MYB and the two have been known to cooperate, a relationship that is important in the context of certain cancers (54).
THAP1 has two discovered motifs, both of which match the known YY1 motif (the first with additional specificity added by an apparent HNF4 motif). To our knowledge, the relationship between THAP1 and YY1 has not been directly observed; however, THAP1 has been known to associate with the coactivator HCF-1 (55), and YY1 and HCF-1 are known to interact (56). Our result suggests that THAP1 and YY1, possibly with the addition of HNF4, may interact at least in the K562 cell line for which we have THAP1 binding data. RAD21_disc3 also matches YY1, suggesting an additional interaction.
NANOG, an important pluripotency TF, has a known motif that is only weakly enriched (1.3-fold) in the bound regions and not discovered by our pipeline. We see much stronger enrichment for the known POU5F1 and POU2F2 motifs, for which we also find similar motifs (NANOG_disc2 and NANOG_disc4, respectively), consistent with their shared roles in pluripotency (57,58). The interaction of these factors is further supported by POU5F1_disc2 matching the known POU2F2 motif. Additionally, NANOG_disc2 and disc3 match the known motifs for TCF7L2 and TCF12, respectively, again consistent with the important role TCF proteins play in stem cells (59).
CTCF plays a variety of vital roles in the organization of chromatin architecture (60) and the motifs we discover matching the known CTCF specificity (RAD21_disc1, SMC3_disc1,2-4, CTCFL_disc1,10, ZBTB7A_disc1,2, SP2_disc3 and RXRA_disc2,5; some weakly) are largely compatible with this role. RAD21 is a highly conserved protein involved in DNA double-strand repair (61) known to co-localize with CTCF (62). Cohesin, of which SMC3 is a subunit, is brought to the chromatin by CTCF (63). Further, although the function of the CTCF paralog CTCFL is not completely known, it does appear to be involved in imprinting through interaction with a histone methyltransferase (64).
Combinations of motifs
A few of the discovered motifs contain additional specificity or have distinct segments matching multiple motifs. For example, EGR1_disc4 appears to be a combination of multiple motifs (EGR1, IKZF1 and a homeobox motif), and SETDB1_disc1 contains the ZNF143 core sequence with significant additional specificity. The appearance of these motifs suggests highly specific ‘grammars’ for these motifs that may require specific spacing and orientation of binding sites for functionality.
We find several additional enrichments of potential interest. PBX3_disc2 matches the known MEIS1 motif, consistent with the known cooperative binding of MEIS1 and PBX (65). TAL1_disc1 matches GATA, with the potential connection that GATA and TAL1 are known to be important in hematopoesis and vascular development (66,67). HSF_disc1 matches the known CEBP motif and has much higher enrichment in HSF data sets (31-fold) compared with the known motifs for HSF (<9-fold). Additionally, EGR1_disc5, HNF4_disc5, NRF1_disc3, PAX5_disc2, RXRA_disc4/PAX5_disc3 and SREBP_disc1 match the known motifs for ZIC, SOX, SP1, PAX2/PAX3, IRF and RFX5, respectively, suggesting additional previously uncharacterized interactions. Lastly, we find some motifs that show more ambiguous matches: SMARC_disc2 shows weak similarity to homeobox TGTAGT motif, NR2C2_disc2–3 weakly matches the known HNF4 motif and EGR1_disc3/SETDB1_disc2 matches the repetitive NRF1 motif.
General factors enriched in cell line-specific key regulators
Factors directly responsible for the establishment of enhancers, chromatin restructuring or polymerase recruitment frequently exhibit binding that is highly cell type specific. Because most of these factors do not have their own sequence specificity, their binding is often correlated with that of regulators important for the specific cell line. We analyze several such factors (BCL, BDP1, CCNT2, EP300, FOXA, HDAC2, HMGN3, TATA, TCF12 and TRIM28) and find that key cell line regulators can be identified by examining enrichments in cell lines-specific data sets.
As a transcriptional coactivator, EP300 interacts with numerous TFs [reviewed in (68)] and has been shown to have binding that can identify tissue-specific enhancers (69). Conversely, FOXA has a DNA binding domain and plays an important role in liver development and function (70) and is a pioneer factor responsible for priming chromatin for the binding of other factors (reviewed in (71)]. Other proteins involved in chromatin restructuring include HDAC2, which transcriptionally represses through histone deacetylation (72) and HMGN3 (73). Further, two factor groups are directly involved in transcription including three RNA Pol3 subunits (BDP1, RPC155 and TFIIIC-110) and CCNT2, which is involved in the elongation of Pol2 (74).
Eight of these ten factor groups have at least one data set in K562 (erythroleukemia cells), and for four of these we discover motifs that match the GATA consensus, which is then enriched specifically in the K562 data sets (BCL_disc5, CCNT2_disc1, HDAC2_disc1 and HMGN3_disc2). GATA has a known important role in K562 (75), and we also have previously found an association with GATA motifs and chromatin state-derived enhancers for K562 cells (76). We also find three additional motifs that have enrichment specific to the factor group’s K562 data set: BDP1_disc1, a 23-nt motif that contains the STAT consensus; HMGN3_disc1, which matches the TRE; and TRIM28_disc2, which matches no known motif and may be associated with an uncharacterized regulator active in this cell line.
Likewise, for GM12878, an EBV-mediated lymphoblastoid cell line, we find three discovered motifs (BCL_disc4, EP300_disc5 and TCF12_disc4) that match the known IRF consensus. IRF4 has been shown to be important in the establishment of these cell lines (77), and the family is an important player in immune cells (78). This enrichment is also consistent with our previous study using epigenetic marks (76), where we found IRF to be the strongest enriched motif in GM12878-specific enhancers. We also find GM12878-specific enrichment for motifs matching NFKB (BCL_disc6) and POU2F2 (TATA_disc9), consistent with the known biology of these factors (79,80).
The motifs we find specifically enriched in HepG2 (liver carcinoma) data sets match the known motifs for FOXA (EP300_disc3, HDAC2_disc2, and TCF12_disc2), HNF4 (FOXA_disc5 and HDAC2_disc5) and CEBP (EP300_disc2,6), three key liver regulators (70,81). We find motifs with enrichments specific to H1-hESC, which include matches to the pluripotency factor POU2F2 (TATA_disc9), the near universally expressed repressor REST (BCL_disc3 and HDAC2_disc4) and key metabolic regulator NRF1 (HDAC2_disc4). We find additional cell line-specific enrichments for FOXA_disc3 (TCF12) in ECC-1, FOXA_disc4 (STAT) in both T-47D and ECC-1 and EP300_disc2,6 (CEBP) and EP300_disc4 (ETS) with enrichment in the HeLa-S3 data set.
Even for these factors, we find motifs that are consistently enriched across assayed cell lines for a given factor. FOXA_disc1, for example, matches the known FOXA motif, indicating that FOXA’s own motif also plays an important role in its specificity. Most of the motifs we identify for RNA Pol2 machinery (TAF1, GTF2B, GTF2F1 and TBP) are enriched in all cell lines, including the known TATAAA motif (TATA_known2). Also, TATA_disc1, disc6 and disc8 have consistent enrichment and match the known motifs for YY1 (which is known to be important in establishing transcription) (82), NFY and ETS. The top discovered motif BCL_disc1 matches the known ETS motif and is also enriched across data sets.
Interestingly, we find that the TRE motif is found and enriched in a cell line-specific manner for several factors, but for different cell lines. For example, HMGN3_disc1 is enriched in K562, BCL_disc2 has the highest enrichment in GM12878, TRIM28_disc1 is only enriched in the HEK2932 and U2OS cell lines and EP300_disc7 has enrichment in the neuroblastoma cell line SK-N-SH-RA and HeLa-S3. This suggests that perhaps AP1 or other factors recognizing TRE are selectively interacting with these proteins depending on the cell line.
Novel motifs raise possibility of unknown regulators
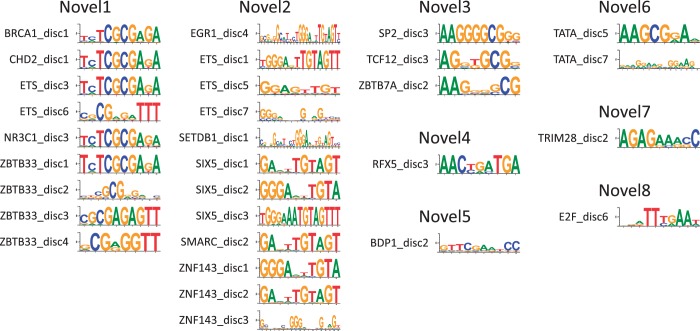
Although we are able to putatively explain the majority of the motifs we discover as either matches to previously known motifs or low complexity sequences, we do identify 30 putative novel motifs (Figure 5). We placed these into eight groups on the basis of their similarity: Novel1 (BRCA1_disc1, CHD2_disc1, ETS_disc3,6, NR3C1_disc3 and ZBTB33_disc1-4), Novel2 (EGR1_disc4, ETS_disc1,5,7, SETDB1_disc1, SIX5_disc1-3, SMARC_disc2 and ZNF143_disc1-3), Novel3 (SP2_disc3, TCF12_disc3 and ZBTB7A_disc2), Novel4 (RFX5_disc3), Novel5 (BDP1_disc2), Novel6 (TATA_disc5,7), Novel7 (TRIM28_disc2) and Novel8 (E2F_disc6).
Figure 5.
Putative novel motifs. We find eight motifs that are not represented in the literature motifs we collected, three of which are found for at least two factor groups. These patterns may represent the binding specificity of the factors for which they are discovered or for other factors that cooperate with them.
Novel1 (using ZBTB33_disc1) is highly enriched in at least one data set for each of the factor groups for which it is found (BRCA1, CHD2, ETS, NR3C1 and ZBTB33). All five factor groups except CHD2 have at least one known motif, and for each of these data sets Novel1 is more enriched in at least one data set than any known motif [the result for NR3C1 is questionable because only one data set has enrichment and that data set has been independently flagged as problematic; see http://www.encodeproject.org/encode/qualityMetrics.html]. The shared role of BRCA1 and CHD2 in DNA damage repair (83,84) suggests that Novel1 may be involved in this or other shared roles for these factors and highlights the utility in shared motif enrichment even outside of motifs directly tied to a factor.
Similarly, for SIX5, we see only weak enrichment of the known SIX5 motif and fail to discover a motif similar to it. However, Novel2 (using SIX5_disc1) shows over 100-fold enrichment for all three data sets (K562, GM12878 and H1-hESC). Novel2 also shows high enrichment in data sets for which it was not rediscovered, including ATF3 (all data sets have >20-fold enrichment with GM12878 having 106-fold) and NRF1 (all data sets have >30-fold enrichment). Moreover, the known ZNF143 motif, which is 4-fold enriched in the one ZNF143 data set, is also not recovered, but Novel2 is 24-fold enriched. The breath of data sets sharing this motif suggests it may be recognized by an important yet unknown or under-characterized regulator.
Like the known ZBTB7A motif, Novel3 (using SP2_disc3) is largely poly-G, which causes us to underestimate its enrichment due to our shuffling process. Despite this, however, it does show enrichment in several data sets, including for the factor groups for which it was identified. This motif shows similarity to other poly-G motifs, such as known SP1 motifs, but appears to be distinct due to its other bases.
Novel4 (RFX5_disc3) shows moderate, but consistent (2- to 6-fold) enrichment across the RFX5 data sets. The consensus is composed of two of the same components as the known motifs (AAC and TGA), but ordered differently. Consequently, it may represent the binding specificity of, for example, an alternative isoform of RFX5. The remaining motifs (Novel5-8), were found for factors that show cell line-specific enrichments. Consequently, these may represent specificities for regulators that are previously unidentified.
Experimental and evolutionary validation of novel motifs
Following the motif discovery and selection of these putative novel motifs, a study released hundreds of new motifs generated using high-throughput SELEX (16). Two of the putative novel motifs described in this section match motifs generated by (16): Novel1 matches the motif for ETV6 and Novel6 matches ZBED1. Although we have incorporated these SELEX motifs into our resource, we continue to include Novel1 and Novel6 as putative novel motifs because they were identified without knowledge of these new specificities and thereby strengthen the evidence for the remaining novel motifs.
Four of these putatively novel motif groups (Novel1–3, 6) match motifs that were previously identified using conservation signals across four mammals (85) (Supplementary Table S5). Therefore, this study provides additional support for these conservation-based motifs and, conversely, the motifs identified here gain comparative evidence. The relatively few distinct novel patterns that are found in this study and the comparative support for many of the few that are found suggests that there may be a limited number of human TF motifs with many instances and which interact with one of the assayed factors that remain unknown.
DISCUSSION
In this article, we provide a systematic and comprehensive collection of motifs for hundreds of human TF binding data sets. TF binding can be complex, with a factor recognizing several or motifs or binding in the apparent absence of any motif [reviewed in (86)]. We also show that it is possible to identify cofactors that may be partially responsible for binding or function.
This motif resource has already been used in several articles while this article was in preparation, demonstrating its value for high-throughput analyses. Our motifs are being matched at low stringency to identify peaks that are void of any motif to understand the mechanism through which motif-less peaks are generated (8). The collection of known motifs and enrichment techniques we present here was also used as a secondary validation of peaks (87). Because having the motifs allows for more precisely determining the bases responsible for binding, these motifs enable analyzes involving population data (88) and for interpreting GWAS data (89). Two other ENCODE articles also perform motif discovery: (90) produce a non-redundant list of discovered motifs but do not perform an extensive analysis of the relationships between factors and (91) use DNaseI footprinting data to identify relevant motifs.
Having a motif catalog is also the first step in identifying high-quality computational targets of factors, which may allow the identification of binding sites that were, for example, not found in the conditions assayed. Two popular strategies are used for this purpose. One is using clustering of motif instances for factors known to cooperate to form cis-regulatory modules (92,93). This resource is well suited for this purpose because it naturally provides sets of motifs that are likely to cooperate.
A second approach is the use of conservation on many closely related species (85,94–97). This can be performed readily on these motif instances because a dense tree of mammalian species has been sequenced readily permitting their alignment and measuring selection of a near-nucleotide level. Because changes in the underlying motif matches are largely responsible for changes in binding across species (98), evolutionary-based approaches on the motif instances may be a means to deal with the high rate of non-functional binding (99–101).
AVAILABILITY
A web interface, along with data files and accompanying software, is available at http://compbio.mit.edu/encode-motifs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [102–110].
FUNDING
National Institutes of Health (NIH) [HG004037, HG007000 and HG006991]. Funding for open access charge: NIH [HG004037, HG007000 and HG006991].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Ewan Birney, Christopher Bristow, Luke Ward, Jason Ernst, Anshul Kundaje, Gerald Quon and other members of the Kellis Laboratory for helpful discussions.
REFERENCES
- 1.Solomon MJ, Larsen PL, Varshavsky A. Mapping proteinDNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 2.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 3.Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 4.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 5.Qi Y, Rolfe A, MacIsaac KD, Gerber GK, Pokholok D, Zeitlinger J, Danford T, Dowell RD, Fraenkel E, Jaakkola TS, et al. High-resolution computational models of genome binding events. Nat. Biotechnol. 2006;24:963–970. doi: 10.1038/nbt1233. [DOI] [PubMed] [Google Scholar]
- 6.Guo Y, Papachristoudis G, Altshuler RC, Gerber GK, Jaakkola TS, Gifford DK, Mahony S. Discovering homotypic binding events at high spatial resolution. Bioinformatics. 2010;26:3028–3034. doi: 10.1093/bioinformatics/btq590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Hendriks CL, et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moorman C, Sun LV, Wang J, deWit E, Talhout W, Ward LD, Greil F, Lu X, White KP, Bussemaker HJ, et al. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2006;103:12027–12032. doi: 10.1073/pnas.0605003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan K-K, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC(R): transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandelin A, Alkema W, Engstrm P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pea-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 17.Hughes JD, Estep PW, Tavazoie S, Church GM. Computational identification of Cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- 18.Liu XS, Brutlag DL, Liu JS. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol. 2002;20:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- 19.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Int. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 20.Pavesi G, Mauri G, Pesole G. An algorithm for finding signals of unknown length in DNA sequences. Bioinformatics. 2001;17:S207–S214. doi: 10.1093/bioinformatics/17.suppl_1.s207. [DOI] [PubMed] [Google Scholar]
- 21.Ettwiller L, Paten B, Ramialison M, Birney E, Wittbrodt J. Trawler: de novo regulatory motif discovery pipeline for chromatin immunoprecipitation. Nat. Methods. 2007;4:563–565. doi: 10.1038/nmeth1061. [DOI] [PubMed] [Google Scholar]
- 22.Che D, Jensen S, Cai L, Liu JS. BEST: binding-site estimation suite of tools. Bioinformatics. 2005;21:2909–2911. doi: 10.1093/bioinformatics/bti425. [DOI] [PubMed] [Google Scholar]
- 23.Romer KA, Kayombya G, Fraenkel E. WebMOTIFS: automated discovery, filtering and scoring of DNA sequence motifs using multiple programs and Bayesian approaches. Nucleic Acids Res. 2007;35:W217–W220. doi: 10.1093/nar/gkm376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun H, Yuan Y, Wu Y, Liu H, Liu JS, Xie H. Tmod: toolbox of motif discovery. Bioinformatics. 2010;26:405–407. doi: 10.1093/bioinformatics/btp681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crooks GE, Hon G, Chandonia J, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bar-Joseph Z, Gifford DK, Jaakkola TS. Fast optimal leaf ordering for hierarchical clustering. Bioinformatics. 2001;17:S22–S29. doi: 10.1093/bioinformatics/17.suppl_1.s22. [DOI] [PubMed] [Google Scholar]
- 27.Bairoch A. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2004;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruitt KD, Maglott DR. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frietze S, Lan X, Jin VX, Farnham PJ. Genomic targets of the KRAB and SCAN domain-containing zinc finger protein 263. J. Biol. Chem. 2010;285:1393–1403. doi: 10.1074/jbc.M109.063032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karin M, Liu Zg, Zandi E. AP-1 function and regulation. Curr. Opin. Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 32.Kawana M, Lee ME, Quertermous EE, Quertermous T. Cooperative interaction of GATA-2 and AP1 regulates transcription of the endothelin-1 gene. Mol. Cell. Biol. 1995;15:4225–4231. doi: 10.1128/mcb.15.8.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 34.Ito T, Yamauchi M, Nishina M, Yamamichi N, Mizutani T, Ui M, Murakami M, Iba H. Identification of SWI.SNF complex subunit BAF60a as a determinant of the transactivation potential of Fos/Jun dimers. J. Biol. Chem. 2001;276:2852–2857. doi: 10.1074/jbc.M009633200. [DOI] [PubMed] [Google Scholar]
- 35.Nateri AS, Spencer-Dene B, Behrens A. Interaction of phosphorylated c-Jun with TCF4 regulates intestinal cancer development. Nature. 2005;437:281–285. doi: 10.1038/nature03914. [DOI] [PubMed] [Google Scholar]
- 36.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 37.Huang Y, Myers SJ, Dingledine R. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat. Neurosci. 1999;2:867–872. doi: 10.1038/13165. [DOI] [PubMed] [Google Scholar]
- 38.Nascimento EM, Cox CL, Macarthur S, Hussain S, Trotter M, Blanco S, Suraj M, Nichols J, Kbler B, Benitah SA, et al. The opposing transcriptional functions of Sin3a and c-Myc are required to maintain tissue homeostasis. Nat. Cell Biol. 2011;13:1395–1405. doi: 10.1038/ncb2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zervos AS, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- 40.Li-Weber M, Davydov I, Krafft H, Krammer P. The role of NF-Y and IRF-2 in the regulation of human IL-4 gene expression. J. Immunol. 1994;153:4122–4133. [PubMed] [Google Scholar]
- 41.Scott E, Simon M, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 42.Villard J, Peretti M, Masternak K, Barras E, Caretti G, Mantovani R, Reith W. A functionally essential domain of RFX5 mediates activation of major histocompatibility complex class II promoters by promoting cooperative binding between RFX and NF-Y. Mol. Cell. Biol. 2000;20:3364–3376. doi: 10.1128/mcb.20.10.3364-3376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu L, Wu Q, Yang CP, Horwitz SB. Coordination of transcription factors, NF-Y and C/EBP beta, in the regulation of the mdr1b promoter. Cell Growth Differ. 1995;6:1505–1512. [PubMed] [Google Scholar]
- 44.Roder K, Wolf S, Larkin K, Schweizer M. Interaction between the two ubiquitously expressed transcription factors NF-Y and Sp1. Gene. 1999;234:61–69. doi: 10.1016/s0378-1119(99)00180-8. [DOI] [PubMed] [Google Scholar]
- 45.Caretti G, Salsi V, Vecchi C, Imbriano C, Mantovani R. Dynamic recruitment of NF-Y and histone acetyltransferases on cell-cycle promoters. J. Biol. Chem. 2003;278:30435–30440. doi: 10.1074/jbc.M304606200. [DOI] [PubMed] [Google Scholar]
- 46.Ivanov VN, Bhoumik A, Krasilnikov M, Raz R, Owen-Schaub LB, Levy D, Horvath CM, Ronai Z. Cooperation between STAT3 and c-jun suppresses fas transcription. Mol. Cell. 2001;7:517–528. doi: 10.1016/s1097-2765(01)00199-x. [DOI] [PubMed] [Google Scholar]
- 47.Choi S, Cho Y, Kim H, Park J. ROS mediate the hypoxic repression of the hepcidin gene by inhibiting C/EBPalpha and STAT-3. Biochem. Biophys. Res. Commun. 2007;356:312–317. doi: 10.1016/j.bbrc.2007.02.137. [DOI] [PubMed] [Google Scholar]
- 48.Sementchenko VI, Watson DK. Ets target genes: past, present and future. Oncogene. 2000;19:6533–6548. doi: 10.1038/sj.onc.1204034. [DOI] [PubMed] [Google Scholar]
- 49.Rothbcher U, Bertrand V, Lamy C, Lemaire P. A combinatorial code of maternal GATA, Ets and beta-catenin-TCF transcription factors specifies and patterns the early ascidian ectoderm. Development. 2007;134:4023–4032. doi: 10.1242/dev.010850. [DOI] [PubMed] [Google Scholar]
- 50.Taylor JM, Dupont-Versteegden EE, Davies JD, Hassell JA, Houl JD, Gurley CM, Peterson CA. A role for the ETS domain transcription factor PEA3 in myogenic differentiation. Mol. Cell. Biol. 1997;17:5550–5558. doi: 10.1128/mcb.17.9.5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Geen H, Lin Y, Xu X, Echipare L, Komashko VM, He D, Frietze S, Tanabe O, Shi L, Sartor MA, et al. Genome-wide binding of the orphan nuclear receptor TR4 suggests its general role in fundamental biological processes. BMC Genomics. 2010;11:689. doi: 10.1186/1471-2164-11-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams B, Drfler P, Aguzzi A, Kozmik Z, Urbnek P, Maurer-Fogy I, Busslinger M. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev. 1992;6:1589–1607. doi: 10.1101/gad.6.9.1589. [DOI] [PubMed] [Google Scholar]
- 53.Fitzsimmons D, Hodsdon W, Wheat W, Maira SM, Wasylyk B, Hagman J. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes Dev. 1996;10:2198–2211. doi: 10.1101/gad.10.17.2198. [DOI] [PubMed] [Google Scholar]
- 54.Dudek H, Tantravahi RV, Rao VN, Reddy ES, Reddy EP. Myb and Ets proteins cooperate in transcriptional activation of the mim-1 promoter. Proc. Natl Acad. Sci. USA. 1992;89:1291–1295. doi: 10.1073/pnas.89.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mazars R, Gonzalez-de-Peredo A, Cayrol C, Lavigne A, Vogel JL, Ortega N, Lacroix C, Gautier V, Huet G, Ray A, et al. The THAP-zinc finger protein THAP1 associates with coactivator HCF-1 and O-GlcNAc transferase: a link between DYT6 and DYT3 dystonias. J. Biol. Chem. 2010;285:13364–13371. doi: 10.1074/jbc.M109.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu H, Mashtalir N, Daou S, Hammond-Martel I, Ross J, Sui G, Hart GW, Rauscher FJR, Drobetsky E, Milot E, et al. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol. Cell. Biol. 2010;30:5071–5085. doi: 10.1128/MCB.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Looijenga LH, Stoop H, deLeeuw HP, deGouveia Brazao CA, Gillis AJ, vanRoozendaal KE, vanZoelen EJ, Weber RF, Wolffenbuttel KP, vanDekken H, et al. POU5F1 (OCT3/4) identifies cells with pluripotent potential in human germ cell tumors. Cancer Res. 2003;63:2244–2250. [PubMed] [Google Scholar]
- 58.Loh Y, Wu Q, Chew J, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 59.Yi F, Merrill BJ. Stem cells and TCF proteins: a role for beta-catenin-independent functions. Stem Cell Rev. 2007;3:39–48. doi: 10.1007/s12015-007-0003-9. [DOI] [PubMed] [Google Scholar]
- 60.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKay MJ, Troelstra C, vander P, Kanaar R, Smit B, Hagemeijer A, Bootsma D, Hoeijmakers JH. Sequence conservation of therad21 Schizosaccharomyces pombeDNA double-strand break repair gene in human and mouse. Genomics. 1996;36:305–315. doi: 10.1006/geno.1996.0466. [DOI] [PubMed] [Google Scholar]
- 62.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 63.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc. Natl Acad. Sci. USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jelinic P, Stehle J, Shaw P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006;4:e355. doi: 10.1371/journal.pbio.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bischof LJ, Kagawa N, Moskow JJ, Takahashi Y, Iwamatsu A, Buchberg AM, Waterman MR. Members of the Meis1 and Pbx homeodomain protein families cooperatively bind a cAMP-responsive sequence (CRS1) from BovineCYP17. J. Biol. Chem. 1998;273:7941–7948. doi: 10.1074/jbc.273.14.7941. [DOI] [PubMed] [Google Scholar]
- 66.Kappel A, Schlaeger TM, Flamme I, Orkin SH, Risau W, Breier G. Role of SCL/Tal-1, GATA, and ets transcription factor binding sites for the regulation of flk-1 expression during murine vascular development. Blood. 2000;96:3078–3085. [PubMed] [Google Scholar]
- 67.Mouthon MA, Bernard O, Mitjavila MT, Romeo PH, Vainchenker W, Mathieu-Mahul D. Expression of tal-1 and GATA-binding proteins during human hematopoiesis. Blood. 1993;81:647–655. [PubMed] [Google Scholar]
- 68.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 69.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Costa RH, Kalinichenko VV, Holterman AL, Wang X. Transcription factors in liver development, differentiation, and regeneration. Hepatology. 2003;38:1331–1347. doi: 10.1016/j.hep.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 71.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johnson CA, Turner BM. Histone deacetylases: complex transducers of nuclear signals. Semin. Cell Dev. Biol. 1999;10:179–188. doi: 10.1006/scdb.1999.0299. [DOI] [PubMed] [Google Scholar]
- 73.Furusawa T, Cherukuri S. Developmental function of HMGN proteins. Biochim. Biophys. Acta. 2010;1799:69–73. doi: 10.1016/j.bbagrm.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peng J, Zhu Y, Milton JT, Price DH. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Partington GA, Patient RK. Phosphorylation of GATA-1 increases its DNA-binding affinity and is correlated with induction of human K562 erythroleukaemia cells. Nucleic Acids Res. 1999;27:1168–1175. doi: 10.1093/nar/27.4.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu D, Zhao L, Del Valle L, Miklossy J, Zhang L. Interferon regulatory factor 4 is involved in Epstein-Barr virus-mediated transformation of human B lymphocytes. J. Virol. 2008;82:6251–6258. doi: 10.1128/JVI.00163-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Paun A, Pitha PM. The IRF family, revisited. Biochimie. 2007;89:744–753. doi: 10.1016/j.biochi.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Corcoran LM, Karvelas M, Nossal GJ, Ye ZS, Jacks T, Baltimore D. Oct-2, although not required for early B-cell development, is critical for later B-cell maturation and for postnatal survival. Genes Dev. 1993;7:570–582. doi: 10.1101/gad.7.4.570. [DOI] [PubMed] [Google Scholar]
- 80.Baeuerle PA, Henkel T. Function and activation of NF-kappa B in the immune system. Annu. Rev. Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 81.Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- 82.Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- 83.Nagarajan P, Onami TM, Rajagopalan S, Kania S, Donnell R, Venkatachalam S. Role of chromodomain helicase DNA-binding protein 2 in DNA damage response signaling and tumorigenesis. Oncogene. 2009;28:1053–1062. doi: 10.1038/onc.2008.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Deng C. Roles of BRCA1 in DNA damage repair: a link between development and cancer. Hum. Mol. Genet. 2003;12:113R–123R. doi: 10.1093/hmg/ddg082. [DOI] [PubMed] [Google Scholar]
- 85.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3[prime] UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Farnham PJ. Insights from genomic profiling of transcription factors. Nat. Rev. Genet. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Spivakov M, Akhtar J, Kheradpour P, Beal K, Girardot C, Koscielny G, Herrero J, Kellis M, Furlong EE, Birney E. Analysis of variation at transcription factor binding sites in Drosophila and humans. Genome Biol. 2012;13:R49. doi: 10.1186/gb-2012-13-9-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2011;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl Acad. Sci. USA. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schroeder MD, Pearce M, Fak J, Fan H, Unnerstall U, Emberly E, Rajewsky N, Siggia ED, Gaul U. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2:e271. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 95.Moses A, Chiang D, Pollard D, Iyer V, Eisen M. MONKEY: identifying conserved transcription-factor binding sites in multiple alignments using a binding site-specific evolutionary model. Genome Biol. 2004;5:R98. doi: 10.1186/gb-2004-5-12-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kheradpour P, Stark A, Roy S, Kellis M. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res. 2007;17:1919–1931. doi: 10.1101/gr.7090407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, et al. Five-vertebrate ChIP-seq Reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono KI, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.MacArthur S, Li X, Li J, Brown J, Chu HC, Zeng L, Grondona B, Hechmer A, Simirenko L, Keranen S, et al. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 2009;10:R80. doi: 10.1186/gb-2009-10-7-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pietrokovski S. Searching databases of conserved sequence regions by aligning protein multiple-alignments. Nucleic Acids Res. 1996;24:3836–3845. doi: 10.1093/nar/24.19.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gray KA, Daugherty LC, Gordon SM, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2013. Nucleic Acids Res. 2013;41:D545–D552. doi: 10.1093/nar/gks1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotech. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Touzet H, Varre J. Efficient and accurate P-value computation for position weight matrices. Algorithms Mol. Biol. 2007;2:15. doi: 10.1186/1748-7188-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wilson EB. Probable Inference, the Law of Succession, and Statistical Inference. J. Am. Stat. Assoc. 1927;22:209–212. [Google Scholar]
- 109.Mahony S, Auron PE, Benos PV. DNA familial binding profiles made easy: comparison of various motif alignment and clustering strategies. PLoS Comput. Biol. 2007;3:e61. doi: 10.1371/journal.pcbi.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sandelin A, Wasserman WW. Constrained binding site diversity within families of transcription factors enhances pattern discovery bioinformatics. J. Mol. Biol. 2004;338:207–215. doi: 10.1016/j.jmb.2004.02.048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.