Abstract
Ten-eleven translocation (TET) family enzymes convert 5-methylcytosine to 5-hydroxylmethylcytosine. However, the molecular mechanism that regulates this biological process is not clear. Here, we show the evidence that PGC7 (also known as Dppa3 or Stella) interacts with TET2 and TET3 both in vitro and in vivo to suppress the enzymatic activity of TET2 and TET3. Moreover, lacking PGC7 induces the loss of DNA methylation at imprinting loci. Genome-wide analysis of PGC7 reveals a consensus DNA motif that is recognized by PGC7. The CpG islands surrounding the PGC7-binding motifs are hypermethylated. Taken together, our study demonstrates a molecular mechanism by which PGC7 protects DNA methylation from TET family enzyme-dependent oxidation.
INTRODUCTION
DNA methylation at the 5 position of the cytosine pyrimidine ring (5mC) in genomic DNA is an important epigenetic mark that regulates gene transcription during numerous physiological and pathological processes (1–5). In particular, during early embryogenesis, DNA methylation is dynamically regulated to precisely control transcription for defining cell fate (6–8). A global DNA demethylation occurs in the paternal pronucleus of a zygote, which is likely to remove epigenetic barriers that restrict gene transcription and developmental potential (9–11). This active DNA demethylation process is mediated by TET family enzymes that oxidize methyl groups at the 5 position of cytosine pyrimidine rings into hydroxymethyl groups (5hmC) in the presence of 2-oxoglutarate (2-OG) and Fe(II) (11–13). Further oxidation, probably catalyzed by these oxygenases, generates 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (14,15). Both 5fC and 5caC are relatively unstable on the chromatin (14,15) and are recognized by thymine-DNA glycosylase (TDG) that excises the base of 5fC or 5caC from the chromatin for the completion of the active demethylation process(14,16).
Among these TET enzymes, TET3, but not TET1 and TET2, is specifically expressed in zygotes (17–21). Thus, it is TET3 that mediates global DNA demethylation in the paternal pronucleus of zygote (17–20). Interestingly, TET3 is located in both maternal and paternal pronucleus although TET3 associates with the paternal pronucleus more tightly than with the maternal pronucleus (19). How can TET3 specifically remove DNA methylation mark in paternal pronucleus but not maternal pronucleus? Recent studies indicate that PGC7, a maternal factor, partially protects DNA methylation status in the maternal pronucleus (19,20). PGC7 is a 159-residue nuclear polypeptide that is mainly expressed in germ cells and pluripotent cells (22–24). Interestingly, the PGC7 gene only exists in mammals and evolved very quickly (24–26). PGC7+/− mice could give birth to PGC7 null offspring without obvious developmental defects. However, oocytes from PGC7−/− mice induce early embryogenesis arrestment at the 4-cell stage following fertilization (26–28). Detailed analysis on the PGC7-deficient zygotes suggests that loss of PGC7 induces TET3-dependent DNA demethylation in the maternal pronucleus, suggesting that PGC7 is an important maternal factor for protecting DNA methylation (19,20). It has been shown that PGC7 binds to dimethylated histone H3K9 (H3K9me2) in the heterochromatin region through its N-terminal tail (19). Interestingly, in this region, PGC7 contains a putative DNA-binding domain (26). Thus, the molecular mechanism by which PGC7 suppresses TET enzyme-induced DNA demethylation could be more complicated. Moreover, a large portion of DNA methylation remains intact even in the absence of PGC7 (28), suggesting that PGC7 only partially protects methylated DNA from TET3-dependent demethylation.
To examine the molecular mechanism of PGC7 in protecting DNA methylation, we analyzed the domains important for the function of PGC7. Although the homology of PGC7 is relatively low among different mammalian species, it contains a conserved DNA-binding domain at the N-terminus (26). Here, we characterized human PGC7 and found that the N-terminal DNA-binding domain of PGC7 interacted with TET2 and TET3. This interaction suppresses the enzymatic activity of TET2 and TET3 both in vitro and in vivo. Moreover, the DNA-binding domain of PGC7 recognizes a consensus DNA sequence and protects the surrounding CpG methylation in the chromatin. These results provide a novel molecular mechanism by which PGC7 suppresses TET family enzymes-induced DNA demethylation.
MATERIALS AND METHODS
Plasmids and antibodies
Full-length complementary DNA (cDNA) of TET1, TET2 and TET3 were cloned into pS-FLAG-SBP (SBP) vector. For protein co-immunoprecipitation experiments, PGC7 and PGC7 deletion mutants were cloned into SBP vector as well as pCMV–Myc vector. TET2 deletion mutants were cloned into SBP vector. TET3CD was subcloned into pCMV–HA vector and SBP vector.
Anti-FLAG (M2) antibody was purchased from Sigma. Anti-HA and anti-Myc monoclonal antibody were purchased from Covance. Anti-5hmC polyclonal antibody was purchased from Zymo Research. Anti-5mC monoclonal antibody was purchased from Calbiochem. Rabbit anti-human PGC7 antibody was raised against full length of human PGC7. Rabbit anti-human TET3 antibody was raised against the a.a. 351–668 of hTET3, rabbit anti-human TET1 antibody was raised against C-terminus (a.a. 1500–2008) of TET1. Rabbit anti-human TET2 antibody was purchased from Abcam (ab94580).
Recombinant proteins
Recombinant proteins were purified from Sf9 insect cells. For generating baculovirus, DNA fragments containing full-length human PGC7, deletion mutants of PGC7, TET2 and TET3CD were subcloned into pFastBac vector with a GST or SBP tag. Baculoviruses were generated in accordance with the manufacturer’s instructions (Invitrogen). After Sf9 cells were infected with baculoviruses for 48 h, the cells were harvested, washed with cold phosphate buffered saline (PBS) three times and lysed with ice-cold NETN100 buffer (20 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 % Nonidet P-40). The soluble fraction was incubated with glutathione-sepharose beads (for GST-tag proteins) or streptavidin-conjugated beads (for SBP-tag proteins) and eluted with glutathione or biotin. For protein binding assay, baculoviruses expressing GST-tagged PGC7 and SBP-tagged TET3CD co-infected Sf9 cells.
Immunofluorescence staining
Cells grown on coverslips were fixed with 3% paraformaldehyde for 20 min and permeabilized with 0.5% Triton X-100 in PBS for 5 min at room temperature. Cells were then washed with cold PBS and incubated with blocking buffer (10% donkey serum, 3% bovine serum albumin in PBS containing 0.1% Triton X-100) before incubation with primary antibodies overnight in a humidified chamber at 4°C. For 5hmC staining, permeabilized cells were denatured with 2 N HCl for 30 min, neutralized with 100 mM Tris–HCl (pH 8.5) before blocking. After three consecutive 5-min washes with PBS, cells were incubated with secondary antibodies for 30 min. The cells were washed again three times with PBS and then mounted onto glass slides and visualized by a fluorescence microscope.
Cell lysis, immunoprecipitation and western blotting
For immunoprecipitation assays, 293 T cells were lysed with ice-cold NETN400 buffer (0.5% NP-40, 50 mM Tris–HCl pH 8.0, 2 mM EDTA and 400 mM NaCl) containing 10 mM NaF and 50 mM β-glycerophosphate. The soluble fractions were collected and diluted to NETN100, then directly subjected to electrophoresis or immunoprecipitation with indicated antibodies followed by western blot analysis with indicated antibodies.
Chromatin immunoprecipitation assay and methylated DNA immunoprecipitation assay
Chromatin immunoprecipitation assays (ChIP) were performed according to the protocol described by Upstate. The 293 T cell DNA was sonicated to an average size between 300 and 600 bp. Solubilized chromatin was immunoprecipitated with the antibody against TET3 and PGC7. Antibody–chromatin complexes were pulled down using protein A-sepharose beads, washed and then eluted. After cross-link reversal and proteinase K treatment, immunoprecipitated DNA was extracted with phenol-chloroform, ethanol-precipitated and treated with RNase. ChIP DNA was qualified using PicoGreen.
For methylated DNA immunoprecipitation (MeDIP), genomic DNA was extracted using Promega Wizard Genomic DNA Purification Kit. After Proteinase K and RNase treatment, the genomic DNA was extracted with phenol-chloroform, precipitated with ethanol and dissolved with Tris-EDTA (TE) buffer. The purified genomic DNA was sonicated using Diagenode Bioruptor 200. The fragment size of DNA was determined by 2% agarose gel. The DNA fragment size was 200–800 bp. The samples were heat-denatured and mixed with 1 µg 5mC antibody. The mixtures were incubated overnight at 4°C and then pulled down by protein A-sepharose beads. The beads were extensively washed and digested by Proteinase K. DNA was further purified by phenol-chloroform extraction and ethanol precipitation. MeDIP DNA was qualified using PicoGreen.
ChIP sequencing
DNA fragments isolated from ChIP were repaired to blunt ends by T4 DNA polymerase and phosphorylated with T4 polynucleotide kinase using the END-IT kit (Epicentre). A single ‘A’ base was added to 3′-end with Klenow. Double-stranded adaptors (75 bp with a ‘T’ overhang) were ligated to the fragments with DNA ligase. Ligation products between 200 and 600 bp and were gel purified to remove unligated adaptors and subjected to 20 polymerase chain reaction (PCR) cycles. Completed libraries were quantified with PicoGreen. The DNA libraries were analyzed by Illumina Hiseq2000 high-throughput sequencing. The read quality of each sample was determined by FastQC software. After pre-filtering the raw data by removing sequence adaptors and low quality reads, the tags were mapped to the human genome (hg19) by Bowtie software. Parameters settings were listed as follows: −v, 3 (reported alignments with at most three mismatches), −5, 3 and −3, 7 (trim three bases from 5′-end and seven from 3′-end to remove low-quality bases). Peak detection was performed using MACS software from Galaxy browser (http://www.galaxy.psu.edu). Parameter settings were as follows: IgG ChIP-Seq aligned reads were used as control file, tag size with 25 bp and band width 300 bp. The peaks obtained from ChIP sequencing (ChIP-Seq) were matched to the annotated reference genome (human hg19) using Cisgenome 2.0. To view the peak density and position, Cisgenome 2.0 was used. The relative accumulations of tags around transcription start sites (TSS) were performed using SEQMINER software. The TSS position of each gene was downloaded from UCSC genome browser. Integrative Genomics Viewer (IGV) was used to analyze the peak distribution in the chromosome level. To obtain the binding motif of PGC7, the online software Peak-motifs (http://rsat.ulb.ac.be/peak-motifs_form.cgi) was used. A set of 30 PCR primer pairs (Supplementary Table S1) were designed to amplify 100–140 bp fragments from genomic regions showing a wide range of signals for TET3 and PGC7 ChIP-Seq as well as MeDIP sequenceing (MeDIP-Seq). ChIP quantitative PCR (ChIP-qPCR) values reflect two independent ChIP assays, and each was evaluated in duplicate by qPCR. For methylation analysis of the aligned reads of MeDIP-Seq, the Batman algorithm has been used (29). Batman algorithm infers the absolute methylation state for 100-bp windows by estimating local sequencing read enrichment for methylation taking into account the varying densities of methylated CpGs across the genome. Batman output is in the form of GFF format, each GFF file represents a score that equals the median of methylation states in a 100-bp window. The score ranges between 0 and 1 according to the level of methylation (percentage of methylated CpGs in the specific region). Methylation states of Group I or Group II genes were summarized, respectively, based on the Batman score at TSS (± 1 kb) (Figure 5B). ChIP-Seq data of O-linked N-acetylglucosamine (GlcNac) transferase (OGT) was according to our previous study (30).
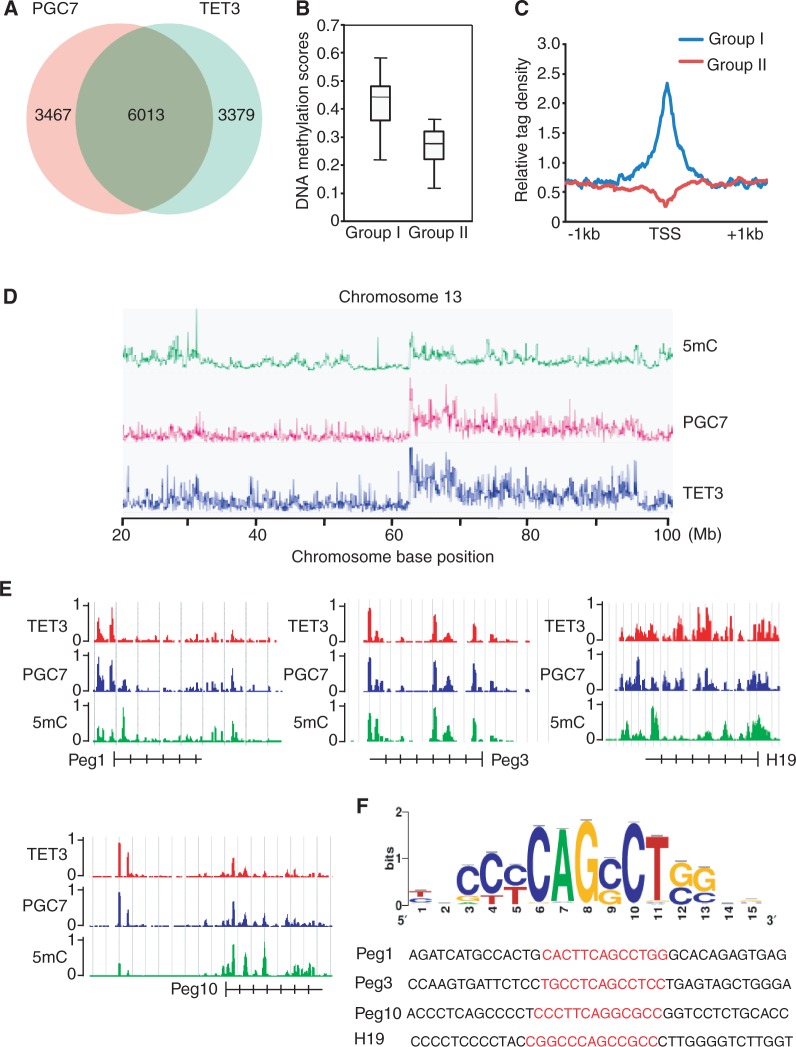
Figure 5.
PGC7 co-localizes with TET3 at a set of genes. (A) Venn diagram shows a significant overlap between PGC7 and TET3 target genes. (B) 5mC level in the genes occupied by PGC7 and TET3 (Group I genes) is significantly higher than that in the genes occupied by only TET3 (Group II genes). (C) Mean distribution of 5mC at TSS (± 1 kb) of Group I or Group II genes. (D) PGC7 and TET3 co-occupancy is associated with 5mC at whole-chromosome level. Significant overlap is shown in chromosome 13. (E) ChIP-Seq results show the co-occupancy of PGC7 and TET3 at methylated DNA region of Peg1, Peg3, Peg10 and H19 loci. (F) The consensus DNA-binding motif of PGC7 is analyzed according to ChIP-Seq result. The binding sequences in Peg1, Peg3, Peg10 and H19 loci are shown in red.
In vitro 5hmC assay
To examine the effect of PGC7 on the activity of TET2 and TET3 in vitro, 0.4 µg double-stranded DNA substrates with 5-methylcytosine replacing cytosine were mixed with 5 µg wild-type recombinant PGC7 or PGC7 mutant proteins as well as TET3CD or TET2CD proteins and incubated with 50 mM HEPES, pH 8, 50 mM NaCl, 2 mM Ascorbic Acid, 1 mM 2-OG, 100 µM FAS (Fe2+), 1 mM DTT and 1 mM ATP for 1 h at 37°C. The samples were subjected to dot blotting assay or PvuRts1I restriction enzyme digestion.
Dot blotting assay
Genomic DNA from 293T cells or the DNA substrate from in vitro assays was denatured by 0.2 N NaOH and dotted on Hybond-N+ nitrocellulose membrane (Amersham Pharmacia Biotech). After drying at 65°C, the membrane was blocked with 10% non-fat milk for 1 h at room temperature followed by 2-h incubation with anti-5hmC antibodies at room temperature. After three consecutive 10-min washes with Tris-buffered saline with Tween®-20 (TBST), the membrane was incubated with horseradish peroxidase (HRP)-conjugated goat-anti-rabbit secondary antibody for 1 h. The membrane was washed again three times with TBST and developed using the Enhanced ChemiLuminescence plus (ECL+) detection system (GE Healthcare).
PvuRts1I restriction enzyme digestion assay
CpGenome 5mC DNA standard (Millipore, Cat. # S8005) was subjected to in vitro 5hmC assay. The DNA from in vitro assay was digested by PvuRts1I restriction enzyme (Active Motif) at 22°C. After 30 min, the DNA samples were subjected to gel electrophoresis or qPCR. PCR primers are listed in Supplementary Table S1.
Bisulfite sequencing
The methylation profiles of Peg1, Peg3, Peg10 and H19 were determined by a slightly modified genomic sequencing technique after bisulfite treatment. Briefly, 500 ng genomic DNA isolated from HBL100, or HBL100 with PGC7 knockdown or TET3 overexpression or both was treated with bisulfite according to the protocol of the Zymo EZ DNA Methylation Kit (Kit # D5001). The bisulfite-converted DNA was subjected to PCR or nesting PCR. The primers are listed in Supplementary Table S1. The PCR products were separated by 1% agarose gel, purified by gel extraction kit and then cloned to a T vector for sequencing.
Retroviral vectors construction, virus production and infection
Retrovirus short hairpin RNA (shRNA) vectors targeting human PGC7 (shPGC7) and Scramble were constructed by inserting short hairpin RNA templates into pMSCV-neo-U6, a RNAi vector that was constructed in Dr Zhao’s lab (31). The short hairpin RNA templates specifically targeting PGC7 were designed, synthesized and annealed as previously reported. The sequences are shown in Supplementary Table S1. The shPGC7 and Scramble vectors were transfected into packaging cell line 293T with two other helper packaging plasmids pMD-MLV-OGP (gag-pol) and pVSV-G (env). After 48 h of transfection, the cell culture medium was harvested and the viral particles were concentrated by ultracentrifugation at 50 000g for 3 h and then resuspended in expansion medium. HBL100 cells were infected with retrovirus at a multiplicity of infection (MOI) of 10 for 4 h in the presence of 8 µg/ml polybrene. After infection, HBL100 cells were allowed to recover for 24 h and then selected in expansion medium with 800 µg/ml G418 (Sigma Aldrich) for 1 week and maintained in the medium with 400 µg/ml of G418.
RESULTS
Recent study suggests that PGC7 plays an important role in protecting DNA methylation during early embryogenesis, especially at several imprinting loci (28). To examine the molecular mechanism of this interesting phenomenon, we hypothesized that PGC7 might interact with DNA methylation and demethylation enzymes, namely DNMT family and TET family enzymes. It has been reported that DNMT1, 3 a, 3 b and TET3 all regulate DNA methylation pattern during early embryogenesis (17,32–35). Thus, we examined the interaction between PGC7 and these enzymes using co-immunoprecipitation (co-IP) assays (Supplementary Figure S1A). Interestingly, we found that PGC7 specifically associated with TET3 but not the DNA methyltransferase (DNMT) family enzymes. In addition to TET3, both TET1 and TET2 can convert 5mC into 5hmC for potential DNA demethylation. Thus, we also examined the interaction between PGC7 and these two TET enzymes and found that only TET2 interacts with PGC7 (Figure 1A). Collectively, these results suggest that PGC7 selectively interacts with TET2 and TET3.
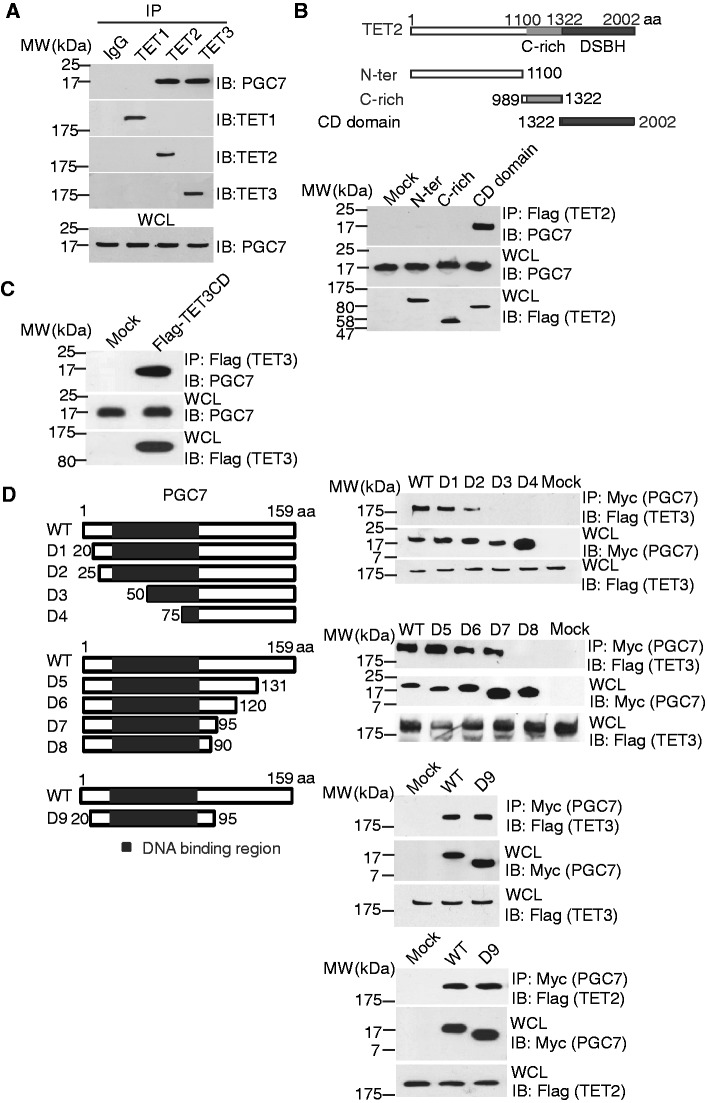
Figure 1.
PGC7 interacts with TET2 and TET3. (A) PGC7 interacts with TET2 and TET3, but not with TET1. Co-IP was performed in 293T cells. PGC7 and TET1-3 were examined with indicated antibodies. The whole cell lysates (WCL) of 293T was used as the input. An irrelevant IgG was used as the IP control. (B) The catalytic DSBH (CD) domain of TET2 interacts with PGC7. Flag-tagged deletion mutants of TET2 were expressed in 293T cells. Co-IP assays were performed with indicated antibodies. Mock transfected cells were used as the control. (C) The CD domain of TET3 also interacts with PGC7. (D) The fragment of PGC7 with a.a. 20–95 interacts with TET3 catalytic domain (TET3CD) and TET2 catalytic domain (TET2CD). Myc-tagged N-terminus or C-terminus deletion mutants of PGC7 were generated and expressed in 293T cells. Co-IP assays were performed with indicated antibodies.
To examine the details of these interactions, we mapped the regions that mediate these interactions. Based on the predicted protein folding, we generated a series of truncation mutants and found that only the C-terminal catalytic DSBH (double strand β-helix) domain (CD domain) of TET2 interacts with PGC7 (Figure 1B). Consistently, the CD domain of TET3 also interacted with PGC7 (Figure 1C), suggesting that the CD domains in TET2 and TET3 are important for the interaction with PGC7. To map the binding region in PGC7, a series of N-terminal and C-terminal truncation mutants were generated. Deletion of the N-terminal a.a. 1–19 (D1) or C-terminal a.a. 96–159 (D7) did not affect the interaction. However, deletion of the N-terminal a.a.1–25 (D2) and C-terminal a.a. 91–159 (D8) significantly decreased or even abolished the interaction with TET3. Moreover, the region encoding a.a. 20–95 was sufficient to interact with TET2 or TET3 (Figure 1D). Interestingly, this region of PGC7 is a putative DNA-binding domain. Thus, it is likely that this domain of PGC7 interacts with TET2 and TET3. In addition, to examine whether PGC7 directly interacts with TET enzymes, we co-infected Sf9 cells with baculoviruses encoding GST-PGC7 and SBP-TET3 CD domain. These two recombinant proteins could be co-purified (Supplementary Figure S1B). Taken together, these results suggest that PGC7 interacts with TET3 in vitro and in vivo.
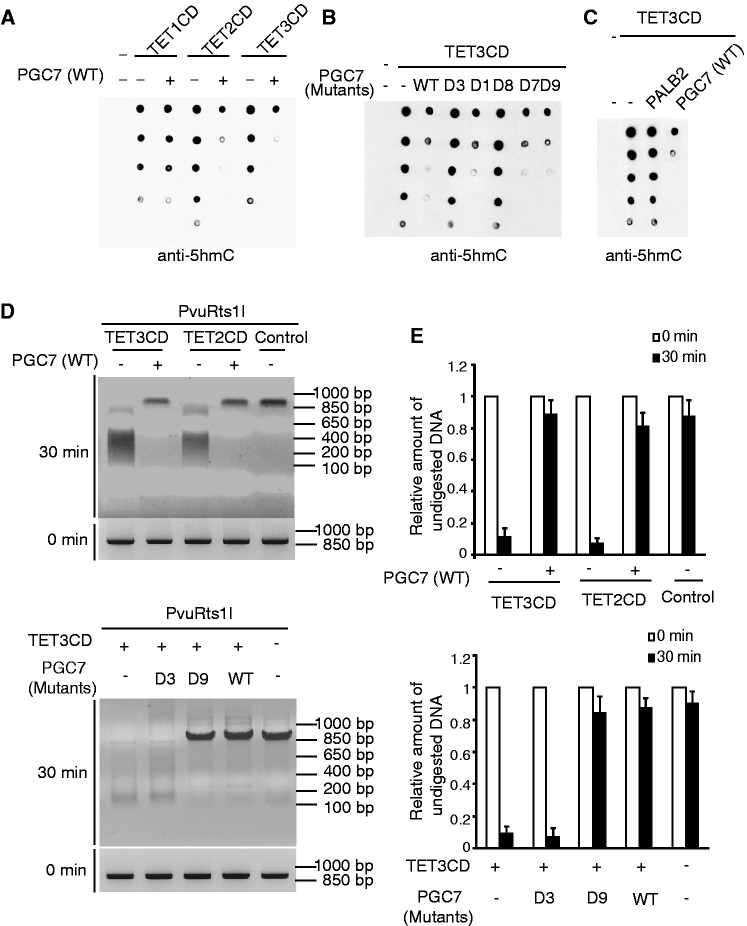
To understand the functional significance of the interaction between PGC7 and TET2 or TET3, we examined whether PGC7 regulated TET2 or TET3-dependent 5mC oxidation. Accumulated evidence shows that TET enzymes oxidize 5mC to 5hmC in the presence of Fe(II) and 2-OG (12,13). Thus, we established the in vitro DNA oxidation assays to detect 5hmC. Using dot blotting assays, we observed that the recombinant CD domains of TET family enzymes converted 5mC to 5hmC. Surprisingly, when the recombinant PGC7 was supplemented into this in vitro DNA oxidation assay, the recombinant PGC7 repressed the enzymatic activity of the CD domains of TET2 and TET3 (Figure 2A and Supplementary Figure S2A). Next, we generated the recombinant D1, D3, D7-9 mutants of PGC7 (Supplementary Figure S2B). As shown in Figure 2B, the D3 and D8 mutants of PGC7 that abolish the interaction with TET3 failed to suppress the enzymatic activity of the CD domain of TET3, whereas the D1, D7 and D9 mutants of PGC7 that still interact with TET3 could suppress the TET3-mediated DNA oxidation. These results suggest that the interaction between PGC7 and TET enzymes is critical for this suppression. Moreover, PGC7 failed to suppress TET1-dependent oxidation of methylated DNA as PGC7 does not interact with TET1 (Figure 2A). The interaction region in PGC7 is a putative DNA-binding domain. To exclude the possibility that the inhibition of TET3 is due to the non-specific DNA-binding of PGC7, which blocks the 5mC epitope recognized by TET3, we included the recombinant PALB2 as the negative control. It has been shown that PALB2 non-specifically binds both single-stranded DNA and double-stranded DNA (36). However, supplement of PALB2 in this in vitro DNA oxidation assay could not block the TET3-dependent oxidation of methylated DNA (Figure 2C). As the putative DNA-binding domain of PGC7 directly interacts with the catalytic domain of TET3, it is likely that the interaction masks the catalytic site of TET3 and thus suppresses TET3-dependent oxidation of methylated DNA. We also used a different approach to confirm the results. PvuRts1I is a restriction enzyme that specifically digests double-stranded DNA containing 5hmC but not 5mC (37). When 5mC was converted to 5hmC by the CD domain of TET2 or TET3, the DNA substrate was digested by PvuRts1I. However, in the presence of PGC7, the enzymatic activity of TET2 or TET3 was suppressed, which failed to convert 5mC into 5hmC. Thus, PvuRts1I could not digest the DNA substrate in the presence of PGC7 (Figure 2D). We also examined the mutants of PGC7 in this assay. Again, the D9 mutant of PGC7 that still interacts with TET3 and suppresses the enzymatic activity of TET3 inhibited the DNA substrate digestion by PvuRts1I. However, with the D3 mutant of PGC7 that does not interact with TET3, PvuRts1I digested the DNA substrate oxidized by TET3 (Figure 2D). Moreover, we quantitatively measured the remaining intact DNA substrate following PvuRts1I treatment by qPCR. The results are consistent with the agarose gel electrophoresis (Figure 2E). Taken together, these results suggest that the direct interaction between PGC7 and TET2 or TET3 suppresses the enzymatic activity of TET2 or TET3.
Figure 2.
PGC7 suppresses the enzymatic activity of TET2 or TET3 in vitro. (A) Recombinant PGC7 suppresses TET2 or TET3, but not TET1, for 5mC to 5hmC conversion in vitro. The in vitro 5hmC assays were performed and described in ‘Materials and Methods’ section. (B) The interaction between PGC7 and TET3 is required for the inhibition of TET3 in vitro. Recombinant PGC7 mutants were generated and examined in the in vitro 5hmC assays. (C) Non-specific DNA-binding of PALB2 does not suppress the enzymatic activity of TET3CD. (D), (E) Wild-type PGC7 and D9 mutant, but not the D3 mutant, suppress TET2CD- or TET3CD-induced 5hmC. 5hmC catalyzed by TET2CD or TET3CD was examined by PvuRts1I digestion assays as described in ‘Materials and Methods’ section. DNA samples were analyzed by gel electrophoresis (D) or qPCR (E). Error bars indicate standard deviation (SD) (n = 3).
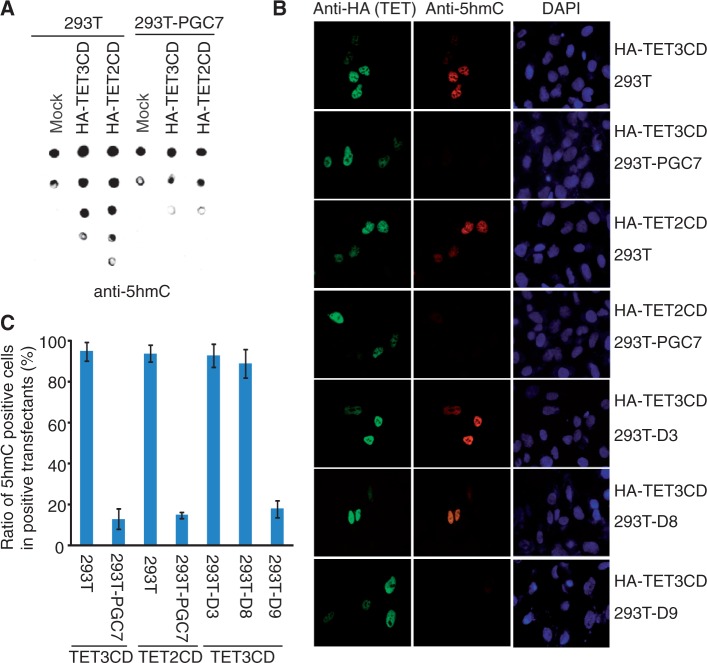
Next, we examined whether PGC7 could suppress TET2- and TET3-dependent DNA oxidation in vivo. As 293T cells express TET1 (Figure 1A), depleting endogenous TET2 and TET3 did not significantly affect the level of 5hmC. Thus, we generated a 293T cell line stably expressing PGC7 and named it as 293T-PGC7 (Supplementary Figure S3A). We then expressed the catalytic domain of TET2 or TET3 in 293T and 293T-PGC7-cells. Compared with that in 293T cells, the level of 5hmC was remarkably reduced in 293T-PGC7 cells when the catalytic domain of TET2 or TET3 was expressed (Figure 3A). Moreover, 5hmC was also examined by immunofluorescence staining. The 5hmC positively-stained population in 293T-PGC7 cells was significantly less than that in 293T cells when the catalytic domains of TET2 or TET3 were expressed (Figure 3B and C; Supplementary Figure S3B). We also established 293T cell lines stably expressing D3, D8 or D9 mutants of PGC7 and named them as 293T-D3, 293T-D8 and 293T-D9, respectively. Again, the D3 and D8 mutants of PGC7 that do not interact with TET2 or TET3 failed to suppress the TET2 CD domain or TET3 CD domain-induced 5hmC, whereas the D9 mutant that still interacts with TET2 or TET3 inhibits the TET2- or TET3-dependent 5hmC generation in vivo (Figure 3B and C). Collectively, these results demonstrate that PGC7 suppresses the enzymatic activity of TET2 and TET3 in vivo.
Figure 3.
PGC7 suppresses the enzymatic activity of TET2CD and TET3CD in vivo. (A) The 5hmC level is examined by dot blotting assays in 293T or 293-PGC7 cells overexpressing TET2CD or TET3CD. (B) Immunofluorescence staining shows that TET2CD- or TET3CD-induced 5hmC is suppressed by wild-type PGC7 or the D9 mutant, but not the D3 or D8 mutant. (C) The ratio of 5hmC positively stained cells in TET2CD or TET3CD positively transfected cells was summarized.
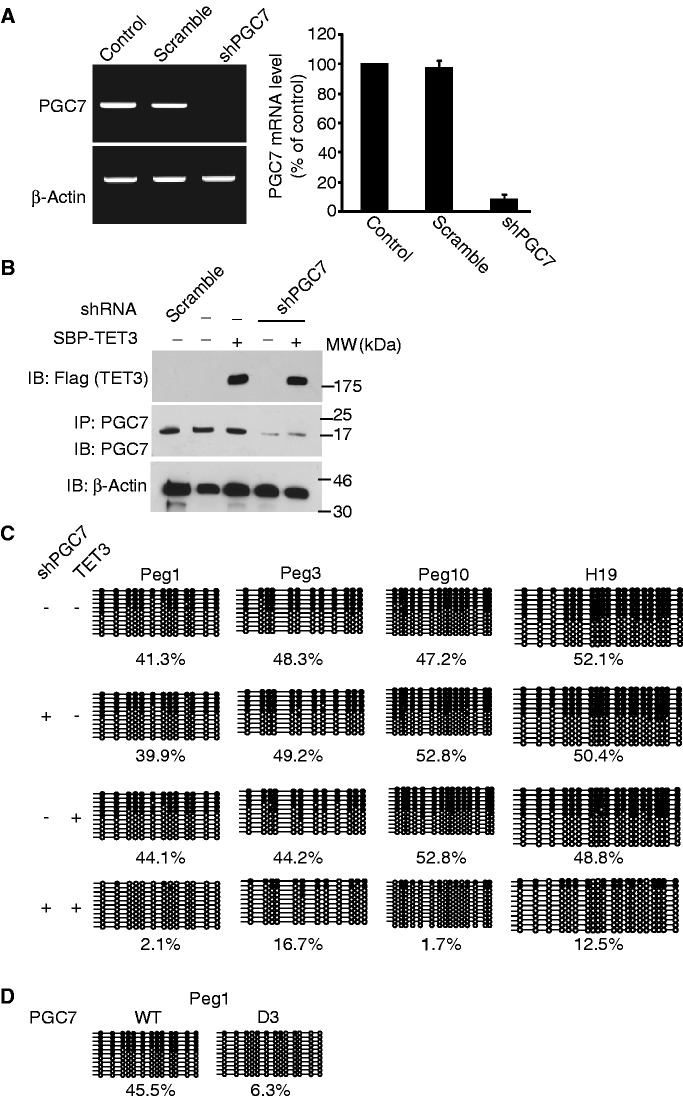
Previous studies reported that PGC7 is important for the protection of DNA methylation mark in the maternal pronucleus from TET3-mediated global DNA demethylation (19). However, not all the methylated CpG loci are protected by PGC7 (28). Lacking PGC7 in oocytes induces loss of DNA methylation mark at certain imprinting loci (28), suggesting that PGC7 might selectively protect methylated CpG loci from TET3-dependent DNA demethylation. To examine whether the interaction between PGC7 and TET3 protects the DNA methylation status of the imprinting loci, we analyzed epithelial HBL100 cell line, a diploid cell line established in vitro from the milk of an apparently healthy woman (38). Unlike in 293T cells, few genetic alterations occur in HBL100 cells. We studied DNA methylation in HBL100 cells. Using shRNA, PGC7 was stably down-regulated in HBL100 cells (Figure 4A and B). Moreover, we also stably expressed TET3 in these cells as the expression of both endogenous TET3 and TET2 in HBL100 is very low (Figure 4B and Supplementary Figure S4A). We examined and compared the DNA methylation status in the parental HBL100 cells and cells with down-regulation of PGC7 and/or up-regulation of TET3. Consistent with PGC7 knockout zygotes, we found that DNA methylation was significantly lost in Peg1, Peg3 and Peg10 loci in HBL100 cells with both down-regulation of PGC7 and up-regulation of TET3 (Figure 4C). Of note, although up-regulation of TET3 significantly reduces the overall 5mC level (Supplementary Figure S4B), it did not drastically change the methylation pattern at the imprinting loci without down-regulation of PGC7 (Figure 4C), suggesting that the limited amount of endogenous PGC7 may selectively protect these imprinting loci, but not global 5mC in HBL-100 cells. As Peg1, Peg3 and Peg10 loci are maternal imprinting loci, we also examined H19 locus, a paternal imprinting locus. Again, the DNA methylation at H19 locus was also lost when cells lacked PGC7 and expressed TET3 (Figure 4C). Moreover, using reverse transcriptase–PCR (RT–PCR), we found that the transcription of these genes significantly increased in the HBL100 cells with down-regulation of PGC7 and up-regulation of TET3 (Supplementary Figure S4C). Thus, these results suggest that PGC7 protects DNA methylation from the TET3-dependent DNA demethylation process. Moreover, we introduced shRNA-resistant PGC7 or the D3 mutant back into the cells lacking PGC7 and expressing TET3. Only wild-type PGC7, and not the D3 mutant, could suppress the loss of DNA methylation at the Peg1 locus (Figure 4D). Taken together, these results demonstrate that the direct interaction between PGC7 and TET3 protects the imprinting loci from TET3-dependent DNA demethylation. Thus, this human cell model recapitulates the DNA methylation protection by PGC7 during early embryo development. Moreover, it demonstrates a novel molecular mechanism by which PGC7 protects methylated CpG from TET enzyme-dependent DNA demethylation.
Figure 4.
PGC7 protects CpG methylation at imprinting loci from TET3-induced DNA demethylation. (A) shRNA treatment specifically down-regulates the mRNA level of PGC7 in HBL100 cells. The level of PGC7 mRNA was examined by RT–PCR (left panel) and RT–qPCR (right panel). Error bars indicate SD (n = 3). Scramble shRNA was used as negative control. (B) SBP-TET3 is stably expressed in HBL100 cells. IP and western blotting were examined by indicated antibodies. (C) Bisulfite sequencing shows methylated CpG at Peg1, Peg3, Peg10 and H19 loci in the presence or absence of PGC7 depletion and/or TET3 overexpression. (D) Expression of RNAi-resistant PGC7, but not the D3 mutant, rescues the loss of DNA methylation at Peg1 locus in HBL100 cells with endogenous PGC7 depletion and TET3 overexpression.
As the region of PGC7 mediating the interaction with TET3 is a putative DNA-binding domain (26), we asked if this putative DNA-binding domain recognizes any specific DNA sequence in those loci analyzed above, thus determining the region under PGC7 protection against TET3-mediated DNA demethylation. We performed ChIP–qPCR assay and found that PGC7 indeed bound these loci. Moreover, like wild-type PGC7, the mutant D9, containing only the DNA-binding domain also associated with these imprinting loci (Supplementary Figure S5). To determine if PGC7 recognizes specific DNA sequence, we performed high-throughput ChIP-Seq to examine the genome-wide localization of PGC7 and TET3 in 293T cells. A total of 9480 PGC7 target genes and 9392 TET3 target genes were identified from the ChIP-Seq analysis. The ChIP-Seq results were validated by using ChIP–qPCR to examine 30 randomly picked loci that represent a broad range of ChIP-Seq fragment counts (Supplementary Figure S6A). We found 63% PGC7 target genes were bound by TET3, whereas 64% target genes of TET3 were bound by PGC7, indicating a significant overlap of the PGC7 and TET3 target genes (Figure 5A). To examine the role of PGC7 in the protection of DNA methylation, we named those genes bound by both PGC7 and TET3 as group I, and the genes only bound by TET3 as group II. We examined the DNA methylation using MeDIP. Comparing group I with group II genes, we found that the DNA methylation level in group I genes was significantly higher than that in group II genes (Figure 5B). TET enzymes have been shown to locate at the TSS (8,30,39,40). Thus, we examined the TSS occupied by TET3. The 5mC levels are significantly higher at TSS in the presence of PGC7 than those without PGC7 (Figure 5C). Moreover, chromosome level analysis further indicates that PGC7 and TET3 co-occupied regions are associated with high levels of 5mC, such as chromosome 13 as shown (Figure 5D). We also examined the localization of PGC7 and TET3 at the Peg1, Peg3, Peg10 and H19 imprinting loci with the high-throughput ChIP-Seq analysis. Again, PGC7 co-localized with TET3 in these gene loci (Figure 5E). To further characterize the DNA-binding of PGC7, we analyzed the DNA-binding sequence that is recognized by PGC7 using Peak-motifs software (http://rsat.ulb.ac.be/peak-motifs_form.cgi). Based on the PGC7-enriched regions, a specific DNA-binding sequence was concluded by the software and this sequence was confirmed in the Peg1, Peg3, Peg10 and H19 imprinting loci, which are occupied by PGC7 (Figure 5F). Electromobility shift assays further confirmed the interaction between PGC7 and DNA with the consensus motif (Supplementary Figure S6B).
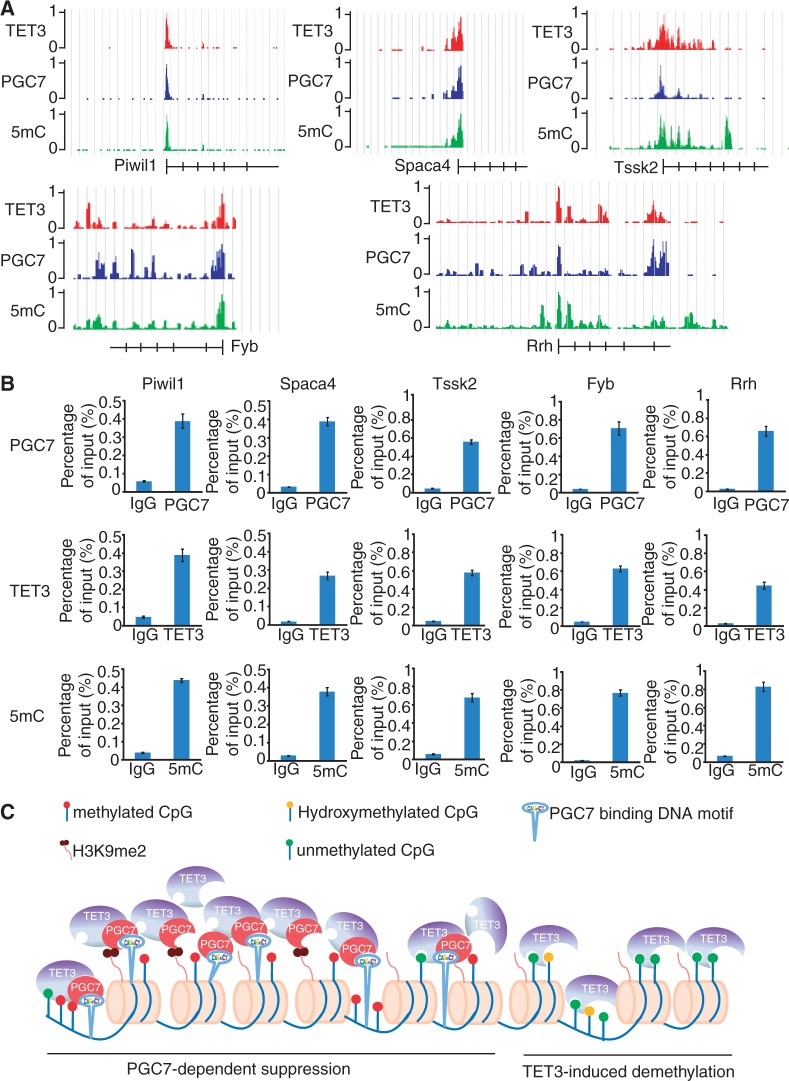
Besides the imprinting genes, PGC7 and TET3 co-localize at many other gene loci containing the consensus DNA sequence recognized by PGC7 (Supplementary Table S2). The ontology analysis classified these genes into different pathways (Supplementary Figure S6C). Interestingly, a number of these genes are tissue-specific genes, such as male germline expressing genes and somatic genes specifically expressed in hematopoietic linages or retina (Supplementary Table S3). Like imprinting loci, it has been reported that DNA methylation status in the promoter region of these genes could be inherited from parental gametes (41). We randomly picked up five genes, including male germline expressing gene Piwil1, Spaca4, Tssk2, hematopoietic gene Fyb and retina-related gene Rrh and examined the localization of PGC7, TET3 and 5mC. Like those imprinting loci, PGC7 co-localizes with TET3 and is associated with the high level of 5mC at these loci (Figure 6A). ChIP–qPCR was performed for these genes to confirm the ChIP-Seq results (Figure 6B). Thus, based on our study, we propose that PGC7 protects DNA methylation from TET enzyme-dependent oxidation by interaction with TET2 and TET3 catalytic domain at a set of gene loci (Figure 6C).
Figure 6.
Analysis of non-imprinting loci co-occupied by PGC7 and TET3. (A) ChIP-Seq results show the PGC7 and TET3 co-occupancy at Piwil1, Spaca4, Tssk2, Fyb and Rrh loci, which is associated with high 5mC level. (B) ChIP–qPCR results confirm the localization of PGC7, TET3 and 5mC in these loci. (C) A model shows that PGC7 suppresses TET3-dependent DNA demethylation.
DISCUSSION
In this study, we show the evidence that PGC7 suppresses the enzymatic activity of TET2 and TET3 and protects 5mC from oxidation. The interaction between PGC7 and TET2 or TET3 is mediated by the DNA-binding domain of PGC7 and the catalytic domain of TET2 or TET3. Thus, it is likely that the DNA-binding domain of PGC7 directly blocks the activation sites of TET2 and TET3, thus suppressing the enzymatic activity of TET2 and TET3. However, the DNA-binding domain of PGC7 does not interact with TET1. Interestingly, the homology between the catalytic domain of TET2 and TET3 is higher than that between these two TET enzymes and TET1. The major difference is in a variable region inside of the catalytic domain (13,42). However, only the variable regions from TET2 and TET3 do not interact with PGC7 (Supplementary Figure S7). It is possible that the variable region alone could not be correctly folded. Future structural analysis will reveal the details of the interaction between PGC7 and TET2/3. Nevertheless, our study suggests that the different catalytic domains of TET enzymes determine the specific interactions with their binding partners. Of note, the interaction between PGC7 and TET2 or TET3 is similar to that between OGT and TET2 or TET3 (30,43,44). Like PGC7, OGT directly interacts with the catalytic domain of TET2 or TET3 (30,43,44). As both PGC7 and OGT interact with the catalytic domain of TET2 and TET3, we examined whether PGC7 and OGT are in one complex. However, we could not detect any interaction between OGT and PGC7 using IP and western blotting (Supplementary Figure S8). We further compared genome-wide distribution of OGT and PGC7, and did not observe the significant overlapping between OGT and PGC7-occupied genes, suggesting that the OGT/TET2 and PGC7/TET2 complexes are functionally independent. It is possible that OGT and PGC7 compete with each other to interact with TET2/TET3. Moreover, OGT has been shown to functionally interact with TET2 for the transcription activation (30,43,44). Here, we found that PGC7 negatively regulates TET2 or TET3 by suppressing the enzymatic activity of TET2 or TET3. Further analysis of TET2 and TET3 complexes will reveal the functional correlation between PGC7 and OGT.
Our analysis suggests that the DNA-binding domain of PGC7 not only blocks the enzymatic sites of TET2 and TET3 but also recognizes a consensus DNA sequence. Thus, the DNA-binding domain of PGC7 has dual functions. In somatic cells or other development stages, low levels of PGC7 bind consensus DNA sequence and protect surrounding CpG loci from TET enzyme-dependent DNA demethylation. It is possible that the relatively high levels of PGC7 in germ cells may not only protect DNA methylation at all the PGC7-binding loci, but additionally block TET3-mediated global DNA demethylation non-specifically after saturating the PGC7-binding sites on the chromatin. This is supported by the interesting phenomenon that the chromatin-associated TET3 is easily eluted from the chromatin at the maternal pronucleus following Triton X-100 treatment in the presence of high levels of PGC7 in zygotes (19). In addition, recent studies indicate that PGC7 recognizes H3K9me2 through its N-terminal tail (19). We examined the role of H3K9me2 in the recruitment of PGC7 to chromatin by reducing H3K9me2 with two different approaches (Supplementary Figure S9). Lacking H3K9me2 only slightly reduces the chromatin-bound PGC7, indicating that both DNA and H3K9me2 function together to recruit PGC7 and the N-terminal tail of PGC7 is important for both DNA-binding and histone binding as well as protecting DNA methylation (Figure 6C).
In this study, we have shown that PGC7 suppresses the enzymatic activity of both TET2 and TET3. Our genome-wide ChIP-Seq analyses indicate that PGC7 and TET3 target genes are largely overlapped. However, a significant portion of PGC7 targets are not occupied by TET3. As PGC7 also interacts with TET2, it is likely that PGC7 adopts a similar mode to protect CpG methylation from TET2-dependent DNA demethylation in these loci. Moreover, PGC7 could not protect all the CpG methylation from TET enzyme-mediated global DNA methylation. In particular, DNA methylation in the repetitive sequence is only mildly reduced and a number of imprinting regions are not affected even lacking PGC7 in zygotes (28). Thus, other mechanisms may exist to protect the maternal pronucleus from TET enzyme-mediated global DNA demethylation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [45].
FUNDING
The University of Michigan Cancer Center and GI Peptide Research Center; the Era of Hope Scholar Award from the Department of Defense (to X.Y.); National Institute of Health [CA132755 and CA130899 to X.Y.]. Funding for open access charge: NIH [R01CA130899].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Qiang Chen and Yibin Chen for technical support, Drs Robert Chunhua Zhao and Alex Bortvin for the reagents. We thank Dr Chao Liu and Andrew Josiah Gorton for proofreading.
REFERENCES
- 1.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 3.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 5.Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Curr. Opin. Cell Biol. 2007;19:266–272. doi: 10.1016/j.ceb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi S, Hong K, Liu R, Inoue A, Shen L, Zhang K, Zhang Y. Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res. 2013;23:329–339. doi: 10.1038/cr.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu H, Zhang Y. Early embryos reprogram DNA methylation in two steps. Cell. Stem Cell. 2012;10:487–489. doi: 10.1016/j.stem.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senner CE, Krueger F, Oxley D, Andrews S, Hemberger M. DNA methylation profiles define stem cell identity and reveal a tight embryonic-extraembryonic lineage boundary. Stem Cells. 2012;30:2732–2745. doi: 10.1002/stem.1249. [DOI] [PubMed] [Google Scholar]
- 10.Seisenberger S, Peat JR, Hore TA, Santos F, Dean W, Reik W. Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philos. Trans. R Soc. Lond. B Biol. Sci. 2013;368:20110330. doi: 10.1098/rstb.2011.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 18.Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl Acad. Sci. USA. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura T, Liu YJ, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, Nakano T. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature. 2012;486:415–419. doi: 10.1038/nature11093. [DOI] [PubMed] [Google Scholar]
- 20.Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 21.Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C, Hu Y, Hu D, Zhao B, Cerovina T, et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell. 2012;151:1200–1213. doi: 10.1016/j.cell.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato M, Kimura T, Kurokawa K, Fujita Y, Abe K, Masuhara M, Yasunaga T, Ryo A, Yamamoto M, Nakano T. Identification of PGC7, a new gene expressed specifically in preimplantation embryos and germ cells. Mech. Dev. 2002;113:91–94. doi: 10.1016/s0925-4773(02)00002-3. [DOI] [PubMed] [Google Scholar]
- 23.Bowles J, Teasdale RP, James K, Koopman P. Dppa3 is a marker of pluripotency and has a human homologue that is expressed in germ cell tumours. Cytogenet. Genome Res. 2003;101:261–265. doi: 10.1159/000074346. [DOI] [PubMed] [Google Scholar]
- 24.Clark AT, Rodriguez RT, Bodnar MS, Abeyta MJ, Cedars MI, Turek PJ, Firpo MT, Reijo Pera RA. Human STELLAR, NANOG, and GDF3 genes are expressed in pluripotent cells and map to chromosome 12p13, a hotspot for teratocarcinoma. Stem Cells. 2004;22:169–179. doi: 10.1634/stemcells.22-2-169. [DOI] [PubMed] [Google Scholar]
- 25.Renfree MB, Papenfuss AT, Deakin JE, Lindsay J, Heider T, Belov K, Rens W, Waters PD, Pharo EA, Shaw G, et al. Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 2011;12:R81. doi: 10.1186/gb-2011-12-8-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Payer B, Saitou M, Barton SC, Thresher R, Dixon JP, Zahn D, Colledge WH, Carlton MB, Nakano T, Surani MA. Stella is a maternal effect gene required for normal early development in mice. Curr. Biol. 2003;13:2110–2117. doi: 10.1016/j.cub.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 27.Bortvin A, Goodheart M, Liao M, Page DC. Dppa3 / Pgc7 / stella is a maternal factor and is not required for germ cell specification in mice. BMC Dev. Biol. 2004;4:2. doi: 10.1186/1471-213X-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- 29.Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat. Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Z, Bian C, Zhou H, Huang S, Wang S, Liao L, Zhao RC. MicroRNA hsa-miR-138 inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells through adenovirus EID-1. Stem Cells Dev. 2011;20:259–267. doi: 10.1089/scd.2010.0072. [DOI] [PubMed] [Google Scholar]
- 32.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 33.Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 34.Takebayashi S, Tamura T, Matsuoka C, Okano M. Major and essential role for the DNA methylation mark in mouse embryogenesis and stable association of DNMT1 with newly replicated regions. Mol. Cell Biol. 2007;27:8243–8258. doi: 10.1128/MCB.00899-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 36.Dray E, Etchin J, Wiese C, Saro D, Williams GJ, Hammel M, Yu X, Galkin VE, Liu D, Tsai MS, et al. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat. Struct. Mol. Biol. 2010;17:1255–1259. doi: 10.1038/nsmb.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szwagierczak A, Brachmann A, Schmidt CS, Bultmann S, Leonhardt H, Spada F. Characterization of PvuRts1I endonuclease as a tool to investigate genomic 5-hydroxymethylcytosine. Nucleic Acids Res. 2011;39:5149–5156. doi: 10.1093/nar/gkr118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaffney EV. A cell line (HBL-100) established from human breast milk. Cell Tissue Res. 1982;227:563–568. doi: 10.1007/BF00204786. [DOI] [PubMed] [Google Scholar]
- 39.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borgel J, Guibert S, Li Y, Chiba H, Schubeler D, Sasaki H, Forne T, Weber M. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 2010;42:1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 42.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vella P, Scelfo A, Jammula S, Chiacchiera F, Williams K, Cuomo A, Roberto A, Christensen J, Bonaldi T, Helin K, et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol. Cell. 2013;49:645–656. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 44.Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kubicek S, O'Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.