Abstract
The anti-silencing function protein 1 (Asf1) is a chaperone that forms a complex with histones H3 and H4 facilitating dimer deposition and removal from chromatin. Most eukaryotes possess two different Asf1 chaperones but their specific functions are still unknown. Trypanosomes, a group of early-diverged eukaryotes, also have two, but more divergent Asf1 paralogs than Asf1 of higher eukaryotes. To unravel possible different functions, we characterized the two Asf1 proteins in Trypanosoma brucei. Asf1A is mainly localized in the cytosol but translocates to the nucleus in S phase. In contrast, Asf1B is predominantly localized in the nucleus, as described for other organisms. Cytosolic Asf1 knockdown results in accumulation of cells in early S phase of the cell cycle, whereas nuclear Asf1 knockdown arrests cells in S/G2 phase. Overexpression of cytosolic Asf1 increases the levels of histone H3 and H4 acetylation. In contrast to cytosolic Asf1, overexpression of nuclear Asf1 causes less pronounced growth defects in parasites exposed to genotoxic agents, prompting a function in chromatin remodeling in response to DNA damage. Only the cytosolic Asf1 interacts with recombinant H3/H4 dimers in vitro. These findings denote the early appearance in evolution of distinguishable functions for the two Asf1 chaperons in trypanosomes.
INTRODUCTION
Anti-silencing function 1 (Asf1) is a chaperone specific for histone H3 and H4 heterodimers. This protein was identified by its capacity to induce expression of telomeric silenced genes in yeast (1,2). More recently, Asf1 was found to participate in the deposition and removal of histone H3 and H4 during several biological processes such as DNA replication, transcription and DNA damage repair (3). Asf1 also plays a role in cell cycle regulation through the interaction with the checkpoint kinase Rad53 in yeast (4,5). Furthermore, Asf1 is the substrate of Tousled-like kinases during cell cycle progression and on DNA damage response (6). In addition, Asf1 is required together with SWI/SNF proteins for the activation of promoters of DNA damage stress responses during transcription (7). When associated with Rtt106 histone acetyltransferase, Asf1 also participates in the control of histone gene transcription during S phase (8,9).
Asf1 proteins display a highly conserved N-terminal region that interacts with histone H3 and H4 (10–12) and with other histone processing molecules, such as the chromatin assembly factor I (13,14) and HIRA chaperone (15). These interactions are required for nucleosome assembly and disassembly (16,17). In contrast, the C-terminal domain of Asf1 is more variable. It is not required for chaperone function but it is associated with regulatory roles of the protein (18). For example, it is necessary for interaction with the chromatin fiber (19), transcriptional silencing and it mediates binding of histones to Rad53 (20). Heterogeneity of the C-terminal tails most likely accounts for different functions of Asf1 proteins in eukaryotes (21,22).
In some species there are two isoforms of Asf1, called Asf1A and Asf1B, mainly distinguishable by differences in the C-terminus (6). The reasons why there are two independent isoforms are not well understood. Asf1B facilitates histone deposition during replication, but does not participate in replication-independent nucleosome deposition, which is mediated by Asf1A and HIRA (23). Interestingly, although both Asf1 proteins are localized in the nucleus, only Asf1B was proposed to be involved in the transport of the H3/H4 heterodimers from the cytosol to the nucleus (24).
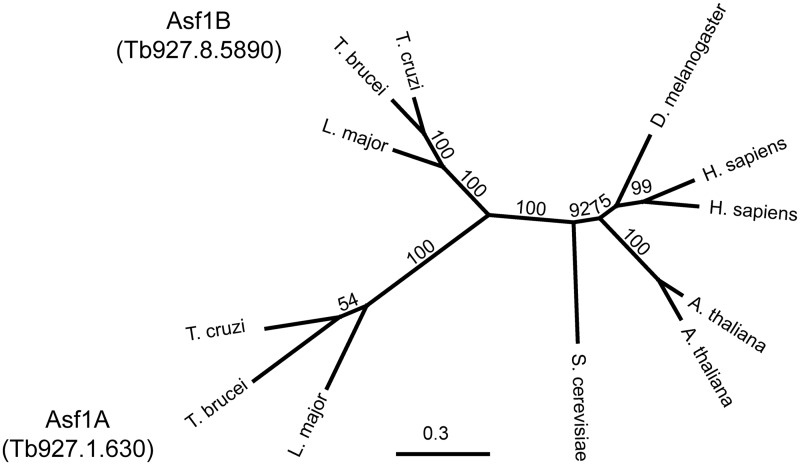
In trypanosomes, a group of protozoan parasites, two Asf1 isoforms (Asf1A and Asf1B) are involved in cell cycle progression and were described as substrates for a Tousled-like kinase (25). Asf1A has been implicated in the control of the exclusive transcription of a single variant surface glycoprotein (VSG) gene out of several hundreds by affecting the chromatin structure in the telomeric expression sites in Trypanosoma brucei. (26). When we compared the two Asf1 isoforms in trypanosomes, we found different C-terminal sequences (Supplementary Figure S1). A phylogenetic tree generated with the protein alignments clearly indicates that Asf1 chaperones in trypanosomes are more divergent than in other organisms (Figure 1). As trypanosomes are early divergent eukaryotes and display unique features regarding the control of gene expression (27,28), DNA replication (29,30) and repair (31,32), we decided to further investigate the functions of these proteins in T. brucei. We confirmed that depletion of the proteins arrest the cells in S phase, although at different moments. More important, we observed strikingly different cellular localizations and functions for these two forms of Asf1.
Figure 1.
Phylogenetic analysis reveals that Asf1 paralogs in trypanosomatids are more divergent compared with those in other eukaryote species. The tree was constructed based on the genetic distance model representing the consensus of 100 bootstrap iterations of the alignment generated by neighbor joining, using the Geneious software and the sequences shown in Supplementary Figure S1. The numbers indicate the consensus support in percentage, and the line indicates the distance scale.
MATERIALS AND METHODS
Parasites
Procyclic form (PCF) of T. brucei strains Lister427 and 29–13 (33) was cultivated in SDM-79 medium (34) in the presence of 10% fetal bovine serum (FBS) at 28°C. hygromycin B (50 µg/ml) and geneticin (15 µg/ml) were added to strain 29–13 cultures. Bloodstream trypomastigotes (BSF) strain 427 was maintained in HMI-9 in the presence of 10% FBS at 37°C (35). Transfections were performed using 5 × 106 PCF resuspended in 200 µl of Zimmerman's Post Fusion Medium (ZPFM) buffer (36) with 5 µg of plasmid DNA in 0.2-mm cuvettes and program U33 of Amaxa Nucleofactor (Lonza). Transfectants were selected and cloned in the same medium containing phleomycin (5 µg/ml) or blasticidin (2.5 µg/ml) according to the used plasmid. For RNA interference (RNAi) induction, 1 µg/ml tetracycline (Tet) was added every day to the culture. To analyze the effect of genotoxic agents, 1 × 106 cells/ml were induced (or not) with Tet and treated either with 0.001% methyl methanesulfonate (MMS, Sigma-Aldrich) or subjected to γ-irradiation (40 Gy). Cell numbers were determined in Neubauer counting chambers in three independent experiments, and statistical analysis was performed with Prisma software. The presented data are results of experiments performed with single clones and correspond to independent experiments performed separately. Nevertheless, similar results were obtained with more than one clone. Non-clonal populations were used in the case of Myc-tags at the N-terminus.
DNA cloning and plasmid constructions
Gene fragments were amplified by polymerase chain reaction (PCR) with DNA extracted from PCF 427 as in (37). The PCR fragments were purified by agarose gel electrophoresis, cloned in pGEM®-T Easy Vector (Promega) and the inserts confirmed by restriction analysis and DNA sequencing before transfer to the final vector. For cloning into pET14b vector (Novagen), Asf1A gene (TriTryp database IDs Tb927.1.630) was amplified using Asf1AFowNde (5′-CATATGAGCATACAACCAATT) and Asf1ARevBamHI (5′-GGATCCTCATCTGGGTTCAAGTGC) primers. Asf1B gene (Tb927.8.5890, respectively) was amplified using Asf1BEcoRIfow (5′-GAATTCACCACAGCCGGTCAG) and the Asf1BNotRev (5′- GCGGCCGCTTAACGGTGGTGC-TTTTCTTTC) primers and inserted in pET28a (Novagen). pZJM-Asf1A fragment was amplified using primers Asf1AHindIIIFow (5′-AAGCTTATGAGCATACAACCAATTG) and Asf1AXhoRev (5′- CTCGAGTCTGGGTTCAAGTGCTTC) and digested with HindIII and XhoI. pZJM-Asf1B fragment was amplified using primers Asf1BHindFow (5′ AAGCTTATGACCACAGCCGGTCAG) and Asf1ASalIFow (5′-GTCGACATGACCACAGCCGGTC) and digested with HindIII and SalI. For cloning into pRP6Myc and pNAT6Myc (38); inserts were generated with Asf1AAvrIIFow (5′-CCTAGGAGCATACAACCAATTGTAC) and Asf1ARevBamHI primers, and Asf1BAvrIIFow (5′-CCTAGGACCACAGCCGGTCAGTC) and Asf1BBglIIRev (5′-AGATCTTTAACGGTGGTGCTTTTC) and digested with AvrII and BamHI (Asf1A) or AvrII and BglII (Asf1B). pNATAsf1A (Asf1B-12)-Myc was prepared using HindIII and XbaI digested PCR products amplified with Asf1AHindFow and Asf1AXbaIRev (5′ TCTAGATCTGGGTTCAAGTGCTTC) or Asf1BXbaRev (5′-TCTAGAACGG-TGGTGCTTTTCTTTC). To generate Asf1 proteins fused to the N-terminus of maltose binding protein (MBP), Asf1A and Asf1B coding sequences were amplified by PCR from genomic DNA using the following primers: Asf1A_for (5′-CGGGATCCATGAGCATACAACCAATTGTACAAC); Asf1A_rev (5′-ACGCGTCGACTCATCTGGGT TCAAGTGCTTC); Asf1B_for (5′-ATGACCACAGCCGGTCAGTC); Asf1B_rev (5′-CCCCAAGCTTTTAACGGTGGTGCTTTTCTTTC), respectively. PCR products were cloned into the expression vector pMAL-c2X (New England BioLabs) digested with BamHI/SalI (for Asf1A) or XmnI/HindIII (for Asf1B).
Antibodies
Antibodies specific for Asf1A and Asf1B were prepared by immunization of rabbits and mice using recombinant proteins obtained in Escherichia coli BL21 DE3 strain after induction with 0.5 mM isopropyl β-D-1-thiogalactopyranoside for 14 h at 28°C. Bacterial pellets were resuspended in Bug Buster Protein Extraction Reagent (Novagen), and soluble proteins were purified using a Ni-Sepharose High Performance column (GE Healthcare Life Sciences) equilibrated in 20 mM Tris–HCl (pH 7.5), 500 mM NaCl and 40 mM imidazole. After extensive washes (500 column volumes), the proteins were eluted in the same buffer with 500 mM imidazole. Affinity-purified antibodies to Asf1A were obtained by incubation of sera with recombinant Asf1A coupled to CNBr-activated Sepharose (GE Healthcare Life Sciences). Specific antibodies to Asf1A were eluted from the column with 0.1 M triethylamine (pH 11.5), and the eluted fractions were neutralized with 1 M Tris–HCl (pH 7.4) and kept at 4°C in the presence of 2 mg/ml BSA. Anti-Trypanosoma cruzi β-tubulin antibodies were prepared as described previously (39). Anti-histone H4 was obtained from Abcam and anti-T. cruzi histone H4 acetylated at lysine 4 was prepared as described previously (40). Anti-histone H3 was obtained from Abcam and anti-acetylated histone H3 from Upstate Biotechnology. The monoclonal anti-Myc 9E10 (41) was used as ascitic fluid.
SDS–PAGE, western blot, immunofluorescence and pulldown assays
Protein samples were separated by 12.5% SDS–PAGE and transferred using the semidry apparatus (Bio-Rad) to nitrocellulose membranes for 20 min at 20 V. Membranes were then treated for 2 h with phosphate-buffered saline (PBS) with 5% non-fat dry milk and 0.1% Tween 20, and incubated in the same buffer with primary antibodies for 1 h. After three washes in PBS with 0.1% Tween 20, bound antibodies were detected with anti-IgG-peroxidase conjugates (Life Technologies) and chemiluminescent peroxidase substrate (Millipore).
For immunofluorescence analysis, exponentially growing cell culture samples were mixed with the same volume of 4% paraformaldehyde in PBS and incubated for 20 min. Cells were washed twice with PBS, resuspended to 1 × 107 cells/ml and added onto glass slides. After 5 min, attached cells were treated with 0.1% Triton X100 in PBS for 5 min. After three washes in PBS, the slides were incubated with 2% FBS in PBS for 1 h, incubated with primary antibodies diluted in blocking solution, washed again in PBS and stained with anti-IgG coupled to Alexa 488 or to Alexa 594 (Life Technologies), diluted in the blocking solution containing 10 µg/ml of 4′,6-diamidino-2-phenylindole (DAPI) and mounted in ProLong Gold anti-fading reagent (Life Technologies). Serial images (0.2 µm) were collected using a 100× objective 1.40 numerical aperture using the Cell^M software (Olympus Europe) in a motorized Olympus IBX61 microscope. Images were processed by blind deconvolution using Autoquant X 2.2 (Media Cybernetics).
For in vitro pulldown assays, Asf1 paralogs in pMAL-c2X were transformed into E. coli Rosetta Blue (Novagen) and cells were grown in selective media (Luria–Bertani medium supplemented with 100 µg/ml ampicillin and 34 µg/ml chloramphenicol) to an OD600 of 0.5–0.6 before induction of protein expression with 1 mM isopropyl β-D-1-thiogalactopyranoside for 1 h at 37°C. Cells were resuspended in lysis buffer (25 mM HEPES, pH 7.5, 150 mM KCl, 10% glycerol, 12.5 mM MgCl2, 1 mM DTT and 1 mM PMSF) and lysed using a Bioruptor (Diagenode). Pulldown assays were performed as described (11) using whole bacteria extracts. Recombinant T. brucei histones H3, H4, H3V and H4V (TriTryp database IDs Tb927.1.2510, Tb927.5.4170, Tb927.10.15350, Tb927.2.2670, respectively) were expressed in E. coli BL21 (manuscript in preparation), purified from inclusion bodies and refolded as described (42). MBP fusion proteins were detected by western blots analysis using a monoclonal rat MBP antibody (gift from E. Kremmer). Histones H3 and H4 (and their variants) were detected using a polyclonal rabbit antibody (43) or a polyclonal peptide antibody (44), respectively. Secondary antibodies [IRDye 800CW goat anti-rabbit and IRDye 680LT goat anti-rat (LI-COR Biosciences)] were detected with an Odyssey infrared imaging system (LI-COR). For pulldown assays with trypanosome whole cell extracts, 2 × 107 PCF were harvested (1300g, 10 min, 4°C) and washed in cold PBS. The cell pellet was resuspended in lysis buffer [25 mM HEPES, pH 7.5, 150 mM KCl, 10% Glycerol, 12.5 mM MgCl2, 1 mM DTT, 1 mM PMSF and Protease inhibitor cocktail (Roche)] and lysed by three freeze and thaw cycles using liquid nitrogen, followed by 10 sonication rounds (30 s on and 30 s off) using a Bioruptor (Diagenode). Cell extracts were cleared by centrifugation (10.000 g, 10 min 4°C) and treated with DNaseI (Thermo Scientific) for 10 min at 37°C. MBP-tagged Asf1c or Asf1n were incubated with the trypanosome cell extract and pulldown assay was performed and monitored as described earlier in the text.
Cell cycle analysis
For cell cycle analysis, parasites were centrifuged for 10 min at 2000 g. The cells were resuspended and washed in PBS and finally resuspended in cold ethanol:PBS (1:1). After 1 h incubation on ice the cells were centrifuged, washed in PBS twice and incubated 30 min with 10 µg/ml RNAse and 1 µg/ml propidium iodide (PI) diluted in PBS. The cells were washed once in PBS and analyzed by flow cytometry using a FACSCalibur (BD Biosciences).
RESULTS
The T. brucei Asf1 have different cellular localizations
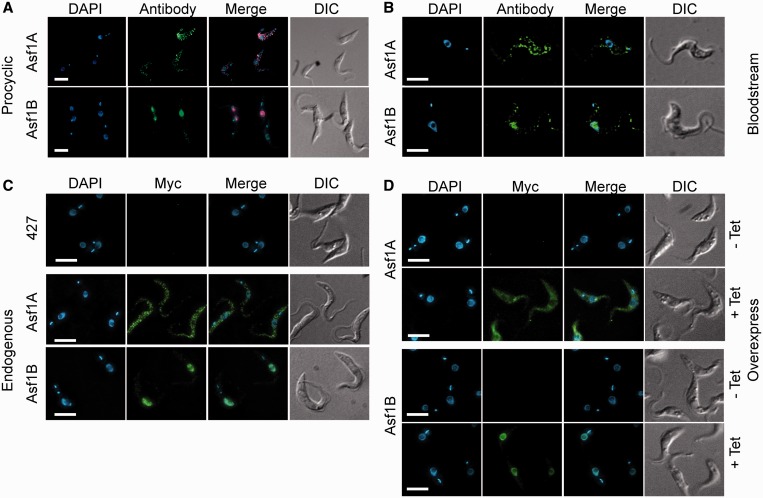
To start the characterization of the different histone chaperones, we analyzed their cellular distribution by indirect immunofluorescence analysis (IFA). Surprisingly, IFA showed that the protein previously called Asf1A in T. brucei (25,26) is mainly localized in the cytosol, whereas Asf1B is mostly found in the nucleus of procyclic and bloodstream forms (Figure 2A and B, respectively). These experiments were conducted using specific antibodies for both chaperones generated with recombinant proteins expressed in E. coli. Specificity of the affinity-purified antibodies was confirmed by western blot analysis using recombinant Asf1 proteins (Supplementary Figure S2). Recombinant Asf1A and Asf1B proteins migrated in SDS–PAGE corresponding to 25 and 40 kDa, respectively, rather than the predicted 19 for Asf1A and 21 kDa for Asf1B. However, accurate expression and purification of both proteins could be confirmed by mass spectrometry analysis (data not shown), indicating that this abnormal migration pattern must be due to SDS-gel artifacts.
Figure 2.
T. brucei Asf1A is nuclear, whereas Asf1B is predominantly cytosolic. Panels (A) and (B) show IFA of procyclic and bloodstream T. brucei, respectively, using specific anti-Asf1A and anti-Asf1B antibodies (antibody), DAPI to stain DNA, merged and DIC images. Panel (C) shows IFA using anti-Myc monoclonal antibody, DAPI staining, merged images and DIC of wild-type procyclics (427) or procyclics expressing the endogenously 12×Myc-tagged versions of Asf1 or Asf1B. Panel (D) shows representative images of procyclics containing the plasmids pRP-Asf1A and pRP-Asf1B, which enable induced overexpression of proteins tagged with a 6×Myc peptide at the N-terminus. The analysis was performed in cells after 48 h without (−Tet) or with tetracycline-induced (+Tet) protein overexpression. Bars = 5 μm.
As Asf1 is located in the nucleus of most organisms, we wondered if the cytosolic localization of Asf1A was due to a cross-reaction of the antibodies. Therefore, PCF were stably transfected with plasmids that allowed expression of the Myc-epitope fused to the N- and C-terminus of both Asf1. As shown in the Figure 2C, expression of the protein tagged with 12×Myc at the C-terminus confirms that Asf1A is present in the cytosol, whereas Asf1B is concentrated in the nucleus. Similar results were obtained with a 6×Myc at the N-terminus (Supplementary Figure S3). Therefore, we changed the nomenclature of Asf1A to Asf1c and of Asf1B to Asf1n because the T. brucei Asf1B is more similar to Asf1A in other organisms.
Asf1c overexpression causes changes in histone H3 and H4 acetylation levels
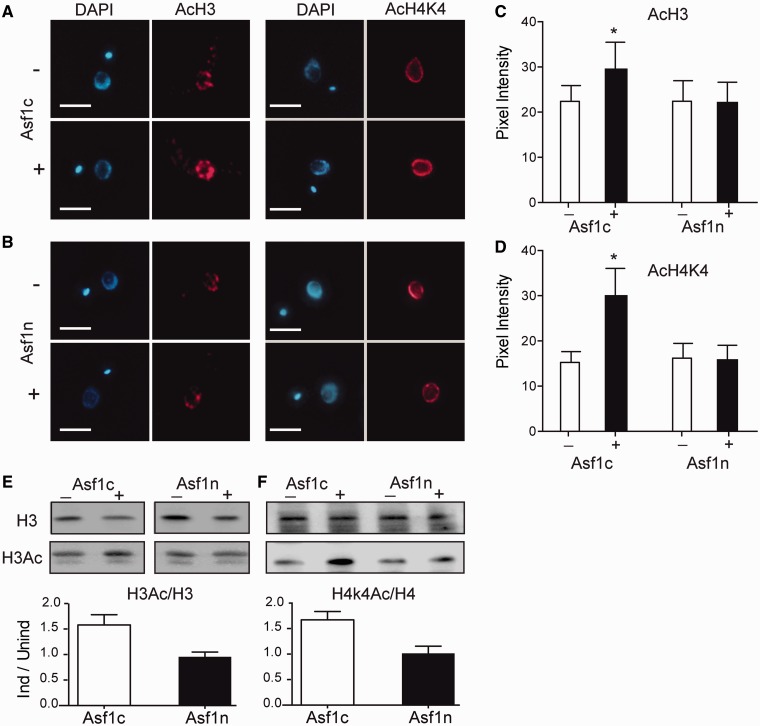
As it has been previously shown that Asf1 controls the acetylation of histone H3 in yeast (45), we examined whether overexpression of Asf1 proteins would also affect histone acetylation levels in T. brucei. Cells overexpressing the cytosolic Asf1c, but not Asf1n, showed an increased labeling for acetylated histone H3 (Figure 3A) and acetylated lysine K4 of histone H4 (H4K4) (Figure 3B) by IFA. A quantitative analysis of the immunofluorescence showed a slight but significant increase in H3 acetylation labeling and a 2.5-fold increase of H4K4 labeling after overexpression of Asf1c (Figure 3C and D). As immunofluorescence labeling depends on antigen accessibility, we performed western blot analyses and confirmed that the acetylated histone levels increased and that this is not a result of increased total amounts of histone H3 or H4 (Figure 3E and F). In summary, these data confirm that only overexpression of cytosolic Asf1 increases the levels of histone H3 and H4K4 acetylation.
Figure 3.
Cytosolic but not nuclear Asf1 overexpressors show increased levels of acetylated forms of histone H3 and H4. Control (–) or parasites overexpressing (+) Asf1c (panel A) or Asf1n (Panel B) were stained with specific antibodies against acetylated histone H3 (right) or acetylated histone H4 (left), both in red. DNA is stained with DAPI. Bars = 3 µm. Panels (C) and (D) show the quantification of the fluorescence intensity (means ± standard deviations) for AcH3 and AcH4K4, respectively, in Asf1-overexpressing cells (n = 35 each). The asterisk indicates significant differences as determined by Student’s t-test (P < 0.05). (C) The same parasites were used for western blot analysis using antibodies specific for histone H3 and H3Ac (E) or histone H4 and H4K4Ac (F). The ratio between acetylated and total histones is shown below the panels. Similar amounts of protein were separated in the gel as assessed by Ponceau S labeling. Similar ratios were obtained in two additional experiments, for each histone.
The two Asf1 act in different phases of the cell cycle
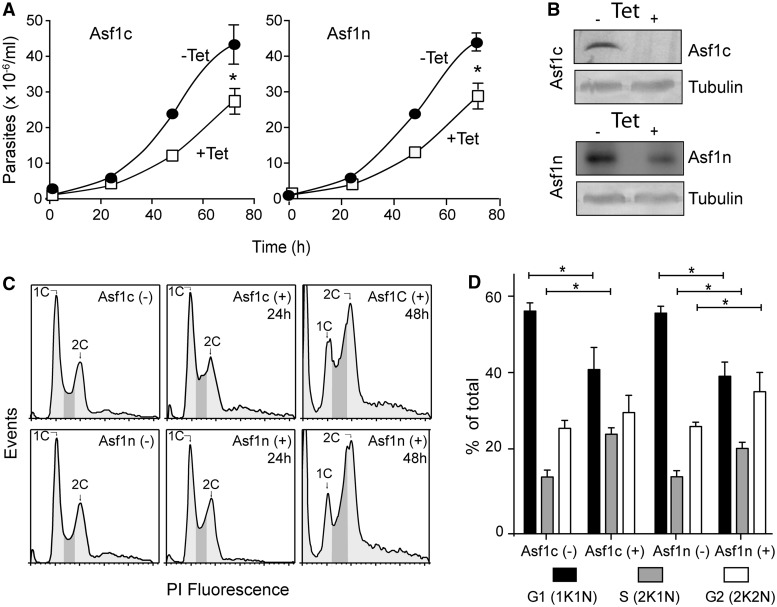
Both T. brucei Asf1 proteins are required for accurate cell cycle progression because knockdown of each one caused cell cycle arrest in S phase (25). When we reexamined the knockdown phenotypes of Asf1 proteins, we observed a decrease in cell growth in both cases (Figure 4A). The levels of both proteins were barely affected 24 h after RNAi induction (data not shown) but decreased substantially after 48 h (Figure 4B). Nevertheless, defects in cell cycle progression observed by flow cytometry analysis could be detected already 24 h after RNAi induction and were more evident after 48 h (Figure 4C). Trypanosomes with depleted Asf1c showed relatively more cells with DNA content compatible with cells in S phase, whereas Asf1n RNAi causes an accumulation of cells with DNA content compatible with cells moving to the G2 phase, as estimated by flow cytometry. As the kinetoplast (K), which consists of thousands of mini-circle DNA that form the mitochondrial genome, divides ahead of the nucleus, it could be used to confirm these findings by microscopic analysis. The results shown in Figure 4D indicate that more cells with 2 kinetoplasts and 1 nucleus (2K1N) are found after Asf1c knockdown compared with Asf1n-depleted cells. Consistently, 2K2N cells are more frequent in Asf1n knockdowns. These observations might suggest that the cytosolic Asf1 is important in earlier stages of the cell cycle compared with the nuclear Asf1.
Figure 4.
Knockdown of each Asf1 arrests the cells at different positions in S phase. (A) Growth curve of parasites after induction (+Tet, open squares) or no induction (−Tet, closed circles) of RNAi targeting Asf1c (left panel) or Asf1n (right panel). The values are means ± standard deviations (n = 4). Asterisks indicate statistically significant differences (P < 0.05). (B) Western blot analysis of whole parasite cell extracts obtained from non-induced (−) or Tet-induced for 48 h (+) incubated with anti-Asf1c and anti-Asf1n antibodies. Tubulin served as a loading control. (C) Representative flow cytometer histograms of 10 000 events of both knockdown cell lines incubated without (−) or with Tet for 24 and 48 h (+). The peaks correspond to single and double genomic DNA content (C1 and C2, respectively), and the dark gray area corresponds to cells in S phase (D) Graphic showing the percentage of cells in different stages of the cell cycle after 48 h of treatment with (+) or without (−) Tet for induction of RNAi (G1 populations with 1 kinetoplast and 1 nucleus (1K1N) are shown in black, S (2K1N) in gray, and G2 (2K2N) in white bars) scored by microscopic examination of cells stained with DAPI. Bars with asterisks indicate statistically significant differences using Student’s t-test P < 0.055 after counting at least 40 cells in each situation.
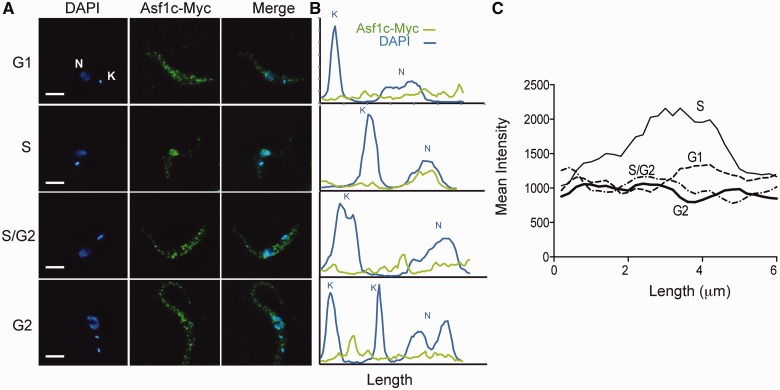
Next, we examined the distribution of the cytosolic Asf1 during cell cycle progression. By using the cell line expressing an endogenous 12×Myc-tagged version of Asf1c, we could detect it clearly in the cytosol of the parasite during most phases of the cell cycle (Figure 5A). However, we noticed that the majority of Asf1c migrates into the nucleus in S phase. This could be better visualized by measuring the pixel intensity along a line from the kinetoplast (K) to the nucleus (N). Examples are shown in Figure 5B. An overlay of the mean fluorescence intensity of several curves across the nucleus and kinetoplast was plotted against the line length for each cell cycle phase (Figure 5C). This plot clearly indicates that cells in S phase have larger amounts of Asf1c near the nucleus compared with nuclei in other cell cycle phases. A similar accumulation of Asf1c in toward the nucleus was observed in non-transfected cells using the anti-Asf1c antiserum (Supplementary Figure S4A), indicating that this effect is not due to the tag incorporation or overexpression of the protein.
Figure 5.
Asf1c accumulates in the nucleus during S phase. Localization of endogenously 12×Myc-tagged Asf1c was analyzed by immunofluorescence with anti-Myc antibodies. Cells in different cell cycle phases were scored by DAPI staining. Panel A shows representative images of G1, S, S/G2 and G2 phase cells and panel B the quantitative analysis of the DAPI and Alexa 488 fluorescence intensity on a line passing through the kinetoplast (K) and nucleus (N). Data analysis was performed on the entire Z-stacking. Panel C shows the mean fluorescent intensity (n = 30) in the nucleus length for cells in G1, S, S/G2 and G2 phase.
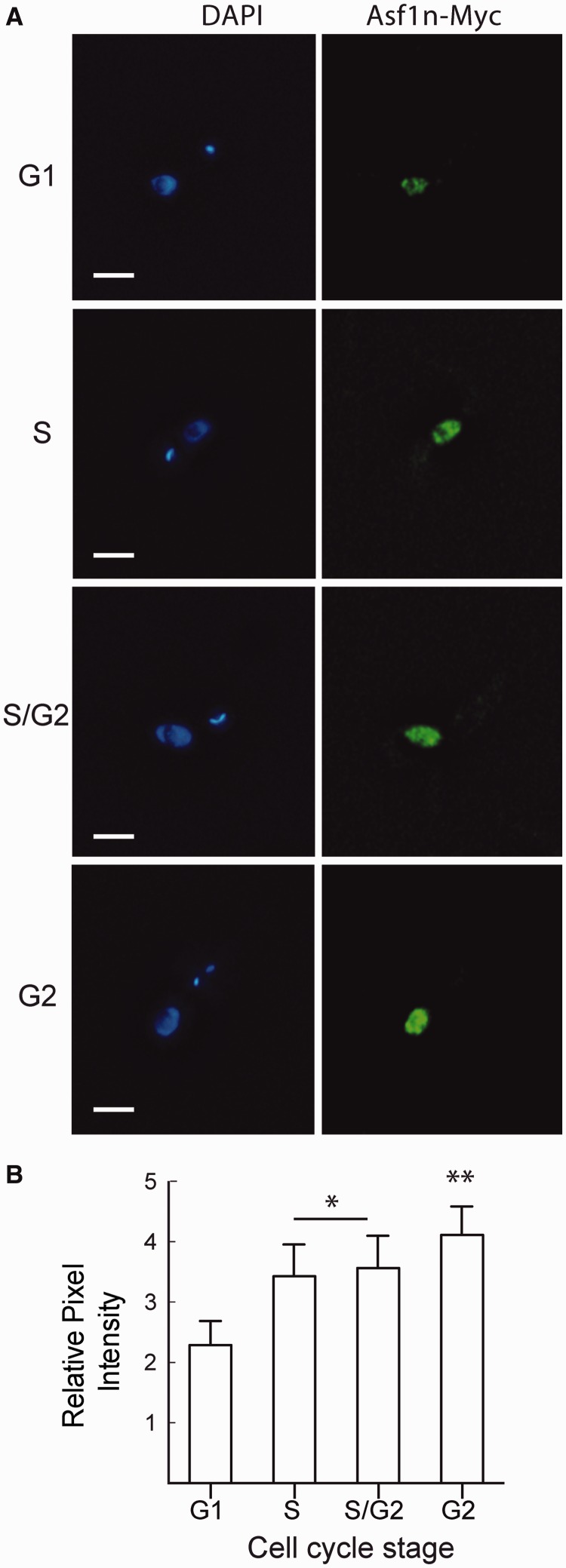
In contrast, Asf1n remains inside the nucleus throughout the cell cycle. Remarkably, we observed an increase in the labeling intensity during cell cycle progression, with higher fluorescence intensity appearing in G2 cells. The intensity of Asf1 signals relative to DAPI staining doubles from G1 to G2 phase and was observed in tagged (Figure 6) and untagged parasites (Supplementary Figure S4B). The fact that Asf1n progressively increases until G2 phase of the cell cycle, whereas Asf1c accumulates in the nucleus in S phase, supports the hypothesis that the cytosolic Asf1 acts in early and the nuclear Asf1 in late S phase.
Figure 6.
Asf1n levels increase during the S phase of the cell cycle. Parasites expressing endogenously 12×Myc-tagged Asf1n were analyzed by immunofluorescence with anti-Myc antibodies throughout the cell cycle. Panel A shows representative images of G1, S, S/G2 and G2 phase cells and Panel B the quantitative analysis of the fluorescence intensity in the nucleus of the entire Z-stacking using ImageJ software. The values are mean ± standard deviation (n = 50). Asterisks indicate significant differences as determined by Kruskall–Wallis test (P < 0.05).
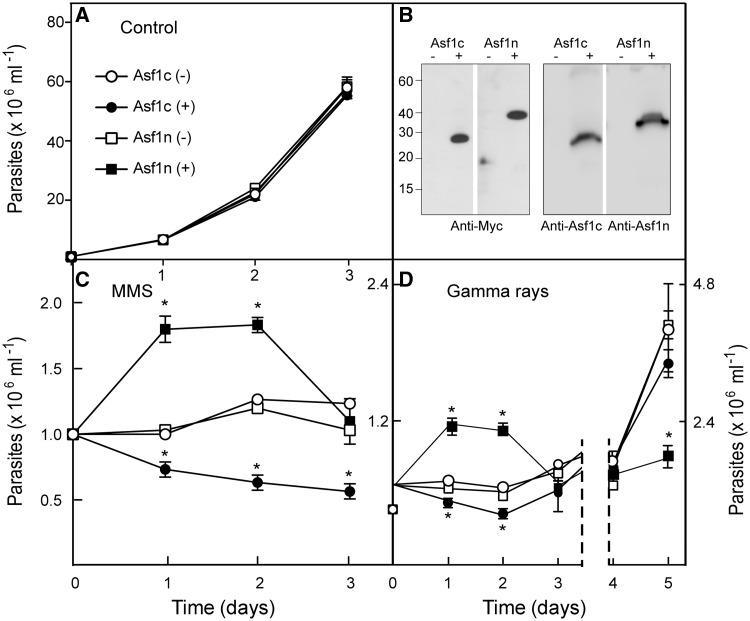
The two Asf1 overexpression have different effects on DNA damage sensitivity
Several studies have demonstrated that Asf1 chaperones play an important role in chromatin remodeling during DNA damage repair (46). To address how T. brucei Asf1 homologs affect DNA repair, we examined the growth of parasites overexpressing Asf1c and Asf1n after γ-irradiation or MMS treatment. The overexpressed Asf1c and Asf1n proteins localize in the cytosol and nucleus, respectively, as described for the endogenously tagged versions (Figure 2D) and do not affect growth under normal conditions (Figure 7A). These proteins are significantly overexpressed. Endogenous protein levels were low and were not detectable in comparable experiments (Figure 7B). Cells with normal Asf1 levels (non-induced) stop growing on treatment with 0.001% MMS (Figure 7C). In contrast, Asf1n overexpressing parasites continue to grow for 24 h before they start to die two days after treatment, as judged by lost of motility and refringence. Overexpression of the cytosolic Asf1 causes initially more cell death. Similarly, the Asf1n overexpressor submitted to γ-irradiation showed a longer initial growth period with a poor recovery after three days of treatment (Figure 7C), suggesting that cells accumulate unrepaired DNA damage. Asf1c overexpressors are initially more sensitive to γ-irradiation, but the cells recover faster. Importantly, overexpressed Asf1c does not accumulate inside the nucleus after γ-irradiation or MMS treatment (data not shown), indicating that its effect occurs in the cytosol. These results suggest that the two isoforms have different functions after DNA damage.
Figure 7.
Overexpression of nuclear Asf1, but not cytosolic Asf1, promotes growth but impairs recovery after DNA damage. Procyclics transfected with plasmids coding for an inducible system to overexpress Asf1c (circles) or Asf1n (squares) were incubated with (dark symbols) or without Tet (empty symbols) for 24 h. (A) The cells were grown without DNA damaging (control). (B) Overexpression of induced proteins were monitored by western blot with anti-Myc (left panels), anti-Asf1c or anti-Asf1n antibodies (right panels) in non-induced (−) or induced (+) cells. The cells were incubated with 0.001% MMS (C) or subjected to γ-irradiation (40 Gy) (D) maintained in the presence or absence of Tet and counted daily. The numbers are mean cell concentrations ± standard deviation of three independent experiments. Asterisk indicates significant differences between induced and non-induced parasites, calculated by Student’s t-test (P < 0.05). We could not detect the 6×Myc-tagged at the endogenous locus by western blot, although it was clearly observed by immunofluorescence analysis. Nevertheless, both Asf1 proteins endogenously tagged with 12×Myc were detected by western blot.
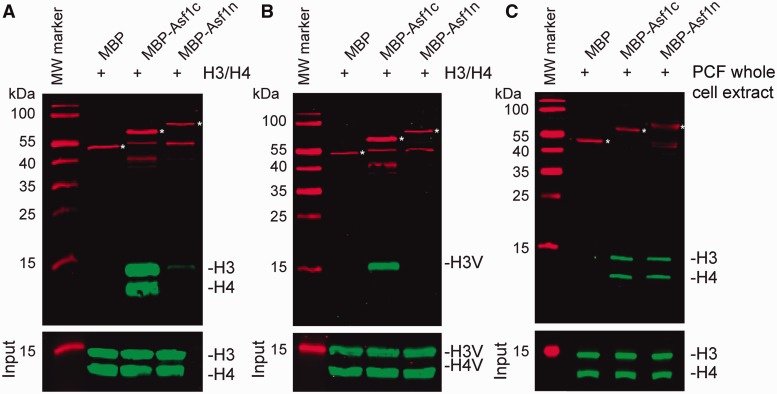
Cytosolic Asf1 binds to H3/H4 dimers in vitro
To gain more insights into the different functions of Asf1 proteins in trypanosomes, we decided to analyze the binding capacity of the chaperones to histone H3/H4 dimers. To elucidate direct interactions, we chose a cell-free system based on recombinant proteins. Asf1c- or Asf1n-MBP fusion proteins were expressed in E. coli, and cell extracts were incubated with recombinant H3/H4 complexes. Interactions of Asf1 proteins were analyzed by western blot after isolating the complexes with amylose-coupled beads. Interestingly, only the cytosolic Asf1 but not the nuclear Asf1 could bind H3/H4 complexes in vitro (Figure 8A). To test for potential specificity of the different Asf1 proteins for histone variants, we repeated this experiment with complexes made with histone H3 and H4 variants (H3V/H4V). Again only Asf1c could bind to recombinant variant histones (Figure 8B). Surprisingly, it was not possible to pulldown histone H4V in this assay. This suggests a direct interaction of the cytosolic Asf1 with histone H3 or H3V. Histone H4V is probably lost in this assay because the variant dimer is less stable than the canonical H3/H4 complex as shown before for the H2Bv/H2AZ variant nucleosome complex (47). The fact that Asf1n does not bind to recombinant histones (PTM) at all might be due to the lack of post-translational protein modifications on histones expressed in E. coli. To test the hypothesis that histone PTMs are a prerequisite for interaction with Asf1n, we repeated the pulldown assay using whole trypanosome cell extracts instead of recombinant proteins (Figure 8C). Interestingly, both chaperones could bind to histones from whole cell extracts with the same efficiency supporting the idea that Asf1n binds specifically modified histone complexes in the nucleus.
Figure 8.
Cytosolic but not the nuclear Asf1 binds to histones in vitro. (A) Bacterial extracts containing MBP-tagged Asf1c or Asf1n were incubated with purified histone H3/H4 complexes. Protein interactions were tested by pulldown assays and monitored by western blot analysis. H3/H4 input samples are shown in the lower panel. MBP was used as control. Full-length fusion proteins are marked with white asterisks (B) Interaction of MBP-tagged Asf1n and Asf1c with histone variant complexes (H3V/H4V) were analyzed as described previously. (C) Purified MBP-tagged Asf1c or Asf1n were incubated with PCF trypanosome whole cell extracts from 3 × 107 cells for each pulldown. Protein interactions were analyzed as described previously.
DISCUSSION
Here we showed different subcellular localization and binding activity, which might be associated with distinct functions for T. brucei Asf1c (Asf1A) and Asf1n (Asf1B). Initially, we demonstrated that the two Asf1 of T. brucei are mainly found in distinct cellular localizations in the two major developmental forms of the parasite. These differences were observed by using specific antibodies and tagged proteins, showing that Asf1A is distributed throughout the cytosol, but accumulates in the nucleus in S phase, whereas Asf1B is found mostly inside the nucleus.
The mainly cytosolic localization of Asf1A is in contrast to the localization of Asf1 in several other eukaryotes (48,49), although small cytosolic fractions of Asf1 were detected in complexes of freshly synthesized histone H3 and H4 (24). Asf1 is also found in the cytosol of rapidly dividing Drosophila embryos, but accumulates in the nucleus and associates with chromatin during each S phase (50). The nuclear localization of Asf1 is assumed to be a consequence of the import of the H3/H4-Asf1 complex carried out via histone recognition (51), which makes the mainly cytosolic localization of Asf1c in T. brucei intriguing. Perhaps in trypanosomes, Asf1c is found free of histone H3/H4 dimers, and it is imported to the nucleus when the histones are synthesized in S phase. The import of Asf1c into the nucleus might occur through binding to H3/H4 dimer and mediated by other proteins as shown in other organisms (52), whereas a nuclear localization signal is present in Asf1n (VSPTKKKARCE and DVKEKDHNGHAAYDDEAKVPEGVSPTKKKAR) predicted by cNLSMapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi). Therefore, we cannot exclude the possibility that T. brucei Asf1c plays other roles in addition to histone binding in the cytosol, at least when cells are not in S phase. Nevertheless, our data support the notion that the cytosolic Asf1 possesses histone chaperone functions. First, it accumulates in the nucleus in S phase. Second, when depleted it arrests the cells in early S phase. Third, when overexpressed, it increases the level of H3/H4 acetylation, known to be dependent of Asf1 and nuclear transport (51). And finally, Asf1c but not Asf1n is able to bind unmodified H3/H4 tetramers. Therefore, it is likely that Asf1c in T. brucei mediates the delivery of newly synthesized H3/H4 complexes to a nuclear envelope transporter. In contrast, Asf1n is able to bind histone if they are modified, as suggested by the immunoprecipitation experiments using whole cell lysates, and, therefore, it would act inside the nucleus. We cannot exclude, however, the possibility that Asf1n itself has to be modified to become active, or additional components might be necessary to form a chaperone/histone complex.
Although Asf1 proteins are highly conserved, there are species differences regarding the acetylation requirements for nuclear import of histones. In yeast, Asf1 stimulates the acetylation of histone H3 by Rtt109 targeting the lysines 9, 23, 56 and 27, the latter dependent on the presence of Vps75, another chaperone, most likely in the cytosol (53–56). In contrast, human Asf1 promotes the cytosolic acetylation of lysines 5 and 12 of H4 by Hat1 (24). In this case, following translation, histone H3 binds to HSC70 chaperones. The histone is then transferred to HSP90, which assembles the H3/H4 heterodimer with the help of co-chaperone tNASP. The heterodimer binds to chaperone sNASP, exposing the C-terminal domain of histone H4 to the RbAp46 protein, allowing the binding of the acetylase Hat1, which acetylates histone H4 at lysines 5 and 12. After acetylation, the heterodimer H3/H4 is delivered to Asf1B and this complex binds to importin-4, which mediates nuclear import. These modifications seem to be required because depletion of Hat1p is lethal for human cells, whereas in yeast mutations of K5 and K12 of the histone H4 does not affect transport and histone incorporation in the nucleus (57). Similar acetylation sites are present in trypanosomes (47,58,59). Additionally, we observed that single mutations in the H4 acetylation sites do not prevent nuclear import and incorporation in the chromatin (manuscript in preparation).
T. brucei Asf1n is always present in the nucleus, and its expression seems to increase during cell cycle progression until cell division. This observation is compatible with a role in chromatin deposition during DNA replication, which involves other histone chaperones and chromatin remodeling factors present in trypanosomes [reviewed in (59)]. It is also in line with the observation that Asf1n gene knockdown causes arrest later in the cell cycle compared with Asf1c, and that there is no effect on H3/H4 acetylation when overexpressed. These results also support the notion that the nuclear Asf1 could exhibit histone chaperone functions during the replication process, using histones derived from disassembled chromatin (60). It is also possible that Asf1n affects replication directly as shown in other organisms in which Asf1 modulates the helicase activity of the minichromosome maintenance complex and the progression of the replication fork (61).
In addition, our results indicate that the presence of an excess of each Asf1 affects the response of cells to DNA damaging agents in different ways. Overexpression of the nuclear Asf1 initially supported growth after damage followed by a poor recover. This effect can be explained by alterations of checkpoint mechanisms, as shown for Asf1 of other organisms (46). In human cells, for example, Asf1 restores the levels of histone H3 acetylation of lysine 56 (H3K56Ac), which decreases after DNA damage. H3K56Ac is a marker of newly synthesized histones and it is required for dephosphorylation of the γ-histone H2AX and for recovery from cell cycle arrest (62). Interestingly, although a homolog of H2AX has been recently reported in T. brucei (63), no acetylation of H3K56Ac, or a similar residue is reported in trypanosomes. Nevertheless, both T. brucei Asf1 proteins are phosphorylated by TLK1 (25), a member of the Tousled-like kinase family, which is involved in cell cycle control and checkpoint signaling (64). When phosphorylated, Drosophila and human Asf1A [ortholog of Asf1n (Asf1B) of T. brucei] show an increased stability (64), which might be involved in checkpoint regulation (6,49,65,66). Therefore, an excess of Asf1n in T. brucei could compensate for its degradation and/or phosphorylation preventing cell cycle arrest after DNA damage, explaining the poor recovery after accumulation of DNA breaks, similar to the situation described in yeast (7). The nature of DNA damage checkpoints are largely unknown in trypanosomes (67), which do not possess homologs of the Rad53 kinase, an enzyme that mediates replication arrest in many organism (68). Similarly, we observed that overexpression of an Asf1A homolog caused an increased sensitivity to genotoxic agents by affecting chromatin remodeling during DNA repair in Leishmania major (69).
In contrast, an excess of Asf1c initially reduces cell growth after DNA damage followed by a normal recover. This effect could be a consequence of the increasing levels of histone acetylation, known to affect the chromatin structure and DNA repair in several eukaryotes (70), including trypanosomes (71). Accordingly, we found that the acetylation of lysine 10 and 14 of histone H4 increases in T. cruzi during repair of DNA double strand breaks caused by γ-irradiation (72). In addition, we can exclude a role of the cytoplasmic Asf1 in the checkpoint, as it did not migrate to the nucleus after DNA damage.
T. brucei telomeres contain several genes coding for the VSG, which forms a surface coat in bloodstream forms of the parasite (73). A single VSG gene is expressed at any given time and when the mammalian host generates antibodies to this VSG, some cells in the population that have switched to a new VSG gene, will escape the host’s immune response (74). One mechanism of changing the VSG is dependent on homologous recombination after a double strand break event near the telomere and requires chromatin-remodeling proteins and histone chaperones (75,76). Furthermore, depletion of the cytosolic Asf1 by RNAi partially activates silent VSGs (26), probably by changing the acetylation levels in the histones.
In conclusion, we show that the two Asf1 proteins in T. brucei acquired distinguishable cellular localizations and functions during evolution of this parasite. This divergence is mainly based on variations in the C-terminus, known to be involved in interactions of Asf1 with regulatory factors (6). A more recent divergence also occurred in vertebrate organisms (77). In the case of trypanosomes, it might be an adaptation to the unique control of gene expression. Further studies with this parasite as a model system can help to understand the multiple functions of Asf1 proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fundação de Amparo à Pesquisa do Estado de São Paulo—FAPESP [2011/51973-3 to S.S. and 2007/59950-7 to B.P.]; Conselho Nacional de Desenvolvimento Científico e Tecnológico—CNPq [477143/2011-3 to S.S. and to Instituto Nacional de Ciência e Tecnologia de Vacinas] from Brazil; and from the Universität Bayern PhD fellowship (to G.D.), the collaborative research center TR5 of the Deutsche Forschungsgemeinschaft (to C.J.J.). Funding for open access charge: Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Claudio Rogério Oliveira and Claudeci Medeiros for technical help. The authors also thank Elisabeth Kremmer for providing antibodies.
REFERENCES
- 1.Le S, Davis C, Konopka JB, Sternglanz R. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 2.Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, Goggin C, Mahowald M, Gottschling DE. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mousson F, Ochsenbein F, Mann C. The histone chaperone Asf1 at the crossroads of chromatin and DNA checkpoint pathways. Chromosoma. 2007;116:79–93. doi: 10.1007/s00412-006-0087-z. [DOI] [PubMed] [Google Scholar]
- 4.Emili A, Schieltz DM, Yates JR, 3rd, Hartwell LH. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell. 2001;7:13–20. doi: 10.1016/s1097-2765(01)00150-2. [DOI] [PubMed] [Google Scholar]
- 5.Hu F, Alcasabas AA, Elledge SJ. Asf1 links Rad53 to control of chromatin assembly. Genes Dev. 2001;15:1061–1066. doi: 10.1101/gad.873201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sillje HH, Nigg EA. Identification of human Asf1 chromatin assembly factors as substrates of Tousled-like kinases. Curr. Biol. 2001;11:1068–1073. doi: 10.1016/s0960-9822(01)00298-6. [DOI] [PubMed] [Google Scholar]
- 7.Minard LV, Lin LJ, Schultz MC. SWI/SNF and Asf1 independently promote derepression of the DNA damage response genes under conditions of replication stress. PLoS One. 2011;6:e21633. doi: 10.1371/journal.pone.0021633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osley MA, Lycan D. Trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol. Cell. Biol. 1987;7:4204–4210. doi: 10.1128/mcb.7.12.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fillingham J, Kainth P, Lambert JP, van Bakel H, Tsui K, Pena-Castillo L, Nislow C, Figeys D, Hughes TR, Greenblatt J, et al. Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol. Cell. 2009;35:340–351. doi: 10.1016/j.molcel.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 10.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 12.Das C, Tyler JK, Churchill ME. The histone shuffle: histone chaperones in an energetic dance. Trends Biochem. Sci. 2010;35:476–489. doi: 10.1016/j.tibs.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 14.Tyler JK, Collins KA, Prasad-Sinha J, Amiott E, Bulger M, Harte PJ, Kobayashi R, Kadonaga JT. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol. Cell. Biol. 2001;21:6574–6584. doi: 10.1128/MCB.21.19.6574-6584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green EM, Antczak AJ, Bailey AO, Franco AA, Wu KJ, Yates JR, 3rd, Kaufman PD. Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 2005;15:2044–2049. doi: 10.1016/j.cub.2005.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adkins MW, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell. 2004;14:657–666. doi: 10.1016/j.molcel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 17.Tang Y, Poustovoitov MV, Zhao K, Garfinkel M, Canutescu A, Dunbrack R, Adams PD, Marmorstein R. Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat. Struct. Mol. Biol. 2006;13:921–929. doi: 10.1038/nsmb1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daganzo SM, Erzberger JP, Lam WM, Skordalakes E, Zhang R, Franco AA, Brill SJ, Adams PD, Berger JM, Kaufman PD. Structure and function of the conserved core of histone deposition protein Asf1. Curr. Biol. 2003;13:2148–2158. doi: 10.1016/j.cub.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 19.Minard LV, Williams JS, Walker AC, Schultz MC. Transcriptional regulation by Asf1: new mechanistic insights from studies of the DNA damage response to replication stress. J. Biol. Chem. 2011;286:7082–7092. doi: 10.1074/jbc.M110.193813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamburini BA, Carson JJ, Linger JG, Tyler JK. Dominant mutants of the Saccharomyces cerevisiae ASF1 histone chaperone bypass the need for CAF-1 in transcriptional silencing by altering histone and Sir protein recruitment. Genetics. 2006;173:599–610. doi: 10.1534/genetics.105.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Umehara T, Chimura T, Ichikawa N, Horikoshi M. Polyanionic stretch-deleted histone chaperone cia1/Asf1p is functional both in vivo and in vitro. Genes Cells. 2002;7:59–73. doi: 10.1046/j.1356-9597.2001.00493.x. [DOI] [PubMed] [Google Scholar]
- 22.Dennehey BK, Noone S, Liu WH, Smith L, Churchill ME, Tyler JK. The C terminus of the histone chaperone Asf1 cross-links to histone H3 in yeast and promotes interaction with histones H3 and H4. Mol. Cell. Biol. 2013;33:605–621. doi: 10.1128/MCB.01053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamburini BA, Carson JJ, Adkins MW, Tyler JK. Functional conservation and specialization among eukaryotic anti-silencing function 1 histone chaperones. Eukaryot Cell. 2005;4:1583–1590. doi: 10.1128/EC.4.9.1583-1590.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campos E, Fillingham J, Li G, Zheng H, Voigt P, Kuo WHW, Seepany H, Gao Z, Day L, Greenblatt J, et al. The program for processing newly synthesized histones H3.1 and H4. Nat. Struct. Mol. Biol. 2010;17:1343–1351. doi: 10.1038/nsmb.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, Gourguechon S, Wang CC. Tousled-like kinase in a microbial eukaryote regulates spindle assembly and S-phase progression by interacting with Aurora kinase and chromatin assembly factors. J. Cell Sci. 2007;120:3883–3894. doi: 10.1242/jcs.007955. [DOI] [PubMed] [Google Scholar]
- 26.Alsford S, Horn D. Cell-cycle-regulated control of VSG expression site silencing by histones and histone chaperones ASF1A and CAF-1b in Trypanosoma brucei. Nucleic Acids Res. 2012;40:10150–10160. doi: 10.1093/nar/gks813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer S. Developmental regulation of gene expression in the absence of transcriptional control: the case of kinetoplastids. Mol. Biochem. Parasitol. 2012;181:61–72. doi: 10.1016/j.molbiopara.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 28.De Gaudenzi JG, Noe G, Campo VA, Frasch AC, Cassola A. Gene expression regulation in trypanosomatids. Essays Biochem. 2011;51:31–46. doi: 10.1042/bse0510031. [DOI] [PubMed] [Google Scholar]
- 29.Calderano SG, de Melo Godoy PD, Motta MC, Mortara RA, Schenkman S, Elias MC. Trypanosoma cruzi DNA replication includes the sequential recruitment of pre-replication and replication machineries close to nuclear periphery. Nucleus. 2011;2:136–145. doi: 10.4161/nucl.2.2.15134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tiengwe C, Marcello L, Farr H, Gadelha C, Burchmore R, Barry JD, Bell SD, McCulloch R. Identification of ORC1/CDC6-interacting factors in Trypanosoma brucei reveals critical features of origin recognition complex architecture. PLoS One. 2012;7:e32674. doi: 10.1371/journal.pone.0032674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Passos-Silva DG, Rajao MA, Nascimento de Aguiar PH, Vieira-da-Rocha JP, Machado CR, Furtado C. Overview of DNA Repair in Trypanosoma cruzi, Trypanosoma brucei, and Leishmania major. J. Nucleic Acids. 2010;2010:840768. doi: 10.4061/2010/840768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alsford S, Horn D, Glover L. DNA breaks as triggers for antigenic variation in African trypanosomes. Genome Biol. 2009;10:223. doi: 10.1186/gb-2009-10-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wirtz E, Clayton C. Inducible gene expression in trypanosomes mediated by a prokaryotic repressor. Science. 1995;268:1179–1183. doi: 10.1126/science.7761835. [DOI] [PubMed] [Google Scholar]
- 34.Brun R, Schönenberger Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- 35.Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- 36.Clayton CE, Fueri JP, Itzhaki JE, Bellofatto V, Sherman DR, Wisdom GS, Vijayasarathy S, Mowatt MR. Transcription of the procyclic acidic repetitive protein genes of Trypanosoma brucei. Mol. Cell. Biol. 1990;10:3036–3047. doi: 10.1128/mcb.10.6.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medina-Acosta E, Cross GAM. Rapid isolation of DNA from trypanosomatid protozoa using a simple ‘mini-prep’ procedure. Mol. Biochem. Parasitol. 1993;59:327–330. doi: 10.1016/0166-6851(93)90231-l. [DOI] [PubMed] [Google Scholar]
- 38.Alsford S, Horn D. Single-locus targeting constructs for reliable regulated RNAi and transgene expression in Trypanosoma brucei. Mol. Biochem. Parasitol. 2008;161:76–79. doi: 10.1016/j.molbiopara.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chung J, Rocha AA, Tonelli RR, Castilho BA, Schenkman S. Eukaryotic initiation factor 5A dephosphorylation is required for translational arrest in stationary phase cells. Biochem. J. 2013;451:257–267. doi: 10.1042/BJ20121553. [DOI] [PubMed] [Google Scholar]
- 40.da Cunha JPC, Nakayasu ES, de Almeida IC, Schenkman S. Post-translational modifications of Trypanosoma cruzi histone H4. Mol. Biochem. Parasitol. 2006;150:268–277. doi: 10.1016/j.molbiopara.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 41.Chan S, Gabra H, Hill F, Evan G, Sikora K. A novel tumour marker related to the c-myc oncogene product. Mol. Cell. Probes. 1987;1:73–82. doi: 10.1016/0890-8508(87)90008-9. [DOI] [PubMed] [Google Scholar]
- 42.Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Method Mol. Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- 43.Gassen A, Brechtefeld D, Schandry N, Arteaga-Salas JM, Israel L, Imhof A, Janzen CJ. DOT1A-dependent H3K76 methylation is required for replication regulation in Trypanosoma brucei. Nucleic Acids Res. 2012;40:10302–10311. doi: 10.1093/nar/gks801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siegel TN, Kawahara T, Degrasse JA, Janzen CJ, Horn D, Cross GA. Acetylation of histone H4K4 is cell cycle regulated and mediated by HAT3 in Trypanosoma brucei. Mol. Microbiol. 2008;67:762–771. doi: 10.1111/j.1365-2958.2007.06079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D'Arcy S, Luger K. Understanding histone acetyltransferase Rtt109 structure and function: how many chaperones does it take? Curr. Opin. Struct. Biol. 2011;21:728–734. doi: 10.1016/j.sbi.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim JA, Haber JE. Chromatin assembly factors Asf1 and CAF-1 have overlapping roles in deactivating the DNA damage checkpoint when DNA repair is complete. Proc. Natl Acad. Sci. USA. 2009;106:1151–1156. doi: 10.1073/pnas.0812578106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siegel TN, Hekstra DR, Kemp LE, Figueiredo LM, Lowell JE, Fenyo D, Wang X, Dewell S, Cross GA. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009;23:1063–1076. doi: 10.1101/gad.1790409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sutton A, Bucaria J, Osley MA, Sternglanz R. Yeast ASF1 protein is required for cell cycle regulation of histone gene transcription. Genetics. 2001;158:587–596. doi: 10.1093/genetics/158.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mello JA, Sillje HH, Roche DM, Kirschner DB, Nigg EA, Almouzni G. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 2002;3:329–334. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moshkin YM, Armstrong JA, Maeda RK, Tamkun JW, Verrijzer P, Kennison JA, Karch F. Histone chaperone ASF1 cooperates with the Brahma chromatin-remodelling machinery. Genes Dev. 2002;16:2621–2626. doi: 10.1101/gad.231202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keck KM, Pemberton LF. Histone chaperones link histone nuclear import and chromatin assembly. Biochim. Biophys. Acta. 2012;1819:277–289. doi: 10.1016/j.bbagrm.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blackwell JS, Jr, Wilkinson ST, Mosammaparast N, Pemberton LF. Mutational analysis of H3 and H4 N termini reveals distinct roles in nuclear import. J. Biol. Chem. 2007;282:20142–20150. doi: 10.1074/jbc.M701989200. [DOI] [PubMed] [Google Scholar]
- 53.Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 55.Burgess RJ, Zhou H, Han J, Zhang Z. A role for Gcn5 in replication-coupled nucleosome assembly. Mol. Cell. 2010;37:469–480. doi: 10.1016/j.molcel.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh MC, Greenblatt JF. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol. Cell. Biol. 2008;28:4342–4353. doi: 10.1128/MCB.00182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma XJ, Wu J, Altheim BA, Schultz MC, Grunstein M. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc. Natl Acad. Sci. USA. 1998;95:6693–6698. doi: 10.1073/pnas.95.12.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Figueiredo LM, Cross GA, Janzen CJ. Epigenetic regulation in African trypanosomes: a new kid on the block. Nat. Rev. Microbiol. 2009;7:504–513. doi: 10.1038/nrmicro2149. [DOI] [PubMed] [Google Scholar]
- 59.Schenkman S, Pascoalino Bdos S, Nardelli SC. Nuclear Structure of Trypanosoma cruzi. Adv. Parasitol. 2011;75:251–283. doi: 10.1016/B978-0-12-385863-4.00012-5. [DOI] [PubMed] [Google Scholar]
- 60.Avvakumov N, Nourani A, Cote J. Histone chaperones: modulators of chromatin marks. Mol. Cell. 2011;41:502–514. doi: 10.1016/j.molcel.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 61.Groth A, Corpet A, Cook AJ, Roche D, Bartek J, Lukas J, Almouzni G. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 62.Battu A, Ray A, Wani AA. ASF1A and ATM regulate H3K56-mediated cell-cycle checkpoint recovery in response to UV irradiation. Nucleic Acids Res. 2011;39:7931–7945. doi: 10.1093/nar/gkr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Glover L, Horn D. Trypanosomal histone gammaH2A and the DNA damage response. Mol. Biochem. Parasitol. 2012;183:78–83. doi: 10.1016/j.molbiopara.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pilyugin M, Demmers J, Verrijzer CP, Karch F, Moshkin YM. Phosphorylation-mediated control of histone chaperone ASF1 levels by Tousled-like kinases. PLoS One. 2009;4:e8328. doi: 10.1371/journal.pone.0008328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Benedetti A. Tousled kinase TLK1B mediates chromatin assembly in conjunction with Asf1 regardless of its kinase activity. BMC Res. Notes. 2010;3:68. doi: 10.1186/1756-0500-3-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carrera P, Moshkin YM, Gronke S, Sillje HH, Nigg EA, Jackle H, Karch F. Tousled-like kinase functions with the chromatin assembly pathway regulating nuclear divisions. Genes Dev. 2003;17:2578–2590. doi: 10.1101/gad.276703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hammarton TC. Cell cycle regulation in Trypanosoma brucei. Mol. Biochem. Parasitol. 2007;153:1–8. doi: 10.1016/j.molbiopara.2007.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gunjan A, Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- 69.Scher R, Garcia JB, Pascoalino B, Schenkman S, Cruz AK. Characterization of anti-silencing factor 1 in Leishmania major. Mem. Inst. Oswaldo Cruz. 2012;107:377–386. doi: 10.1590/s0074-02762012000300013. [DOI] [PubMed] [Google Scholar]
- 70.Hunt CR, Ramnarain D, Horikoshi N, Iyenger P, Pandita RK, Shay JW, Pandita TK. Histone modifications and DNA double-strand break repair after exposure to ionizing radiations. Radiat. Res. 2013;179:383–392. doi: 10.1667/RR3308.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garcia-Salcedo JA, Gijon P, Nolan DP, Tebabi P, Pays E. A chromosomal SIR2 homologue with both histone NAD-dependent ADP-ribosyltransferase and deacetylase activities is involved in DNA repair in Trypanosoma brucei. EMBO J. 2003;22:5851–5862. doi: 10.1093/emboj/cdg553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nardelli SC, da Cunha JP, Motta MC, Schenkman S. Distinct acetylation of Trypanosoma cruzi histone H4 during cell cycle, parasite differentiation, and after DNA damage. Chromosoma. 2009;118:487–499. doi: 10.1007/s00412-009-0213-9. [DOI] [PubMed] [Google Scholar]
- 73.Marcello L, Barry JD. Analysis of the VSG gene silent archive in Trypanosoma brucei reveals that mosaic gene expression is prominent in antigenic variation and is favored by archive substructure. Genome Res. 2007;17:1344–1352. doi: 10.1101/gr.6421207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Horn D, McCulloch R. Molecular mechanisms underlying the control of antigenic variation in African trypanosomes. Curr. Opin. Microbiol. 2010;13:700–705. doi: 10.1016/j.mib.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kawahara T, Siegel TN, Ingram AK, Alsford S, Cross GA, Horn D. Two essential MYST-family proteins display distinct roles in histone H4K10 acetylation and telomeric silencing in trypanosomes. Mol. Microbiol. 2008;69:1054–1068. doi: 10.1111/j.1365-2958.2008.06346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Povelones M, Gluenz E, Dembek M, Gull K, Rudenko G. Histone H1 plays a role in heterochromatin formation and VSG expression site silencing in Trypanosoma brucei. PLoS Pathog. 2012;8:e1003010. doi: 10.1371/journal.ppat.1003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abascal F, Corpet A, Gurard-Levin ZA, Juan D, Ochsenbein F, Rico D, Valencia A, Almouzni G. Subfunctionalization via adaptive evolution influenced by genomic context: the case of histone chaperones ASF1a and ASF1b. Mol. Biol. Evol. 2013;30:1853–1866. doi: 10.1093/molbev/mst086. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.