Abstract
Many chloroplast transcripts are protected against exonucleolytic degradation by RNA-binding proteins. Such interactions can lead to the accumulation of short RNAs (sRNAs) that represent footprints of the protein partner. By mining existing data sets of Chlamydomonas reinhardtii small RNAs, we identify chloroplast sRNAs. Two of these correspond to the 5′-ends of the mature psbB and psbH messenger RNAs (mRNAs), which are both stabilized by the nucleus-encoded protein Mbb1, a member of the tetratricopeptide repeat family. Accordingly, we find that the two sRNAs are absent from the mbb1 mutant. Using chloroplast transformation and site-directed mutagenesis to survey the psbB 5′ UTR, we identify a cis-acting element that is essential for mRNA accumulation. This sequence is also found in the 5′ UTR of psbH, where it plays a role in RNA processing. The two sRNAs are centered on these cis-acting elements. Furthermore, RNA binding assays in vitro show that Mbb1 associates with the two elements specifically. Taken together, our data identify a conserved cis-acting element at the extremity of the psbH and psbB 5′ UTRs that plays a role in the processing and stability of the respective mRNAs through interactions with the tetratricopeptide repeat protein Mbb1 and leads to the accumulation of protected sRNAs.
INTRODUCTION
In the chloroplast, post-transcriptional steps play a major role in the control of gene expression. Many chloroplast genes are part of polycistronic transcription units, and RNA maturation is complex. It involves endonucleolytic and exonucleolytic processing at the 5′-end, the 3′-end and in intergenic spacers, intron splicing and in plants, RNA editing (1–4). These maturation events in turn influence messenger RNA (mRNA) translation (5–7). RNA maturation, RNA stability and translation are regulated by developmental programs and by environmental factors such as light or nutrient availability (6,8,9). Numerous nucleus-encoded factors are imported in the chloroplast where they govern these post-transcriptional events (4,10). Most of these factors are highly specific and generally target only one or a few genes. A prominent example for such RNA-binding proteins is the members of the helical-repeat protein super-family, which fulfill various tasks for the maturation of organellar RNAs and include pentatricopeptide repeat (PPR), octotricopeptide repeat (OPR) or TPR/HAT (tetratricopeptide repeat/half a tetratricopeptide repeat) proteins (11–16). The prototypical example of helical-repeat proteins is Pumilio, where each repeat is composed of three alpha-helices that interact to provide a super-helical scaffold. Each repeat presents specific amino acid residues that bind to 1 nt of the RNA substrate (17). The OPR family has expanded during the evolution of Chlamydomonas reinhardtii, whereas it is the PPR family that is most prominent in the higher-plant lineages (12–14). Members of these helical-repeat protein families can protect chloroplast RNAs against exonucleolytic degradation by tightly binding sequences in the UTRs and thus increase the stability of their substrates (18–24). A well-studied example is the binding of PPR10 to its target sequences, which impedes the progression of both 5′- and 3′-exonucleases and can thus protect either the downstream or the upstream RNA, respectively (22,23). Intriguingly, PPR10 as well as other helical repeat proteins generate short, 15–30-nt-long sRNAs simply by protecting the bound RNA segment, i.e. the footprint, against exonucleolytic degradation (22–24). Almost 100 such sRNAs are found in plant chloroplast transcriptomes, many corresponding to ends of transcripts and to known or presumed protein binding sites (25–27). Consequently, sRNAs can be used as a proxy to identify binding sites of RNA-binding proteins. To date, sRNA data sets have been presented for different species of angiosperms, namely, barley, maize, Arabidopsis and Chinese cabbage (25,26,28). Whether sRNAs are present in chloroplasts of other lineages is at present unclear, although this is suggested by the wide evolutionary distribution of chloroplast-targeted helical-repeat proteins.
Here, we identify chloroplast sRNAs of the green alga C. reinhardtii in public data sets from high-throughput RNA-sequencing experiments. We show that some sRNAs co-localize with transcript ends and can be detected by RNA gel blot analysis. To investigate their biological significance, we focus on two sRNAs that map to the psbB/psbT/psbH gene cluster, which is transcribed as a unit and processed to give rise to the monocistronic psbB and dicistronic psbB/T mRNAs, as well as to several forms of psbH RNA, all of which encode subunits of PSII (29,30). The nucleus-encoded factor Mbb1 is specifically required for the stable accumulation of all the transcripts from this cluster (29,31). The analysis of reporter constructs has shown that Mbb1 acts through the 5′ untranslated region (5′ UTR) of psbB, and there is genetic evidence that it also acts directly on psbH (19).
Mbb1 is one of the rare RNA-binding proteins in chloroplasts for which an ortholog can be identified in higher plants. This ortholog, named HCF107 (high chlorophyll fluorescence 107), is required for expression of psbB and psbH (32,33). In the hcf107 mutant, RNA processing upstream of psbH is deficient and its translation is impaired. Translation of psbB is also defective in this mutant (32), even though the pattern of psbB transcripts appears normal. In vitro assays have demonstrated that recombinant HCF107 binds the 5′-end of the psbH transcript and can protect it against exonucleolytic degradation from either the 5′ or the 3′ side (24). An sRNA representing the footprint of HCF107 is detected in vivo. Thus, the TPR/HAT protein HCF107 seems to act similarly to PPR proteins like PPR10, in line with their similar predicted helical-repeat structure (34).
Here we show that the sRNAs mapping to the ends of the 5′ UTRs of psbH and psbB are missing in the mbb1 mutant, suggestive of a direct functional link between Mbb1 and these short RNA segments. Using chloroplast site-directed mutagenesis, we demonstrate the importance of the corresponding sequence elements for mRNA stability in vivo by a systematic genetic survey of the entire psbB 5′ UTR and of conserved sequences in the psbH 5′ UTR. Association of Mbb1 with this RNA sequence element is demonstrated by in vitro binding assays.
MATERIALS AND METHODS
Strains and media
The C. reinhardtii mbb1-222E mutant strain was described previously (29). For phenotypic analysis of mutant strains (spot-tests), 2 ml of culture was grown overnight in Tris acetate phosphate medium in the dark, and then 15 μl aliquots were spotted on agar plates containing Tris acetate phosphate or high salt minimum (35). Biolistic transformation of C. reinhardtii and selection on spectinomycin were described previously (19).
RNA and protein analysis
RNA was extracted using TriReagent (Sigma-Aldrich) and analyzed by agarose gel electrophoresis, capillary transfer to nylon membranes and hybridization with 32P-labeled probes as described previously (36). The psbB probe was a 1.1-kb NcoI—EcoRI fragment of p38ANco. The psaA exon 3 probe was a 2.2-kb EcoRI—HindIII fragment of R17EH4 (37). The psbH probe was a 250-bp polymerase chain reaction (PCR) fragment amplified with the two oligonucleotides 5′ PSBH and 3′ PSBH. The 5′ psbH probe was a 309-bp PacI—XmnI fragment, and the 3′ psbH probe was a 484-bp AatII—EcoRI fragment. Probes were stripped in 0.5% sodium dodecyl sulphate at 60°C for 1 h, and the membranes were checked for residual signal using a phosphor screen. The autoradiographs shown in Figure 1 were all obtained using one membrane that was repeatedly stripped and reprobed.
Figure 1.
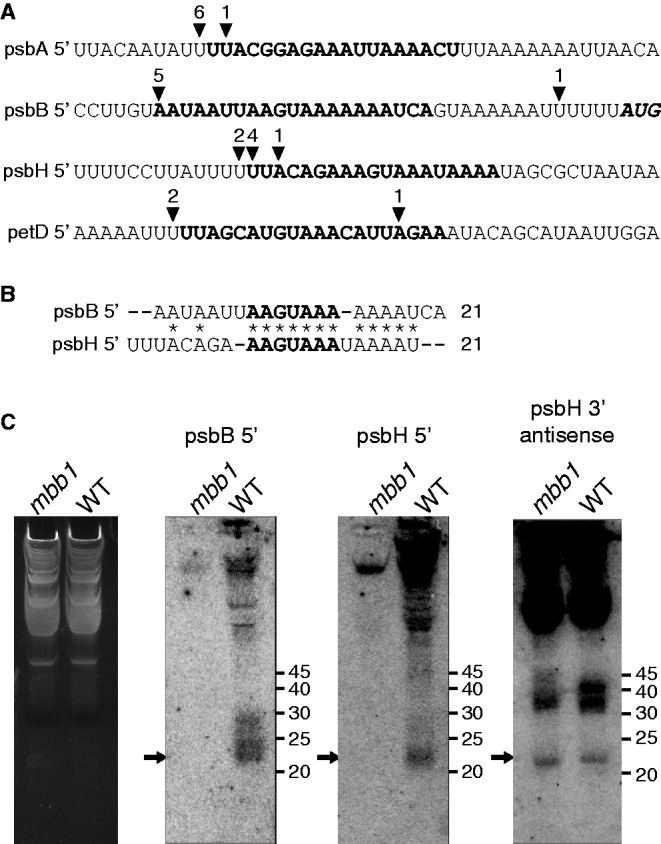
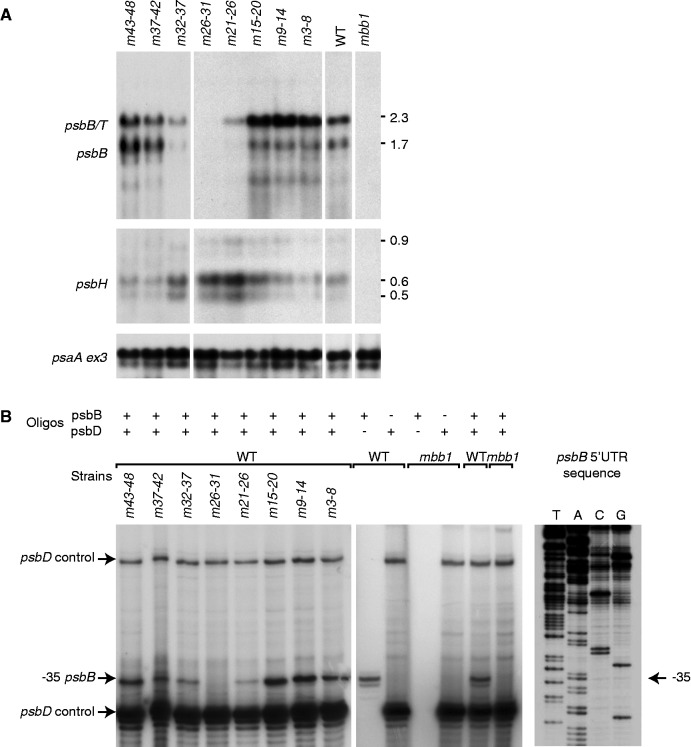
sRNAs correspond to the 5'-ends of chloroplast mRNAs in Chlamydomonas. (A) The sequences corresponding to four chloroplast sRNAs (Supplementary Table S1) are highlighted in bold. The position of the 5'-ends as mapped by RACE is shown with arrowheads. The numbers above the arrowheads indicate the number of 5'-RACE clones that end at the respective sites. Most clone ends coincide with sRNA 5'-ends. (B) Sequence alignment of the psbB 5' and psbH 5' sRNAs. Asterisks indicate conserved positions. (C) Autoradiograms of an RNA blot of chloroplast RNA from the WT and the mbb1 mutant hybridized with the radioactive probes indicated above the panels. The arrows indicate the positions of the sRNAs. Low molecular weight RNA was enriched from total RNA and separated on a 15% sodium dodecyl sulphate-polyacrylamide gel electrophoresis containing 8 M urea. A fluorescence image of the gel lanes stained with ethidium bromide is shown as loading control in the leftmost panel. The probes were single-stranded DNA oligonucleotides (≤25 nt) antisense to the sRNAs. The same membrane was repeatedly probed and stripped as described in Materials and Methods section.
Primer extension mapping of the psbB and psbH 5′ UTR was performed with 10 μg of RNA, 32P-labeled oligonucleotides and M-MLV reverse transcriptase (Promega) at 50°C essentially as described (38).
Chlamydomonas sRNA data were retrieved from Ibrahim et al. (39), available from the National Center for Biotechnology Information as GEO Series GSE17815, including reads from four different libraries: two libraries had been prepared from a knockout line of the terminal nucleotidyltransferase and two libraries from the knockdown of a tryptophan synthase beta subunit. Three further libraries from the Comparative Sequencing of Plant Small RNAs project are available as GEO Series GSE32457. They had been prepared from a wild-type (WT) Chlamydomonas strain grown under normal conditions, under phosphate starvation or under sulfate starvation. sRNA sequences represented by >1 read per sample were mapped to the chloroplast genome (NC_005353) using CLC Genomics Workbench (version 6.0.1) allowing one mismatch in the core sequence. sRNAs with >15 reads were extracted.
sRNAs were enriched according to (40) from total RNA prepared as described by (41). RNA was resolved on 15% polyacrylamide gels containing 8-M urea and transferred to Hybond-N nylon membranes (GE Healthcare). Membrane-bound RNAs were chemically cross-linked as described by (42). DNA oligonucleotides were end-labeled using polynucleotide kinase (Thermo Scientific) and γ32P-ATP (Hartmann-Analytics) and used as probes.
The 5′ and 3′ RACE experiments were carried out as described (25), with primers described in Supplementary Table S1.
Protein samples were prepared and analyzed by immunoblotting as described previously (36).
Site-directed mutagenesis
The mutations were introduced in a transformation vector [p38ANco; (19)] containing an aadA cassette, which confers spectinomycin resistance as a selectable marker for chloroplast transformation (43). The marker was inserted at position −320 and was transcribed in the opposite direction relative to psbB/T/H (Supplementary Figure S1). In this vector, the two nucleotides preceding the ATG start codon were changed to create an NcoI restriction site; this change did not measurably affect psbB/T/H mRNA expression but reduced the translation of the PSII subunits and the growth rate of the strains [Supplementary Figure S2; (19)]. Restriction sites used for these constructs are shown in Supplementary Figure S1. The mutant psbB 5′ UTR fragments (m3-8 to m32-37) were generated by PCR with the sense primer StuNde and the respective antisense primer (Apa-3 to Apa-32). The PCR products were cloned in the SmaI site of pBluescriptKS(+), from which they were excised with NdeI and NcoI. For m37-42 and m43-48, the WT 325-bp NdeI—NcoI fragment inserted in pBluescriptKS(+) was used as a template for PCR around the entire plasmid with 5′ phosphorylated oligonucleotides containing half of the ApaI at their 5′-ends: s-37>-15 with as-40>-69, or s-45>-15 with as-45>-74, respectively. The PCR product was ligated and cloned, and the mutant UTR fragments were excised with NdeI and NcoI. The sequences of the mutant fragments were verified by sequencing. The NdeI—NcoI fragments were inserted in the vector P38.NcoΔ0 (19) digested with the same enzymes. Finally, the spectinomycin resistance cassette from pUCatpXaadA (43) was inserted as a 1.9-kb EcoRV SmaI fragment into the StuI site to obtain the mutant transformation vectors.
The psbH transformation vector p41A was prepared as follows. A 3216-bp BamHI—ScaI genomic fragment was cloned in the pBluescriptKS(+) vector digested with EcoRV and BamHI (plasmid p41). Then the atpXaadA cassette was inserted as a 1.9-kb EcoRV—SmaI fragment in the MscI site, in the same orientation as psbH (p41A). The mutant psbH 5′ UTR fragments were generated with the antisense primer Sta-as and the respective sense primers ApaS, ComS and DelS. These were cloned in pCRII-Topo (Invitrogen), verified by sequencing, excised with PacI and AfeI and inserted into p41A digested with the same enzymes to obtain the mutant transformation vectors.
For psbB transformation, the C. reinhardtii recipient hosts were obtained as follows. A vector (pANcoΔ3) was constructed by replacing the NcoI–SacII fragment of p38ANco (19) with the recyclable aadA cassette (44) excised with SacI and KpnI from pKS483aadA483. This vector was used to transform C. reinhardtii (WT or mbb1-222E), and homoplasmic strains were obtained by repeated selection on spectinomycin. Further growth in absence of spectinomycin allowed intrachain recombination between the 483-bp repeats (44) and obtention of homoplasmic psbB 5′ UTR deletion strains (non-photosynthetic, spectinomycin-sensitive), which were verified by Southern blotting. One such deletion strain (Δ3) was used as a host for transformations with the linker-scan mutations of psbB in vector p38ANco to ensure that the desired mutations were incorporated together with the selectable marker (Supplementary Figure S1).
For psbH transformation, the C. reinhardtii recipient host was obtained as follows. A vector (p41K7R) was constructed from p41 by replacing the XmnI—MscI segment with the recyclable aadA cassette (44). This was done in two steps: first inserting the cassette (amplified from pKS483aadA483 with phosphorylated M13 and M13 reverse primers) in the MscI site, in the same orientation as psbH; in the second step, the 1.8-kb ApaI fragment (one site in the plasmid vector, one site at the edge of the cassette) was replaced with a 0.7-kb ApaI—XmnI fragment to obtain p41K7R. This vector was used as described above to transform C. reinhardtii and obtain homoplasmic psbH deletion strains (non-photosynthetic, spectinomycin-sensitive), which were verified by PCR. Such a deletion strain (ΔH) was used as a host for transformations with the psbH mutations in the p41A vector, so as to ensure that the mutations were inserted together with the selectable marker, similar to what is described earlier and in Supplementary Figure S1 for psbB mutants.
Electrophoretic mobility shift assay
Protein crude extracts were prepared from a strain expressing a version of Mbb1 tagged with a triple hemagglutinin (HA) epitope obtained by rescuing the mbb1-222E mutant with the Mbb1 gene fused to a triple HA tag at the 3′-end (31), and as a negative control a strain expressing the gene coding HA-tagged sedoheptulose-1,7-bisphosphatase from Dunaliella salina.
A total of 1 × 108 cells in exponential phase were collected and re-suspended in 1 ml of extraction buffer (80 mM NaCl, 10 mM Tris HCl, 1 mM ethylenediaminetetraacetic acid, 2 mM dithiothreitol, 5% glycerol, pH 8.0, and 1× Sigma protease inhibitors). The cells were lysed by adding 500 µl of glass beads (diameter 0.3 mm) and vigorous agitation for 2 ×1 min. The lysate was centrifuged for 12 min at 13 000 g to pellet cell debris. Partial purification of Mbb1-HA and Sbp-HA was performed using an anti-HA affinity matrix (Roche) according to the recommendations of the manufacturer, using a synthetic HA-peptide (Genescript Inc., USA) for the elution.
For the preparation of the RNA probes, templates corresponding to the WT psbB 5′ UTR and the m26-31 mutant psbB 5′ UTR were prepared by PCR using as templates the p38ANco and the p38ANcoApa-26 plasmids that were used for site-directed mutagenesis. The template was amplified with the sense primer T7–35 (which contains the T7 promoter) and the corresponding antisense primer psbB-rev3-probe. RNAs were in vitro transcribed in the presence or absence of 32P-UTP using T7 RNA polymerase (Promega). The probes obtained extend from the psbB processing site at −35 to position +64 relative to the AUG. The probes were verified by gel electrophoresis and quantified by scintillation counting (32P-labeled RNA) or absorbance at 260 nm (competitor RNA).
Binding reactions contained 150 mM NaCl, 50 mM Tris-HCl (pH 7.5), 4 mM dithiothreitol, 0.04 mg/mL bovine serum albumin, 0.5 mg/mL heparin, 15% glycerol, 10 U of RNAsin (Promega), 0.5% Triton X100, 15 pM radiolabeled RNA and protein amounts as indicated. Reactions were incubated for 20 min at 25°C and resolved on 5% native polyacrylamide gels. The radioactive signals were detected with a phosphorimager (BioRad Molecular Imager FX).
RESULTS
Identification of chloroplast sRNAs within Chlamydomonas RNA-seq data sets
Small RNAs such as small interfering RNAs and micro RNAs can be discovered by high-throughput sequencing of their cDNAs. Such approaches have been carried out in a number of autotrophic organisms including Chlamydomonas (39). Starting from total cellular RNA, short RNAs were separated by gel electrophoresis, eluted and used to prepare a template library for high-throughput sequencing. These data sets include short RNAs derived from all three genomes in the nucleus, the chloroplasts and the mitochondria.
We pooled reads from seven libraries originally designed to analyze micro RNAs and small interfering RNAs in Chlamydomonas (39) and from the Comparative Sequencing of Plant Small RNAs project (http://smallrna.udel.edu). For each library, identical reads were combined and called sequences. A total of 17 400 sequences were mapped to the chloroplast genome. This corresponds to 2.2% of all sequences in the seven libraries. Such sequences can represent up to hundreds of individual reads. The identified sequences are scattered across the entire chloroplast genome with distinct local enrichments. Genes known to be highly expressed like the rrn operon, or the psbA and rbcL mRNAs show the expected higher sequence densities compared with the rest of the genome. For further analysis, we excluded such extended regions of high sequence density, as we were interested in isolated and narrow peaks of sequence density—a hallmark of sRNAs (25). We focused on non-coding regions as prime targets of RNA-stabilizing proteins and disregarded all sequence clusters within coding sequences. Furthermore, only clusters of at least 3 overlapping sequences, and representing >10 reads, were retained. The core of each cluster was defined as those nucleotides represented in 50% of all sequences within the cluster. A set of 61 of such clusters, which we henceforth call sRNAs, was identified (Supplementary Table S2).
The mbb1 mutant fails to accumulate two sRNAs from its target UTRs
Angiosperm chloroplast sRNAs in non-coding regions show a bias in location toward transcript ends—consistent with the idea that they are generated by the protective action of helical-repeat proteins. In our data set, we find seven sRNAs to map immediately down- or upstream to known transcript ends (Supplementary Table S2). We confirmed the ends of four of these mRNAs by rapid amplification of cDNAs (RACE, Figure 1A). Two of these sRNAs are located in the psbB and psbH 5′ UTRs, which are the proposed target regions of Mbb1. To test whether Mbb1 is functionally linked to the accumulation of these sRNAs, we probed total RNA from the mbb1 null mutant by RNA gel blot hybridization (Figure 1C). We could detect neither of these two sRNAs in the mutant RNA extract, while both sRNAs accumulated in WT RNA preparations. The specificity of the loss of psbB and psbH sRNAs in mbb1 was confirmed by the presence of the psbH 3′-antisense sRNA (Figure 1C). It is striking that Mbb1 is required for the accumulation of the psbB 5′ and the psbH 5′ sRNAs, as both correspond to mRNAs that depend on the presence of Mbb1 for their stable accumulation (29). A comparison of the psbB and psbH 5′ UTRs showed that they share an identical sequence, AAGUAAA, at the same positions relative to their respective processing sites (Figure 1B). This sequence is at the core of both sRNAs, and will be referred to as ‘the S-box’. In the following, we analyzed the functional significance of both S-box sequence elements by mutational analyses in vivo using chloroplast transformation.
A cis-acting element required for psbB mRNA accumulation
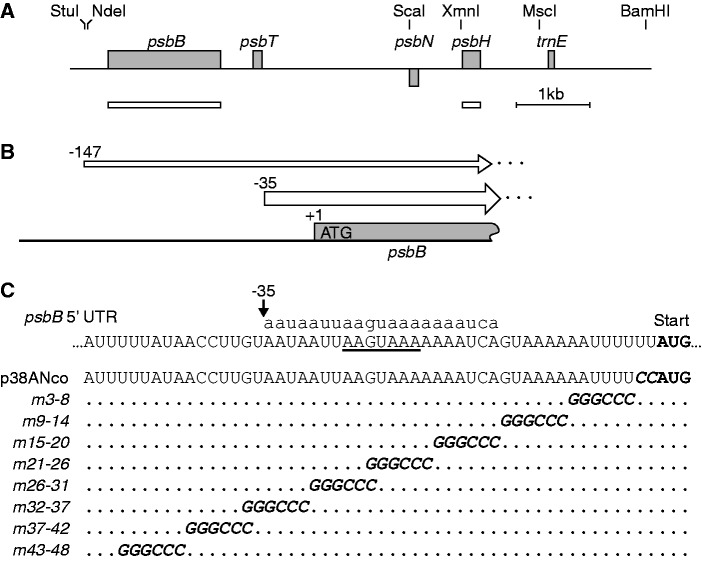
To systematically investigate the function of cis-acting elements in the psbB 5′ UTR, and in particular the short form beginning at −35, which had not been previously investigated, we designed a series of mutations. In a sequential manner, six bases of the sequence were replaced by GGGCCC, the recognition site for the restriction enzyme ApaI, a strategy dubbed linker-scan mutagenesis (Figure 2). The successive mutations spanned the region −3 to −48 relative to the translation start codon, thus extending 13 nt upstream of −35, the 5′-end of the major form of the psbB 5′ UTR. Some of the mutations overlapped so that every base in the sequence had been mutated in at least one of the constructs. To facilitate the recovery of homoplasmic mutant lines, we first generated a recipient strain that carried a substitution upstream of the ATG of psbB in the chloroplast genome (Supplementary Figure S1). This ensured that all transformants with the aadA insertion also carried the desired mutations and excluded the possibility that WT psbB 5′ UTR sequences be retained in the mutant lines because of homologous recombination between the selectable marker and the mutation. The mutated vectors were used for biolistic transformation of the chloroplast genome and selection on spectinomycin-containing media. After several rounds of sub-culturing on spectinomycin, the selected lines were homoplasmic and harbored the expected mutations as shown by PCR amplification followed by ApaI digestion (Supplementary Figure S1B).
Figure 2.
Mutational analysis of the psbB 5' UTR. (A) Map of the psbB/psbT/psbH locus. The gray bars represent the open reading frames of psbB, psbT and psbH, and below the line of the putative psbN, which would be transcribed from the opposite strand. Restriction sites relevant to this work are indicated at the top, and the probes used for RNA blot hybridization are shown below as white bars. (B) The 5' UTRs of the psbB transcripts. The major form of the psbB 5' UTR begins at −35 relative to the start codon. A minor form, which is two orders of magnitude less abundant, begins at −147 (19). (C) Linker-scan mutagenesis of the psbB 5' UTR. The sequence of the WT is shown at the top with the translation start codon of psbB highlighted in bold and the S-box underlined. The corresponding sRNA (psbB 5') is shown in lowercase above the sequence. The black arrow shows the positions of the major 5'-end at −35. The sequences of the chloroplast transformation vector p38ANco and of the eight linker-scan mutants are aligned below, with the ApaI sites in italics and with dots representing unchanged bases.
To investigate the effect of the mutations on mRNA accumulation, we analyzed the transcripts of the psbB/T/H cluster. RNA blots were hybridized with a probe specific for psbB to reveal the monocistronic form as well as the dicistronic transcript psbB/T (Figure 3A). In the WT MBB1 background, the most dramatic reduction of the psbB mRNA and of the dicistronic psbB/T mRNA was with the mutation between positions −26 and −31 (m26-31), for which the mRNAs were not detectable. For the flanking mutations m21-26 and m32-37, the levels were approximately one-third of the WT (Figure 3A). In the other mutants, the amounts of these mRNAs were in the WT range. It is noteworthy that the psbB 5′ sRNA coincides with the region where mutants display reduced RNA accumulation.
Figure 3.
RNA analysis of the psbB 5' UTR mutants. (A) RNA blot hybridization analysis of the psbB/T/H transcripts. Total RNA was analyzed by agarose gel electrophoresis and blot hybridization using the probes indicated to the left of each panel. A probe for exon 3 of psaA was used as a control. (B) Primer extension analysis of the psbB RNAs. Total RNA was used as a template for reverse transcription using oligonucleotide primers specific for psbB, psbD or both as indicated at the top of the panel. The psbD primer serves as an internal control. Owing to the abundance of psbD mRNA, background precludes the detection of the longer form of psbB RNA starting at −147, which is two orders of magnitude less abundant than the form at −35 (19). The panel on the right shows sequencing reactions of the psbB 5' UTR with the psbB primer.
The RNA blots were then hybridized with a probe for psbH, which detected two transcripts that differ by the length of their 3′ UTR (30). In the WT background, the psbH transcripts were detectable at normal levels in all the mutants, and their accumulation was even increased in the strains where psbB mRNA was less abundant (Figure 3A). This suggests that psbB and psbH may compete for a common limiting factor that confers RNA stability (see later).
Mutant psbB transcripts are accurately processed at −35
In certain mutants, a normal pattern of psbB RNAs was present as determined by RNA blotting, but the possibility remained that in some mutants, the 5′-end of the transcripts could be changed to a different position. To address this question, we determined by primer extension the end of the 5′ UTR of the psbB transcripts (Figure 3B). We found that the differences of accumulation of the transcripts ending at -35 among the mutated strains correlated well with the levels detected by RNA blot analysis for the psbB and psbB/T RNAs. Moreover, the length of the 5′ UTR was not affected by any of the mutations for which RNA is present, even for the mutation m32-37, which alters the sequence at the position of the major 5′-end (−35). This shows that processing does not require the recognition of a specific sequence at the exact site corresponding to the end of the mature product. A similar situation was observed for the psbD 5′ UTR, where substitutions of sequences around the processing site (−47) do not alter the position of processing (20). Likewise, substitution of four bases at the processing site in the 5′ UTR of psbA did not change the position or extent of processing (45).
We also determined the effects of the mutations on growth and on the accumulation of the psbB gene product CP47 (Supplementary Figure S2). The mutant strains were compared with the vector control (p38ANco), which accumulates normal levels of psbB/T mRNAs (19), but has an NcoI restriction site engineered at the ATG start codon, which reduces translation efficiency and photosynthetic growth. Phototrophic growth of the strains whose mutations are located upstream of the −35 end (m37-42 and m43-48) was similar to the vector control (p38ANco). The m32-37 mutant, where mRNA levels were strongly reduced (Figure 3A), grew photoautotrophically, albeit more slowly than the vector control. It was previously reported for other chloroplast mRNAs that their abundance under normal conditions is not limiting for translation (46). All mutations within the short leader (strains m3-8 to m26-31) further reduced phototrophic growth below vector-control levels (p38ANco). The levels of CP47 in the mutant strains were consistent with their capacity for photosynthetic growth (Supplementary Figure S2), suggesting that the mutations within the short 5′ UTR may affect translation efficiency.
The psbH 5′ UTR contains a copy of the S-box, which is required for processing and translation
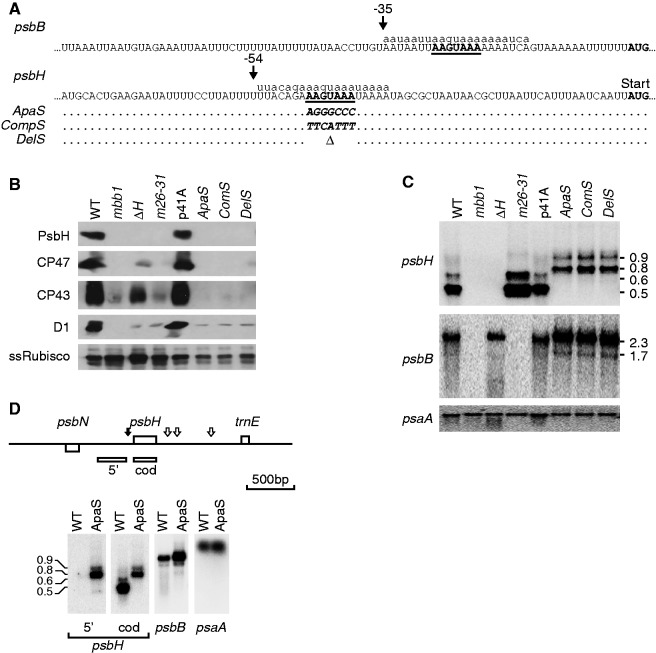
To analyze the importance of the sequence element represented by the psbH 5′ sRNA for the maturation and stability of psbH mRNA in vivo, we mutated its conserved core motif, dubbed the S-box. Three different mutations of the S-box were introduced in the chloroplast genome (Figure 4A), with the S-box replaced by an ApaI site (ApaS), by the complementary sequence (CompS) or deleted (DelS).
Figure 4.
Mutational analysis of the conserved S-box in the psbH 5' UTR. (A) Mutagenesis of the psbH 5' UTR. The sequence of the psbB 5' UTR is shown at the top for comparison, and the sequence of the psbH 5' UTR below it, with the translation start codons in bold. The conserved ‘S-box’ is highlighted in bold and underlined. The sequences of the sRNAs are shown in lowercase above the respective sequences. The black arrows show the positions of the major 5'-ends (see Figure 2 and Supplementary Figure S3). The transformation vector (p41A) carries a selectable aadA spectinomycin resistance cassette inserted downstream of psbH (Materials and Methods). The Chlamydomonas host strain (ΔH) carries a substitution of the psbH gene and its 5' UTR to ensure that transformants carrying the aadA marker also bear the desired mutation. (B) Immunoblot analysis of PSII components. Total protein extracts were analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis and immunoblotting with the antibodies indicated on the left. From left to right, the samples are the WT, the mbb1-222E mutant (mbb1), the ΔH transformation host (ΔH), the psbB m26-31 mutant (m26-31), the transformation host rescued with a WT psbH vector (p41A) and the three mutants shown in (A). (C) RNA blot hybridization analysis of the psbB/T/H transcripts. Total RNA was analyzed by agarose gel electrophoresis and RNA blot hybridization using the psbH and psbB probes as indicated on the left. A probe for psaA was used as a control. (D) Mapping of the psbH transcripts. The schematic map shows the positions of the 5'-end at −54/−53 with a black arrow and of the 3' termini with white arrows [according to (30)]. The upstream probe (5') and coding sequence probe (cod) are depicted below the map as open bars. Total RNA from the WT and a representative mutant (ApaS) was analyzed by agarose gel electrophoresis and RNA blot hybridization using the probes indicated at the bottom of each panel.
The three S-box mutants failed to grow on minimal medium, and immunoblotting showed that they accumulated no detectable PsbH or CP47, and only low levels of D1 and CP43, other subunits of PSII (Figure 4B), as compared with the Rubisco control. This probably reflected a lack of PsbH translation and subsequent proteolytic degradation of the other core subunits of the PSII complex (47). The psbH transcripts were examined by RNA blotting (Figure 4C). In the WT, several forms of psbH mRNAs were present, which were previously determined to differ in the length of their 3′ UTRs (30). However, in the S-box mutants, these were absent and longer forms accumulated. To determine how these longer forms differed from the WT, we compared RNA blots hybridized with probes for the region upstream of the psbH 5′ UTR or for the coding sequence (Figure 4D). The upstream probe detected the longer transcripts in an S-box mutant (ApaS), but not in the WT. This indicated that the longer transcripts in the mutant had 5′ extensions and was confirmed by mapping the psbH RNAs by primer extension (Supplementary Figure S3). In the WT, we observed the mature psbH 5′- ends at position −54, but we did not find an end at position −78 as previously reported (30). In the ApaS S-box mutant, no transcripts ending at −54 were detected, but a major band was found at −617. In the WT, a band migrated to the same position, but was present in lower amounts. The results of primer extension were consistent with those of RNA blotting, and showed that psbH transcripts with a 5′ extension accumulate in the S-box mutants. These data indicate that the S-box is important for processing of psbH RNA at −54, and/or for the stability of the shorter transcripts. Interestingly, the psbB transcripts accumulated to higher levels in the psbH S-box mutants (Figure 4C), again suggesting a competition of these transcripts for a common factor.
Mbb1 associates with the psbB and psbH 5′ UTRs in vitro
It has been shown that Mbb1 acts through the 5′ UTR of psbB to allow stable accumulation of the corresponding transcript and may also play a direct role in the accumulation of psbH (19). The identification of a conserved sequence present on both the psbB and psbH 5′ UTRs, the accumulation of sRNAs encompassing these elements and the effect of mutations of these elements on the maturation of the corresponding transcripts suggest that these may be the target sites of Mbb1. To confirm this hypothesis, we ran electrophoretic mobility shift assays to determine whether Mbb1 associates with the mRNAs. Because our attempts to obtain recombinant Mbb1 expressed in Escherichia coli in soluble form were not successful, we used immunoprecipitation to enrich Mbb1::HA from Chlamydomonas extracts. Immunopurified proteins were incubated with in vitro transcribed RNAs corresponding to the psbB 5′ UTR. As shown in Figure 5A, the psbB 5′ UTR is bound by the Mbb1-HA immune-precipitate but not by the Sbp-HA control (sedoheptulose-1,7-bisphosphatase, an enzyme of the Calvin–Benson cycle). The assays performed with increasing protein amounts resulted in increased shifting of the radiolabeled probe. The specificity of RNA-binding was investigated by comparing competition of the radiolabeled probe with increasing concentration of unlabeled WT probe or a mutant probe carrying the m26-31 substitution that abolishes psbB stability in vivo (Figure 5B). A 10-fold excess of the WT psbB 5’ UTR was sufficient to completely compete the signal, whereas competition with the m26-31 probe required a 100-fold excess. These results indicate that Mbb1 associates with psbB mRNA, and that this interaction involves the sequence element (S-box) that is essential for psbB mRNA stability in vivo. This association could be direct, but could also involve other proteins that are present in the immunoprecipitate.
Figure 5.
Electrophoretic mobility shift assay with Mbb1-HA immunoprecipitates. (A) Binding of Mbb1-HA to the 5' UTR of psbB. A radiolabeled probe corresponding to the mature psbB 5' UTR was incubated with increasing amounts of Mbb1-HA immunoprecipitate (0.1, 1, 2.5 and 4.5 µg) and resolved by native polyacrylamide gel electrophoresis. Bound (B) and unbound (U) probes are marked. A similar experiment was performed with increasing amounts of Sbp-HA (sedoheptulose 1,7 bisphophatase) as a negative control. (B) Competition of RNA binding with WT or mutant psbB 5' UTR. One microgram of Mbb1-HA protein extract was incubated with a radiolabeled probe corresponding to the WT psbB 5' UTR and an excess (10-, 50- or 100-fold) of an unlabeled competitor corresponding either to the m26-31 psbB 5' UTR (left) or the WT psbB 5' UTR (right). (C) Binding of Mbb1-HA to the 5' UTR of psbH. A radiolabeled probe corresponding to the mature psbH 5' UTR was incubated with increasing amounts of Mbb1-HA immunoprecipitate as in (A). (D) Competition of RNA binding with WT or mutant psbH 5' UTR. Competition was performed as in (B) with WT or mutant (ApaS) psbH 5' UTR unlabeled RNA.
In a parallel set of experiments, we used a radiolabeled probe corresponding to the 5′UTR of psbH, and also observed increased shifting with increasing amounts of the Mbb1-HA immunoprecipitate (Figure 5C). The specificity of RNA-binding was tested by comparing competition with increasing concentration of unlabeled WT probe or a mutant form carrying the ApaS substitution, which affects psbH processing in vivo (Figure 5D). The WT probe competed much more efficiently than the mutant version. These results indicate that Mbb1 also associates with psbH mRNA and that the interaction requires the S-box that is essential for proper RNA processing in vivo. A closely related sequence is also found upstream of psbH in higher plants (AAGUcAA, −36 to −30), and is bound in vitro by recombinant HCF107, the plant ortholog of Mbb1 (24). Thus, it is likely that Mbb1 also interacts directly rather than indirectly with its two RNA targets in Chlamydomonas.
DISCUSSION
sRNAs help the identification of cis-acting elements important for RNA processing and stability in the Chlamydomonas transcriptome
We have screened the Chlamydomonas transcriptome for sRNAs as a proxy to identify possible target sites of chloroplast RNA-binding proteins. We found several sRNAs that map to UTRs of mRNAs, adjacent to known transcript ends (Supplementary Table S1). Among RNA-binding proteins, helical-repeat proteins are known to generate sRNAs by binding to transcripts and protecting their footprints against exonucleolytic degradation (2). Other RNA-binding proteins may also contribute to the generation of footprints. For example, we found sRNAs that may represent binding sites of Rbp63, Nac2 and Mcd1 in the psbA, psbD and petD 5′ UTRs, respectively, as these proteins are known to interact with the UTR regions of their target mRNAs (18,48,49). The interaction of these proteins with their RNAs awaits fine-mapping, possibly aided by sRNA location, similar to what we have shown here for Mbb1. It will be interesting to investigate whether there are further links between sRNAs, previously determined functional cis-elements for RNA processing and RNA-binding proteins. For example, there are RNA stabilizing elements in the psbD 5′ UTR, upstream and downstream of the −47 processing site (20). Determinants of petD and rbcL RNA stability reside in sequences adjacent to the 5′-end of the mature mRNA (13,50,51). There are still numerous genes encoding helical-repeat proteins with unassigned functions and targets in the Chlamydomonas genome (13,14,52), and sRNAs could be valuable tools to identify their binding sites. In general, the large set of unassigned sRNAs identified here could be a potential source for not only the identification of novel cis-acting elements, but also a source of baits to biochemically fish for the associated RNA-binding proteins required for RNA stabilization and processing.
Cis-acting targets of Mbb1 required for RNA stabilization
Previous work had shown that both the psbB and psbH RNAs are targets of the nucleus-encoded protein Mbb1, and that the psbB RNA is subject to 5′ exonucleolytic degradation in the absence of Mbb1 (19). Our genetic analysis identifies a cis-acting element in the 5′ UTR, which is essential for the stable accumulation of the psbB and psbB/T transcripts. These RNAs were completely absent in m26-31, and reduced to about one-third of WT levels in the flanking m21-26 and m32-37 mutants. The mutations in m21-26 and m26-31 span a sequence of seven bases (S-box: AAGUAAA), which is also found at position −43 to −49 in the short 5′ UTR of psbH (Figure 4A) and is present at the core of both sRNAs that originate from the two UTRs. Here we show that the S-box is essential for proper psbH mRNA maturation in vivo. We observe that Mbb1 associates with the psbB and psbH 5′ UTR in vitro, and that the interaction can be competed with the WT RNA 10 times more efficiently than with the mutant RNAs carrying a substitution of the S-box, indicating that these interactions are dependent on the S-box. We also detect in vivo small RNAs centered on the S-boxes of both psbB and psbH, whose presence is dependent on Mbb1. Taken together, the data indicate that Mbb1 associates with the 5′ UTRs of psbB and psbH through stable interactions with their respective S-boxes and that these are important for the maturation and stability of the two transcripts.
It is interesting to note that both Mbb1 and its target sequence have been conserved during evolution, as the ortholog HCF107 binds a sequence in maize and Arabidopsis whose 7-nt core (AAGUcAA) is closely related to the S-box (AAGUAAA) (24). Assuming a mode of RNA–protein interaction where one TPR/HAT repeat recognizes one base (53), it is expected that the binding site of Mbb1, which harbors 10 repeats (31), is longer than the S-box. This is compatible with our observation that the levels of psbB RNA are affected in three consecutive linker-scan mutants. While the S-box, defined as the sequence conserved in psbB and psbH, is at position −22 to −28, the loss of psbB mRNA is strongest in the mutant affecting −26 to −31, with a weaker effect in the mutant affecting −21 to −26. A longer target sequence of Mbb1 would also be compatible with the length of the sRNA that we identified (21 nt).
In those mutants that still accumulate measurable levels of psbB RNA, our primer extension analysis showed that the transcripts end at −35, even when the actual sequence around that site is altered (m32-37). Processing of the psbA and psbD RNAs is also independent of the sequence at the processing site (20,45). We have previously presented evidence that a 5′–3′ exonucleolytic activity efficiently degrades the longer −147 transcript both in the WT and in mbb1-222E (19). Thus, the most likely model for the role of Mbb1 in stabilization of psbB RNA is that it stably binds the RNA through the S-box and arrests the 5′–3′ processing activity at position −35, thus creating and protecting the mature 5′-end, possibly in association with other accessory factors. Evidence for a similar mechanism has previously been presented for petA and Mca1, psbD and Nac2, as well as for petD and Mcd1 in Chlamydomonas (20,21,54), and for several plant chloroplast transcripts, including psbH, which is protected by HCF107 at its 5′-end (24). Mbb1 and its plant ortholog HCF107, as well as Nac2 (which protects psbD in Chlamydomonas), belong to the TPR family of helical repeat proteins, and more specifically to its HAT variant (15). There is strong evidence that members of the PPR family act in a similar manner by stably binding to specific chloroplast transcripts and protecting them from exonucleolytic degradation (22,23,55). In some cases, it is remarkable that by binding in the intergenic region of a polycistronic transcription unit, the same PPR protein can protect both the upstream transcript from 3′ degradation and the downstream transcript from 5′ degradation (22,56).
Different functions of Mbb1 for its two target mRNAs
When the S-box was mutated in the psbH leader, accumulation of the respective transcripts was also affected: the mature forms of the mRNA were missing, and longer transcripts with a 5′ extension accumulated (Figure 4). This was not the case with the mutants of the psbB 5′ UTR for which we did not observe an over-accumulation of the longer precursor (Figure 3). Apparently, the role of Mbb1 in allowing the proper maturation of the psbH mRNA is somewhat different than with psbB. To explain the accumulation of the long forms of the transcripts in the S-box mutants of psbH, one possibility is that association of Mbb1 with the S-box contributes to the recruitment of an endonuclease. This could occur through protein–protein interaction, or by a change in the RNA structure on Mbb1 binding that would expose a susceptible site upstream of the Mbb1 binding site. Thus, Mbb1 would play a dual role in recruiting an endonuclease and in protecting the mature psbH RNA against exonucleolytic degradation. In higher plants, it was suggested that RNAse J could account for both endo- and 5′ exo-nuclease processing activities (22,57,58).
We observed in the psbB S-box mutants, where the amounts of psbB and psbB/T RNAs are strongly reduced, that the levels of the psbH RNAs are enhanced compared with the WT. Conversely, in the mutants of the S-box in psbH, the levels of psbB transcripts are elevated. A possible interpretation of these results could be that the psbB and psbH transcripts compete for a common factor, possibly Mbb1, that would enhance the stability of the RNAs and be present in limiting amounts. Experiments with transgenic tobacco chloroplasts have offered evidence for limiting trans-acting factors involved in RNA maturation. Over-expression of a chimeric marker with the 5′ UTR of clpP led to reduced processing (59). Likewise, ectopic over-expression of an RNA editing substrate resulted in reduced editing of the endogenous transcript (60,61). However, our attempts to introduce by chloroplast transformation an additional ectopic copy of the Mbb1 binding site did not result in a measurable decrease in the levels of psbB RNA.
In conclusion, we have identified a conserved cis-acting element that is essential for processing of the chloroplast transcripts psbB/T and psbH and is a target of the RNA-binding protein Mbb1. Chloroplast reverse genetics corroborated the biological significance of the sRNAs that were mapped to the 5′-end of these transcripts.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online including [19,30,48,49,62–69].
FUNDING
The Swiss National Fund for Scientific Research [31003A_133089 to M.G-C.], the European Framework Program 7 Knowledge-Based Bio-Economy project “SUNBIOPATH” [GA 245070 to M.G-C.], and the German Science foundation [SCHM-1698/2-1 to C.S-L.]; supported by the China Scholarschip Council (CSC) with a scholarship (to Y.Q.). Funding for open access charge: University of Geneva and Humboldt University of Berlin.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Uri Pick for providing the gene for HA-tagged sedoheptulose bisphosphatase, Dr Francis-André Wollman for stimulating discussions, Dr Jean-David Rochaix for his critical comments on the manuscript and Nicolas Roggli for preparing the figures.
REFERENCES
- 1.Herrin DL, Nickelsen J. Chloroplast RNA processing and stability. Photosynth. Res. 2004;82:301–314. doi: 10.1007/s11120-004-2741-8. [DOI] [PubMed] [Google Scholar]
- 2.Barkan A. Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 2011;155:1520–1532. doi: 10.1104/pp.110.171231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stern DB, Goldschmidt-Clermont M, Hanson MR. Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 2010;61:125–155. doi: 10.1146/annurev-arplant-042809-112242. [DOI] [PubMed] [Google Scholar]
- 4.Barkan A, Goldschmidt-Clermont M. Participation of nuclear genes in chloroplast gene expression. Biochimie. 2000;82:559–572. doi: 10.1016/s0300-9084(00)00602-7. [DOI] [PubMed] [Google Scholar]
- 5.Zerges W. Translation in chloroplasts. Biochimie. 2000;82:583–601. doi: 10.1016/s0300-9084(00)00603-9. [DOI] [PubMed] [Google Scholar]
- 6.Marin-Navarro J, Manuell A, Wu J, Mayfield S. Chloroplast translation regulation. Photosynth. Res. 2007;94:359–774. doi: 10.1007/s11120-007-9183-z. [DOI] [PubMed] [Google Scholar]
- 7.Lyska D, Meierhoff K, Westhoff P. How to build functional thylakoid membranes: from plastid transcription to protein complex assembly. Planta. 2013;237:413–428. doi: 10.1007/s00425-012-1752-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raynaud C, Loiselay C, Wostrikoff K, Kuras R, Girard-Bascou J, Wollman FA, Choquet Y. Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast. Proc. Natl Acad. Sci. USA. 2007;104:9093–9098. doi: 10.1073/pnas.0703162104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoppel R, Lezhneva L, Schwenkert S, Torabi S, Felder S, Meierhoff K, Westhoff P, Meurer J. Recruitment of a ribosomal release factor for light- and stress-dependent regulation of petB transcript stability in Arabidopsis chloroplasts. Plant Cell. 2011;23:2680–2695. doi: 10.1105/tpc.111.085324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maier UG, Bozarth A, Funk HT, Zauner S, Rensing SA, Schmitz-Linneweber C, Borner T, Tillich M. Complex chloroplast RNA metabolism: just debugging the genetic programme? BMC Biol. 2008;6:36. doi: 10.1186/1741-7007-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Johnson X, Wostrikoff K, Finazzi G, Kuras R, Schwarz C, Bujaldon S, Nickelsen J, Stern DB, Wollman FA, Vallon O. MRL1, a conserved Pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell. 2010;22:234–248. doi: 10.1105/tpc.109.066266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahire M, Laroche F, Cerutti L, Rochaix JD. Identification of an OPR protein involved in the translation initiation of the PsaB subunit of photosystem I. Plant J. 2012;72:652–661. doi: 10.1111/j.1365-313X.2012.05111.x. [DOI] [PubMed] [Google Scholar]
- 15.Preker PJ, Keller W. The HAT helix, a repetitive motif implicated in RNA processing. Trends Biochem Sci. 1998;23:15–16. doi: 10.1016/s0968-0004(97)01156-0. [DOI] [PubMed] [Google Scholar]
- 16.Shikanai T, Fujii S. Function of PPR proteins in plastid gene expression. RNA Biol. 2013 doi: 10.4161/rna.25207. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 18.Drager RG, Girard-Bascou J, Choquet Y, Kindle KL, Stern DB. In vivo evidence for 5′–>3′ exoribonuclease degradation of an unstable chloroplast mRNA. Plant J. 1998;13:85–96. doi: 10.1046/j.1365-313x.1998.00016.x. [DOI] [PubMed] [Google Scholar]
- 19.Vaistij FE, Goldschmidt-Clermont M, Wostrikoff K, Rochaix JD. Stability determinants in the chloroplast psbB/T/H mRNAs of Chlamydomonas reinhardtii. Plant J. 2000;21:469–482. doi: 10.1046/j.1365-313x.2000.00700.x. [DOI] [PubMed] [Google Scholar]
- 20.Nickelsen J, Fleischmann M, Boudreau E, Rahire M, Rochaix JD. Identification of cis-acting RNA leader elements required for chloroplast psbD gene expression in Chlamydomonas. Plant Cell. 1999;11:957–970. doi: 10.1105/tpc.11.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loiselay C, Gumpel NJ, Girard-Bascou J, Watson AT, Purton S, Wollman FA, Choquet Y. Molecular identification and function of cis- and trans-acting determinants for petA transcript stability in Chlamydomonas reinhardtii chloroplasts. Mol. Cell. Biol. 2008;28:5529–5542. doi: 10.1128/MCB.02056-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfalz J, Bayraktar OA, Prikryl J, Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009;28:2042–2052. doi: 10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prikryl J, Rojas M, Schuster G, Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl Acad. Sci. USA. 2011;108:415–420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammani K, Cook WB, Barkan A. RNA binding and RNA remodeling activities of the half-a-tetratricopeptide (HAT) protein HCF107 underlie its effects on gene expression. Proc. Natl Acad. Sci. USA. 2012;109:5651–5656. doi: 10.1073/pnas.1200318109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruwe H, Schmitz-Linneweber C. Short non-coding RNA fragments accumulating in chloroplasts: footprints of RNA binding proteins? Nucleic Acids Res. 2012;40:3106–3116. doi: 10.1093/nar/gkr1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhelyazkova P, Sharma CM, Forstner KU, Liere K, Vogel J, Borner T. The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell. 2012;24:123–136. doi: 10.1105/tpc.111.089441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haili N, Arnal N, Quadrado M, Amiar S, Tcherkez G, Dahan J, Briozzo P, Colas des Francs-Small C, Vrielynck N, Mireau H. The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Res. 2013;41:6650–6663. doi: 10.1093/nar/gkt337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Yu X, Wang H, Lu YZ, de Ruiter M, Prins M, He YK. A novel class of heat-responsive small RNAs derived from the chloroplast genome of Chinese cabbage (Brassica rapa) BMC Genomics. 2011;12:289. doi: 10.1186/1471-2164-12-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monod C, Goldschmidt-Clermont M, Rochaix JD. Accumulation of chloroplast psbB RNA requires a nuclear factor in Chlamydomonas reinhardtii. Mol. Gen. Genet. 1992;231:449–459. doi: 10.1007/BF00292715. [DOI] [PubMed] [Google Scholar]
- 30.Johnson CH, Schmidt GW. The psbB gene cluster of the Chlamydomonas reinhardtii chloroplast: sequence and transcriptional analyses of psbN and psbH. Plant Mol. Biol. 1993;22:645–658. doi: 10.1007/BF00047405. [DOI] [PubMed] [Google Scholar]
- 31.Vaistij FE, Boudreau E, Lemaire SD, Goldschmidt-Clermont M, Rochaix JD. Characterization of Mbb1, a nucleus-encoded tetratricopeptide-like repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA. 2000;97:14813–14818. doi: 10.1073/pnas.97.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felder S, Meierhoff K, Sane AP, Meurer J, Driemel C, Plucken H, Klaff P, Stein B, Bechtold N, Westhoff P. The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell. 2001;13:2127–2141. doi: 10.1105/TPC.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sane AP, Stein B, Westhoff P. The nuclear gene HCF107 encodes a membrane-associated R-TPR (RNA tetratricopeptide repeat)-containing protein involved in expression of the plastidial psbH gene in Arabidopsis. Plant J. 2005;42:720–730. doi: 10.1111/j.1365-313X.2005.02409.x. [DOI] [PubMed] [Google Scholar]
- 34.Howard MJ, Lim WH, Fierke CA, Koutmos M. Mitochondrial ribonuclease P structure provides insight into the evolution of catalytic strategies for precursor-tRNA 5′ processing. Proc. Natl Acad. Sci. USA. 2012;109:16149–16154. doi: 10.1073/pnas.1209062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris EH. The Chlamydomonas Sourcebook. San Diego, CA: Academic Press, Inc; 1989. [Google Scholar]
- 36.Perron K, Goldschmidt-Clermont M, Rochaix JD. A factor related to pseudouridine synthases is required for chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 1999;18:6481–6490. doi: 10.1093/emboj/18.22.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choquet Y, Goldschmidt-Clermont M, Girard-Bascou J, Kuck U, Bennoun P, Rochaix JD. Mutant phenotypes support a trans-splicing mechanism for the expression of the tripartite psaA gene in the C. reinhardtii chloroplast. Cell. 1988;52:903–913. doi: 10.1016/0092-8674(88)90432-1. [DOI] [PubMed] [Google Scholar]
- 38.Ausubel FA, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York, NY: John Wiley & sons; 1998. [Google Scholar]
- 39.Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc. Natl Acad. Sci. USA. 2010;107:3906–3911. doi: 10.1073/pnas.0912632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu C, Meyers BC, Green PJ. Construction of small RNA cDNA libraries for deep sequencing. Methods. 2007;43:110–117. doi: 10.1016/j.ymeth.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 41.Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- 42.Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 43.Goldschmidt-Clermont M. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker of site-directed transformation of chlamydomonas. Nucleic Acids Res. 1991;19:4083–4089. doi: 10.1093/nar/19.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fischer N, Stampacchia O, Redding K, Rochaix JD. Selectable marker recycling in the chloroplast. Mol. Gen. Genet. 1996;251:373–380. doi: 10.1007/BF02172529. [DOI] [PubMed] [Google Scholar]
- 45.Bruick RK, Mayfield SP. Processing of the psbA 5′ untranslated region in Chlamydomonas reinhardtii depends upon factors mediating ribosome association. J. Cell. Biol. 1998;143:1145–1153. doi: 10.1083/jcb.143.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eberhard S, Drapier D, Wollman FA. Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 2002;31:149–160. doi: 10.1046/j.1365-313x.2002.01340.x. [DOI] [PubMed] [Google Scholar]
- 47.Summer EJ, Schmid VH, Bruns BU, Schmidt GW. Requirement for the H phosphoprotein in photosystem II of Chlamydomonas reinhardtii. Plant Physiol. 1997;113:1359–1368. doi: 10.1104/pp.113.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ossenbuhl F, Hartmann K, Nickelsen J. A chloroplast RNA binding protein from stromal thylakoid membranes specifically binds to the 5′ untranslated region of the psbA mRNA. Eur. J. Biochem. 2002;269:3912–3919. doi: 10.1046/j.1432-1033.2002.03057.x. [DOI] [PubMed] [Google Scholar]
- 49.Nickelsen J, van Dillewijn J, Rahire M, Rochaix JD. Determinants for stability of the chloroplast psbD RNA are located within its short leader region in Chlamydomonas reinhardtii. EMBO J. 1994;13:3182–3191. doi: 10.1002/j.1460-2075.1994.tb06617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suay L, Salvador ML, Abesha E, Klein U. Specific roles of 5′ RNA secondary structures in stabilizing transcripts in chloroplasts. Nucleic Acids Res. 2005;33:4754–4761. doi: 10.1093/nar/gki760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Higgs DC, Shapiro RS, Kindle KL, Stern DB. Small cis-acting sequences that specify secondary structures in a chloroplast mRNA are essential for RNA stability and translation. Mol. Cell. Biol. 1999;19:8479–8491. doi: 10.1128/mcb.19.12.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eberhard S, Loiselay C, Drapier D, Bujaldon S, Girard-Bascou J, Kuras R, Choquet Y, Wollman FA. Dual functions of the nucleus-encoded factor TDA1 in trapping and translation activation of atpA transcripts in Chlamydomonas reinhardtii chloroplasts. Plant J. 2011;67:1055–1066. doi: 10.1111/j.1365-313X.2011.04657.x. [DOI] [PubMed] [Google Scholar]
- 53.Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, Small I. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8:e1002910. doi: 10.1371/journal.pgen.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drager RG, Higgs DC, Kindle KL, Stern DB. 5′ to 3′ exoribonucleolytic activity is a normal component of chloroplast mRNA decay pathways. Plant J. 1999;19:521–531. doi: 10.1046/j.1365-313x.1999.00546.x. [DOI] [PubMed] [Google Scholar]
- 55.Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell. 2003;15:1480–1495. doi: 10.1105/tpc.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barkan A, Walker M, Nolasco M, Johnson D. A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 1994;13:3170–3181. doi: 10.1002/j.1460-2075.1994.tb06616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hotto AM, Schmitz RJ, Fei Z, Ecker JR, Stern DB. Unexpected diversity of chloroplast noncoding RNAs as revealed by deep sequencing of the arabidopsis transcriptome. G3. 2011;1:559–570. doi: 10.1534/g3.111.000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luro S, Germain A, Sharwood RE, Stern DB. RNase J participates in a pentatricopeptide repeat protein-mediated 5′ end maturation of chloroplast mRNAs. Nucleic Acids Res. 2013;41:9141–9151. doi: 10.1093/nar/gkt640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuroda H, Maliga P. The plastid clpP1 protease gene is essential for plant development. Nature. 2003;425:86–89. doi: 10.1038/nature01909. [DOI] [PubMed] [Google Scholar]
- 60.Chateigner-Boutin AL, Hanson MR. Cross-competition in transgenic chloroplasts expressing single editing sites reveals shared cis elements. Mol. Cell. Biol. 2002;22:8448–8456. doi: 10.1128/MCB.22.24.8448-8456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaudhuri S, Carrer H, Maliga P. Site-specific factor involved in the editing of the psbL mRNA in tobacco plastids. EMBO J. 1995;14:2951–2957. doi: 10.1002/j.1460-2075.1995.tb07295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choquet Y, Vallon O. Synthesis, assembly and degradation of thylakoid membrane proteins. Biochimie. 2000;82:615–634. doi: 10.1016/s0300-9084(00)00609-x. [DOI] [PubMed] [Google Scholar]
- 63.Adam Z. Chloroplast proteases: Possible regulators of gene expression? Biochimie. 2000;82:647–654. doi: 10.1016/s0300-9084(00)00612-x. [DOI] [PubMed] [Google Scholar]
- 64.Baena-Gonzalez E, Aro EM. Biogenesis, assembly and turnover of photosystem II units. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2002;357:1451–1459. doi: 10.1098/rstb.2002.1141. discussion 1459–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Erickson JDR, Goldschmidt-Clermont M, Merchant S. 1998. Assembly of Photosystem II. In Rochaix, J.-D., Goldschmidt-Clermont, M., Merchant S. (eds), The Molecular Biology of Chloroplast and Mitochondria in Chlamydomonas. Kluwer Academic Publishers, Dordrecht, pp. 255–285. [Google Scholar]
- 66.Markham NR, Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sturm NR, Kuras R, Buschlen S, Sakamoto W, Kindle KL, Stern DB, Wollman FA. The petD gene is transcribed by functionally redundant promoters in Chlamydomonas reinhardtii chloroplasts. Mol. Cell. Biol. 1994;14:6171–6179. doi: 10.1128/mcb.14.9.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Silk GW, Wu M. Posttranscriptional accumulation of chloroplast tufA (elongation factor gene) mRNA during chloroplast development in Chlamydomonas reinhardtii. Plant. Mol. Biol. 1993;23:87–96. doi: 10.1007/BF00021422. [DOI] [PubMed] [Google Scholar]
- 69.Drapier D, Suzuki H, Levy H, Rimbault B, Kindle KL, Stern DB, Wollman FA. The chloroplast atpA gene cluster in Chlamydomonas reinhardtii. Functional analysis of a polycistronic transcription unit. Plant Physiol. 1998;117:629–641. doi: 10.1104/pp.117.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.