Abstract
To investigate the principles driving recognition between proteins and DNA, we analyzed more than thousand crystal structures of protein/DNA complexes. We classified protein and DNA conformations by structural alphabets, protein blocks [de Brevern, Etchebest and Hazout (2000) (Bayesian probabilistic approach for predicting backbone structures in terms of protein blocks. Prots. Struct. Funct. Genet., 41:271–287)] and dinucleotide conformers [Svozil, Kalina, Omelka and Schneider (2008) (DNA conformations and their sequence preferences. Nucleic Acids Res., 36:3690–3706)], respectively. Assembling the mutually interacting protein blocks and dinucleotide conformers into ‘interaction matrices’ revealed their correlations and conformer preferences at the interface relative to their occurrence outside the interface. The analyzed data demonstrated important differences between complexes of various types of proteins such as transcription factors and nucleases, distinct interaction patterns for the DNA minor groove relative to the major groove and phosphate and importance of water-mediated contacts. Water molecules mediate proportionally the largest number of contacts in the minor groove and form the largest proportion of contacts in complexes of transcription factors. The generally known induction of A-DNA forms by complexation was more accurately attributed to A-like and intermediate A/B conformers rare in naked DNA molecules.
INTRODUCTION
Interactions between proteins and DNA are essential for molecular processes of replication, transcription, gene regulation or chromosome packaging. Despite an extensive effort to understand the principles governing protein/DNA recognition, no simple and general rules have been found. The paradigm of molecular biology, DNA self-recognition via Watson–Crick base pairing, has probably no analogy in protein/DNA recognition. According to Matthews, there is no simple ‘code of recognition’ between amino acids and nucleotides (1), and the reason might be that the interaction between these two structurally complicated molecules has too many degrees of freedom (2).
Proteins recognize specific DNA sequences by two strategies commonly referred to as ‘direct’ and ‘indirect’ readout (3). However useful, this classification is artificial, and all protein/DNA high-affinity interactions depend on the conformational flexibility of the binding partners. Intrinsic conformational flexibility is more frequent in protein regions binding to DNA than in regions that do not bind to DNA (4). DNA is also known to conformationally adapt to its binding partner, e.g. by varying double helical groove widths, the helical twist, other base-pair parameters and the backbone conformations (3). The knowledge accumulated about modulations of DNA structure and electrostatics has complicated the idea of straightforward sequence-dependent readout by hydrogen-bonding patterns (5) and ultimately led to understanding that proteins recognize sequence-dependent flexibility or deformability rather than the sequence by direct readout (6). Such a complex nature of protein/DNA interactions requires elaborate functional and structural analysis of complexes (7) that has led to identification of specific rules of recognition for various families of protein/DNA complexes. An algorithm revealing likely sequences of potential transcription factors has been published soon after their first structures had been solved (8). Later, with many more experimental structures available, protein structural, physicochemical characteristics and thermodynamic properties have been examined to determine the rules of residue conservation in DNA-binding proteins (9,10); other studies analyzed the structural principles governing protein/DNA recognition (11) and classified protein motifs that bind to DNA (12). Rules determining recognition of DNA by some protein motifs, e.g. zinc fingers (13–15), or helix-turn-helix (16,17), have been discovered. These studies provide evidence that diverse structural descriptors have to be considered to describe origins of the binding specificity for different protein families.
Analysis of structural and physicochemical properties of the protein/DNA interface and of atom–atom interactions has demonstrated that amino acid and base compositions are correlated (18–20). The interface is formed mostly by positive and polar amino acids forming hydrogen bonds with bases and phosphates; the interface is more polar than basically lipophilic protein/protein interfaces (18,21); and contacts are often water-mediated. The importance of interactions between charged phosphate groups and charged or polar amino acid for the stability of complexes points to a key role of electrostatics in protein/DNA recognition, and modeling of electrostatic potentials has been used to predict DNA-binding sites (22–24). Another specific type of interaction, hydrogen bonding, has also attracted a considerable attention: networks of hydrogen bonds have been correlated to recognition of DNA by transcription factors (25) and direct amino acid—base contacts have been statistically analyzed (26). More specific types of interactions such as CH…O interactions (27) or pi/H-bond stacking motifs (28) have also been studied. Both proteins and DNA are heavily hydrated molecules, and an importance of water and of other solvent species for the binding has been recognized from early days of DNA structural research (29) and later recapitulated in several reviews (30–32).
The growing availability of structures of protein/DNA complexes has facilitated purely bioinformatics approaches to protein/DNA recognition. Many of these studies emphasize the active role of proteins in the recognition process, e.g. in graph representation of the interactions (33,34), or in structural classification of the interfaces from over a hundred protein/DNA structures (35). Structural alignment of interfacial protein and DNA residues has revealed surprising similarities between proteins of different folds (36). Similarly, surprising results have been obtained by using 11 structural descriptors that classify protein/DNA interfaces of 62 crystal complexes (37), concluding that DNA-binding proteins with the same binding motif (such as zinc-finger) may belong to different structural and functional classes. A recent work (4) has investigated local conformational changes at the interfaces of DNA-binding proteins classifying protein conformations by a protein structural alphabet but not distinguishing between different subfamilies of protein binding motifs and using subjective and coarse classification of DNA conformations.
In this work, we present a novel bioinformatics analysis of protein/DNA interactions. Both protein and DNA structures were classified using a well-established concept of structural alphabet (38–43). To characterize local conformations of proteins, we used the Protein Blocks (PBs) (44,45) that consist of 16 folding patterns of five consecutive amino acid residues; DNA local conformations were described at the dinucleotide (ntC) level (46). We then determined counts of mutually interacting PBs and ntCs, which form the protein/DNA interface, and compared their populations with numbers of non-interacting PBs and ntCs. The scope of over a thousand analyzed protein/DNA complexes and simultaneous objective classification of protein and DNA conformations offer a detailed insight into the protein/DNA interactions.
MATERIALS AND METHODS
Selection of protein/DNA structures
Protein/DNA complexes were retrieved from the Nucleic Acid Database (47) and the Protein Data Bank (PDB) (48). X-ray structures were selected containing protein and DNA longer than 6 nt, not RNA, and with crystallographic resolution better than 3.3 Å. The resolution limit of 3.3 Å was used to include as many functionally different complexes as possible. Short nucleotides were excluded for their low information content. The resulting 1475 structures are listed in Supplementary Table S1. Locally installed MolProbity suite (49,50) was used to add hydrogens, utilizing the option to flip oxygens and nitrogens in asparagine, glutamine and histidine residues.
Elimination of sequence identities and similarities
Sequence redundancy among 1475 structures was treated at two levels of stringency leading to two different datasets—Que and Umb. A list of selected structures is given in Supplementary Table S1.
Que–data set containing 339 complexes with sequentially unique proteins. Close evolutionary relationships among the protein sequences were avoided by removing structures with 50% or larger protein sequence identity. From two redundant structures, the one with higher crystallographic resolution was retained. If the resolution between two structures differed by <0.2 Å, structure with lower MolProbity score (49) was selected.
Umb–data set containing 1018 complexes with unique interfaces. This selection was based only on the identity of DNA sequences. Two complexes were considered unique when they differed at least by two (for strands shorter than 24 nt) or by three (for strands longer than 25 nt) nucleotides. The rationale for this less stringent selection based primarily on DNA sequences lies in the fact that we studied the structural features of the protein/DNA interfaces, not the protein or DNA behavior per se. A larger size of the Umb data set allowed an additional classification of structures by a protein functional class and by crystallographic resolution.
Protein classification
In addition to Que and Umb data sets, data sets containing proteins with more specific functions were analyzed. Structures were divided into broad categories consisting of enzymes (Enz), proteins regulating transcription (TrF) and structural proteins (Str). Structures containing enzymes were further classified as nucleases (Nuc) and polymerases (Pol). Other groups of structures such as DNA complexes with DNA repair proteins (Air), proteins operating on DNA topology (Top) and histone particles (His) were created, but they were not large enough to perform statistically reliable analysis. Functional classification of proteins was based primarily on the Pfam database (51); ∼15% of structures with missing Pfam annotations were classified manually based on the information in their original articles.
Because many structural features depend on the crystallographic resolution, the complexes were analyzed in three resolution bins: high-resolution structures up to 1.9 Å (labeled R1), middle-resolution structures between 1.9 and 2.8 Å (labeled R2) and low-resolution structures between 2.8 and 3.3 Å (labeled R3). Abbreviations and counts of structures in various functional groups and resolution bins are summarized in Table 1.
Table 1.
Number of analyzed structures
| Group of structures |
Crystallographic resolution |
||||
|---|---|---|---|---|---|
| Description | Code | R1: up to 1.90 Å | R2: 1.90–2.80 Å | R3: 2.80–3.30 Å | |
| All | Unique interface | Umb | 200 | 636 | 182 |
| Subsets of structures | Enzymes | Enz | 121 | 351 | 80 |
| Regulatory | TrF | 71 | 255 | 90 | |
| Structural | Str | 8 | 32 | 18 | |
| Nuclease | Nuc | 46 | 101 | 20 | |
| Polymerase | Pol | 32 | 133 | 22 | |
| Repair | Air | 28 | 82 | 20 | |
| Topology | Top | 3 | 31 | 22 | |
| Histone | His | 2 | 14 | 1 | |
| Sequentially unique | Que | 100 | 205 | 34 | |
Shown are the numbers of structures in the considered groups as a function of crystallographic resolution. Umb, ‘Unique interfaces’ represent the largest analyzed group, all others are just subsets.
Modified nucleotides and amino acids
Modified amino acid residues were not excluded from the analysis because they are rare, chemically homogeneous (mostly phosphorylated serines) and most of them occur outside the contact area with DNA. The identity of the modified amino acids was assigned to the parent natural amino acid.
On the other hand, chemically modified nucleotides occur more frequently and their modifications may be significant. Hence, we analyzed chemical structure of all modified nucleotide residues individually; of all 84 types of chemically modified nucleotides, 38 were judged chemically close to their parent residues and sterically not too different from the natural nucleotides, so they were included in the analyzed sample, and the other 46 were excluded. The list of all modified residues and PDB IDs of structures where they occur is given in the Supplementary Table S2.
Protein/DNA contacts
Nucleotide and amino acid residues in contact define the protein/DNA interface. We calculated direct and water-mediated protein/DNA contacts using in-house scripts using the Visual Molecular Dynamics (VMD) program (52). A nucleotide and amino acid residues were considered in a direct contact if any of their non-hydrogen atoms were closer than 3.40 Å. The direct contacts were classified as polar between polar atoms and as van der Waals between non-polar atoms. Water-mediated protein/DNA contacts were assigned to nucleotide and amino acid atoms that were connected by water oxygen no further than 3.40 Å. Direct and water-mediated contacts were assigned independently, i.e. an atom may be involved in both. All contacts were determined considering the crystallographic symmetry.
Classification of local conformations
Protein blocks
PBs are pentapeptide conformers defined by five pairs of the Φ, Ψ peptidic dihedral angles. The 16 local prototypes of the alphabets labeled from a to p were obtained by an unsupervised classification similar to Kohonen Maps and hidden Markov models of 342 non-homologous protein structures (44). This structural alphabet allows a reasonable approximation of local protein 3D structures with a root-mean-square deviation evaluated to be 0.42 Å, and is currently the most widely used structural alphabet (53). The PBs were assigned to all protein chains in the analyzed set of complexes according to the published procedure (54). A brief qualitative description of PB conformations and their occurrence at and outside the protein/DNA interface are given in Table 2.
Table 2.
PBs (44) assigned to proteins in the 1018 analyzed protein/DNA complexes with unique interface (Umb) and their occurrence
| PB label | Brief characterization | Occurrencea |
|
|---|---|---|---|
| At the interface | Outside the interface | ||
| a, b, c | N-terminus of β-strand | 4465 | 74 544 |
| d | Center of β-strand | 5163 | 78 833 |
| e, f | C-terminus of β-strand | 3097 | 38 039 |
| g, h, i, j | Coil, various forms | 2241 | 22 072 |
| k, l | N-terminus of α-helix | 5884 | 50 877 |
| m | Center of α-helix | 7978 | 174 348 |
| n, o, p | C-terminus of α-helix | 1561 | 40 357 |
aNumber of PBs identified at and outside the protein/DNA interface in 1018 analyzed structures.
Assignment of DNA conformer classes (ntC)
A DNA structural alphabet characterizing local conformations of ntC units was developed by Svozil et al. (46). In the present work, we critically consolidated a larger set of originally published conformers into a group of 18 letters. Three Z-DNA conformers were assigned but not further analyzed, and an additional ntC (referred to as ‘NN’) was designated to conformations that could not be assigned to any of the existing classes. NtCs were assigned to DNA steps using a modified version of a k-nearest neighbor algorithm (55). The ntC classes are briefly characterized in Table 3 and their backbone torsions are summarized in the Supplementary Table S3. After the assignment, three conformers with χ angle in the syn region (χ < 180°), ntCs 119, 121 and 122, were pooled into one ntC labeled 155. Together with structurally diverse ntC class NN, we analyzed 14 DNA conformational classes.
Table 3.
Nucleotide conformers (ntC) used for the conformational assignment (55) of 57 797 DNA steps in the 1018 analyzed protein/DNA complexes (the Umb data set)
| ntCa | Symbolb | Characterization | Occurrencec |
|
|---|---|---|---|---|
| At the interface | Outside the interface | |||
| 8 | A | The most frequent A-DNA | 1242 | 354 |
| 13 | A | A-DNA, BI-like χ | 727 | 202 |
| 19 | A | A-DNA, α+1/γ+1 crank | 573 | 205 |
| 41 | A2B | A-to-B, δ > C3′−, δ+1 C2′-endo | 2014 | 724 |
| 32 | BI2A | BI-to-A, δ+1 O4′-endo | 1574 | 909 |
| 109 | BII2A | BII-to-A, δ+1 > C3′-endo | 333 | 106 |
| 110 | BII2A | as 109 plus α+1/γ+1 crank, high β+1 | 457 | 267 |
| 54 | BI | The most frequent BI variant | 9261 | 7529 |
| 50 | BI | BI variant | 3677 | 2073 |
| 86 | BII | the most frequent BII variant | 2805 | 2820 |
| 96 | BII | BII variant | 1620 | 1133 |
| 116 | BI | BI, α+1/γ+1 crank, α/γ normal | 2431 | 1935 |
| 155 | BIsyn | orig. 119: 5′-mismatches, BI, χ syn, α/γ crank | 254 | 188 |
| 155 | BIsyn | orig. 121: 3′-mismatches, δ O4′-endo, χ+1 syn | ||
| 155 | BIsyn | orig. 122: as 121 plus α+1/γ+1 crank | ||
| NN | Unassigned conformers | 3421 | 2854 | |
aNumerical label of the nucleotide conformers as in (46). Torsion angle values of all ntCs are given in Supplementary Table S3.
bSymbol of a conformation family.
cNumber of ntCs identified at and outside the protein/DNA interface in the Umb data set.
Statistical analysis of structural features of the interface
Statistical analyses were performed to compare the distributions of the following descriptors at and outside the protein/DNA interface: amino acid and nucleotide residues, PBs and ntCs and protein secondary structure elements. The differences between the descriptors involved in the interaction and not involved in the interaction were measured by the logodds ratios, P(i, j), that represented the propensity of descriptor’s elements i and j to interact. Values of P(i, j) were calculated using the following formula:
where fc(i,j) was the observed number of pairs i, j in contact between i (DNA descriptor) and j (protein descriptor); fe(i, j) was the expected number of interacting pairs of i, j between protein and DNA if there were no contacts between them. The expected number was calculated from the following formula:
where fnc(i) was the frequency of the descriptors of type i not in contact. The fnc(i) was calculated as N(i)nc/Nnc and fnc(j) as N(j)nc/Nnc, where N(i)nc was number of non-interacting descriptor i and Nnc was the total number of non-interacting descriptors.
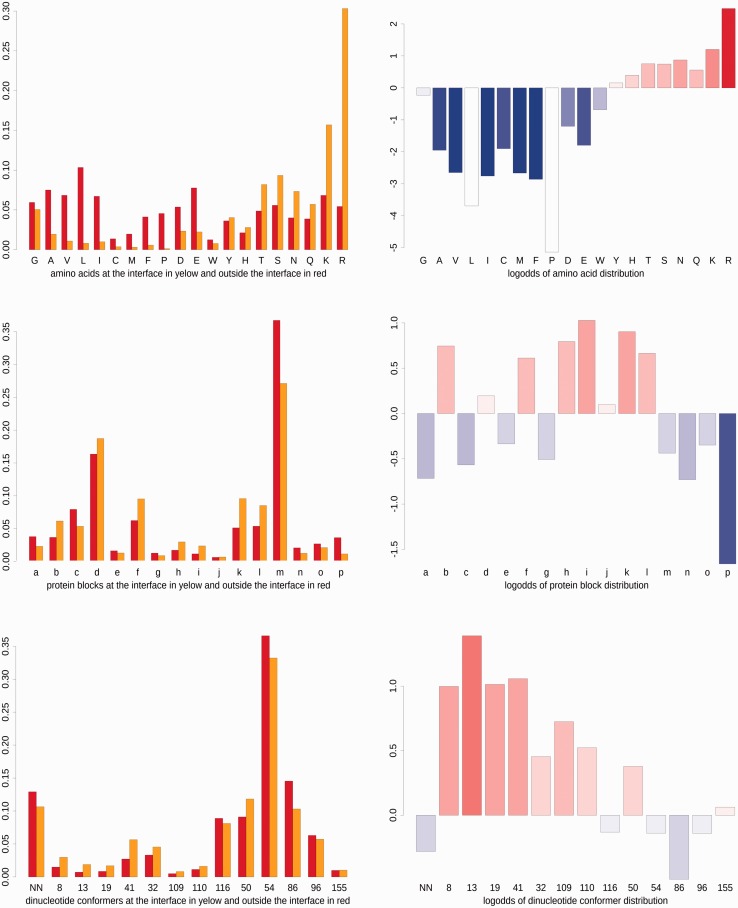
For example, the data set Umb-R2 contains 4082 PBs m in contact with DNA and 15 550 of all PBs in contact with DNA, so that f(m)c = 4082/15550 = 0.26251. The number of PB m not in contact with DNA is 83 694 and there are 225 348 of all PBs, f(m)e = 83694/225348 = 0.37140. Logodd value of PB m in Umb-R2 is then P(m) = log2(0.26251/0.37140) = −0.50060, the value plotted in the right side histogram of Figure 1.
Figure 1.
Occurrence of protein and DNA structural descriptors at and outside the protein/DNA interface for the group of structures Umb-R2 (636 complexes with crystallographic resolution between 1.90 and 2.80 Å). Histograms show distributions of amino acid residues (top), PBs (center) and ntCs (bottom) involved in direct polar contacts. Histograms on the left show the relative frequencies at the interface (in yellow) and outside the interface (in red). Histograms on the right show logodds of these frequencies, with underpopulation indicated by blue and overpopulation by red; hue indicates the significance of the effect. PBs are labeled by their one-letter codes (Table 2) and ntC by their numbers as defined in Table 3. Histograms for other groups of complexes are given in Supplementary Figure S1.
RESULTS AND DISCUSSION
In this section, we compare statistics for direct polar and water-mediated contacts between proteins and DNA, and briefly describe differences between contacts to the DNA minor and major grooves, and phosphate atoms. Finally, we compare general features of the protein/DNA interface and in two particular groups of structures: transcription factors (TrF) and polymerases (Pol). The structures are divided into three groups based on their crystallographic resolution; the middle-resolution bin R2 comprising structures between 1.90 and 2.80 Å contains most structures (Table 1), so we primarily concentrate on the analysis of this bin.
Statistics of contacts for selected classes of structures
Table 4 shows selected statistics of direct polar contacts for selected groups of structures in the three resolution bins; a more detailed account of various statistical measures of the interactions can be found in Supplementary Table S4. In the high-resolution bin R1, only enzyme complexes are numerous enough to be analyzed as a separate subgroup. On the other hand, in the medium-resolution bin R2, we could also analyze transcription factors, nucleases and polymerases (TrF, Nuc and Pol) individually.
Table 4.
Protein/DNA contacts
| Structuresa |
Residues in polar contactsb |
Atom-to-atom polar contacts per residuec |
HOH/polard |
||||
|---|---|---|---|---|---|---|---|
| Code | Number | aa | nt | aa | nt | aa | nt |
| Umb-R1 | 200 | 3764 | 2445 | 1.33 | 1.81 | 1.31 | 1.13 |
| Enz-R1 | 121 | 2399 | 1491 | 1.29 | 1.81 | 1.17 | 1.04 |
| TrF-R1 | 71 | 1238 | 866 | 1.38 | 1.80 | 1.54 | 1.25 |
| Pol-R1 | 32 | 562 | 378 | 1.24 | 1.42 | 0.90 | 1.05 |
| Nuc-R1 | 46 | 1166 | 678 | 1.33 | 2.09 | 1.10 | 0.95 |
| Que-R2 | 205 | 3707 | 2803 | 1.32 | 1.69 | 0.76 | 0.66 |
| Umb-R2 | 636 | 14 869 | 10 039 | 1.35 | 1.71 | 0.78 | 0.70 |
| Enz-R2 | 351 | 8342 | 5312 | 1.33 | 1.73 | 0.74 | 0.70 |
| TrF-R2 | 255 | 5594 | 4056 | 1.35 | 1.68 | 0.90 | 0.74 |
| Str-R2 | 32 | 975 | 699 | 1.45 | 1.65 | 0.48 | 0.47 |
| Nuc-R2 | 101 | 2746 | 1726 | 1.34 | 1.91 | 0.98 | 0.81 |
| Pol-R2 | 133 | 2843 | 1902 | 1.35 | 1.53 | 0.66 | 0.69 |
| Umb-R3 | 182 | 4156 | 2997 | 1.32 | 1.63 | ||
aStatistics for selected groups of structures, for abbreviations see Table 1.
bThe columns list the total number of amino acids (aa) and nucleotides (nt) in direct polar contacts in selected groups of structures.
cThe columns show how many protein (‘aa’) or DNA (‘nt’) atoms forming direct polar contacts interact per residue.
d'‘HOH/polar’ show the number of water-mediated contacts divided by the number of direct polar contacts for protein (‘aa’) or DNA (‘nt’) atoms.
Table 4 shows that polar contacts are, on average, mediated by 1.3 atoms in amino acid residues, and by 1.7 atoms in nucleotides. For amino acids, these numbers are remarkably similar within all groups of structures, and slightly more variable for nucleotides. Water-mediated contacts are as common as direct polar contacts as demonstrated by numbers under the ‘HOH/polar’ column in Table 4, and their role is discussed in greater detail in ‘The role of water-mediated contacts’.
To test the robustness of the observed features of the large Umb group (group with sequentially unique interfaces), we compared them with the features of the Que group (sequentially unique proteins). Descriptors given in Table 4 show virtually identical values for Que-R2 and Umb-R2 data sets, and other descriptors analyzed in this work also demonstrate similar-to-identical characteristics of these two groups in all resolution bins (see also Supplementary Table S4).
Protein structure elements
Neither type of interactions (direct polar, water-mediated, van der Waals) nor resolution changes the general pattern of protein binding characteristics. As expected (18,19,26), most contacts to DNA are formed by arginine and lysine followed by other polar and/or charged amino acids (Figure 1, Supplementary Figure S1). Positively charged arginine is overpopulated at the negatively charged DNA surface regardless of the structural type or resolution, and lysine is overpopulated in most groups. Lipophilic amino acids, namely, leucine, valine, isoleucine, methionine and phenylalanine, have low occurrence at the polar interface and are statistically underrepresented. Strong underrepresentation of proline at the interface likely originates in its structural rather than lipophilic properties. In contrast to large differences in the presence of individual amino acids at and outside the interface, protein secondary structural elements do not show any preferences for the interface (not shown). In other words, no secondary structural element can be identified as a key building block for DNA recognition.
As Figure 1 shows, PBs have a larger discriminatory power in identifying structural elements recognizing DNA than secondary structure elements. PBs overpopulated at the interface are N-termini of α-helix and β-sheet (PBs k, l, b) and coil blocks (PBs h, j), and PBs underpopulated are central and especially C-terminal parts of α-helix (PBs p and n). We observed no real differences in the occurrence of these PBs between direct polar and water-mediated interactions.
Description of the protein local structure by PBs allowed observing differences between the general protein structure and structural features observed at the interface with DNA. Coil-related PB g, the second least frequent PB (56) associated with flexible regions, is even less present at the interface. Underrepresentation was also observed for some frequent sequences of PBs classified by de Brevern (57) as ‘Structural Words’, e.g. mnopac.
DNA structure elements
The dominant DNA form, BI-DNA, is represented here by ntCs 54 and 50. It is the most common form at the protein/DNA interface in all groups of structures. What distinguishes interacting DNA from unbound DNA is a larger relative occurrence of the A-forms in protein/DNA complexes (25,58–60). We observed an increased occurrence of the ‘canonical’ A-form (ntC 8), but owing to our finer classification of DNA conformers, also of deformed A-like and especially of mixed A/B conformers. The population of ntC 13 is notably increased. The occurrences ntCs 41 and 19 are also increased. NtC 41 with the A-like backbone but B-like values of the glycosidic torsion angle χ preserves perpendicular orientation of the base pairs relative to the helical axis; ntC 19 is an A-form with α and γ torsions switched from the 300°/60° canonical values to the 150°/180° combination (‘crankshaft’ motion). Although the most common BII-form (ntC 86) is disfavored at the interface, other BII conformers rare in naked DNA (ntCs 109 and 110) are well represented in protein/DNA complexes.
Unclassified nucleotides (ntC NN) representing extreme structural variations are not significantly enriched at the interface. Apparently, the interaction of proteins with DNA does not induce any novel DNA local conformers, but it stabilizes A (ntC 13) and A/B forms (ntCs 41, 32, 109, 110) that appear more often at the interface than in uncomplexed DNA. Some of these conformers (namely ntC 32) exhibit values of torsion δ, which defines sugar pucker, between 90° and 100° indicating high C3′-endo or even O4′-endo pucker. Large number of these conformers at the interface (especially in high-resolution structures) refutes doubts about the existence of the O4′-endo sugar pucker in DNA and demonstrates a smooth deformation of the deoxyribose ring from the C3′-endo to C2′-endo pucker via the O4′-endo observed in high-resolution small nucleoside and nucleotide structures (61,62). In this context, virtual absence of the O4′-endo pucker in RNA structures (63) may be more a consequence of the force fields used to refine RNA structures than reflection of the actual distribution of sugar puckers.
Binding statistics in the group of low-resolution structures
Distributions of direct polar and van der Waals contacts for structures at the lowest resolution bin R3 (2.80–3.30 Å) show the same general features as distributions of structures at the higher resolution bins (Table 4 and Supplementary Table S4). What discriminates low-resolution structures is a larger number of unclassified ntC NN that may be attributed to refinement difficulties with poorly resolved electron density maps and incorrectly fitted nucleotide conformations. Unexpected is a high frequency of ntC 116, rare BI-form with alpha/gamma crankshaft compensation. The low number of observed water molecules in low-resolution structures does not allow analysis of water-mediated contacts.
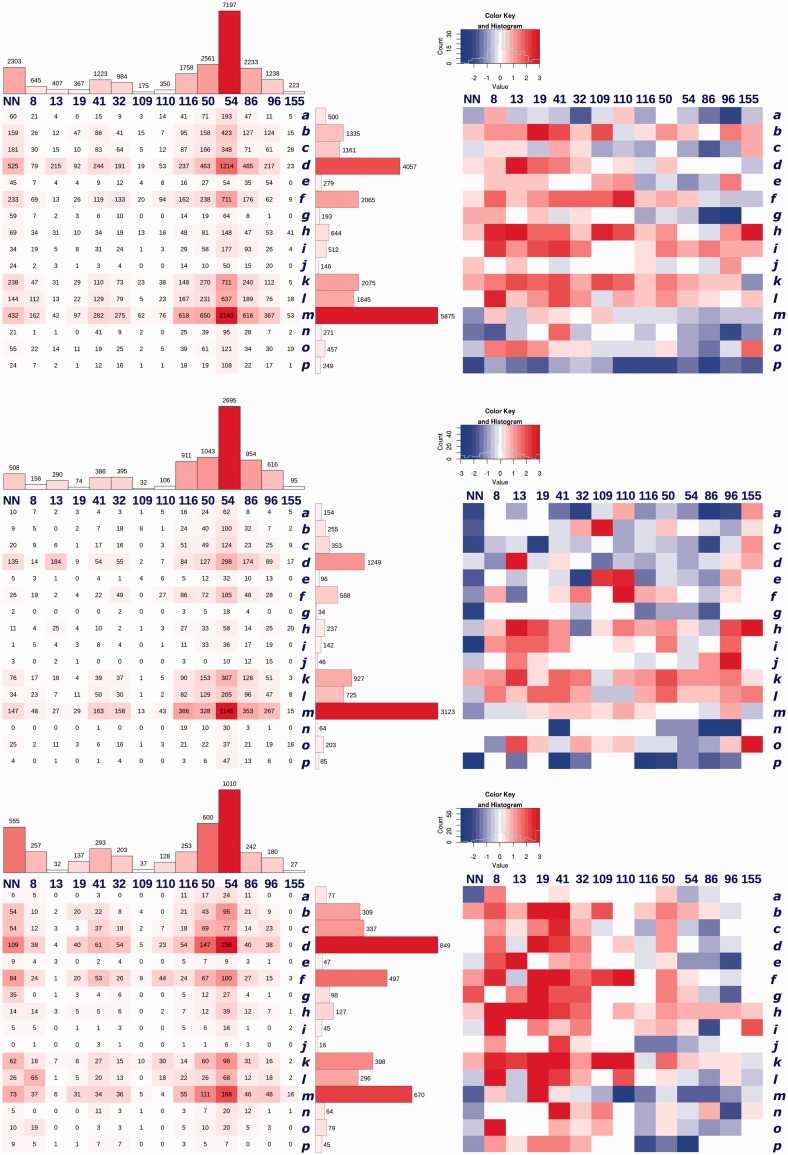
Interaction matrices: correlations between interacting PBs and ntCs
The counts of mutually interacting PBs and ntCs are presented in a form of ‘interaction matrices’ that show how many protein and nucleotide conformers of certain type interact and reflect therefore the local geometry of the interface. Figure 2 shows interaction matrices for direct polar contacts in the medium-resolution group of structures Umb-R2, and its subgroups TrF-R2 and Nuc-R2. Interaction matrices for direct polar (Figure 2), water-mediated (Supplementary Figure S2a) and van der Waals (not shown) contacts are similar. Moreover, most observations made for the medium resolution structures are also valid for the high-resolution data set Umb-R1 (Supplementary Figure S2b).
Figure 2.
Interaction matrices for direct polar contacts of the three groups of structures with crystallographic resolution between 1.90 and 2.80 Å (bin R2). Top: 636 protein/DNA complexes, Umb-R2. Center: 255 complexes of transcription factors, TrF-R2. Bottom: 101 complexes of nucleases, Nuc-R2. The matrices on the left show how many peptide blocks, PBs, interact with nucleotide conformers, ntCs, the highest populations are highlighted in red. The matrices on the right show how much are the interactions statistically different from their expected frequencies estimated by the logodd analysis. Higher-than-expected populations (overrepresentation) are indicated by red, underrepresentation by blue, hue indicates intensity of the deviation from the neutral distribution. PBs are plotted vertically, ntCs horizontally, their symbols and characterization are given in Tables 2 and 3. Supplementary Figure S2 shows more interaction matrices, always for groups of structures Umb, TrF and Nuc: for water-mediated contacts and for direct polar contacts in the minor groove, major groove and phosphates in the medium resolution bin R2 and also for direct polar contacts in the high-resolution bin R1.
The most frequent interactions occur between the main architectural units of proteins and DNA, DNA BI form ntC 54 and protein α-helical PB m and β-strand PB d, which form 15 and 12% of all contacts, respectively. However, according to the logodds analysis, neither m54 nor d54 combination prefers or avoids the interface. Combinations of conformers that characterize the interface (occur at the interface with higher than expected frequency and are therefore ‘statistically overrepresented’) are A and mixed A/B DNA forms (mainly ntCs 8, 13, 19) associated with β-sheet (PBs b, d) and coil (PBs h, i, j). Strongly overrepresented are also interactions between less populated B-to-A ntCs 109 and 110 and PBs e (C-terminus of β-strand), h (coil) and k (N-terminus of α-helix). In contrast, conformers that avoid the interface are BII forms (ntCs 86, 96) and the C-terminal segments of the α-helix (PBs n, o and especially p). The most negatively correlated associations are BII forms with the coil PB g and the N-terminal β-sheet PB a. The described pattern is similar for medium- as well as high-resolution structures and for direct polar and water-mediated contacts.
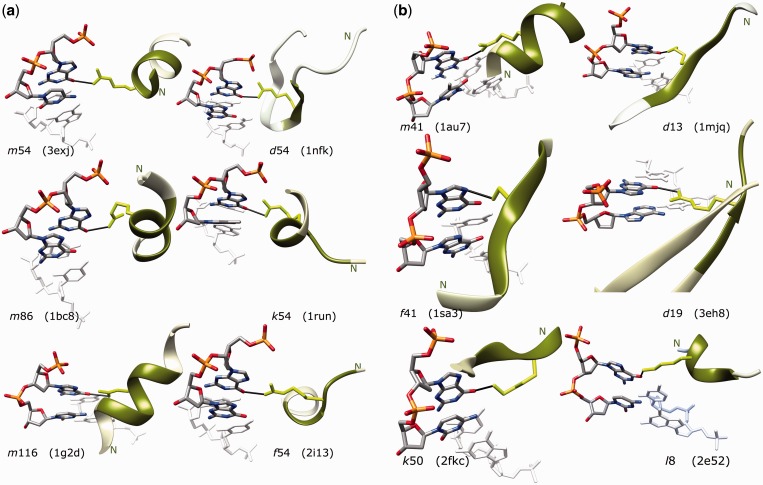
Figure 3a depicts examples of the most frequent PB/ntC interaction partners. The dominant BI form (ntC 54) participates frequently in contacts with α-helical (m54, k54) as well as β-sheet (d54, f54) PBs. The BII ntC 86 is common at the interface (even when statistically underrepresented) and its contacts with the main α-helical PB m are frequent (motif m86 in Figure 3a). A comparison of the three binding motifs between the α-helical PB m and three B-DNA conformers, 54, 86 and 116 (less-populated BI conformer), shows variability of the mutual orientation between the B-DNA major groove and α-helix. Arginine contacting the major groove guanine O6 is, in most cases, in its extended rotamer, but it can also accommodate more compact rotameric forms as in motifs m86 and k54.
Figure 3.
Examples of the common protein/DNA interactions. Interacting motifs are labeled by the codes of the interacting PB and ntC (Tables 2 and 3, respectively) and by PDB id of structures in which they were identified. Interacting PBs are drawn as green cartoon with atoms of the central amino acid in light green and the nucleotide step as a stick model using commonly used ‘chemical’ colors; the contacts (black sticks) are directed to the major groove edge of guanines in the right-handed double helical DNA. The 5'-end phosphates are on the left top of each motif. The N-ends of the PBs are labeled; the complementary DNA strand and amino acids adjacent to the depicted PB are in light gray. Molecular graphics was created by program Chimera (64). (a) Motifs common to all types of structures approximately in order of their occurrence in the group of all 1018 structures. All contacts shown are between the guanine atom O6 and the arginine NH observed in crystal structures 3exj (65), 1nfk (66), 1bc8 (67), 1run (68), 1g2d (69) and 2i13 (70). (b) Motifs m41 and d13 are highly populated in transcription factors (TrF-R2) and underrepresented in nucleases (Nuc-R2), motifs f41, d19, k50 and l8 are highly populated in Nuc-R2 and less in TrF-R2. They appear in crystal structures 1au7 (71), 1mjq (72), 1sa3 (73), 3eh8 (74), 2fkc (75) and 2e52 (76). The motifs m41, d13 and d19 show interaction between the guanine O6 and arginine NH, k50 and l8 between the guanine O6 and lysine NZ, and f41 between the guanine N7 and serine OG.
While motifs drawn in Figure 3a are common in all types of complexes, Figure 3b depicts motifs typical for complexes of transcription factors TrF-R2 (m41 and d13), and for nucleases Nuc-R2 (f41, d19, k50 and l8). Complexes of transcription factors have interaction matrices similar to the matrices of the whole data set Umb-R2 with dominating BI-DNA and α-helical conformers. In contrast, complexes of nucleases (Nuc-R2) use a wider spectrum of conformers at the interface, dominance of BI ntC 54 is visibly weaker and more contacts are actually formed by β-strand PB d than by otherwise more populated α-helical PB m; many contacts are also formed by β-strand PB f. Preference for the A-like forms measured by logodds is much stronger than in Umb or TrF data sets, especially in combinations with β-strand f, coil h and N-terminal α-helical PBs k and l. The population of undefined nucleotides NN is surprisingly high. The BII forms are infrequent and statistically disfavored. Conformational diversity of DNA/nuclease interactions is underscored by their larger chemical variability when fewer contacts are formed by arginine; we show interacting lysine side chains (k50, l8) and also a serine motif f41.
Contacts to the DNA minor groove, major groove and phosphate
Protein interactions to DNA constituents, the minor groove (mig), the major groove (MAG), the phosphate (PH) and deoxyribose, are distributed unevenly. The phosphate atoms OP1 and OP2 form a large part of all polar contacts to protein atoms, more than a half. On the other side, deoxyribose atoms O4′, O5′ and O3′ together form ∼5% of the contacts and are not important for protein binding. The proportion for direct polar contacts is mig:MAG:PH = 1:2:9 in the Umb-R1 data set, and comparable 1:3:15 in Umb-R2 (data for other datasets are given in Supplementary Table S5). Water-mediated contacts are distributed more evenly, and the corresponding ratios for water-mediated contacts are 1:2:6 and 1:2:7, respectively. Lower relative number of water-mediated contacts at phosphates shows that water molecules are better localized in the grooves than around more accessible phosphates.
Interaction matrices of the minor groove contacts have distinct patterns, and also other statistics of contacts to mig differ from matrices constructed for MAG and PH (Supplementary Figure S2c versus S2d and S2e). The interaction matrices are formed by more β-sheet than α-helix contacts and also BI dominance is much lower than for contacts to MAG or PH. The second most populated nucleotide conformer is ntC NN that strongly correlates with β-sheet PB d; we do not have explanation for this observation. The differences observed between interaction matrices of TrF and Nuc for all contacts are more pronounced in mig; despite the lower counts in the mig matrices, it seems clear that these interactions disfavor the BI-form, may induce unusual DNA conformers (ntC NN) and generally prefer β-sheet over α-helix.
Water-mediated contacts to the minor groove show fewer of these extreme features, and their interaction matrices resemble the interaction matrices of major groove and phosphates. A notable overall feature of the minor groove atoms is that they actually form more water-mediated than direct polar contacts, 1.5 times more in the medium-resolution structures (Umb-R2), the corresponding ratios are 1.1 in MAG, and 0.7 in PH. High-resolution structures (Umb-R1) show the same trend. Interaction of the narrow mig with proteins, therefore, requires either its substantial deformations or alleviation of the steric constraints by water-mediated contact.
Distribution of protein contacts to the grooves and phosphates is in some groups of structures different from the average values given above. Extreme behavior was observed for transcription factors (TrF) that have direct polar and water-mediated contacts distributed similarly between mig, MAG and PH, and for polymerases (Pol) with different distributions (ratios are listed in Supplementary Table S5). Because polymerases distribute fewer water contacts per residue than transcription factors (0.66 versus 0.90, Table 4), their interface is ‘more’ dehydrated than the interface of transcription factors, and this dehydration of polymerases is most pronounced for phosphate atoms.
The role of water-mediated contacts
The number of residues linked by direct polar contacts and by water bridges is comparable even for the medium-resolution structures (Umb-R2) where 20 000 amino acids contact DNA directly and 16 000 via water. The last two columns of Table 4 (‘HOH/polar’) show that the number of water-mediated contacts divided by the number of direct polar contacts varies between various groups of structures. The highest proportion of water-mediated contacts was observed for complexes of transcription factors and nucleases, the lowest for polymerases (extremely low value for Str-R2 may be skewed by histone complexes). High proportion of water-mediated contacts in transcription factors in both relevant resolution bins, TrF-R1 and TrF-R2, is perhaps surprising, especially in the light of the fact that polymerases with arguably less stringent demand for specificity of interaction have their proportion of water contacts lower.
High proportion of water-mediated contacts in all complexes, and especially in complexes of transcription factors, suggests that these structured water molecules play an active role in the process of protein/DNA recognition and do not serve as mere fillers of cavities formed at imperfectly matching protein and DNA molecular surfaces as has been sometimes suggested (77). Similarity of the PB/ntC interaction matrices for direct polar and water-mediated contacts (Figure 2 and Supplementary Figure S2a) further demonstrates that interaction by direct polar contacts and via the interface waters has similar conformational constraints on both protein and DNA partners and indirectly points again to the active role of water to the recognition.
On complexation, heavily hydrated surfaces of protein and DNA molecules release a large number of water molecules and ions increasing entropy of the interaction and thus compensating for the entropy loss caused by the complex formation (32,78–80). Around the naked DNA double helices, water and cations lie in spatially localized hydration sites (81–83) that coincide largely with protein interaction sites (84). The waters trapped at the interface represent the remains of the first-shell waters and cations that have specific physical properties (79,85–87), and become an ‘integral part’ (29) of the protein/DNA interface (30). The packing of atoms at protein–DNA interfaces is as high as in the protein interior, and cavities at the interface are filled with water more frequently than the protein interior (88). Therefore, it is plausible to state that water contributes significantly to the protein/DNA recognition (84,89) and participates in protein/DNA interactions (90,91).
Stabilization of the A-forms at the interface
High relative occurrence of A- and A/B DNA forms at the protein/DNA interface observed in the interaction matrices can be interpreted as remodeling of the B-form to the A-form. Almost continuous plastic deformation from B-to-A state through several minor conformational states (46) is accompanied by bending of the duplex that modifies the widths of the major and minor grooves and changes the exposition of the base pairs, deoxyribose and mainly phosphate atoms (59). The narrowing of the major groove of the protein-induced A and A/B conformers could provide one mechanism for forming specific contacts to a protein-binding motif preserving the essential stacking interactions of the base pairs (18). In some complexes, binding requires a high degree of DNA distortion (92,93), and a shift in the distribution of conformers from naked to complexed DNA suggests that conformational deformability and flexibility of DNA are essential for the recognition (94–96). The tendency to induce A-like conformers at the interface is accompanied by a shift from the C2′-endo sugar pucker typical for B-forms toward the C3′-endo pucker family, the effect described as the ‘sugar switching’ that facilitates hydrophobic recognition in the minor groove (97,98).
The driving force of the A-to-B transformation in naked DNA, partial dehydration of the DNA surface, is well known (99) (100) so that partial dehydration of DNA on complexation with proteins works in accord with the aforementioned steric reasons, and may contribute to the relative preference of the A- over the B-forms at the interface. The fact that the A-like structures are similarly overrepresented at the interface for direct polar and water-mediated contacts does not directly confirm or exclude such possibility, and in our opinion, the A and A/B conformers are induced in the protein/DNA complexes likely by a combination of two factors, the partial dehydration required by the complexation and the ability of DNA to adjust its conformation to protein (58,59) and in a broader sense, to reflect the environment (101,102).
CONCLUSIONS
We analyzed structural features of the protein/DNA interface and compared them with the features of non-interacting parts of proteins and DNA. Structures of proteins and DNA were classified by structural alphabets. Protein local conformers were classified into 16 pentapeptide PBs (44,53), and DNA into 14 ntCs (46,55). These structural alphabets describe biopolymer conformations at greater detail than elements of protein secondary structure and than crude and sometimes subjective DNA structural types such as A, BI and BII. Direct polar and water-mediated protein–DNA contacts were analyzed in >1000 protein/DNA crystal structures in three bins of crystallographic resolution. The counts of mutually interacting PBs and ntCs were assembled into ‘interaction matrices’ that serve as comprehensive description of structural features of the interface. The matrices demonstrate that minor DNA conformers are often significantly enriched at the interface so that the ability of DNA to adopt non-canonical conformers rare in naked DNA is clearly essential for the recognition by proteins. Rare DNA forms introduce significant deformations to the DNA regular structure and the occurrence of these rare forms was characterized here enabling better understanding of the role of non-B-DNA structures for genetic instability and evolution (103).
The well-known tendency of DNA to adopt A-like forms on protein binding (58,59) should be understood as a relative preference because the BI forms are the most frequent even at the interface (Figures 1 and 2). Our detailed structural classification of DNA conformers allowed a specific characterization of A-like forms enriched at the interface. We showed that the interaction with proteins induces more gradual deformations of the B form into B-A, A-B and exotic A conformations rather than solely into the canonical A-DNA. Importantly, unclassified conformers (ntC NN) representing rare or incorrectly refined conformers are not overpopulated at the interface so that interactions with proteins do not induce conformations unseen in naked DNA but only stabilize the less stable forms. The relative stabilization of the A-like forms at the interface is likely facilitated by synergy of the steric accommodation to the interacting protein and dehydration occurring during the interaction that also stabilizes the A-form.
The interaction matrices of direct polar and water-mediated contacts are remarkably similar, and water-mediated contacts are nearly as numerous as direct polar ones. Water molecules trapped at the interface are important for the binding by alleviating steric incompatibility between protein and DNA so that the interacting peptide and nucleotide fragments can remain in their energetically low-lying conformations. An important role of water molecules for the recognition is further underscored by their high occurrence at the interfaces with transcription factors (Table 4, column HOH/polar).
Both features characterizing protein/DNA binding, i.e. reduction of the mutual steric incompatibility by water bridges and induction of the B-to-A transition, are best visible in interaction matrices constructed for contacts to the narrow minor groove. They are conspicuously different from the matrices constructed for contacts to the major groove and phosphate atoms. Remarkably, water-mediated interactions form more than a half of all the contacts in the minor groove, while the proportion of ordered waters around the major groove and especially phosphate atoms is lower.
Interaction matrices counting contacts between protein and DNA residues classified into structural alphabets represent robust and comprehensive description of the interface and contribute to the understanding of principles underlying protein/DNA recognition.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Czech-France collaboration Barrande [MEB021032]; BIOCEV CZ.1.05/1.1.00/02.0109 from the ERDF, [P305/ 80 12/1801] from the Czech Science Foundation, and institutional [AV0Z50520701]; supported by [MSM 6046137302 to D.S. and P.Č.]; supported by the Ministry of Research (France), University Paris Diderot, Sorbonne Paris Cité (France), National Institute of Blood Transfusion (INTS, France), National Institute of Health and Medical Research (INSERM, France) and ‘Investissements d’avenir’, Laboratories of Excellence GR-Ex (France) (to J.C.G. and A.G.dB.). Funding for open access charge: Czech Science Foundation and Academy of Sciences of the Czech Republic.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
This work is dedicated to Prof. Helen M. Berman (RCSB PDB, Rutgers University), a great tutor and true aficionado of structural biology.
REFERENCES
- 1.Matthews BW. No code for recognition. Nature. 1988;335:294–295. doi: 10.1038/335294a0. [DOI] [PubMed] [Google Scholar]
- 2.Pabo CO, Nekludova L. Geometric analysis and comparison of protein-DNA interfaces: why is there no simple code for recognition? J. Mol. Biol. 2000;301:597–624. doi: 10.1006/jmbi.2000.3918. [DOI] [PubMed] [Google Scholar]
- 3.Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sunami T, Kono H. Local conformational changes in the DNA interfaces of proteins. PLoS One. 2013;8:e56080. doi: 10.1371/journal.pone.0056080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seeman NC, Rosenberg JM, Rich A. Sequence specific recognition of double helical nucleic acids by proteins. Proc. Natl Acad. Sci. USA. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickerson R. In: Oxford Handbook of Nucleic Acid Structure. Neidle S, editor. Oxford: Oxford University Press; 1999. pp. 145–198. [Google Scholar]
- 7.Stormo GD, Zhao Y. Determining the specificity of protein-DNA interactions. Nat. Rev. Genet. 2010;11:751–760. doi: 10.1038/nrg2845. [DOI] [PubMed] [Google Scholar]
- 8.Sarai A, Takeda Y. Lambda repressor recognizes the approximately 2-fold symmetric half-operator sequences asymmetrically. Proc. Natl Acad. Sci. USA. 1989;86:6513–6517. doi: 10.1073/pnas.86.17.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarai A, Kono H. Protein-DNA recognition patterns and predictions. Annu. Rev. Biophys. Biomol. Struct. 2005;34:379–398. doi: 10.1146/annurev.biophys.34.040204.144537. [DOI] [PubMed] [Google Scholar]
- 10.Ahmad S, Keskin O, Sarai A, Nussinov R. Protein-DNA interactions: structural, thermodynamic and clustering patterns of conserved residues in DNA-binding proteins. Nucleic Acids Res. 2008;36:5922–5932. doi: 10.1093/nar/gkn573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pabo CO, Sauer RT. Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 12.Luscombe NM, Austin SE, Berman HM, Thornton JM. An overview of the structures of protein-DNA complexes. Genome Biol. 2000;1:1–37. doi: 10.1186/gb-2000-1-1-reviews001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki M, Gerstein M, Yagi N. Stereochemical basis of DNA recognition by Zn fingers. Nucleic Acids Res. 1994;22:3397–3405. doi: 10.1093/nar/22.16.3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choo Y, Klug A. Physical basis of a protein-DNA recognition code. Curr. Opin. Struct. Biol. 1997;7:117–125. doi: 10.1016/s0959-440x(97)80015-2. [DOI] [PubMed] [Google Scholar]
- 15.Mandel-Gutfreund Y, Margalit H. Quantitative parameters for amino acid-base interaction: implications for prediction of protein-DNA binding sites. Nucleic Acids Res. 1998;26:2306–2312. doi: 10.1093/nar/26.10.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki M, Gerstein M. Binding geometry of alpha-helices that recognize DNA. Proteins. 1995;23:525–535. doi: 10.1002/prot.340230407. [DOI] [PubMed] [Google Scholar]
- 17.McLaughlin WA, Berman HM. Statistical models for discerning protein structures containing the DNA-binding helix-turn-helix motif. J. Mol. Biol. 2003;330:43–55. doi: 10.1016/s0022-2836(03)00532-1. [DOI] [PubMed] [Google Scholar]
- 18.Jones S, van Heyningen P, Berman HM, Thornton JM. Protein-DNA interactions: a structural analysis. J. Mol. Biol. 1999;287:877–896. doi: 10.1006/jmbi.1999.2659. [DOI] [PubMed] [Google Scholar]
- 19.Luscombe NM, Thornton JM. Protein-DNA interactions: amino acid conservation and the effects of mutations on binding specificity. J. Mol. Biol. 2002;320:991–1009. doi: 10.1016/s0022-2836(02)00571-5. [DOI] [PubMed] [Google Scholar]
- 20.Lejeune D, Delsaux N, Charloteaux B, Thomas A, Brasseur R. Protein-nucleic acid recognition: statistical analysis of atomic interactions and influence of DNA structure. Proteins. 2005;61:258–271. doi: 10.1002/prot.20607. [DOI] [PubMed] [Google Scholar]
- 21.Nadassy K, Wodak SJ, Janin J. Structural features of protein-nucleic acid recognition sites. Biochemistry. 1999;38:1999–2017. doi: 10.1021/bi982362d. [DOI] [PubMed] [Google Scholar]
- 22.Jones S, Shanahan HP, Berman HM, Thornton JM. Using electrostatic potentials to predict DNA-binding sites on DNA-binding proteins. Nucleic Acids Res. 2003;31:7189–7198. doi: 10.1093/nar/gkg922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuchiya Y, Kinoshita K, Nakamura H. Structure-based prediction of DNA-binding sites on proteins using the empirical preference of electrostatic potential and the shape of molecular surfaces. Proteins. 2004;55:885–894. doi: 10.1002/prot.20111. [DOI] [PubMed] [Google Scholar]
- 24.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandel-Gutfreund Y, Schueler O, Margalit H. Comprehensive analysis of hydrogen bonds in regulatory protein DNA-complexes: in search of common principles. J. Mol. Biol. 1995;253:370–382. doi: 10.1006/jmbi.1995.0559. [DOI] [PubMed] [Google Scholar]
- 26.Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandel-Gutfreund Y, Margalit H, Jernigan RL, Zhurkin VB. A role for CH•••O interactions in protein-DNA recognition. J. Mol. Biol. 1998;277:1129–1140. doi: 10.1006/jmbi.1998.1660. [DOI] [PubMed] [Google Scholar]
- 28.Biot C, Wintjens R, Rooman M. Stair motifs at protein-DNA interfaces: nonadditivity of H-bond, stacking, and cation-pi interactions. J. Am. Chem. Soc. 2004;126:6220–6221. doi: 10.1021/ja049620g. [DOI] [PubMed] [Google Scholar]
- 29.Westhof E. Water: an integral part of nucleic acid structure. Annu. Rev. Biophys. Chem. 1988;17:125–144. doi: 10.1146/annurev.bb.17.060188.001013. [DOI] [PubMed] [Google Scholar]
- 30.Schwabe JW. The role of water in protein-DNA interactions. Curr. Opin. Struct. Biol. 1997;7:126–134. doi: 10.1016/s0959-440x(97)80016-4. [DOI] [PubMed] [Google Scholar]
- 31.Berman HM, Schneider B. In: Oxford Handbook of Nucleic Acid Structure. Neidle S, editor. Oxford: Oxford University Press; 1999. pp. 295–312. [Google Scholar]
- 32.Jayaram B, Jain T. The role of water in protein-DNA recognition. Annu. Rev. Biophys. Biomol. Struct. 2004;33:343–361. doi: 10.1146/annurev.biophys.33.110502.140414. [DOI] [PubMed] [Google Scholar]
- 33.Ponomarenko JV, Bourne PE, Shindyalov IN. Building an automated classification of DNA-binding protein domains. Bioinformatics. 2002;18(Suppl. 2):S192–S201. doi: 10.1093/bioinformatics/18.suppl_2.s192. [DOI] [PubMed] [Google Scholar]
- 34.Sathyapriya R, Vijayabaskar MS, Vishveshwara S. Insights into protein-DNA interactions through structure network analysis. PLoS Comput. Biol. 2008;4:e1000170. doi: 10.1371/journal.pcbi.1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biswas S, Guharoy M, Chakrabarti P. Dissection, residue conservation, and structural classification of protein-DNA interfaces. Proteins. 2009;74:643–654. doi: 10.1002/prot.22180. [DOI] [PubMed] [Google Scholar]
- 36.Siggers TW, Silkov A, Honig B. Structural alignment of protein—DNA interfaces: insights into the determinants of binding specificity. J. Mol. Biol. 2005;345:1027–1045. doi: 10.1016/j.jmb.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 37.Prabakaran P, Siebers JG, Ahmad S, Gromiha MM, Singarayan MG, Sarai A. Classification of protein-DNA complexes based on structural descriptors. Structure. 2006;14:1355–1367. doi: 10.1016/j.str.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 38.Unger R, Harel D, Wherland S, Sussman JL. A 3D building blocks approach to analyzing and predicting structure of proteins. Proteins. 1989;5:355–373. doi: 10.1002/prot.340050410. [DOI] [PubMed] [Google Scholar]
- 39.Bystroff C, Baker D. Prediction of local structure in proteins using a library of sequence-structure motifs. J. Mol. Biol. 1998;281:565–577. doi: 10.1006/jmbi.1998.1943. [DOI] [PubMed] [Google Scholar]
- 40.Kolodny R, Koehl P, Guibas L, Levitt M. Small libraries of protein fragments model native protein structures accurately. J. Mol. Biol. 2002;323:297–307. doi: 10.1016/s0022-2836(02)00942-7. [DOI] [PubMed] [Google Scholar]
- 41.Guyon F, Camproux AC, Hochez J, Tuffery P. SA-Search: a web tool for protein structure mining based on a Structural Alphabet. Nucleic Acids Res. 2004;32:W545–W548. doi: 10.1093/nar/gkh467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fourrier L, Benros C, de Brevern AG. Use of a structural alphabet for analysis of short loops connecting repetitive structures. BMC Bioinform. 2004;5:58. doi: 10.1186/1471-2105-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benros C, de Brevern AG, Etchebest C, Hazout S. Assessing a novel approach for predicting local 3D protein structures from sequence. Prot. Struct. Funct. Bioinform. 2006;62:865–880. doi: 10.1002/prot.20815. [DOI] [PubMed] [Google Scholar]
- 44.de Brevern AG, Etchebest C, Hazout S. Bayesian probabilistic approach for predicting backbone structures in terms of protein blocks. Prots. Struct. Funct. Genet. 2000;41:271–287. doi: 10.1002/1097-0134(20001115)41:3<271::aid-prot10>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 45.Offmann B, Tyagi M, de Brevern AG. Local protein structures. Curr. Bioinform. 2007;2:165–202. [Google Scholar]
- 46.Svozil D, Kalina J, Omelka M, Schneider B. DNA conformations and their sequence preferences. Nucleic Acids Res. 2008;36:3690–3706. doi: 10.1093/nar/gkn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berman HM, Westbrook J, Feng Z, Iype L, Schneider B, Zardecki C. The Nucleic Acid Database. Acta Crystallogr. D Biol. Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003487. [DOI] [PubMed] [Google Scholar]
- 48.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, et al. The Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 2002;58:889–898. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 49.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38, 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 53.Joseph AP, Agarwal G, Mahajan S, Gelly J-C, Swapna LS, Offmann B, Cadet F, Bornot A, Tyagi M, Valadié H, et al. A short survey on protein blocks. Biophys. Rev. 2010;2:137–145. doi: 10.1007/s12551-010-0036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tyagi M, Sharma P, Swamy CS, Cadet F, Srinivasan N, de Brevern AG, Offmann B. Protein Block Expert (PBE): a web-based protein structure analysis server using a structural alphabet. Nucleic Acids Res. 2006;34:W119–W123. doi: 10.1093/nar/gkl199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cech P, Kukal J, Cerny J, Schneider B, Svozil D. Automatic workflow for the classification of local DNA conformations. BMC Bioinform. 2013;14:205. doi: 10.1186/1471-2105-14-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Brevern AG. New assessment of a structural alphabet. In Silico Biol. 2005;5:283–289. [PMC free article] [PubMed] [Google Scholar]
- 57.de Brevern AG, Valadie H, Hazout S, Etchebest C. Extension of a local backbone description using a structural alphabet: a new approach to the sequence-structure relationship. Protein Sci. 2002;11:2871–2886. doi: 10.1110/ps.0220502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shakked Z, Rabinovich D, Kennard O, Cruse WBT, Salisbury SA, Viswamitra MA. Sequence-dependent conformation of an A-DNA double helix. The crystal structure of the octamer d(G-G-T-A-T-A-C-C) J. Mol. Biol. 1983;166:183–201. doi: 10.1016/s0022-2836(83)80005-9. [DOI] [PubMed] [Google Scholar]
- 59.Lu X-J, Shakked Z, Olson WK. A-form conformational motifs in ligand-bound DNA structures. J. Mol. Biol. 2000;300:819–840. doi: 10.1006/jmbi.2000.3690. [DOI] [PubMed] [Google Scholar]
- 60.Steffen NR, Murphy SD, Lathrop RH, Opel ML, Tolleri L, Hatfield GW. The role of DNA deformation energy at individual base steps for the identification of DNA-protein binding sites. Genome Inform. 2002;13:153–162. [PubMed] [Google Scholar]
- 61.Taylor R, Kennard O. Molecular Structures of Nucleosides and Nucleotides. 2. orthogonal coordinates for standard nucleic acid base residues. J. Am. Chem. Soc. 1982;104:3209–3212. [Google Scholar]
- 62.Gelbin A, Schneider B, Clowney L, Hsieh S-H, Olson WK, Berman HM. Geometric parameters in nucleic acids: sugar and phosphate constituents. J. Am. Chem. Soc. 1996;118:519–528. [Google Scholar]
- 63.Richardson JS, Schneider B, Murray LW, Kapral GJ, Immormino RM, Headd JJ, Richardson DC, Ham D, Hershkovits E, Williams LD, et al. RNA backbone: consensus all-angle conformers and modular string nomenclature (an RNA Ontology Consortium contribution) RNA. 2008;14:465–481. doi: 10.1261/rna.657708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 65.Malecka KA, Ho WC, Marmorstein R. Crystal structure of a p53 core tetramer bound to DNA. Oncogene. 2009;28:325–333. doi: 10.1038/onc.2008.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ghosh G, van Duyne G, Ghosh S, Sigler PB. Structure of NF-kappa B p50 homodimer bound to a kappa B site. Nature. 1995;373:303–310. doi: 10.1038/373303a0. [DOI] [PubMed] [Google Scholar]
- 67.Mo Y, Vaessen B, Johnston K, Marmorstein R. Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins. Mol. Cell. 1998;2:201–212. doi: 10.1016/s1097-2765(00)80130-6. [DOI] [PubMed] [Google Scholar]
- 68.Parkinson G, Gunasekera A, Vojtechovsky J, Zhang X, Kunkel TA, Berman H, Ebright RH. Aromatic hydrogen bond in sequence-specific protein DNA recognition. Nat. Struct. Biol. 1996;3:837–841. doi: 10.1038/nsb1096-837. [DOI] [PubMed] [Google Scholar]
- 69.Wolfe SA, Grant RA, Elrod-Erickson M, Pabo CO. Beyond the “recognition code”: structures of two Cys2His2 zinc finger/TATA box complexes. Structure. 2001;9:717–723. doi: 10.1016/s0969-2126(01)00632-3. [DOI] [PubMed] [Google Scholar]
- 70.Segal DJ, Crotty JW, Bhakta MS, Barbas CF, III, Horton NC. Structure of Aart, a designed six-finger zinc finger peptide, bound to DNA.J. Mol. Biol. 2006;363:405–421. doi: 10.1016/j.jmb.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 71.Jacobson EM, Li P, Leon-del-Rio A, Rosenfeld MG, Aggarwal AK. Structure of Pit-1 POU domain bound to DNA as a dimer: unexpected arrangement and flexibility. Genes Dev. 1997;11:198–212. doi: 10.1101/gad.11.2.198. [DOI] [PubMed] [Google Scholar]
- 72.Garvie CW, Phillips SE. Direct and indirect readout in mutant Met repressor-operator complexes. Structure. 2000;8:905–914. doi: 10.1016/s0969-2126(00)00182-9. [DOI] [PubMed] [Google Scholar]
- 73.Xu QS, Kucera RB, Roberts RJ, Guo HC. An asymmetric complex of restriction endonuclease MspI on its palindromic DNA recognition site. Structure. 2004;12:1741–1747. doi: 10.1016/j.str.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 74.Takeuchi R, Certo M, Caprara MG, Scharenberg AM, Stoddard BL. Optimization of in vivo activity of a bifunctional homing endonuclease and maturase reverses evolutionary degradation. Nucleic Acids Res. 2009;37:877–890. doi: 10.1093/nar/gkn1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Horton JR, Zhang X, Maunus R, Yang Z, Wilson GG, Roberts RJ, Cheng X. DNA nicking by HinP1I endonuclease: bending, base flipping and minor groove expansion. Nucleic Acids Res. 2006;34:939–948. doi: 10.1093/nar/gkj484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watanabe N, Takasaki Y, Sato C, Ando S, Tanaka I. Structures of restriction endonuclease HindIII in complex with its cognate DNA and divalent cations. Acta Crystallogr. D Biol. Crystallogr. 2009;65:1326–1333. doi: 10.1107/S0907444909041134. [DOI] [PubMed] [Google Scholar]
- 77.Sonavane S, Chakrabarti P. Cavities in protein-DNA and protein-RNA interfaces. Nucleic Acids Res. 2009;37:4613–4620. doi: 10.1093/nar/gkp488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anderson CF, Record MT., Jr Polyelectrolyte theories and their applications to DNA. Annu. Rev. Phys. Chem. 1982;33:191–222. [Google Scholar]
- 79.Rau DC, Parsegian VA. Direct measurement of the intermolecular forces between counterion-condensed DNA double helices. Biophys. J. 1992;61:246–259. doi: 10.1016/S0006-3495(92)81831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chalikian TV, Sarvazyan AP, Plum GE, Breslauer KJ. The influence of base composition, base sequence, and duplex structure on DNA hydration: apparent molar volumes and apparent molar adiabatic compressibilities of synthetic and natural DNA duplexes at 25 oC. Biochemistry. 1994;33:2394–2401. doi: 10.1021/bi00175a007. [DOI] [PubMed] [Google Scholar]
- 81.Schneider B, Berman HM. Hydration of the DNA bases is local. Biophys. J. 1995;69:2661–2669. doi: 10.1016/S0006-3495(95)80136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schneider B, Patel K, Berman HM. Hydration of the phosphate group in double helical DNA. Biophys. J. 1998;75:2422–2434. doi: 10.1016/S0006-3495(98)77686-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schneider B, Kabelac M. Stereochemistry of binding of metal cations and water to a phosphate group. J. Am. Chem. Soc. 1998;120:161–165. [Google Scholar]
- 84.Woda J, Schneider B, Patel K, Mistry K, Berman HM. An analysis of the relationship between hydration and protein-DNA interactions. Biophys. J. 1998;75:2170–2177. doi: 10.1016/S0006-3495(98)77660-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anderson CF, Record MT., Jr Ion distributions around DNA and other cylindrical polyions: theoretical descriptions and physical implications. Annu. Rev. Biophys. Chem. 1990;19:423–465. doi: 10.1146/annurev.bb.19.060190.002231. [DOI] [PubMed] [Google Scholar]
- 86.Leikin S, Parsegian VA, Rau DC. Hydration forces. Annu. Rev. Phys. Chem. 1993;44:369–395. doi: 10.1146/annurev.pc.44.100193.002101. [DOI] [PubMed] [Google Scholar]
- 87.Chalikian TV, Breslauer KJ. Thermodynamic analysis of biomolecules: a volumetric approach. Curr. Opin. Struct. Biol. 1998;8:657–664. doi: 10.1016/s0959-440x(98)80159-0. [DOI] [PubMed] [Google Scholar]
- 88.Nadassy K, Tomas-Oliveira I, Alberts I, Janin J, Wodak SJ. Standard atomic volumes in double-stranded DNA and packing in protein–DNA interfaces. Nucleic Acids Res. 2001;29:3362–3376. doi: 10.1093/nar/29.16.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reddy CK, Das A, Jayaram B. Do water molecules mediate protein-DNA recognition? J. Mol. Biol. 2001;314:619–632. doi: 10.1006/jmbi.2001.5154. [DOI] [PubMed] [Google Scholar]
- 90.Otwinowski Z, Schevitz RW, Zhang R-G, Lawson CL, Joachimiak A, Marmorstein RQ, Luisi BF, Sigler PB. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988;335:321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- 91.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 92.Winkler FK, Banner DW, Oefner C, Tsernoglou D, Brown RS, Heathman SP, Bryan RK, Martin PD, Petratos K, Wilson KS. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993;12:1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Horton NC, Perona JJ. Role of protein-induced bending in the specificity of DNA-recognition: Crystal structure of EcoRV endonuclease complexed with d(AAAGAT) + d(ATCTT) J. Mol. Biol. 1998;277:779–787. doi: 10.1006/jmbi.1998.1655. [DOI] [PubMed] [Google Scholar]
- 94.Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 95.Dickerson RE, Chiu TK. Helix bending as a factor in protein/DNA recognition. Biopolymers. 1997;44:361–403. doi: 10.1002/(SICI)1097-0282(1997)44:4<361::AID-BIP4>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 96.Kono H, Sarai A. Structure-based prediction of DNA target sites by regulatory proteins. Proteins. 1999;35:114–131. [PubMed] [Google Scholar]
- 97.Tolstorukov MY, Jernigan RL, Zhurkin VB. Protein-DNA hydrophobic recognition in the minor groove is facilitated by sugar switching. J. Mol. Biol. 2004;337:65–76. doi: 10.1016/j.jmb.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 98.Locasale JW, Napoli AA, Chen S, Berman HM, Lawson CL. Signatures of protein-DNA recognition in free DNA binding sites. J. Mol. Biol. 2009;386:1054–1065. doi: 10.1016/j.jmb.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Saenger W, Hunter WN, Kennard O. DNA conformation is determined by economics in the hydration of phosphate groups. Nature. 1986;324:385–388. doi: 10.1038/324385a0. [DOI] [PubMed] [Google Scholar]
- 100.Tolstorukov MY, Ivanov VI, Malenkov GG, Jernigan RL, Zhurkin VB. Sequence-dependent B<–>A transition in DNA evaluated with dimeric and trimeric scales. Biophys. J. 2001;81:3409–3421. doi: 10.1016/S0006-3495(01)75973-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shakked Z, Guerstein-Guzikevich G, Eisenstein M, Frolow F, Rabinovich D. The conformation of the DNA double helix in the crystal Is dependent on its environment. Nature. 1989;342:456–460. doi: 10.1038/342456a0. [DOI] [PubMed] [Google Scholar]
- 102.Shakked Z. The influence of the environment on DNA structures determined by X-ray crystallography. Curr. Opin. Struct. Biol. 1991;1:446–451. [Google Scholar]
- 103.Zhao J, Bacolla A, Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability and evolution. Cell. Mol. Life Sci. 2010;67:43–62. doi: 10.1007/s00018-009-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.