Abstract
The addition of uridine nucleotide by the poly(U) polymerase (PUP) enzymes has a demonstrated impact on various classes of RNAs such as microRNAs (miRNAs), histone-encoding RNAs and messenger RNAs. Cid1 protein is a member of the PUP family. We solved the crystal structure of Cid1 in complex with non-hydrolyzable UMPNPP and a short dinucleotide compound ApU. These structures revealed new residues involved in substrate/product stabilization. In particular, one of the three catalytic aspartate residues explains the RNA dependence of its PUP activity. Moreover, other residues such as residue N165 or the β-trapdoor are shown to be critical for Cid1 activity. We finally suggest that the length and sequence of Cid1 substrate RNA influence the balance between Cid1's processive and distributive activities. We propose that particular processes regulated by PUPs require the enzymes to switch between the two types of activity as shown for the miRNA biogenesis where PUPs can either promote DICER cleavage via short U-tail or trigger miRNA degradation by adding longer poly(U) tail. The enzymatic properties of these enzymes may be critical for determining their particular function in vivo.
INTRODUCTION
Cytoplasmic messenger RNA (mRNA) homeostasis is a complex phenomenon in which the balance between mRNA stability and mRNA degradation is tightly monitored. Eukaryotic species, from plants to mammals, rely heavily on the poly(A) tail to control the mRNA degradation process (1–2). Recently, a long-known family of proteins, named poly(U) polymerases (PUPs), was shown to be an unexpected player in mRNA homeostasis in various eukaryotes (3–6). The PUPs add poly(U) tails to mRNAs, which appear to modulate the rates and/or the directionality of the mRNA degradation process (4,7–9). Furthermore, members of the PUP family also regulate the production of histone mRNAs, microRNAs, siRNAs or U6 snRNA in the nucleus (6,10–17).
The PUP enzymes contain a nucleotidyl transferase domain, which has been characterized structurally (18–23). These structures show that the nucleotide triphosphate and the substrate RNA are bound in a space between the catalytic domain and the central domain (21,24). The PUP Cid1 protein was identified in Schizosaccharomyces pombe and was shown to have the capacity to use both uridine triphosphate (UTP) and adenosine triphosphate (ATP) as substrates for polymerization in vitro, while apparently being specific for UTP in vivo (25–26). This is surprising, as other members of the PUP family were depicted to be highly specific for UTP (18–20,27–28). Residue H336 found in the nucleotide recognition motif (NRM) loop has been shown to be mainly responsible for determining the specificity of the nucleotide recognized in vitro and in vivo (21–23). Additional charged residues were shown to be involved in substrate RNA association, although their exact function was not entirely characterized (21–22).
To further understand the mechanism behind RNA recognition and elongation, we determined the structure of Cid1 bound to a non-hydrolyzable nucleotide UMPNPP and bound to its minimal pseudo-product ApU. These structures revealed new key residues for the substrate/product recognition. In particular, one of the three catalytic aspartate residues interacts with the 2′-OH group of the pseudo-product-bound dinucleotide, thus explaining the RNA specificity displayed by this class of enzymes. Moreover, residue N165 interacts with the adenosine base in our pseudo-product-bound crystal structure. This interaction is essential for the enzymatic reaction. Finally, we demonstrate that the polymerizing capacities of Cid1 are modulated by a highly flexible loop named the β-trapdoor (residues 310–322) and by the residue K144 when UTP is used as a substrate (22). Our study thus highlights the structural basis behind several unique properties of the PUP enzyme family: properties mediated by a combination of specific residues and conserved secondary structures. We speculate that the specific type of activity that PUP enzymes display may determine their involvement in particular biochemical pathways and require factors to regulate their polymerizing activity.
MATERIALS AND METHODS
Expression and purification
Schizosaccharomyces pombe Cid1 protein (residues 40–377) was expressed and purified as described previously (21). Briefly, Cid1 was expressed in Escherichia coli BL21 Star cells (Novagen) as a His9-MBP fusion protein and purified using HiTrap FF crude column (GE Healthcare). After cleavage with the Tobacco Etch Virus protease (ratio of 1/50) at 16°C, the sample was reloaded onto the HiTrap column to remove the tag, the protease and the contaminants. The purified protein was finally applied on a Superdex 200 column (GE Healthcare), in a buffer containing 20 mM Hepes (pH 7.5), 150 mM NaCl, 1 mM β-mercaptoethanol and 10% (v/v) glycerol. Fractions containing the protein were pooled and concentrated (30 000 MWCO Amicon) to 10 mg/ml and stored at −80°C. All protein samples eluted from the gel filtration column as monomers. The same expression and purification procedure was performed for all the Cid1 mutants.
Mutagenesis
Cid1 point mutants were prepared by site-directed mutagenesis using the Quick-Change mutagenesis kit from Stratagene according to manufacturer’s instructions. The Cid1 Δ310–322 deletion mutant was prepared by Nested-polymerase chain reaction. All the constructs were verified by DNA sequencing. The oligonucleotides used for the mutations are shown in Supplementary Table S1.
Protein crystallization and X-ray data collection
D160A mutant protein was used at 10 mg/ml. Crystals were produced using the sitting-drop vapor diffusion technique at 18°C, with a reservoir solution containing 0.1 M imidazole/MES (equal ratio) (pH 6.1), 12–20% glycerol, 6–10% polyethylene glycol (PEG) 4000, 126 mM of a halogen salt (NaI, NaBr, NaF in equal ratio) and 10 mM TCEP [tris(2-carboxyethyl)phosphine] as an additive. Crystals appeared in 2 days and grew to a maximum size of around 0.25 × 0.25 × 0.2 mm in 1 week. Crystals of D160A mutant were soaked for 16 h with 2.5 mM UMPNPP and UTP or with 2.5 mM of ApU and ATP, in both cases supplemented with 5 mM of MgCl2. Crystals were passed into the crystallization solution supplemented with 10% glycerol before flash freezing in liquid nitrogen.
Diffraction data for the UMPNPP-soaked and ApU-soaked protein crystals were collected using the Beamline ID14–4 at the European Synchrotron Radiation Facility (ESRF, Grenoble). Complete data sets for UMPNPP- and ApU-soaked crystals were collected to 1.9 and 1.94 Å resolution, respectively, and the statistics are reported in Table I.
Table 1.
Data collection and refinement statistics
| Data collection | ||
| Crystal | UMPNPP-soaked Cid1 | ApU-soaked Cid1 |
| Wavelength (Å) | 0.9394 | 0.9394 |
| Temperature (K) | 100 | 100 |
| Maximum resolution (Å) | 45.0–1.90 | 45.3–1.94 |
| Space group | P21 | P21 |
| Unit cell dimensions (Å) | a = 53.8, b = 77.3, c = 82.1, α = γ = 90.0, β = 90.9 | a = 54.0, b = 77.1, c = 81.8, α = γ = 90.0, β = 90.7 |
| Number of observations | 260 839 | 246 855 |
| Number of unique reflections | 52 778 | 49 569 |
| Redundancy | 5.0 | 5.0 |
| Data completeness (%) | 99.5 (99.7) | 99.6 (99.2) |
| I/σ(I) | 28.47 (13.2) | 13.14 (2.3) |
| Rmeas | 4.4 (16.7) | 9.3 (87.8) |
| Refinement | ||
| Resolution (Å) | 19.8–1.90 | 45.3–1.94 |
| Number of atoms | 5688 | 5569 |
| Protein | 5112 | 5129 |
| Ligand | 58 (UMPNPP) | 78 (ApU) |
| Magnesium ions | 4 | 3 |
| Brome ions | 3 | 3 |
| Water molecules | 511 | 356 |
| Rwork (%) | 17.94 | 17.92 |
| Rfree (%) | 20.81 | 22.57 |
| RMSD from ideal geometry | ||
| Bond lengths (Å) | 0.008 | 0.008 |
| Bond angles (degrees) | 1.082 | 1.077 |
| Mean B (Å2) | ||
| Protein chain | 22.6 | 33.9 |
| Nucleotide/ions | 26.1 | 35.2 |
| Water | 30.6 | 41.3 |
| Residues in favored region of the Ramachandran plot (%) | 97.52 | 96.72 |
| Residues in allowed region of the Ramachandran plot (%) | 2.15 | 2.63 |
Structure determination and refinement
The structures of the D160A mutant in complex with UMPNPP or in complex with ApU were determined by molecular replacement with the Cid1 protein structure as a search model (PDB code 4EP7). The data sets were indexed with the XDS package (29). Phasing was done using the program Phaser from the CCP4 package (30) and the models were refined using the routine phenix.refine from the Phenix program (31). A large unidentified density was visible in both 2Fo – Fc and the Fo – Fc electron density maps where an UMPNPP or an ApU molecule was built (Supplementary Figure S1A and B). Atomic coordinates for the UMPNPP and ApU molecules were taken from the HIC-Up database (32) and fitted in the empty electron density map using the program COOT (33). The final Cid1/UMPNPP and Cid1/ApU models were refined to a resolution of 1.9 and 1.94 Å, respectively. Both crystal structures contain two molecules of Cid1 in the asymmetric unit. The Cid1/UMPNPP structure consists of residues 38–377 from the Cid1 protein, 2 UMPNPP compounds, 4 magnesium ions and 3 bromide ions present in the crystallization condition. The Cid1/ApU model contains the same Cid1 sequence (residues 38–377 for molecule A and residues 39–377 for molecule B), 2 ApU dinucleotides, 3 magnesium ions and 3 bromide ions. For both models, loops containing residues 109–115 and 309–322 were not visible in our electron density maps.
Poly(U) polymerase assays
RNA primers (U15, A15 or ApU) were labeled using T4 polynucleotide kinase (NEB) and γ-32P-ATP using manufacturer’s protocol. Labeled RNA substrates (10 pmoles) were incubated with 5 or 15 pmoles of Cid1 proteins [wild-type (WT) or mutant] in 25 µl of 10 mM Tris-Cl (pH 7.9) containing 50 mM NaCl, 10 mM MgCl2, 1 mM DTT and 0.5 mM UTP (or ATP when indicated) for 40 min at 37°C. Reactions were stopped with an equal volume of 100 mM EDTA and 2% sodium dodecyl sulfate. Extended primers were extracted with phenol-chloroform, precipitated with ethanol and resuspended in 90% formamide loading dye. Products were denatured at 95°C for 2 min and separated by gel electrophoresis in a 12% (or 25% for the ApU substrate) polyacrylamide/7 M urea gel.
Electrophoretic mobility shift assay
The indicated amount of proteins was mixed with the labeled RNAs in 20 mM Hepes (pH 7.5) containing 150 mM NaCl, 5 mM MgCl2, 1 mM β-mercaptoethanol, 10% glycerol and incubated at 37°C for 10 min. Following incubation, 2 µl of loading dye (0.1% bromophenol blue, 0.1% xylene cyanol, 180 mM Tris-boric acid and 50% glycerol) was added, and the samples were immediately loaded on a 10% native polyacrylamide gel in 0.5 × TBE buffer (45 mM Tris-boric acid, 1 mM EDTA). The gel was then run at 300 V for 120 min at 4°C and exposed in a cassette with an X-ray film (Fujifilm) for 30 to 45 min. Following quantification with ImageQuant software (GE Healthcare), affinity constants were calculated using the non-linear regression with one site-specific binding option from GraphPad Prism 6 program (GraphPad).
For comparing directly the RNA binding efficiency of various mutated Cid1 proteins, we used a unique concentration of protein (WT or mutant) and measured complex formation with a given probe (U15 or A15). Then, we quantified and normalized the amount of complex obtained with a given mutant protein by the one obtained with the WT enzyme for the same probe. Protein concentrations were set to 1.5 µM for the U15 probe and 11 µM for the A15 RNA. We used these two fixed concentrations of protein to maximize complex formation based on WT Cid1 Kd value for a given probe and to take into account the lower solubility observed with mutant proteins K144A and F332A. After loading and running the sample as indicated previously, we quantified our signal, normalized it against WT signal and plotted it as histogram. Individual binding reactions were performed three times in triplicates.
RESULTS AND DISCUSSION
Overall structure of the Cid1/UMPNPP and Cid1/ApU complexes
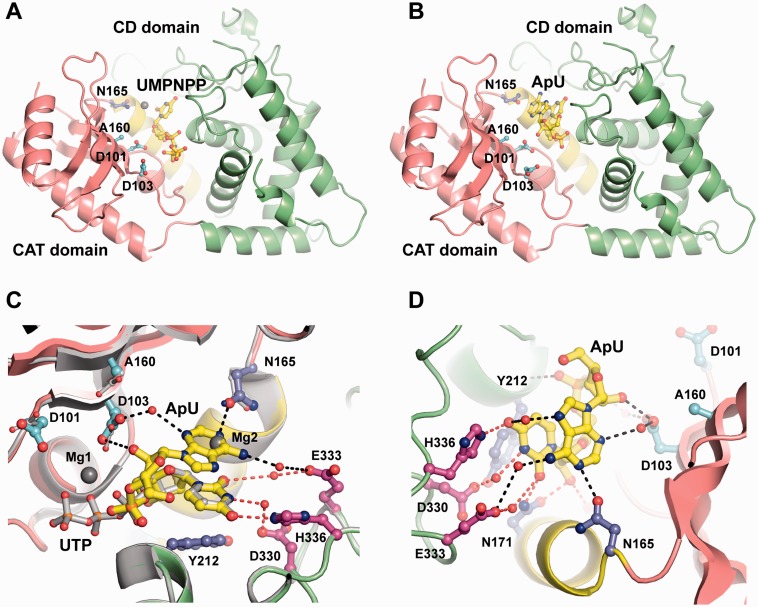
We mutated one residue from the catalytic triad (residue D160) in an attempt to stabilize the UMPNPP- or the ApU-bound Cid1 complexes as successfully done in the poly(A) polymerase (PAP)/ATP/RNA ternary complex structure (24). Here, we report the atomic structures of the D160A Cid1 mutant in complex with the non-hydrolyzable UMPNPP and the D160A Cid1 mutant in complex with a minimal pseudo-product (ApU) (Figure 1A and B; see X-ray statistics in Table I). Both ligands were built using unbiased Fo-Fc Fourier difference electron density map calculated with the protein model only after a round of simulated annealing (Supplementary Figure S1).
Figure 1.
Crystal structure of Cid1 in complex with its minimal pseudo-product. (A) Cartoon representation of D160A Cid1 bound to the non-hydrolyzable UMPNPP. The UMPNPP, the catalytic aspartic acids and D160A residue are shown as ball and sticks and colored according to atom type (carbon, yellow or cyan; nitrogen, blue; oxygen, red; phosphate, orange). (B) Cartoon representation of D160A Cid1 bound to ApU. The ApU molecules are shown and colored as in A. The catalytic (CAT) and central (CD) domains of Cid1 are colored in salmon and green, respectively, with the ‘ratchet’ helix four colored in yellow in A and B. (C) Side view of the active site of the Cid1/UTP and D160A/ApU superposed structures. Cid/UTP atomic model is colored in gray. Residues from the NRM loop are shown as ball and sticks and colored as in A (carbon atom in magenta). Hydrogen bonds within 2.4–3.6 Å distance are shown as black dash lines for adenine stabilization and as red dash lines for uridine stabilization. Amino acids from the Cid1/UTP structure are shown with thin stick radius compared with the one of the Cid1/ApU model. (D) Top view of the D160A showing ApU interactions. Atoms, residues and hydrogen bonds are colored as C. Magnesium ions are illustrated as gray spheres in A and C. All the structural panels here and in the following figures were generated with PyMol (34).
UMPNPP mode of binding is similar to our Cid1/UTP complex (21) confirming that D160A mutation does not induce any conformational changes in the overall protein fold or in the substrate nucleotide recognition by the protein. The only difference is the absence of the first magnesium ion necessary for the nucleolytic attack, as previously observed in the PAP/ATP/RNA crystal structure (Figure 1A) (24).
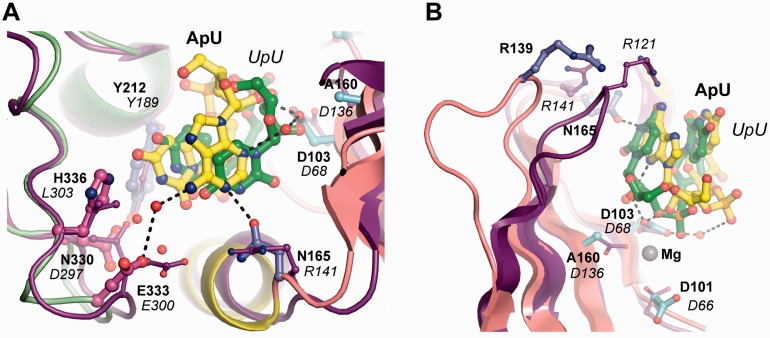
The bound ApU molecule should be considered as a mimic of the post-catalysis reaction state as defined by Stagno et al. (27). As such, ApU is considered as a pseudo-product of the Cid1-mediated reaction. The pseudo-product ApU is representative of the in vivo situation encountered by Cid1 protein as, in S. pombe, Cid1 natural mRNA substrates are polyadenylated (Figure 1B) (26). In the ApU-bound Cid1 structure, the uridine nucleotide is recognized as in the UMPNPP and the UTP-bound Cid1 structures (21–23) (Figure 1C). The adenosine base of the bound ApU molecule is stacked on the uridine base, a situation reminiscent of the TUT4/UpU structure (Figure 1C) (27). Several Cid1 residues are involved in the stabilization of the adenosine nucleotide. First, its ribose 2′-OH group establishes a direct hydrogen bond with the catalytic residue D103 (Figures 1C and D). Although Cid1 was previously classified as an RNA-specific nucleotidyl transferase, the basis for such specificity was not described (8). We suggest that the catalytic residue D103 stabilizes the proper type of substrate near the catalytic site by detecting the presence of a 2′-hydroxyl group (Figure 1D). This mechanism of RNA detection is probably shared across the entire family, as a similar interaction is found in the TUT4/UpU crystal structure (27). Second, residue D103 and the ApU molecule have another water-mediated interaction with the N3 position of the adenine, likely used to stabilize the incoming RNA substrate before the nucleotidyl transfer reaction (Figure 1C and D). The adenine is further stabilized via additional water-mediated interaction between its N6 position and residue E333 (Figure 1C and D). None of these water-mediated interactions confer nucleotide specificity on the 3′-end of the substrate RNA, as expected for a template-independent polymerase (8). In the ApU complex structure, the N1 position of the bound adenine is recognized via a specific hydrogen bond with the OD1 position of residue N165. In Trypanosoma brucei TUT4/UpU structure, the equivalent residue is an arginine (R141), which cannot contact the uracil base because it forms a salt bridge with residue E300 (E333 in Cid1) (27). However, if we superpose the TUT4/UpU and Cid1/ApU structures, the N3 atom of the uracil base in the trypanosomal structure is located at nearly the same place as the N1 atom of the adenosine in our structure (Figure 2A). Therefore, we suggest that N165 is a key residue for binding to the 3′-end of the RNA substrate either before or after the Cid1 uridylation reaction. This proposal is valid whether this substrate RNA contains a 3′-end adenosine nucleotide, as expected for the native poly(A)-containing substrates in S. pombe, or whether the RNA went through at least one round of polyuridylation (Figure 2A). Our proposal can be extrapolated to any RNA 3′-end base, although it will require the nucleotide tautomeric form with a hydrogen-bonded N1 atom to establish a proper bond with N165 OD1 position. Finally, the TUT/PUP superposition also shows that residue R139 in Cid1 is not in contact with the pseudo-product ApU (Figure 2B). We presume that the two enzyme groups may have evolved toward slightly different RNA recognition/stabilization modes, most likely reflecting their specific enzymatic properties.
Figure 2.
Structural comparison between the Cid1/ApU and the TbTUT4/UpU complex structures. (A) Close view of the NRM loop TbTUT4/UpU (PDB code 2Q0G) and Cid1/ApU showing related active site pocket architecture. TbTUT4/UpU structure is shown in cartoon representation and colored in violet. The main interacting amino acids and the UpU molecule are shown as ball and stick model colored in violet or green, respectively. Cid1/ApU model, residues and atoms are shown and colored as in Figure 1. (B) View from the top of the active site highlighting the differential position of the loop between β-sheets four and five. This loop contains residue R139 (R121 in TbTUT4). TbTUT4 residues are labeled in italic and the radius of their ball and stick representations is reduced compared with the one used for Cid1 residues. Magnesium ions are illustrated as gray spheres in all panels. The black dash lines represent hydrogen bonds within 2.4–3.6 Å distance.
RNA binding properties
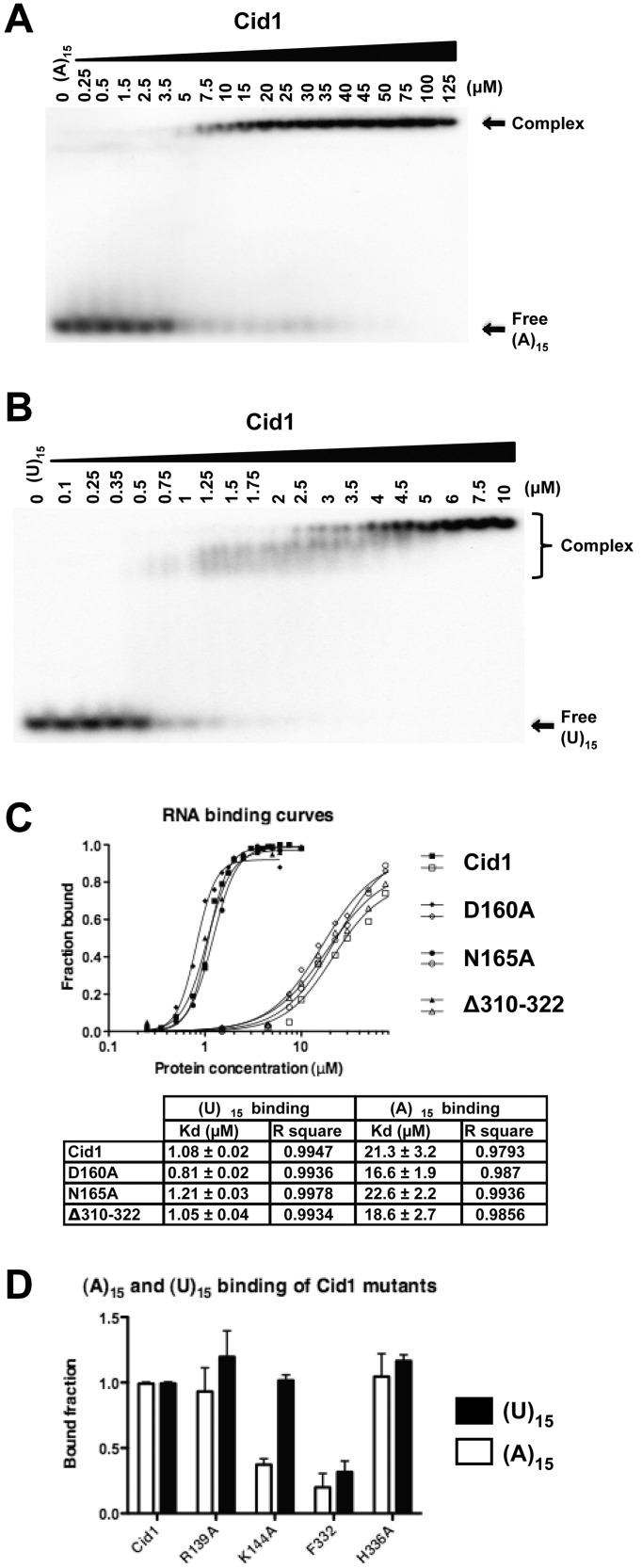
Cid1 has been classified as a non-canonical template-independent nucleotidyl transferase (3,7–8,21–23,35). Previous studies have also shown that the overall activity of Cid1 is unexpectedly high when presented with U15 (26). To further understand the Cid1 enzymatic properties, we generated various Cid1 mutants driven by the analysis of our Cid1/ApU crystal structure. We then measured their binding properties for two different substrates (A15 and U15) in the absence of any polymerizing activity to avoid interference from the newly synthesized tail (Figure 3 and Supplementary Figure S2).
Figure 3.
Characterization of U15 and A15 RNA binding properties of WT and mutant Cid1 protein by EMSA. (A) EMSA showing the interaction of Cid1 with an A15 RNA. (B) Same experiments with a U15 RNA. In A and B, the individual reactions contain either no protein or the indicated amounts. (C) RNA binding curves and calculated Kd from quantified EMSA experiments performed with Cid1 WT, D160A, N165A and Δ310–322 mutants for both A15 and U15. Individual binding experiments were repeated at least three times and representative EMSA experiments are shown in the Supplementary Figure S2. (D) Histogram representation of the interaction measured between various Cid1 mutants with either an U15 or an A15 RNA probe. Bound fractions correspond to the amount of formed complex and were normalized by the one observed between Cid1 WT protein and either the U15 or the A15 probe. Experiments were repeated at least three times and done in triplicate.
We first compared the capacities of the WT protein and of our mutant D160A to bind to the two selected substrates—mimics of a short poly(A) tail (A15) or a newly added U-tail (U15; Figure 3A and B; Supplementary Figure S2A and B). The affinity constant calculated between the Cid1 WT and the U15 probe was ∼1 and 20 µM with the A15 probe (Figure 3C). These values were identical for the D160A mutant, reinforcing the fact that this mutation does not alter the enzyme’s RNA binding capacity. Overall, we observe a 20-fold difference between the affinity constants of the various tested Cid1 proteins for a 15-mer homouridine as compared with the A15 (Figure 3C). Cid1’s intrinsically higher affinity for homouridine polymer probably explains why Cid1 exhibits a more efficient polymerizing activity on U15 RNA (26).
We also mutated residue N165 into an alanine (or an aspartate) and assessed the mutated Cid1 enzyme’s binding capacity (Supplementary Figure S2C and D). The affinity constant for the N165A mutant did not differ from that of the WT protein (Figure 3C). These values are comparable or slightly lower than the reported values for nucleotidyl transferase enzymes (23,27,36). The binding experiments agree with our observation that both A and U nucleotide would be forming a stable hydrogen bond with N165 residue (Figures 2A and 3C). We then tested residue R139, which is found in the loop between β-strands 4 and 5 (Figure 2B). In the TbTUT4, the equivalent amino acid R121 is involved in the primer stabilization as well as in the catalytic activity (Figure 2B) (19,27). The association between the two RNA substrates and the R139A mutant is similar to that of the WT protein (Figure 3D). These binding assays corroborate our structural data, which did not show any interaction with R139 residue (Figure 2B). We concluded that this residue is not important on its own for the recognition of the substrate RNA in contrast to TbTUT4 (Figure 3D). Furthermore, it has been suggested that the β-sheet structure on top of the active site, named β-trapdoor, plays a role during UTP exchange, although its role in RNA association has not been assessed (22). We could not observe any significant difference between the affinity constants measured with Cid1 truncation of residues 310–322 (Δ310–322) for both RNA substrates suggesting that the β-trapdoor does not determine the specificity of the associated RNA per se (Figure 3C).
Several residues that have been shown to be important for the Cid1 activity such as F332, K144 or H336 were finally tested for their potential effect on the RNA binding properties. Strikingly, the F332 mutant had a reduced capacity for binding RNAs (Figure 3D). We previously proposed that F332 was critical for the UTP clamping mechanism mediated by the ratchet helix 4 (21). We suggest that the importance of the domain movement during the nucleotide transfer likely extends to RNA template trapping mechanism as well (Figure 3D) (21). We then looked into the RNA binding capacity of the H336A mutant protein for either of the RNA substrates, which was unimpaired, thereby confirming that H336 is only important for the UTP selectivity (Figure 3D) (21). When we tested the RNA binding capacity of the K144A mutant protein, we observed that its association with A15 was strongly reduced, whereas it was unchanged for a 15-mer U stretch (Figure 3D). Therefore, K144 residue seems important for the stabilization of our short incoming substrate RNA. These results are slightly different from the data published by Gilbert’s laboratory, possibly due to the differences in the RNA used for the binding studies (a 40-mer RNA in their study) (22). The poly(A) polymerase equivalent residue K145 is found to contact the 2′-OH group of the nucleotide -2 in the PAP/ATP/RNA crystal structure (24). Therefore, the Cid1 protein may use a substrate stabilization strategy closer to the one of the PAP enzymes. Altogether, it appears that Cid1 enzyme is highly specialized with residues critical for the nucleotide triphosphate association (H336), residues specialized for the RNA primer association (K144) and residues involved in the nucleotidyl transferase enzymatic cycle (β-trapdoor, D103, N165 or F332).
Polymerization activity of Cid1 mutants
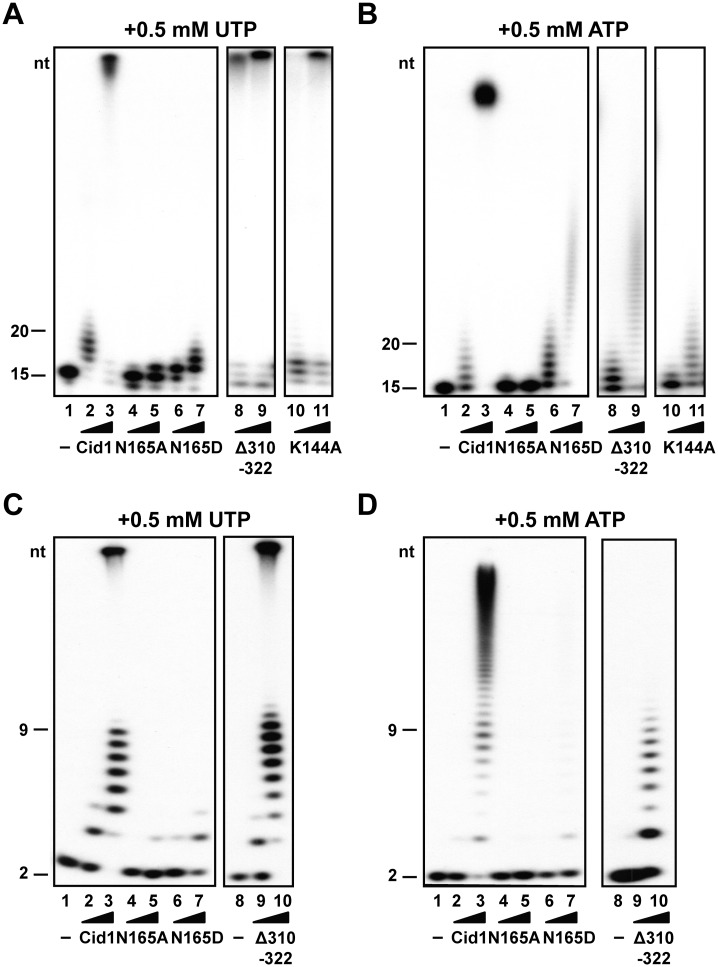
To complete our RNA binding study, we measured the polymerization activities of the same set of mutants. As we observed significant differences between Cid1 affinity for a U-tail as compared with an A-tail, we looked at the polymerizing activity in the presence of either UTP or ATP. The N165A mutant is unable to add more than one nucleotide per substrate regardless of the provided nucleotide triphosphate (UTP or ATP) even though its RNA binding capacity is unimpaired, at least for the 15-mer primer (Figures 3C, 4A and B). If N165 residue is changed into a charged residue (N165D), the enzyme is able to add up to 3–4 uridines or up to 20 adenines to the 15-nt RNA template but is still almost unable to use the ApU substrate (Figure 4, mutant N165D). Overall, these experiments demonstrate the critical role of N165 residue in the polymerizing reaction. Being located at the beginning of helix 4, N165 is likely important during the swivel motion occurring throughout the catalytic cycle of the enzyme (21). The rotation of the catalytic domain around the helix 4 can open the space between the domains. We suggest that N165 residue is involved in pulling on the RNA primer after the addition of the UMP. On displacement of the RNA primer, the UTP binding cavity is emptied allowing a new UTP molecule to take its place, and a new cycle to start.
Figure 4.
Poly(U) and poly(A) polymerization assays with two different RNA probes. (A) Polymerization assay in the presence of UTP with an A15 primer. Lane 1 shows the RNA primer after incubation without enzyme. The RNA after elongation with WT Cid1 (lanes 2–3), N165A (lanes 4–5), N165D (lanes 6–7), Δ310–322 deleted protein (lanes 8–9) and K144A mutants are loaded in the respective lanes. (B) Polymerization assay in the presence of ATP with an A15 as substrate. Similar mutants were used and reactions were loaded as in A. (C) Polymerization assay with UTP and the substrate ApU. The lane 1 shows the ApU substrate after incubation without enzyme. The polymerizing reaction in the presence of WT Cid1, N165A, N165D and Δ310–322 mutant proteins is shown in lanes 2–3, 4–5, 6–7 and 9–10, respectively. (D) Polymerization assays with ATP and an ApU substrate. Similar mutants were used and reactions were loaded as in C.
Another region of interest near the NRM loop is the β-trapdoor (22). Deletion of the β-trapdoor residues does not affect the overall PUP activity of the enzyme for either of the RNA primers as long as we use UTP as a substrate (Figure 4A and C). We can observe the typical biphasic profile with, on one hand, short products of 7–9 added nucleotides corresponding to a distributive activity and, on the other hand, highly elongated products that did not enter the gel (Figure 4A and B). However, when we look at activity of the Δ310–322 mutant protein in the presence of ATP, we do not observe any larger products suggesting that Cid1 without the β-trapdoor is almost exclusively a distributive PAP enzyme (Figure 4B and D). This is even more striking with the short substrate ApU (Figure 4D). Because these observed differences between PAP and PUP activities cannot be linked to a difference in the RNA binding properties of the truncated protein (Figure 3C; Supplementary Figure S2E and F), we conclude that the β-trapdoor has an important role during the catalytic cycle. This may be to favor NTP exchange after each polymerization cycle by promoting domain movements (22).
Finally, we tested the polymerizing activity of the K144A mutant. This single-point mutant has a PUP activity comparable with the WT protein, but its PAP activity is drastically reduced as demonstrated by the almost complete absence of extended products (compare Figure 4A and B). As observed for the Δ310–322 deleted protein, its activity is now almost exclusively distributive when presented with ATP (Figure 4A and B). As the K144A mutant has an impaired A15 RNA association, we propose that the change in the polymerizing profile is linked with the poly(A) RNA binding capacities of the protein.
CONCLUSIONS
Our atomic models provide missing information regarding the association of RNAs with the uridylating enzyme Cid1. We identify multiple determinants behind the RNA binding properties of Cid1 and decipher the roles played by several residues or protein fragments during Cid1’s polymerization activity. When the 3′-end of the template RNA is not a uridine, Cid1 has a low RNA binding affinity and therefore a highly distributive activity reflecting its classification as a template-independent nucleotidyl transferase (8). Our binding tests show that on addition of 6–7 uridine nucleotides, the enzyme's affinity for the 3′-end of its template becomes at least 20 times higher as shown by our binding tests. Following this switch in its overall affinity, Cid1’s activity becomes highly processive. We show that residue K144 is involved in this switch of activity. The Cid1 catalytic cycle is complex and its completion critically depends on several key residues: (i) the β-trapdoor controls at least partially the UTP exchange after the reaction; (ii) N165 residue, which is hydrogen bonded to the 3′-penultimate nucleotide in our atomic model, may be involved in the displacement of the elongated RNA product after UMP addition. Finally, the overall catalytic cycle depends on the previously proposed domain movement around helix 4 and residue F332 to proceed (21).
Until now, the length of the characterized poly(U) tail due to PUP activities in vivo is generally short (between 1 and 5 nt) (4,5,37,38). We propose that Cid1 enzymatic activity is influenced by its higher affinity for the homouridine-containing tails. To prevent Cid1 from generating long U-tails that can trigger RNA degradation by exonucleases like Dis3L2 (9,39), we suggest that factors are used to control the local activity, like the targeting factor Lin28 (11,40).
Lastly, as shown by the differential use of particular protein residues like residue R139 or K144, TUT and PUP enzymes have evolved differently (19,21). It has recently been proposed that DSS1 protein, a uridine-specific exonuclease, plays a critical role during the guide RNA maturation process in trypanosome, a process where RET1 TUT enzyme is critical (41,42). As our present results shed new light on the properties of the polyuridylating enzyme Cid1, it will be exciting to see whether a similar switch exists in the RET1 enzyme (43–45).
ACCESSION NUMBERS
Atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 4NKT for the Cid1-UMPNPP complex and 4NKU for the Cid1-ApU complex.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation [31003A_140924 and 31003A_124909 to S.T.]. Funding for open access charge: Swiss National Science Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the staff of ID14-4 at the ESRF for developing excellent beamlines and supporting optimal data collection. They also thank all the members of Stephane Thore Lab for their critical reading, helpful discussion and technical advice. They thank the University of Geneva for gratefully allowing the installation of the biostructural platform together with the generous financial support of the Boninchi foundation, the Schmidheiny foundation and the Swiss National Science Foundation R'equip grant (316030-128787).
REFERENCES
- 1.Eckmann CR, Rammelt C, Wahle E. Control of poly(A) tail length. Wiley Interdiscip. Rev. RNA. 2011;2:348–361. doi: 10.1002/wrna.56. [DOI] [PubMed] [Google Scholar]
- 2.Schuster G, Stern D. RNA polyadenylation and decay in mitochondria and chloroplasts. Prog. Mol. Biol. Transl. Sci. 2009;85:393–422. doi: 10.1016/S0079-6603(08)00810-6. [DOI] [PubMed] [Google Scholar]
- 3.Rissland OS, Norbury CJ. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat. Struct. Mol. Biol. 2009;16:616–623. doi: 10.1038/nsmb.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sement FM, Ferrier E, Zuber H, Merret R, Alioua M, Deragon JM, Bousquet-Antonelli C, Lange H, Gagliardi D. Uridylation prevents 3′ trimming of oligoadenylated mRNAs. Nucleic Acids Res. 2013;41:7115–7127. doi: 10.1093/nar/gkt465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lapointe CP, Wickens M. The nucleic acid-binding domain and translational repression activity of a Xenopus terminal uridylyl transferase. J. Biol. Chem. 2013;288:20723–20733. doi: 10.1074/jbc.M113.455451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt MJ, West S, Norbury CJ. The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA. 2011;17:39–44. doi: 10.1261/rna.2252511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwak JE, Wickens M. A family of poly(U) polymerases. RNA. 2007;13:860–867. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin G, Keller W. RNA-specific ribonucleotidyl transferases. RNA. 2007;13:1834–1849. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malecki M, Viegas SC, Carneiro T, Golik P, Dressaire C, Ferreira MG, Arraiano CM. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 2013;32:1842–1854. doi: 10.1038/emboj.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Sinha K, Perumal K, Reddy R. Effect of 3′ terminal adenylic acid residue on the uridylation of human small RNAs in vitro and in frog oocytes. RNA. 2000;6:1277–1288. doi: 10.1017/s1355838200000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trippe R, Sandrock B, Benecke BJ. A highly specific terminal uridylyl transferase modifies the 3′-end of U6 small nuclear RNA. Nucleic Acids Res. 1998;26:3119–3126. doi: 10.1093/nar/26.13.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA. 2006;12:1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- 17.Zhao YY, Yu Y, Zhai JX, Ramachandran V, Dinh TT, Meyers BC, Mo BX, Chen XM. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr. Biol. 2012;22:689–694. doi: 10.1016/j.cub.2012.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng J, Ernst NL, Turley S, Stuart KD, Hol WG. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei. EMBO J. 2005;24:4007–4017. doi: 10.1038/sj.emboj.7600861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stagno J, Aphasizheva I, Rosengarth A, Luecke H, Aphasizhev R. UTP-bound and Apo structures of a minimal RNA uridylyltransferase. J. Mol. Biol. 2007;366:882–899. doi: 10.1016/j.jmb.2006.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stagno J, Aphasizheva I, Bruystens J, Luecke H, Aphasizhev R. Structure of the mitochondrial editosome-like complex associated TUTase 1 reveals divergent mechanisms of UTP selection and domain organization. J. Mol. Biol. 2010;399:464–475. doi: 10.1016/j.jmb.2010.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munoz-Tello P, Gabus C, Thore S. Functional implications from the Cid1 poly(U) polymerase crystal structure. Structure. 2012;20:977–986. doi: 10.1016/j.str.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 22.Yates LA, Fleurdepine S, Rissland OS, De Colibus L, Harlos K, Norbury CJ, Gilbert RJ. Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase. Nat. Struct. Mol. Biol. 2012;19:782–787. doi: 10.1038/nsmb.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lunde BM, Magler I, Meinhart A. Crystal structures of the Cid1 poly (U) polymerase reveal the mechanism for UTP selectivity. Nucleic Acids Res. 2012;40:9815–9824. doi: 10.1093/nar/gks740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balbo PB, Bohm A. Mechanism of poly(A) polymerase: structure of the enzyme-MgATP-RNA ternary complex and kinetic analysis. Structure. 2007;15:1117–1131. doi: 10.1016/j.str.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang SW, Toda T, MacCallum R, Harris AL, Norbury C. Cid1, a fission yeast protein required for S-M checkpoint control when DNA polymerase delta or epsilon is inactivated. Mol. Cell. Biol. 2000;20:3234–3244. doi: 10.1128/mcb.20.9.3234-3244.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rissland OS, Mikulasova A, Norbury CJ. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol. Cell. Biol. 2007;27:3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stagno J, Aphasizheva I, Aphasizhev R, Luecke H. Dual role of the RNA substrate in selectivity and catalysis by terminal uridylyl transferases. Proc. Natl Acad. Sci. USA. 2007;104:14634–14639. doi: 10.1073/pnas.0704259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ringpis GE, Stagno J, Aphasizhev R. Mechanism of U-insertion RNA editing in trypanosome mitochondria: characterization of RET2 functional domains by mutational analysis. J. Mol. Biol. 2010;399:696–706. doi: 10.1016/j.jmb.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kabsch W. Xds. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleywegt GJ. Crystallographic refinement of ligand complexes. Acta Crystallogr. D. Biol. Crystallogr. 2007;63:94–100. doi: 10.1107/S0907444906022657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Stevenson AL, Norbury CJ. The Cid1 family of non-canonical poly(A) polymerases. Yeast. 2006;23:991–1000. doi: 10.1002/yea.1408. [DOI] [PubMed] [Google Scholar]
- 35.DeLano WL. The PyMol User's Manual. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 36.Balbo PB, Toth J, Bohm A. X-ray crystallographic and steady state fluorescence characterization of the protein dynamics of yeast polyadenylate polymerase. J. Mol. Biol. 2007;366:1401–1415. doi: 10.1016/j.jmb.2006.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. Mono-Uridylation of Pre-MicroRNA as a key step in the biogenesis of group II let-7 MicroRNAs. Cell. 2012;151:521–532. doi: 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 38.Knouf EC, Wyman SK, Tewari M. The human TUT1 nucleotidyl transferase as a global regulator of microRNA abundance. PloS One. 2013;8:e69630. doi: 10.1371/journal.pone.0069630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ustianenko D, Hrossova D, Potesil D, Chalupnikova K, Hrazdilova K, Pachernik J, Cetkovska K, Uldrijan S, Zdrahal Z, Vanacova S. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA. 2013;19:1632–1638. doi: 10.1261/rna.040055.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 41.Mattiacio JL, Read LK. Roles for TbDSS-1 in RNA surveillance and decay of maturation by-products from the 12S rRNA locus. Nucleic Acids Res. 2008;36:319–329. doi: 10.1093/nar/gkm690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Penschow JL, Sleve DA, Ryan CM, Read LK. TbDSS-1, an essential Trypanosoma brucei exoribonuclease homolog that has pleiotropic effects on mitochondrial RNA metabolism. Eukaryot. Cell. 2004;3:1206–1216. doi: 10.1128/EC.3.5.1206-1216.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aphasizhev R, Aphasizheva I, Simpson L. A tale of two TUTases. Proc. Natl Acad. Sci. USA. 2003;100:10617–10622. doi: 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aphasizheva I, Ringpis GE, Weng J, Gershon PD, Lathrop RH, Aphasizhev R. Novel TUTase associates with an editosome-like complex in mitochondria of Trypanosoma brucei. RNA. 2009;15:1322–1337. doi: 10.1261/rna.1538809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panigrahi AK, Schnaufer A, Carmean N, Igo RP, Jr, Gygi SP, Ernst NL, Palazzo SS, Weston DS, Aebersold R, Salavati R, et al. Four related proteins of the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 2001;21:6833–6840. doi: 10.1128/MCB.21.20.6833-6840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.