Abstract
RNA contains various chemical modifications that expand its otherwise limited repertoire to mediate complex processes like translation and gene regulation. 25S rRNA of the large subunit of ribosome contains eight base methylations. Except for the methylation of uridine residues, methyltransferases for all other known base methylations have been recently identified. Here we report the identification of BMT5 (YIL096C) and BMT6 (YLR063W), two previously uncharacterized genes, to be responsible for m3U2634 and m3U2843 methylation of the 25S rRNA, respectively. These genes were identified by RP-HPLC screening of all deletion mutants of putative RNA methyltransferases and were confirmed by gene complementation and phenotypic characterization. Both proteins belong to Rossmann-fold–like methyltransferases and the point mutations in the S-adenosyl-l-methionine binding pocket abolish the methylation reaction. Bmt5 localizes in the nucleolus, whereas Bmt6 is localized predominantly in the cytoplasm. Furthermore, we showed that 25S rRNA of yeast does not contain any m5U residues as previously predicted. With Bmt5 and Bmt6, all base methyltransferases of the 25S rRNA have been identified. This will facilitate the analyses of the significance of these modifications in ribosome function and cellular physiology.
INTRODUCTION
Ribosomes are the molecular assemblies of RNA and proteins that translate the genetic information. Structural and functional analyses of ribosomes have revealed that it is the rRNA that makes the structural framework of ribosomes and catalyze the joining of amino acids (peptidyl transfer) during translation, hence making ribosome a ribozyme (1,2). However, the chemical composition of RNA, on the other hand, seems to be rather insufficient to provide the structural complexity and flexibility that RNA requires to catalyze the chemical reactions and coordinate the molecular events during protein synthesis. Interestingly, the composition analysis of rRNA has shown that rRNA contains various chemical modifications that apparently expand its stereochemical possibilities to mediate sophisticated processes like translation (3,4). This is well supported by the fact that most of the chemical modifications cluster in the functionally conserved regions of the ribosomes, like peptidyl transferase center and intersubunit bridges (5).
The rRNA molecules contain three different types of chemical modifications, pseudouridylations, ribose methylations and methylation of nitrogenous bases. Most of these modifications are added posttranscriptionally, whereas some are added co-transcriptionally (6). In Saccharomyces cerevisiae, the 18S rRNA of small subunit contains 14 pseudouracils, 17 ribose methylations and 4 base methylations, whereas the 25S rRNA of large subunit contains 30 pseudouracils, 37 ribose methylations and 8 base methylations (7,8).
Unlike, pseudouridylations and ribose methylations, which are catalyzed by H/ACA and C/D box snoRNPs, respectively, the base methylations are performed by specific base methyltransferases. Surprisingly, many of the base methyltransferases of rRNA remained elusive for long time. It is important to note that the identification of these enzymes is crucial to understand the significance of these modifications in the functioning of ribosomes and in cellular physiology.
We have been interested in exploring base methylations of the rRNA and identifying novel methyltransferases catalyzing these modifications. In S. cerevisiae, all presently known rRNA base methyltransferases use reactive methyl group of the S-adenosyl-l-methionine (SAM) for the methylation reaction and belong to either class I or class IV of methyltransferases (9). The majority of the known RNA methyltransferases belong to class I and contain characteristic Rossmann-like fold SAM binding domain (9,10). The methyltransferases belonging to class IV are characterized by an α/β knot structure or SPOUT domain (11). Using rDNA genetics and RP-HPLC approaches, we have been able to identify novel sites of base methylation in the rRNA and have investigated all single-gene deletion mutants of putative uncharacterized and characterized RNA methyltransferases to identify the enzymes responsible for these methylation (8,12,13).
The 18S rRNA in S. cerevisiae contains four base methylations, m1Ψ1191, m62A1781, m62A1782 and m7G1575, catalyzed by enzymes Nep1, Dim1 and Bud23, respectively (14–17). The 25S rRNA of S. cerevisiae was reported to contain eight base methylations (two m1A, two m5C, two m3U and two m5U), the enzymes for which remained elusive (7,8). Recently, we identified Rrp8 and Bmt2 as m1A methyltransferases responsible for the methylation of adenine 645 and 2142 of the 25S rRNA, respectively (12,13). Furthermore, we discovered that Rcm1 methylates C5 of cytosine (m5C) 2278, whereas Nop2 catalyzes the methylation of C5 of recently identified m5C residue at position 2870, located in the peptidyl transferase center (8). The analyses of these modifications revealed that the base methylations play a significant role in 60S biogenesis and in antibiotic sensitivity. Surprisingly, most of rRNA modifications are dispensable for cell viability although the enzymes are essential for growth and ribosome biogenesis. This clearly highlights a dual functionality of these enzymes in ribosome biogenesis. Additionally, many of these methyltransferases are highly conserved. The human homolog of Rrp8, Nucleomethylin, is involved in regulation of rRNA synthesis and is an integral component of eNOSc complex, whereas the human homolog of Nop2 acts as a tumor marker (18,19). Interestingly, the human homolog of Rcm1 is completely deleted in Williams–Beuren syndrome, which is a complex developmental disorder (20). The substrates of most of the human homologs remained to be identified. As the positions of most of these modifications are also highly conserved, with the knowledge of yeast it is now possible to assign the substrate specificities to these human homologs, which are otherwise difficult to analyze.
In the present study, we analyzed the remaining base modifications of the 25S rRNA, the methylation of uridine residues, m3U (N3 methyl uridine) and m5U (C5 methyl uridine). Bmt5 (Yil096c) and Bmt6 (Ylr063w), two previously uncharacterized proteins, turned out to be the base methyltransferases responsible for m3U2634 and m3U2843 base methylation, respectively. The genes were identified by RP-HPLC screening of all deletion mutants of putative RNA methyltransferase and were validated by gene complementation and phenotypic characterization. Surprisingly, in contrast to previous predictions for the presence of m5U methylations, our analysis exhibited that 25S rRNA of S. cerevisiae does not contain any m5U methylation (21,22).
MATERIALS AND METHODS
Yeast strains and plasmids
The strains and plasmids used in the present study are listed in Supplementary Table S1. The polymerase chain reaction primers used for the construction of the plasmids are listed in Supplementary Table S2. A detailed protocol for construction of all plasmids will be provided on request.
Growth conditions and media
Yeast strains were grown at 30°C in YPD (1% yeast extract, 2% peptone, 2% glucose) or in synthetic dropout medium (0.5% ammonium sulphate, 0.17% yeast nitrogen base, 2% glucose). For serial dilution growth assays, yeast cells were grown overnight in YPD medium and diluted to an OD600 of 1 followed by 1:10 serial dilutions. From the diluted cultures, 5 µl were spotted onto YPD plates and incubated at 30 or 19°C.
Northern hybridization and RNA extraction
For northern hybridization, the RNA was prepared by phenol/chloroform extraction as previously described (13). Northern blotting was also performed as described previously (13). 25S rRNA for the RP-HPLC analysis was isolated from the 60S subunits, separated by sucrose gradient centrifugation as described previously (8).
Reversed phase−high performance liquid chromatography
RP–HPLC was performed as previously described with following adjustment (13,23). 25S rRNA (70 µg) was digested to nucleosides by nuclease P1 and alkaline phosphatase. Nucleosides were analyzed by RP-HPLC on a Supelcosil LC-18-S HPLC column (25 cm × 4.6 mm, 5 µm) equipped with a precolumn (4.6 × 20 mm) at 30°C on an Agilent 1200 HPLC system. For optimum separation of m3U residues, we changed the elution conditions as described previously to an isocratic mode using 50% buffer A (2.5% methanol) and 50% buffer B (20% methanol) (24).
Mung bean nuclease assay
The mung bean nuclease protection assay was performed exactly as described previously (13). Complementary synthetic deoxyoligonucleotides were used for hybridization and protection of specific sequence of isolated 25S rRNA. One thousand pico moles of the synthetic deoxyoligonucleotides complementary to yeast 25S rRNA were incubated with 100 pmoles of total rRNA and 1.5 µl of DMSO in 0.3 volumes of hybridization buffer (250 mM HEPES, 500 mM KCl, pH 7.0). The fragments were digested by mung bean nuclease and 0.02 µg/µl RNase A (Sigma-Aldrich) and were separated on a 13% polyacrylamide gel containing 7 M urea. RNA was extracted from the gel using D-Tube™ Dialyzers according to the manufacturer’s protocol for electroelution (Novagen®).
Polysome profiles
The polysome profiles were performed exactly as described previously (8). The yeast strains were grown in YPD medium (100 ml) at 30°C to early logarithmic phase (OD600 = 1.0). Before cell disruption, cycloheximide was added to the final concentration of 100 µg/ml. A 100-ml YPD culture (OD600 = 1.0) was harvested by centrifugation at 4°C. Cells were then washed twice with 10 ml of polysome buffer A (20 mM HEPES, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EGTA and 1 mM DTT), resuspended in 0.5 ml of buffer A and disrupted by vortexing with an equal volume of glass beads. Equivalent amounts of absorbing material were layered on a 10−50% (w/v) sucrose gradient in buffer A. The gradient was made using Gradient Master 107 (Biocomp). The samples were then centrifuged at 19 000 rpm for 17 h at 4°C in a SW40 rotor using Beckman ultracentrifuge (L-70: Beckman). Gradients were fractionated in an ISCO density gradient fractionar and the absorbance profile at 254 nm was analyzed in ISCO UA-5 absorbance monitor.
Immunoprecipitation
Affinity purification was performed exactly as described previously (25). Bmt6 TAP-tagged strain (Thermo Fischer) was inoculated in 1 l of YPD and was grown to an OD600 2–3. Instead of TEV elution, IgG bound proteins were eluted with 0.2 M Glycine (pH 2.5). Bmt6-TAP tag was detected using peroxidise–anti-peroxidase (PAP) antibody, whereas Nop2 was detected using anti-Nop2 (EnCor Biotechnology, Florida, USA), followed by anti-mouse IgG-conjugated horseradish peroxidase (Bio-Rad; 1:10 000 dilution).
Western blot analysis
For western blotting, 30 µg of total protein from each sample were separated with 12% sodium dodecyl sulphate (SDS) polyacrylamide gel and blotted on a PVDF membrane (Millipore). The membrane was blocked with 5% nonfat dry milk and the protein was detected with anti-His monoclonal antibody (Roche; 1:1000 dilution), followed by anti-mouse IgG-conjugated horseradish peroxidase (Bio-Rad; 1:10 000 dilution).
Protein localization
The plasmids pSH22 (GFP-Bmt5) and pSH28 (GFP-Bmt6) were constructed using pUG35 and pUG36 plasmids, respectively (EUROSCARF). Plasmids were then transformed in a strain containing a gene encoding for ScNop56-mRFP (26). Transformants carrying plasmid were grown in synthetic medium lacking uracil at 30°C. GFP-fused Bmt5 and Bmt6 were visualized using a Leica TCS SP5. The RFP-fused Nop56 was used as reference for nucleolar localization.
Primer extension
Primer extension analysis was carried out following the published protocol with some modifications (13). Ten pico moles of the DNA primer PE-2634-GATTTCTGTTCTCCATGAGC and PE 2843-GGAAGAGCCGACATCGAAGAATC, complementary to the positions 2663–2682 and 2863–2885 of 25S rRNA, respectively, were 32P-5′terminally labeled by incubation in the final volume of 40 µl with 50 µCi γ-[32P] ATP and 20 U of polynucleotide kinase in 2 µl of polynucleotide kinase buffer (Fermentas). Reaction was incubated at 37°C for 30 min and then stopped by incubation at 90°C for 2 min. The reaction mixture was next purified using Roche columns (11814419001) to get rid of free γ-[32P] ATP.
The extension reaction premix contained 6 µl of water, 2 µl of dNTP mix (10 µM dGTP, 10 µM dATP, 10 µM dTTP and 10 µM dCTP), 6 µl of RNase-free water and 1 µl (10 U) of Superscript III reverse transcriptase (Invitrogen) per reaction. Fourteen microliters of the premix were added to the reaction tube with the annealed primer/RNA complex. Samples were incubated at 55°C for 15 min. Following incubation, the RNA was precipitated using 3 M sodium acetate, pH 5.2, and 100% ethanol, followed by a washing step with 75% ethanol. After complete removal of supernatant and air-drying of the tubes for 2–3 min. The pellets were resuspended in 6 µl of formamide loading dye (Sambrook et al. 1989). We loaded 1 µl of the sample on to the sequencing gel (Model S2, Biometra) and let it run till the phenol blue dye reached the bottom of the gel. The gel was transferred to Whatman 3MM paper, dried and exposed on a phosphoimager screen. The screen was scanned using Typhoon 9400 (GE) with Storage Phosphor acquisition mode.
Overexpression and immobilized metal affinity chromatography
Plasmids pSH26 and pSH25 carrying N terminal 6xHis-Smt3–tagged Bmt5 and Bmt6, respectively, were constructed using pPK591 by gap repair. For overexpression, BL21 (DE3) cells were transformed with isolated plasmids and grown in LB medium containing 100 mg/ml ampicillin at 37°C to an OD600 of 0.6. Isopropyl-b-d-thiogalactopyranoside at 0.5 mM was added to the culture, and the incubation continued for 4 h. Cells were recovered by centrifugation, washed with water.
His-tagged Bmt5 and Bmt6 were purified by Nickel-NTA metal affinity chromatography. Briefly, cell pellets were thawed on ice using 10 ml of Lysis buffer with 100 μl of PMSF (0.1 M), 20 μl of Benzamidine (0.5 M) and 7 μl of 2-mercaptoethernol (14.3 M). Cells were lysed by sonication. The lysate was treated with 10 μl of RNase A (10 mg/ml) and 10 μl of DNase (10 mg/ml) on ice for 15 min. Cell debris was removed by centrifuging the lysate for 30 min at 15 000 rpm in a JA-20 Beckmann rotor. His-tagged protein in the supernatant fraction was allowed to bind to the Ni-NTA column (Protino® Ni-NTA agarose, MACHEREY-NAGEL), preequilibrated with lysis buffer. Unbound proteins were removed by washing the resin three times with washing buffer. Protein was eluted with 5 ml of lysis buffer containing 150 mM imidazole, collected in 500 -µl fractions, and then analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). The protein concentration was estimated using Bradford’s reagent (27).
To cleave the 6× His-Smt3 tag from the protein, Ulp1 enzyme (1/20th of the amount of eluted protein) was added and the sample was incubated at 37°C for 1 h. Cation exchange column (SP Sepahrose High performance, GE Life Sciences) was used to separate the 6× His-Smt3 tag from the protein. Elution was made with phosphate buffered saline containing different concentration of NaCl from 400 to 1200 mM
Isothermal titration calorimetry
All isothermal titration calorimetry (iTC) experiments were performed with an LLC iTC 200 (Micro Cal. Northampton, MA, USA) at 293.15 K. The sample cell was filled with 50–100 µM protein and the titrations were carried out using 1 mM SAM in phosphate buffered saline buffer. Injection volume of 0.2 μl with an interval of 0.4 s was injected. The time delay to allow equilibration between successive injections was 180 s. Stirring speed (1000 rpm) with reference power of 10 mcal/s was used during the titration. Control experiments were performed to correct the data for the enthalpy of dilution of ligand and buffer mixing. The heat signals obtained from iTC were analyzed using Origin software supplied by Micro Cal yielding the stoichiometry (N), the equilibrium association constant Ka as well as the enthalpy (ΔH) and the entropy (ΔS) of binding. The ΔG value was then used to calculate the dissociation constant (Kd) of SAM binding for Bmt5.
RESULTS
RP-HPLC screening of putative RNA methyltransferases revealed m3U methyltransferases
25S rRNA of S. cerevisiae has been reported to contain eight base methylations, two m1A, two m5C, two m3U and two m5U (8,28). Recently, except uridine methylations, enzymes for both adenine (m1A) and cytosine (m5C) methylation have been identified (8,12,13).
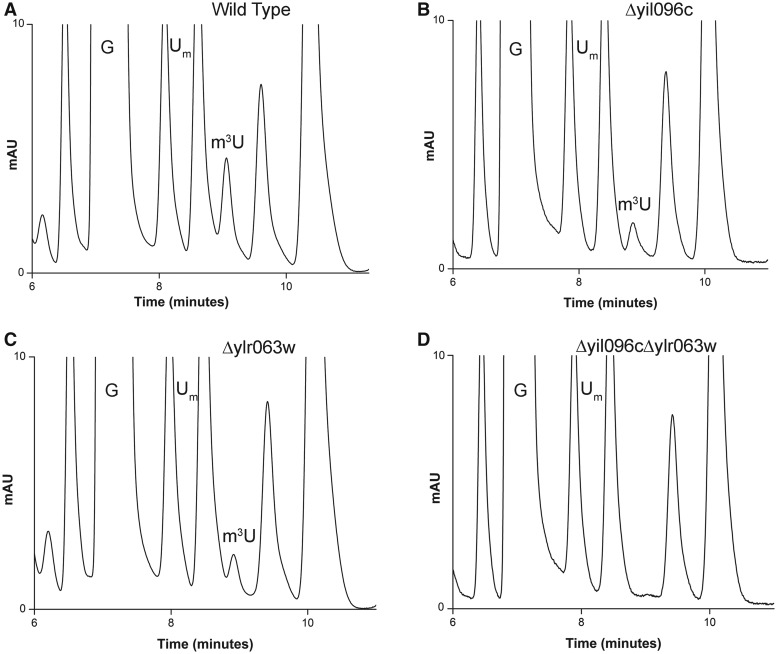
To identify the enzymes responsible for remaining m3U and m5U methylations, we biochemically analyzed single-gene deletion mutants of all uncharacterized RNA methyltransferases. For the analysis, 25S rRNA from each deletion mutant was isolated as described in ‘Materials and Methods’ section and their composition was analyzed using RP-HPLC. Interestingly, as seen in Figure 1, in the deletion mutant of Δyil096c and Δylr063w, the peak corresponding to the m3U reduced to half and disappeared completely in the double mutant Δyil096cΔylr063w, suggesting that Yil096c and Ylr063w are involved in the methylation of the m3U residues.
Figure 1.
RP-HPLC screening of the mutants for identification of m3U methyltransferase. The 25S rRNA from mutants and isogenic wild type were digested to nucleosides using P1 nuclease and alkaline phosphatase. Nucleosides obtained after digestion was then analyzed by RP-HPLC. For optimum separation of m3U residues, we changed the elution conditions to an isocratic mode using 50% buffer A (2.5% methanol) and 50% buffer B (20% methanol). RP-HPLC chromatogram from the wild type (A), Δyil096c (B), Δylr063w (C) and Δyil096cΔylr063w (D) mutants. The peak corresponding to the m3U with a retention time of ∼9 min reduces to half in both Δyil096c and Δylr063w mutants and disappears in double mutant Δyil096cΔylr063w, highlighting the involvement of these two methyltransferases in m3U methylations of the 25S rRNA.
Mung bean nuclease assay for the identification of the targets of Yil096c and Ylr063w
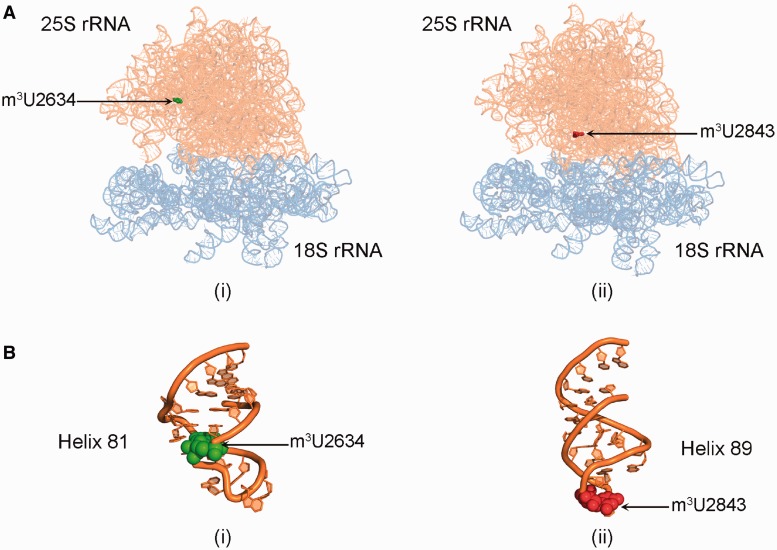
25S rRNA of yeast contains two m3U residues at position 2634 in the helix 81 and at position 2843 in the helix 89 (Figure 2). Therefore, to determine the nucleotide position at which the m3U modification was missing in Δylr063w and Δyil096c mutants, a mung bean nuclease protection assay was performed.
Figure 2.
Location of m3U methylations in the ribosomal RNA. (A) 3D cartoon of the rRNA structure of ribosome. The 18S rRNA is colored light blue, whereas the 25S rRNA is shown in orange. The m3U2634 residue is displayed in green spheres (i) and the residue m3U2843 is highlighted in red spheres (ii). (B) Cartoon representing the helix 81 (i) and 85 (ii) of the 25S rRNA. The RNA is colored in orange and the modified bases U2634 and U2843 are shown as green and red spheres, respectively. The cartoon was made using PyMol software (PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC) with PDB files, 3U5B.pdb and 3U5D.pdb.
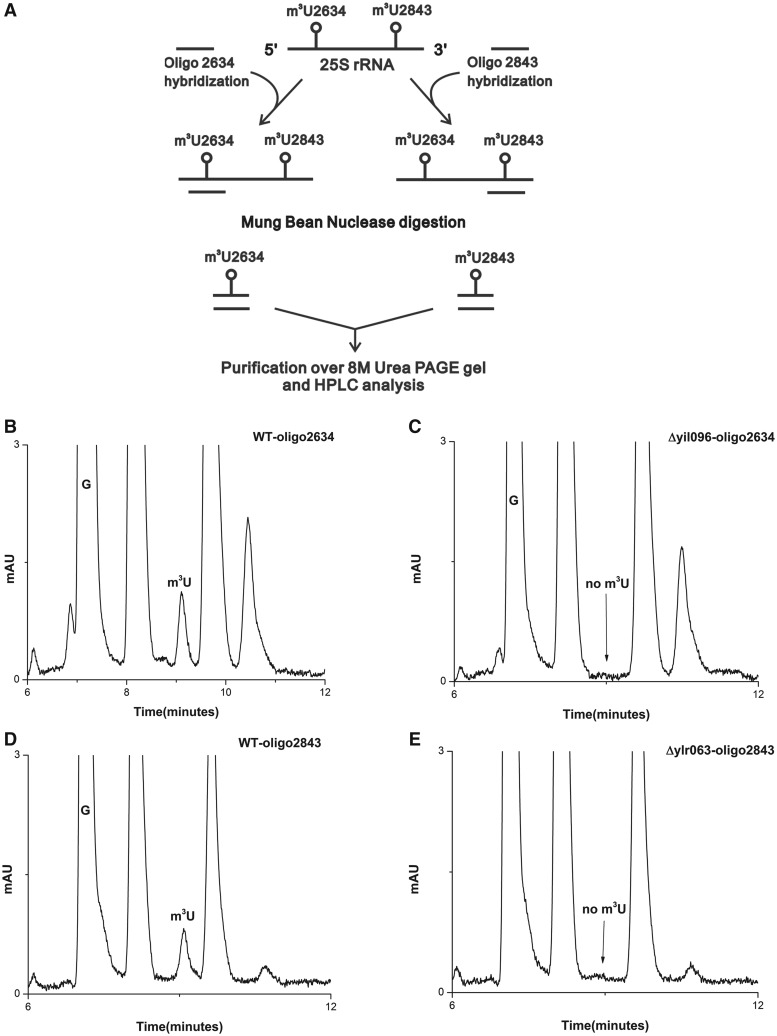
Synthetic oligonucleotides complementary to nucleotides 2613–2657 (Oligo-2634) and 2820−2866 (Oligo-2843), corresponding to position 2634 and 2843, respectively, were designed (Figure 3A). The 25S rRNA from Δyil096c and Δylr063w, and wild-type cells were next hybridized to these oligonucleotides and subjected to nuclease digestion. Six independent experiments using oligonucleotides corresponding to two different m3U positions were performed. Because only these fragments were protected due to double-stranded conformation, both m3U modifications could be separated and addressed independently. These protected RNA-DNA hybrid fragments were then separated from rest of the debris and from each other using 8M-urea PAGE gels.
Figure 3.
Mung bean nuclease protection assay. (A) Graphic representation of Mung bean nuclease protection assay used in the present study for the analysis of specific position of modified m3U nucleoside in the Δyil096c and Δylr063w mutants. RP-HPLC chromatogram of the nucleosides derived from protected RNA fragments. Specific fragments of the 25S rRNA from wild type, Δyil096c and Δylr063w corresponding to both m3U2634 and m3U2843 were isolated. The status of m3U residue in these fragments were then analyzed by RP-HPLC. (B) RP-HPLC chromatogram of the fragment corresponding to m3U2634 isolated from the wild type and (C) from Δyil096c. (D) RP-HPLC chromatogram of the fragment corresponding to m3U2843, isolated from the wild type and (E) from Δylr063w. As evident from the chromatograms, the m3U2634 methylation is absent in Δyil096c, whereas m3U2843 is missing in Δylr063w. This clearly demonstrated that Yil096c and Ylr063w are involved in the methylation of m3U2634 and m3U2843, respectively.
These RNA fragments were next subjected to RP-HPLC analysis for the presence of m3U methylation. As expected, in the wild-type cells, m3U methylation was present in both fragments corresponding to m3U2634 and m3U2843 (Figure 3B and D). However, in the Δyil096c mutant, only m3U2634 methylation was missing (Figure 3C). The m3U2843 methylation in Δyil096c remained unaffected (data not shown). Interestingly, in Δylr063w mutant, only m3U2843 methylation was absent (Figure 3D), whereas the m3U2634 methylation remained unaffected (data not shown). Our results clearly exhibited the involvement of Yil096c in m3U2634 and Ylr063w in m3U2843 methylation.
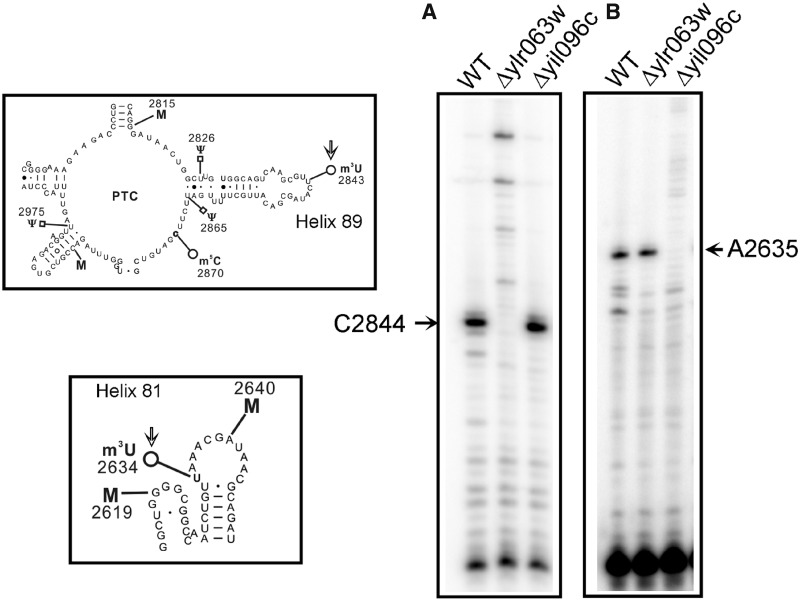
Primer extension analysis of the m3U2634 and m3U2843 methylations of the 25S rRNA
The methylation of the uridine at N3 blocks the Watson–Crick base pairing, and results in a strong stop in the primer extension analysis. Therefore, to further augment the role of Yil096c and Ylr063w in the methylation of m3U residues and validate the positions of the m3U modifications in the 25S rRNA, we performed the primer extension analysis with the 25S rRNA isolated from the Δyil096c and Δylr063w deletion mutants. As evident in Figure 4, the presence of m3U at position 2843 and 2634 in the 25S rRNA from the wild-type cells led to a strong stop at position 2844 and 2635. However, the absence of stop at position 2844 in deletion mutant Δylr063w (Figure 4A) and at position 2635 in deletion mutant Δyil096c (Figure 4B) demonstrated that Ylr063w and Yil096c are involved in the methylation of m3U2843 and m3U2634, respectively. Our primer extension analysis further validated the substrate specificities of Yil096c and Ylr063w in vivo as revealed by mung bean nuclease assay and verified the position of the m3U methylations in the 25S rRNA.
Figure 4.
Primer extension analysis. To further substantiate the role of Yil096c and Ylr063w in the methylation of m3U residues and validate the positions of the m3U modifications in the 25S rRNA, we performed the primer extension analysis with the 25S rRNA isolated from Δyil096c and Δylr063w deletion mutants along with isogenic wild type. The methylation of the uridine at N3 blocks the Watson–Crick base pairing, and results in a strong stop in the primer extension analysis (A) The presence of m3U at position 2843 in the 25S rRNA from the wild type and Δyil096c cells led to a strong stop at position 2844. However, this stop was absent in Δylr063w deletion mutant. (B) Similarly, owing to presence of an m3U residue at position 2634, a strong stop at position 2635 was observed in wild type and Δylr063w but was missing in Δyil096c deletion mutant. The positions of the methylated residues were determined accurately from running side by side a sequencing ladder (data not shown), is given on the sides of the gels, and the bands of interest are shown with arrows. This clearly validated the specific involvement of Ylr063w and Yil096c in the methylation of m3U2843 and m3U2634, respectively. Both helices 81 and 89, carrying m3U residues (marked with an arrow) are displayed on left of the panel A and B.
Once, it became apparent that Yil096c and Ylr063w are the proteins involved in the base methylation at U2634 and U2843 of 25S rRNA. We decided to name gene and YIL096C as BMT5 (Base methyltransferase of twenty five S rRNA 5) and YLR063w as BMT6 (Base methyltransferase of twenty five S rRNA 6).
Bmt5 and Bmt6 are predicted to be Rossmann-fold–like methyltransferase
The amino acid sequence analysis along with 3D modeling of both Bmt5 and Bmt6 demonstrated that both proteins belong to Rossmann-fold–like methyltransferases characterized by a central seven-stranded β-sheet that is flanked by three helices on each side (Supplementary Figure S1) (29).
As far as the conservation of the Bmt5 and Bmt6 is concerned, our searches of the nonredundant sequence database at NCBI using PSI-BLAST indicated that both Bmt5 and Bmt6 orthologs are conserved in members of lower eukaryotes (Supplementary Figures S2 and S3).
Bmt5 (Yil096c) and Bmt6 (Ylr063w) methylate m3U2634 and m3U2843, respectively, in vivo
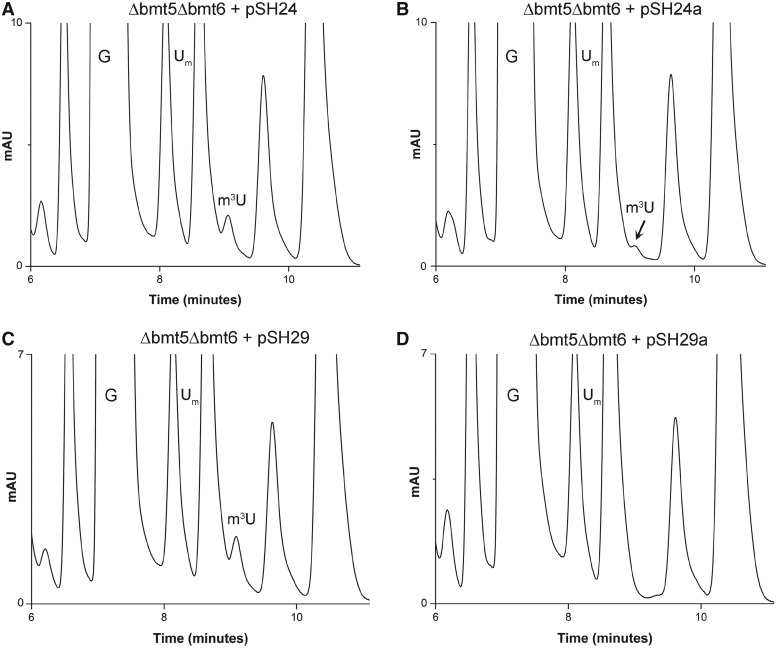
To confirm Bmt5 and Bmt6 to be methyltransferase (MTases) involved in performing m3U methylations, gene complementation analysis was conducted. The plasmids pSH24 and pSH29 carrying C-terminally heptahistidine-tagged Bmt5 and Bmt6, respectively, were transformed into Δbmt5Δbmt6 double mutant. The 25S rRNA from these transformed strains were then isolated and the composition of 25S rRNA was analyzed with RP-HPLC. As seen in Figure 5A and C, episomally expressed Bmt5 as well as Bmt6 were able to perform the m3U modification in vivo, seen as appearance of m3U residues in the double mutant.
Figure 5.
Gene complementation and analysis of methyltransferase dead mutants of Bmt5 and Bmt6. To confirm Bmt5 and Bmt6 to be MTases involved in performing m3U methylations, gene complementation analysis was conducted. The plasmids pSH24 and pSH29 carrying C-terminally heptahistidine-tagged Bmt5 and Bmt6, respectively, were transformed into Δbmt5Δbmt6 double mutant. The 25S rRNA from these transformed strains were then isolated and composition of 25S rRNA was analyzed with RP-HPLC. (A) Chromatogram of the 25S rRNA isolated from Δbmt5Δbmt6 strain carrying C-terminally heptahistidine-tagged Bmt5 (pSH24). (C) Chromatogram of the 25S rRNA isolated from Δbmt5Δbmt6 carrying N-terminally heptahistidine-tagged Bmt6 (pSH29). Both episomally expressed Bmt5 and Bmt6 could methylate their respective targets in vivo, seen as appearance of m3U peak in the RP-HPLC chromatogram. Furthermore, to corroborate the direct involvement of Bmt5 and Bmt6 in performing the m3U modifications, we created mutant alleles for Bmt5 and Bmt6, where we exchanged amino acids in the highly conserved SAM binding domain of both proteins. (B) RP-HPLC chromatogram from the nucleosides derived from 25S rRNA of Δbmt5Δbmt6 strain carrying mutant bmt5-G182R protein, expressed from plasmid pSH24a. (D) RP-HPLC chromatogram from the nucleosides derived from the 25S rRNA of Δbmt5Δbmt6 carrying mutant protein bmt6-G294R, expressed from plasmid pSH29a. The substitution of G182R in Bmt5 resulted in significant reduction in the amount of m3U residues, whereas the exchange of G294R in Bmt6 completely abolished the catalytic function of Bmt6.
To corroborate direct involvement of Bmt5 and Bmt6 in performing m3U modifications, we created mutant alleles for Bmt5 and Bmt6, where we exchanged the amino acids in the highly conserved SAM binding domain of both proteins. The point mutations were generated on plasmid pSH24 and pSH29, so that the expression of the mutant proteins could be analyzed with a western blot using anti-His antibodies. The plasmids pSH24-a (bmt5-G182R) and pSH29-b (bmt6-G294R) carrying mutant allele for Bmt5 and Bmt6, respectively, were then transformed into the Δbmt5Δbmt6 double mutant. The composition of the 25S rRNA from these transformed strains were next analyzed by RP-HPLC. Interestingly, as seen in Figure 5B, exchange of G182R in Bmt5 resulted in drastic reduction in the amount of m3U residues, and the exchange of G294R in Bmt6 completely abolished the m3U methylation (Figure 5D). The western blot exhibited no differences in the protein expression of the mutant proteins compared with wild-type Bmt5 and Bmt6 (Supplementary Figure S4). Apart from bmt5-G182R and bmt6-G294R, we also tested other point mutants of Bmt5 and Bmt6, bmt5-D138A and bmt6-G283R for expression analysis. The point mutations in SAM binding motif for both Bmt5 and Bmt6 did not affect the expression of the respective mutant proteins. Taken together, our results confirmed that Bmt5 and Bmt6 catalyzes the m3U methylation of the 25S rRNA.
Bmt5 binds SAM in vitro
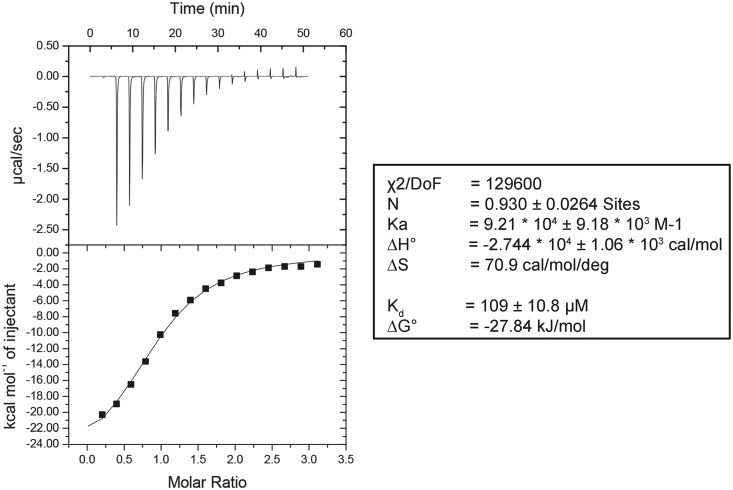
To demonstrate that Bmt5 and Bmt6 bind to SAM in vitro, both BMT5 and BMT6 genes were cloned into pPK591 and were expressed heterologously in Escherichia coli as explained in ‘Materials and Methods’ section. The recombinant DNA obtained codes for an N-terminal 6xHis-Smt3-tag fusion protein, which allowed easy purification by metal affinity chromatography. As seen in Supplementary Figure S5, the Bmt5 protein was purified to homogeneity by Ni-NTA affinity chromatography followed by cation exchange chromatography. The purified Bmt5 was then analyzed for SAM binding assay, using iTC as described in ‘Materials and Methods’ section. Interestingly, as evident in the Figure 6, the purified Bmt5 bind SAM in vitro with Kd value of 109±10.8 μM. Unfortunately, although successfully expressed in E. coli, Bmt6 precipitated during the purification step (data not shown) and we failed to retrieve sufficient amount of protein for the subsequent analysis.
Figure 6.
Bmt5 binds SAM in vitro. To demonstrate SAM binding of Bmt5, the heterologously expressed Bmt5 was purified to homogeneity by Ni-NTA and cation exchange chromatography and its binding was analyzed by iTC. The left panel display titration of SAM into the Bmt5 protein (sample cell). The upper panel shows the baseline-corrected titration data, and the lower panel shows the binding isotherm fit to a model for a single SAM binding site. The right panel shows different parameters calculated from the titration analysis. The heat signals obtained from iTC were analyzed using Origin software supplied by Micro Cal yielding the stoichiometry (N), the equilibrium association constant Ka as well as the enthalpy (ΔH) and the entropy (ΔS) of binding. Bmt5 binds SAM with a Kd of 109 ± 10.8 µM.
Once it became clear that heterologously expressed Bmt5 binds to SAM in vitro, we next examined Bmt5 for in vitro methylation reaction using SAM 510™ methyltransferase kit (G-Biosciences) and 60S subunits (isolated from Δbmt5 strain by sucrose gradient centrifugation) as substrate. Unexpectedly, we failed to observe any methyltransferase activity of the heterologous Bmt5 in vitro. We conjecture that this might be due to different conformation and composition of mature 60S subunit in the cytoplasm, used in the reaction as compared with the pre-60S subunit in nucleolus where Bmt5 catalyzes the m3U2634 methylation.
Cellular localization of Bmt5 and Bmt6
The subcellular localization of the enzymes can provide important information about their biological functions. The cellular localization of both proteins were previously analyzed in a high-throughput analysis (26). To further validate the cellular localization of Bmt5 and Bmt6, plasmids pSH22 (Bmt5-GFP) and pSH28 (Bmt6-GFP) were constructed, where Bmt5 and Bmt6 were expressed as GFP fusion protein. The plasmids were then transformed into a strain where Nop56, a nucleolar protein, was expressed as a RFP fusion protein. The cells expressing GFP-fusion protein were next observed with the help of a confocal Leica TCS SP5 microscope. Intriguingly, as evident in Supplementary Figure S6, Bmt5 localizes exclusively in the nucleolus, whereas Bmt6 localizes predominantly in cytoplasm with seemingly few molecules in nucleus. To rule out any mislocalization of Bmt5 and Bmt6 because of being expressed as GFP fusion proteins, we tested the functionality of both Bm5-GFP and Bmt6-GFP fusion proteins for performing m3U methylation in vivo. Both Bmt5-GFP and Bmt6-GFP fusion proteins turned out to be capable of catalyzing the m3U methylations in vivo (data not shown).
We next analyzed both Bmt5 and Bmt6 for their association with ribosomal or preribosomal particles by sucrose gradient centrifugation. Bmt5 and Bmt6 TAP-tagged strains (Thermo-Fischer) were used for the analysis of co-sedimentation with ribosomal and preribosomal particles. Polysome profiles from the respective strains were made and the fractions corresponding to free pool, 40S, 60S, 80S, preribosomes and polysomes were collected and precipitated for western blotting using PAP antibody. Interestingly, as observed in Supplementary Figure S6C, both Bmt5 and Bmt6 co-sediments with the ribosomal/preribosomal particles. For Bmt6, a significant amount of protein was observed to be present in the free cytoplasmic free pool. Minor bands of both Bmt5 and Bmt6 TAP-tagged proteins were also detected at positions corresponding to complexes of ∼40S and lower, suggesting that both proteins not only interact with ribosomal particle but also with other smaller complexes. Similar sedimentation patterns have also been observed for other base methyltransferases and some of the trans-acting factors involved in ribosome biogenesis (13,30).
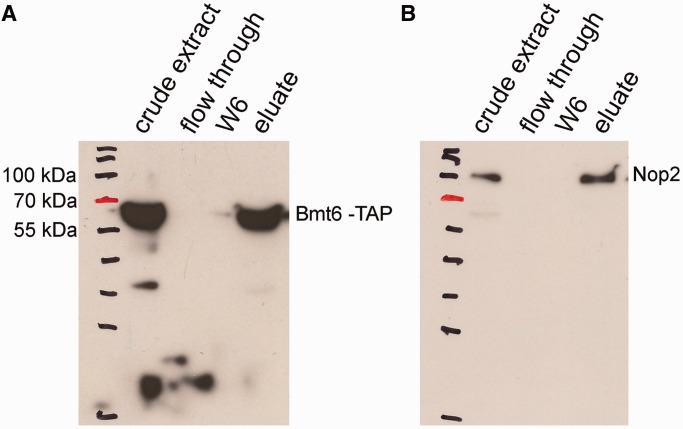
Bmt6 TAP co-immunoprecipitates Nop2
To further demonstrate the interaction of Bmt6 with the preribosomal particles and its minor, yet significant, nuclear localization, we immunoprecipitated the total cell extract from Bmt6-TAP–tagged strain using IgG sepharose and analyzed its interaction for Nop2, a nucleolar protein by western blotting. The reason for selecting Nop2 was its already well-characterized nucleolar localization and its active involvement in ribosome biogenesis, especially 60S subunit. Moreover, since the residues catalyzed by Bmt6 (U2843) and Nop2 (C2870) are in close proximity, we hypothesized that if Bmt6 is present in the pre-60S particle, there is a high chance of Bmt6 and Nop2 to be present in the same complex. Remarkably, as evident in Figure 7, Bmt6-TAP specifically immunoprecipitated Nop2, suggesting that like Nop2, Bmt6 is also a part of pre-60S particle and seemingly catalyze the methylation of U2843 during early maturation steps of 60S biogenesis. Interestingly, Planta’s lab has previously demonstrated that both uridine methylations are performed early during the maturation of 60S subunits in nucleus (4).
Figure 7.
Bmt6 interacts with Nop2 in vivo. To demonstrate the interaction of Bmt6 with the preribosomal particles and its minor nuclear localization, we immunoprecipitated the total cell extract from Bmt6-TAP–tagged strain using IgG sepharose and analyzed its interaction for Nop2 by western blotting. (A) Western blot with PAP antibodies. (B) Western blot with anti-Nop2 (EnCor Biotechnology, Florida, USA) followed by anti-mouse IgG-conjugated horseradish peroxidase (Bio-Rad; 1:10 000 dilution). The samples in the different lanes of the gels are crude extract (total cell extract), flow through (unbound cell extract) and wash fraction 6 (the beads were washed six times with IPP-150 buffer) and eluate.
Growth, polysome profiles and rRNA processing analysis of Δbmt5 and Δbmt6
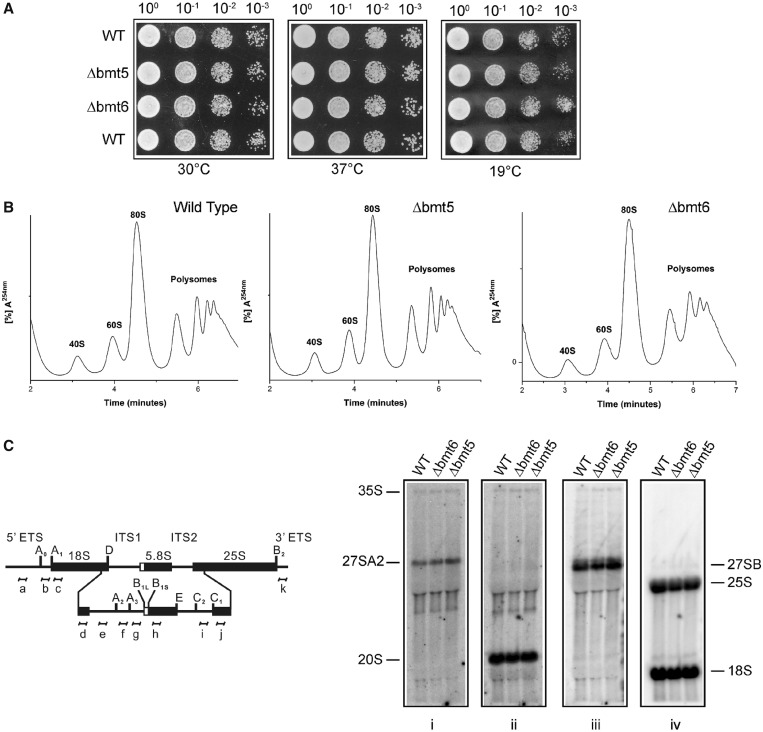
In our previous studies with the adenine and cytosine methylations, we discovered that the loss of methylation influences growth, antibiotic sensitivity and ribosome biogenesis. To investigate any such influence of the m3U modifications on cellular physiology, we initially analyzed the growth of single deletion mutants at different temperatures. Surprisingly, as shown in Figure 8A, loss of both bmt5 and bmt6 did not impair the growth of the cells at 30, 37 and 19°C on YPD media. We next analyzed the antibiotic sensitivity for both Δbmt5 and Δbmt6. The loss of bmt5 and bmt6 did not exhibit any hypersensitivity to paromomycin and anisomycin (data not shown). We also performed the same analyses with Δbmt5 Δbmt6 double mutant and did not observe any growth defects and antibiotic hypersensitivity (data not shown).
Figure 8.
Growth, polysome and rRNA processing analysis for Δbmt5 and Δbmt6 mutants. (A) Ten-fold serial dilutions of the strains were spotted onto solid YPD plates and were incubated at different temperatures. (B) Polysome profile of isogenic wild type (WT), Δbmt5 and Δbmt6 mutant. (C) Illustration for the 35S primary transcript. 35S rRNA contains 18, 5.8 and 25S rRNA sequences separated by ITS1 and ITS2. The processing of the 35S precursor to mature rRNA involves endonucleolytic and exonucleolytic steps at specific sites. (D) Northern blot analysis of the Δbmt5 and Δbmt6 mutant. The membrane was hybridized with radioactive (32P) labeled probes for ITS1, (f) for panel (i) and (e) for panel (ii), for ITS2, (i) for panel (iii) and to oligonucleotides specific to 18S (d) and 25S rRNA (j) for panel (iv). Loss of m3U methylations does not influence the growth and rRNA processing.
To analyze the effect of loss of these modifications on ribosome synthesis and translation, we examined the polysome profiles of both deletion mutants. As observed in Figure 8B, absence of these modifications did not disturb ribosomal stoichiometry and affect translation. We also analyzed rRNA processing in both Δbmt5 and Δbmt6 by northern blotting. Parallel to polysome profiles, we did not observe any change in the pattern of rRNA processing in Δbmt5 and Δbmt6 (Figure 8C).
Yeast 25S rRNA does not contain any m5U residue
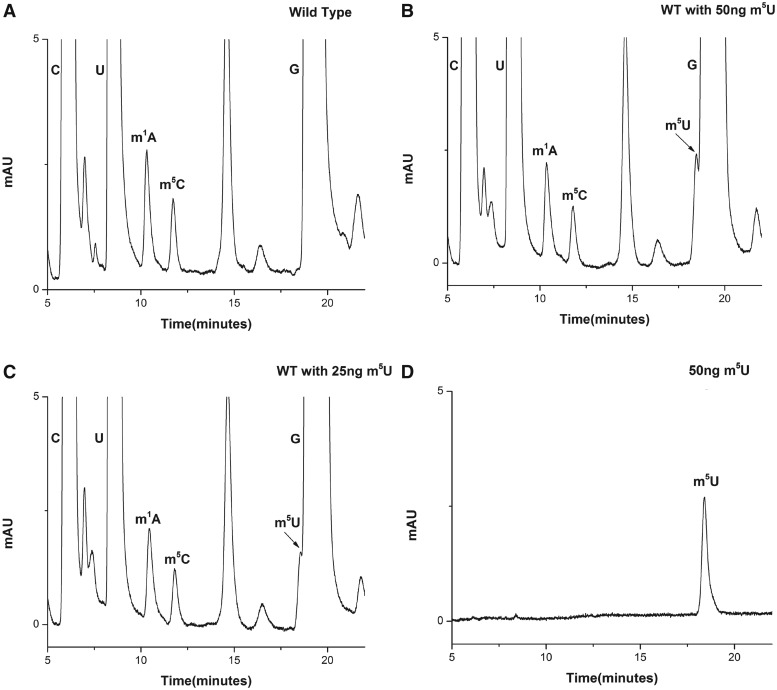
Yeast 25S rRNA was reported to contain two m5U residues at positions 956 and 2924 in the helices 37 and 92, respectively (Supplementary Figure S7). To characterize and identify enzymes associated with these m5U base methylations of 25S rRNA, we analyzed the composition of 25S rRNA isolated from the yeast wild-type strain. Surprisingly, as evident from Figure 9A, we could not detect any m5U nucleosides, which was in contrast to previous reports (21,22) and as mentioned at database (3D Ribosomal Modification Maps Database, http://people.biochem.umass.edu/fournierlab/3dmodmap/main.php). Initially, the absence of m5U modification was seen as a technical limitation. Previous studies have shown that similar retention time of Guanosine (G) and m5U make it difficult to separate the m5U residues from G (31). Moreover, since the amount of G derived from any given amount of 25S rRNA is higher than m5U residues, there is always a possibility of m5U being hidden under the G peak. Therefore, to rule out any technical limitation, we analyzed the resolution of our RP-HPLC chromatogram in terms of separating G and m5U residues. For the resolution check, we made use of pure m5U residues (Berry and Associates Inc. Dexter, MI, USA). Since, 25S rRNA was predicted to contain two m5U residues, we next calculated the amount of two m5U residues expected to be derived from 70 µg of 25S rRNA, using m1A as a control. The reason for selecting m1A, as a control was similar extinction coefficient of adenosine and uridine at 254 nm and equal number of residues expected to be present in the 25S rRNA (32). The amount turned out to be ∼50 ng for two m5U residues.
Figure 9.
25S rRNA of yeast does not contain any m5U residue. (A) RP-HPLC chromatogram of 25 S rRNA displaying modified bases, m1A, m5C along with C, G and U. Surprisingly no m5U residue was detected in the chromatogram. (B) RP-HPLC chromatogram of yeast 25 S rRNA with 50 ng of m5U residues. (C) RP-HPLC chromatogram of yeast 25S rRNA with 25 ng of m5U residues. (D) RP-HPLC chromatogram of pure 50 ng m5U residues. As evident from chromatograms (B) and (C) the m5U peak could be resolved and detected in our RP-HPLC setup. This clearly demonstrates absence of m5U residues from the yeast 25S rRNA.
To check the retention time and peak area of purified m5U, 50 ng of m5U was loaded on to the column. As seen in Figure 9B, m5U carried the retention time of ∼18 min, close to G (∼19.5 min). Also, the peak area of 50 ng of m5U was comparable with the peak area of two m1A residues (derived from 70 µg of 25S rRNA).
Next, we added 50 and 25 ng of pure m5U residues to the nucleosides mixture derived from the 70 µg of 25S rRNA and performed the RP-HPLC analysis. Surprisingly, as seen in Figure 9C and D, we could not only resolve and detect a peak corresponding to m5U modification, but could also rule out the presence of any m5U modified base in the 25S rRNA. Had there been any modification, we should have seen a doubling of the peak area on adding additional 25 and 50 ng pure m5U residues. We also performed similar analysis with the 25S rRNA isolated from the cells grown to different growth phases (exponential, mid-stationary and stationary) and did not observe any m5U residues (data not shown). To further validate the absence of m5U from the rRNA we also analyzed the 25S rRNA by mass spectrometry. The mass spectrometry analysis further validated the absence of m5U residues (data not shown). Our results clearly exhibited that the 25S rRNA of S. cerevisiae does not contain any m5U methylation.
DISCUSSION
Owing to the absence of easily discernible phenotypes, the base methyltransferases of the 25S rRNA remained elusive for a long time. Only recently, we have been able to identify most of these base methyltransferases by using RP-HPLC screening of the deletion mutants of putative methyltransferases. In the present study, we analyzed the m3U and m5U methylations of the 25S rRNA and demonstrated that yeast 25S rRNA contains two m3U residues at positions 2634 and 2843 and does not contain any m5U residues, as shown by Rudi Panta’s lab in early 1973 (3). Mapping of m3U residues on the mature 60S subunit revealed that both residues are located in the functionally conserved region of the 25S rRNA. The m3U2634 is located in the helix 81 and the m3U2843 is located in the helix 89 (Figure 2). Both helices 81 and 89 constitute the domain V of the 25S rRNA, responsible for catalytic function of ribosome (33).
In here we identified Yil096c and Ylr063w, two previously uncharacterized proteins to be the enzymes responsible for methylation of uridine residues at N-3 atom (m3U). We named these two ORFs as Bmt5 and Bmt6, YIL096C as BMT5 and YLR063w as BMT6.
Methylation is the most prevalent modification in rRNA. Base methylation fosters base stacking by increasing the hydrophobicity and expanding the polarizability. Methylation also influences the structure by increasing steric hindrance, blocking hydrogen bonds (e.g. at Watson–Crick positions) and providing positive charge to the nucleosides, which in turn effect the hydrogen bonding (34). We demonstrated by primer extension analysis that m3U indeed hinders the Watson–Crick base pairing. Therefore, it is tempting to speculate that methylation of specific uridines at N3 atoms prevent undesirable base pairing in rRNA that might disturb the conformation of RNA, especially near the catalytic center.
Intriguingly, until now the methylation of uridine at N3 atom (m3U) has only been discovered in the rRNA. rRNA from all three kingdoms of life contain m3U residues. The human 28S rRNA has also been reported to contain a single m3U residue at position 4513. Similarly, a single m3U residue in the 23S rRNA of an archaea Sulfolobus solfataricus and a single m3U in the 16S rRNA of E. coli have also been identified. Surprisingly, apart from E. coli, m3U methyltransferases from other organisms remained elusive. RsmE, the only hitherto known m3U methyltransferase, is a SPOUT methyltransferase that catalyzes the methylation of m3U1498 in the 16S rRNA of E. coli. Contrastingly, the amino acid sequence of Bmt5 and Bmt6 revealed that both proteins belong to Rossmann-like fold protein family. Supportingly, the in silico 3D model of Bmt5 and Bmt6 further illustrated that both proteins contain characteristic central seven-stranded β-sheet, flanked by three helices on each side characteristic of Rossmann-like fold protein family (Supplementary Figures S1 and S2). Moreover, in the present study, we also demonstrated that the substitution of highly conserved glycine residues in the SAM binding motif of both Bmt5 and Bmt6 with arginine abolishes the catalytic function of the protein. Proteins with highly significant similarity also exist in other lower eukaryotes including yeasts like Schizosaccharomyces pombe, Neurospora crassa and Kluyveromyces lactis (Supplementary Figures S2 and S3). The Bmt5 and Bmt6 homologs are likely performing the same function in these organisms.
Ribosome biogenesis is a complex multistep process, where modifications are introduced both co-transcriptionally and posttranscriptionally on to the rRNA molecules, before its assembly into subunits (6). The synthesis of some modifications, however, requires specific conformations of pre-RNA available only in fully or partial assembled subunits. This might also be the reason why we failed to observe any significant methyltransferase activity of heterologous Bmt5 using mature 60S subunits. Proteomics and structure analysis (rRNA) of preribosomal particles have revealed that precursors of ribosomal subunits differ both in composition and conformation as compared with mature subunits (35,36). Also, as ribosome biogenesis is a highly dynamic process with several fleeting interaction of different trans-acting factors, identification of the in vitro substrates for the base methyltransferases is a major challenge.
In the present study, we also provide some evidences for the site and timing of m3U methylations. The nucleolar localization of Bmt5 along with its preribosomal association made it apparent that the Bmt5-mediated m3U2634 methylation is performed early during the synthesis of 60S subunits. Likewise, even though Bmt6 is predominantly located in the cytoplasm, the Nop2 Bmt6 interaction suggests that Bmt6-catalyzed m3U2843 methylation is apparently performed in the nucleus, suggesting that Bmt6 is a nucleocytoplasmic protein. Interestingly, previous studies have also assigned a similar chronology to the m3U methylations (4). As far as nucleocytoplasmic localization of Bmt6 is concerned, further studies are required to decipher any role of Bmt6 in cytoplasm.
The base methylations influences the antibiotic sensitivity and also impacts the synthesis of 60S subunits (8,13). Surprisingly, for these two uridine methylations, we could not discover any such phenotypes. Therefore, the question, what are the significances of these modifications in ribosomal function, remained open.
The existence of m5U was dubious right from the beginning, where Bakin et al. only speculated the presence of a possible m5U modification at positions 956 and 2924 of the 25S rRNA (21). The assumption was solely based on the chemical reactivity to l-cyclohexyl-3-(2-morpholinoethyl)-carbodiimide metho-p-toluenesulfonate (CMC)-OH without any further chemical characterization by RP-HPLC or mass spectrometry. Similarly, Lapeyre et al. also could not validate the presence of any m5U modifications (22). On the contrary, Planta’s lab showed explicitly that the 25S rRNA of the yeast does not contain any m5U methylation (3).
Previous studies have proposed that the RNA m5U methyltransferases appear to have appeared in bacteria and were then dispersed by horizontal transfer of an rlmD-type gene to the Archaea and Eukaryota (37). As far as eukaryotes are concerned, m5U residues have been only observed in tRNAs, which is in consent with the presence of only one m5U methyltransferase, Trm2 (in yeast and its homologs), responsible for m5U modification of tRNAs at position 54 (31). Up till now, Trm2 has not been observed to display any interaction with the rRNA in yeast. We also analyzed the 25S rRNA of Candida albicans and S. pombe and like S. cerevisiae we could not detect any m5U residues (data not shown). Similarly, rRNA modification analysis of higher eukaryotes performed elsewhere also did not identify any m5U residues in the large or small rRNA (38). Taken together, this would apparently suggest that during the course of evolution, m5U methylations have disappeared in rRNA of eukaryotes.
In summary, we showed that Bmt5 and Bmt6 are the Rossmann-fold–like methyltransferases that catalyze the m3U2634 and m3U2843 methylations of the 25S rRNA, respectively. Furthermore, we demonstrated that the 25S rRNA of yeast does not contain any m5U methylation as previously predicted. With these two methyltransferases, the enzymes for all currently known base methylation of the 25S rRNA have been identified. The knowledge of these base methyltransferases along with their substrate specificities will foster the future structural and functional analysis of the significance of these modifications in the functionality of the ribosomes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: DFG: Deutsche Forschungsgemeinschaft [En134-9].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Karl-Dieter Entian would like to dedicate this study to late Prof. Dieter Mecke, a great biochemist and mentor. Sunny Sharma would like to thank DAAD for the award of Ph.D scholarship. We would like to thank Benjamin Weiss for helping with sequencing gels and all the members of Entian’s lab for fruitful discussions. We would also like to acknowledge Prof. Dr Mark Helm and Dr Stefanie Kellner for mass spectrometry analysis and Prof. Dr Jens Wöhnert for stimulating discussions and suggestions.
REFERENCES
- 1.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 2.Lilley DM. The ribosome functions as a ribozyme. Chembiochem. 2001;2:31–35. doi: 10.1002/1439-7633(20010105)2:1<31::AID-CBIC31>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 3.Klootwijk J, Planta RJ. Analysis of the methylation sites in yeast ribosomal RNA. Eur. J. Biochem. 1973;39:325–333. doi: 10.1111/j.1432-1033.1973.tb03130.x. [DOI] [PubMed] [Google Scholar]
- 4.Brand RC, Klootwijk J, Van Steenbergen TJ, De Kok AJ, Planta RJ. Secondary methylation of yeast ribosomal precursor RNA. Eur. J. Biochem. 1977;75:311–318. doi: 10.1111/j.1432-1033.1977.tb11531.x. [DOI] [PubMed] [Google Scholar]
- 5.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 6.Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piekna-Przybylska D, Decatur WA, Fournier MJ. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res. 2008;36:D178–D183. doi: 10.1093/nar/gkm855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma S, Yang J, Watzinger P, Kötter P, Entian K-D. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 2013;41:9062–9076. doi: 10.1093/nar/gkt679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin JL, McMillan FM. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002;12:783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- 11.Taylor AB, Meyer B, Leal BZ, Kotter P, Schirf V, Demeler B, Hart PJ, Entian KD, Wohnert J. The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site. Nucleic Acids Res. 2008;36:1542–1554. doi: 10.1093/nar/gkm1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peifer C, Sharma S, Watzinger P, Lamberth S, Kotter P, Entian KD. Yeast Rrp8p, a novel methyltransferase responsible for m1A 645 base modification of 25S rRNA. Nucleic Acids Res. 2013;41:1151–1163. doi: 10.1093/nar/gks1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma S, Watzinger P, Kötter P, Entian K-D. Identification of a novel methyltransferase, Bmt2, responsible for the N-1-methyl-adenosine base modification of 25S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2013;41:5428–5443. doi: 10.1093/nar/gkt195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer B, Wurm JP, Kotter P, Leisegang MS, Schilling V, Buchhaupt M, Held M, Bahr U, Karas M, Heckel A, et al. The Bowen-Conradi syndrome protein Nep1 (Emg1) has a dual role in eukaryotic ribosome biogenesis, as an essential assembly factor and in the methylation of 1191 in yeast 18S rRNA. Nucleic Acids Res. 2011;39:1526–1537. doi: 10.1093/nar/gkq931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wurm JP, Meyer B, Bahr U, Held M, Frolow O, Kotter P, Engels JW, Heckel A, Karas M, Entian KD, et al. The ribosome assembly factor Nep1 responsible for Bowen-Conradi syndrome is a pseudouridine-N1-specific methyltransferase. Nucleic Acids Res. 2010;38:2387–2398. doi: 10.1093/nar/gkp1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lafontaine D, Delcour J, Glasser AL, Desgrès J, Vandenhaute J. The DIM1 gene responsible for the conserved m6(2)Am6(2)A dimethylation in the 3′-terminal loop of 18 S rRNA is essential in yeast. J. Mol. Biol. 1994;241:492–497. doi: 10.1006/jmbi.1994.1525. [DOI] [PubMed] [Google Scholar]
- 17.White J, Li Z, Sardana R, Bujnicki JM, Marcotte EM, Johnson AW. Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre-40S subunits. Mol. Cell. Biol. 2008;28:3151–3161. doi: 10.1128/MCB.01674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khanna-Gupta A. p120 nucleolar-proliferating antigen is a direct target of G-CSF signaling during myeloid differentiation. J. Leukoc. Biol. 2006;79:1011–1021. doi: 10.1189/jlb.0205066. [DOI] [PubMed] [Google Scholar]
- 19.Yang L, Song T, Chen L, Kabra N, Zheng H, Koomen J, Seto E, Chen J. Regulation of SirT1-NML binding by rRNA coordinates ribosome biogenesis with nutrient availability. Mol. Cell. Biol. 2013;33:3838–3848. doi: 10.1128/MCB.00476-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doll A, Grzeschik KH. Characterization of two novel genes, WBSCR20 and WBSCR22, deleted in Williams-Beuren syndrome. Cytogenet. Cell Genet. 2001;95:20–27. doi: 10.1159/000057012. [DOI] [PubMed] [Google Scholar]
- 21.Bakin A, Lane BG, Ofengand J. Clustering of pseudouridine residues around the peptidyltransferase center of yeast cytoplasmic and mitochondrial ribosomes. Biochemistry. 1994;33:13475–13483. doi: 10.1021/bi00249a036. [DOI] [PubMed] [Google Scholar]
- 22.Lapeyre B, Purushothaman SK. Spb1p-directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing stage. Mol. Cell. 2004;16:663–669. doi: 10.1016/j.molcel.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 23.Gehrke CW, Kuo KC. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography. J. Chromatogr. 1989;471:3–36. doi: 10.1016/s0021-9673(00)94152-9. [DOI] [PubMed] [Google Scholar]
- 24.Gehrke CW, Kuo KCT. Analytical Methods for Major and Modified Nucleosides - HPLC, GC, MS, NMR, UV and FT-IR. 1989. Elsevier; Distributors for the U.S. and Canada Elsevier Science Pub. Co Amsterdam, New York, NY, USA. [Google Scholar]
- 25.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 26.Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 27.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 28.Veldman GM, Klootwijk J, de Regt VC, Planta RJ, Branlant C, Krol A, Ebel JP. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981;9:6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 30.Fatica A, Cronshaw AD, Dlakić M, Tollervey D. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell. 2002;9:341–351. doi: 10.1016/s1097-2765(02)00458-6. [DOI] [PubMed] [Google Scholar]
- 31.Nordlund ME, Johansson JO, von Pawel-Rammingen U, Byström AS. Identification of the TRM2 gene encoding the tRNA(m5U54)methyltransferase of Saccharomyces cerevisiae. RNA. 2000;6:844–860. doi: 10.1017/s1355838200992422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cavaluzzi MJ. Revised UV extinction coefficients for nucleoside-5′-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004;32:e13. doi: 10.1093/nar/gnh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenner L, Melnikov S, de Loubresse NG, Ben-Shem A, Iskakova M, Urzhumtsev A, Meskauskas A, Dinman J, Yusupova G, Yusupov M. Crystal structure of the 80S yeast ribosome. Curr. Opin. Struct. Biol. 2012;22:759–767. doi: 10.1016/j.sbi.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 34.Yi C, Pan T. Cellular dynamics of RNA modification. Acc. Chem. Res. 2011;44:1380–1388. doi: 10.1021/ar200057m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bradatsch B, Leidig C, Granneman S, Gnädig M, Tollervey D, Böttcher B, Beckmann R, Hurt E. Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat. Struct. Mol. Biol. 2012;19:1234–1241. doi: 10.1038/nsmb.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez-Fernandez J, Roman A, Las Rivas,De J, Bustelo XR, Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell. Biol. 2007;27:5414–5429. doi: 10.1128/MCB.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auxilien S, Rasmussen A, Rose S, Brochier-Armanet C, Husson C, Fourmy D, Grosjean H, Douthwaite S. Specificity shifts in the rRNA and tRNA nucleotide targets of archaeal and bacterial m5U methyltransferases. RNA. 2011;17:45–53. doi: 10.1261/rna.2323411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maden B. Locations of methyl groups in 28 S rRNA of Xenopus laevis and man: clustering in the conserved core of molecule. J. Mol. Biol. 1988;201:289–314. doi: 10.1016/0022-2836(88)90139-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.