Abstract
The bacterial Rcs phosphorelay signals perturbations of the bacterial cell envelope to its response regulator RcsB, which regulates transcription of multiple loci related to motility, biofilm formation and various stress responses. RcsB is unique, as its set of target loci is modulated by interaction with auxiliary regulators including BglJ. The BglJ–RcsB heteromer is known to activate the HNS repressed leuO and bgl loci independent of RcsB phosphorylation. Here, we show that BglJ–RcsB activates the promoters of 10 additional loci (chiA, molR, sfsB, yecT, yqhG, ygiZ, yidL, ykiA, ynbA and ynjI). Furthermore, we mapped the BglJ–RcsB binding site at seven loci and propose a consensus sequence motif. The data suggest that activation by BglJ–RcsB is DNA phasing dependent at some loci, a feature reminiscent of canonical transcriptional activators, while at other loci BglJ–RcsB activation may be indirect by inhibition of HNS-mediated repression. In addition, we show that BglJ–RcsB activates transcription of bgl synergistically with CRP where it shifts the transcription start by 20 bp from a position typical for class I CRP-dependent promoters to a position typical for class II CRP-dependent promoters. Thus, BglJ–RcsB is a pleiotropic transcriptional activator that coordinates regulation with global regulators including CRP, LeuO and HNS.
INTRODUCTION
RcsB is the response regulator of the Rcs phosphorelay which is conserved in Enterobacteriaceae. RcsB plays a pleiotropic role in the control of biofilm formation and motility as well as in the general stress response (1,2). Correspondingly, RcsB controls transcription initiation at multiple loci. It does so either as homodimer or together with auxiliary transcriptional regulators including RcsA, BglJ, GadE, MatA and others (1,3–6). Interaction of RcsB with the auxiliary factors extends its regulatory repertoire and represents a unique mechanism of modulation of transcriptional control in bacteria, where heteromeric transcription factors are rare.
RcsB and its auxiliary regulators all belong to the same family of transcriptional regulators carrying a characteristic C-terminal helix-turn-helix DNA-binding domain of the FixJ-type (also named LuxR, NarL or UhpA-type) (7,8). The N-terminal domain of RcsB corresponds to a receiver domain typical of two-component response regulators, which becomes phosphorylated by the Rcs phosphorelay in response to perturbations of the outer membrane and peptidoglycan layer (2,9). Perception of these extracellular signals requires the lipoprotein RcsF and the sensory and transmitter proteins RcsC and RcsD (10,11). The N-terminal domains of the auxiliary regulators RcsA (11) and BglJ are related to a receiver domain, but their alignment suggests that BglJ lacks a aspartic acid phosphorylation site.
Genetic and two-hybrid analyses conducted for RcsA and BglJ, as well as molecular analyses for GadE, suggest that RcsB interacts with these auxiliary proteins and possibly forms heterodimers or heterooligomers (1,3,4). Accordingly, the DNA-binding motifs of RcsB homodimers, RcsB–RcsA heteromers and BglJ–RcsB heteromers are similar in only one half, which may be bound by RcsB (4,12,13). RcsA and BglJ are completely dependent on RcsB (1,4). The activity of the heteromeric RcsA–RcsB depends on phosphorylation of RcsB, while BglJ–RcsB is active independently of RcsB phosphorylation (4). An additional level of control of the RcsB-heteromers is based on the regulation of the genes encoding the auxiliary proteins. Expression of rcsA and bglJ is repressed by HNS, a global repressor and nucleoid structuring protein (14,15). Regulation of the gadE gene is part of the complex acid stress response network and involves multiple regulators (16).
In this study, we focused on the RcsB auxiliary protein BglJ. BglJ is an RcsB-dependent transcriptional regulator which was initially identified as activator of the bgl (aryl-β,d-glucoside) operon in Escherichia coli (17). At the bgl locus, the BglJ–RcsB binding site was mapped to a position ∼100 bp upstream of the promoter (4). The bgl promoter is repressed by HNS and StpA, and binding of BglJ–RcsB within the regulatory region of the promoter abrogates repression by HNS (4,18,19). More recent studies demonstrated that BglJ–RcsB also activates the transcription of the leuO gene encoding the pleiotropic transcriptional regulator and HNS antagonist LeuO (20–22). Expression of the leuO gene is likewise repressed by HNS and StpA (20,23). The leuO gene has two promoters: the distal leuO P1 promoter becomes fully active in an hns stpA double mutant, whereas the proximal leuO P2 promoter is activated by BglJ–RcsB. The BglJ–RcsB binding site is situated ∼70 bp upstream of this P2 promoter (20). Furthermore, activation of the leuO P2 promoter by BglJ–RcsB is inhibited by LeuO acting as negative autoregulator (20). As both of the characterized BglJ–RcsB activated loci, bgl and leuO, are repressed by HNS and StpA, BglJ–RcsB is a presumptive HNS and StpA antagonist. Beyond that, microarray data suggest that BglJ–RcsB regulates ∼30 additional loci (20).
Here, we analyzed the activation of 10 target loci by BglJ–RcsB and we define a consensus DNA-binding motif. Our data suggest that BglJ–RcsB activates transcription of HNS repressed and non-HNS repressed loci by several mechanisms. At some promoters the mechanism corresponds to that of ‘classical’ bacterial activators, which bind upstream of the core promoter and interact with RNA polymerase. In addition, BglJ–RcsB is a transcriptional activator that can function as HNS antagonist. Furthermore, we found that BglJ–RcsB at the bgl operon activates a P2 promoter which is located 20 bp upstream of the known CRP-dependent bgl promoter. Both BglJ–RcsB and CRP are required for transcription activation of the newly identified P2bgl. The data suggests that BglJ–RcsB acts synergistically with CRP resulting in a shift of the transcription start site and abrogation of HNS mediated repression of bgl.
MATERIALS AND METHODS
Bacterial strains and plasmids
Strains and plasmids were constructed applying standard techniques. Details on their genotype and relevant structure, respectively, as well as their construction are given in the supplement (see Supplementary Table S1 for strains, Supplementary Table S2 for plasmids and Supplementary Table S3 for oligonucleotides). Briefly, for cloning fragments were amplified by PCR using primers carrying restriction sites at their 5′ ends (Supplementary Table S3). The integrity of the constructed plasmids was analyzed by sequencing of the cloned fragments and by restriction analyses of the overall plasmid structure. Site-specific mutations were introduced by PCR based methods using either combined chain reaction (24) or overlapping PCR fragments, as described earlier (4,20). Promoter lacZ reporter constructs were integrated into the phage Lambda attB site using a plasmid based attB integrations system, as described (25,26). Genomic mutations and deletions were introduced by Red-Gam mediated recombination (27) or transferred by transduction using phage T4GT7 (28). All mutations and insertions were validated by PCR using primers flanking the respective alleles. Antibiotics were added for selection to final concentrations of 25 µg/ml kanamycin, 50 µg/ml ampicillin, 50 µg/ml spectinomycin and 12.5 µg/ml tetracycline, where necessary.
Expression analysis
For expression analysis of promoter lacZ reporter fusions, β-galactosidase assays were performed of exponential cultures grown in LB medium to an optical density of 0.5 at 600 nm, as described (20,29). The assay was repeated at least three times from independent cultures, and the standard deviation of the values (in arbitrary Miller Units) was <15% unless otherwise indicated. For qRT-PCR analyses RNA was isolated from cultures grown in LB medium to the exponential phase (optical density 0.5 at 600 nm) using the bacterial RNAprotect and RNeasy MiniKit system (Qiagen). Then cDNA was synthesized with the SuperScript III First Strand Synthesis Kit (Invitrogen) using 1 mg of RNA and random hexameric oligonucleotides as primers. Subsequently, quantitative PCR was carried out with gene specific oligonucleotide primers (Supplementary Table S3), SYBR Green I and a C1000 touch thermal cycler with optical reaction module CFX96 (Bio-Rad), as described (20). Data were normalized to rpoD, encoding the housekeeping sigma 70 subunit, as reference gene using the CFX Manager Software 2.1 (Bio-Rad) and applying a ΔΔCt algorithm. For each locus, the relative expression was calculated as the ratio of the value obtained for the RNA isolated from mutant strains (T1166, T1048) as compared to the data obtained for RNA isolated from the reference strain (T75).
Mapping of transcription start sites by 5′-RACE
5′ RACE analysis was performed, as described (20,30). Briefly, RNA was isolated from strains T75, T1166 and T1048, as described earlier. Six microgram of the RNA was treated with tobacco acid pyrophosphatase (TAP; Epicentre Biotechnologies). After phenol-chloroform-isoamyl alcohol (25:24:1) extraction and ethanol precipitation the RNA adapter oligonucleotide T268 was ligated to the TAP-treated RNA and to the same amount of untreated RNA using RNA ligase (New England Biolabs) premixed with RNase inhibitor SuperaseIn (Ambion). The RNA adapter ligations were incubated overnight at 17°C, the samples were extracted with phenol-chloroform-isoamyl alcohol (25:24:1), the RNA was precipitated with ethanol, and resuspended in 20 µl H2O. Half of the RNA samples were used for cDNA synthesis using random hexameric oligonucleotide primers and the SuperScript III First Strand Synthesis Kit (Invitrogen). The cDNA was amplified by PCR using the RNA adapter specific DNA primer T265, gene specific primers (Supplementary Table S3) and PlatinumTaq Polymerase (Invitrogen). PCR products were digested with EcoRI and XbaI, separated by agarose gel electrophoresis, and cloned into the EcoRI and XbaI digested vector pUC12. The fragments of several clones were sequenced for mapping of the primary transcription start sites.
RESULTS
Regulation of the presumptive target promoters by BglJ–RcsB
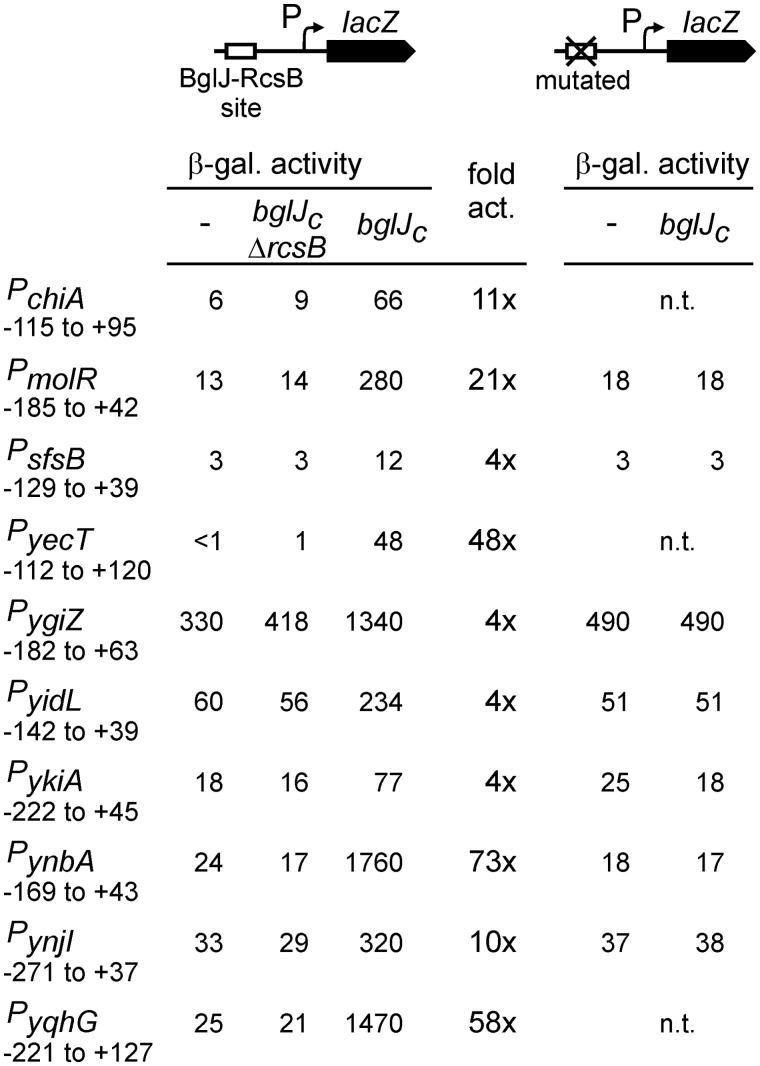
BglJ–RcsB is a presumptive pleiotropic activator, as in a microarray analysis roughly 30 loci were found to be upregulated upon ectopic expression of BglJ in a rcsB+ background but not in an isogenic ΔrcsB mutant (20). To analyze whether transcription of these putative target loci is indeed activated by BglJ–RcsB, in a first step, we constructed lacZ transcriptional fusions of the promoters of 10 putative target loci. In these constructs. the cloned fragments (ranging in size from 168 to 348 bp) encompass the intergenic region in between the target gene and the gene mapping upstream to include the promoter and possible regulatory sites (Figure 1; Supplementary Table S2). The selected loci include chiA encoding endochitinase, yehH (molR) presumably encoding molybdate metabolism regulator, and the loci ykiA, ynbA, yqhG, sfsB, ynjI, yidL, ygiZ and yecT. The latter genes encode proteins of unknown or predicted function (Table 1). The promoter lacZ fusions were integrated into the genome of strain T314 carrying deletion ΔyjjP-yjjQ-bglJ that encompasses bglJ. In addition, isogenic strains carrying a constitutively expressed bglJ allele (bglJC) were constructed. Constitutive expression of allele bglJC is driven by a miniTn10 insertion upstream of bglJ (31). As a further control expression was analyzed in an isogenic bglJC ΔrcsB background. Furthermore, all strains that were constructed carry a lacZ deletion and a deletion of the BglJ–RcsB activated leuO gene encoding the pleiotropic regulator and HNS antagonist LeuO (20,21), although we found later that the presence of leuO has little influence on the BglJ–RcsB targets (data not shown). The results of expression analyses in the bglJ deletion and bglJC strain backgrounds as well as in the bglJC ΔrcsB background demonstrate that all 10 promoter lacZ fusions are specifically activated in the presence of BglJ, ranging from a 4- to 73-fold activation, and that the activation by BglJ is RcsB dependent (Figure 1).
Figure 1.
Specific activation of target gene promoters by BglJ–RcsB. Left panel: The expression level of chromosomal promoter lacZ fusions (coordinates given relative to the transcription start site) was determined in the bglJ deletion background (allele ΔyjjP-yjjQ-bglJ) (−), in isogenic bglJC strains expressing BglJ constitutively, and in isogenic bglJC ΔrcsB mutant derivatives. The factor of activation by BglJ–RcsB (fold act.) was calculated by dividing the β-galactosidase activity (β-gal. activity) determined in the bglJC strain by the activity determined in the absence of BglJ (−). Right panel: BglJ–RcsB binding sites were mutated (mutations given in Figure 2), and the expression level of the mutant promoter lacZ fusions was determined in the absence (−) and presence of BglJ (bglJC) using isogenic bglJ deletion (ΔyjjP-yjjQ-bglJ) and bglJC strains. For the seven promoters analyzed, the mutation of the binding site abrogated their activation by BglJ–RcsB. Cultures were grown in LB medium to the exponential phase and harvested at OD600 = 0.5; β-galactosidase activities are given in arbitrary units (29); n.t. is not tested.
Table 1.
Function of analyzed BglJ–RcsB target loci
| Gene(s) | Function | References |
|---|---|---|
| chiA | Endochitinase (897 aa) | (32) |
| ynbAB | YnbA predicted diacylglycerol choline-phosphotransferase (201 aa)YnbB inner membrane protein (298 aa) | (33,34) |
| ynjI | Inner membrane protein (346 aa) | (34) |
| yecT | Unknown function (162 aa) | (33) |
| yqhGH | YqhG unknown function (308 aa)YqhH predicted lipoprotein (85 aa) | (33) |
| ygiZ | Inner membrane protein (110 aa) | (34) |
| sfsB | SfsB/Nlp transcriptional regulator, Ner-like protein (92 aa) | (33,35) |
| ykiA | Unknown function (113 aa) | (33) |
| yidL | Predicted AraC-type transcription regulator (307 aa) | (33) |
| molR (yehH) | Molybdate metabolism regulator (disrupted in K12) | (33) |
Mapping of the promoters activated by BglJ–RcsB
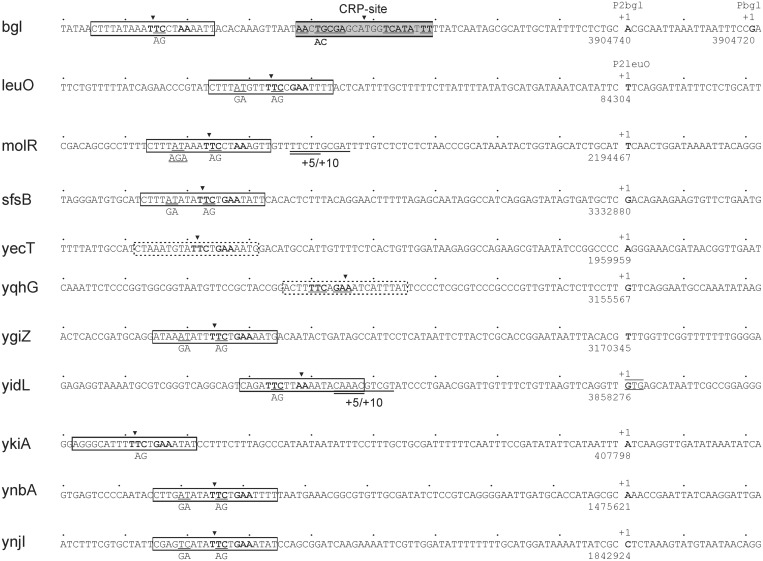
To further characterize the regulation of the presumptive BglJ–RcsB target promoters, we mapped the transcription start sites of the 10 loci by 5′–RACE analyses of primary transcript. For this we isolated RNA from strain T75, carrying the bglJ deletion (allele ΔyjjP-yjjQ-bglJ) and of strain T1166 carrying the constitutive bglJC allele. In the presence of BglJ (expressed from allele bglJC) a specific 5′-RACE product was obtained for each of the 10 loci, as analyzed by agarose gel electrophoresis. In the absence of BglJ the respective 5′-RACE product was apparent but very weak. These results support the conclusion that the analyzed loci are activated by BglJ–RcsB within their chromosomal context. To precisely map the transcription start sites, the 5′-RACE products obtained of the bglJ deletion and the bglJC strains were cloned, and for each locus several independent clones were sequenced. For chiA the previously mapped transcription start site at genome position 3467918 (coordinates as in E. coli K12 MG1655 reference genome sequence NC_000913) (32) was confirmed. The map positions of the newly identified transcription start sites of the other nine loci are summarized in Figure 2.
Figure 2.
Sequence of BglJ–RcsB activated promoters and their BglJ–RcsB binding sites. The transcription starts sites (+1) were mapped by 5′-RACE of RNA isolated from strain T1166 constitutively expressing BglJ, as well as of RNA isolated from strain T75 carrying a bglJ deletion (allele ΔyjjP-yjjQ-bglJ). The positions of the transcription start sites refer to the MG1655 genome sequence NC_000913. For 5′-RACE analysis of bgl also RNA of the Δhns stpA ΔyjjP-yjjQ-bglJ mutant strain T1048 was used. This revealed the transcription start site mapping at position 3904720 in the bglJ deletion background (T75, ΔyjjP-yjjQ-bglJ) and Δhns stpA ΔyjjP-yjjQ-bglJ (T1048), i.e. the absence of BglJ, while the transcription start site at position 3904740 is used in the BglJ positive strain T1166. The BglJ–RcsB activated leuO P2 promoter was mapped previously (20). BglJ–RcsB binding sites marked by boxes with solid lines were mapped by mutagenesis in this work (molR, sfsB, yecT, ygiZ, yidL, ykiA, ynbA and ynjI) or previously [bgl, leuO (4,20)]. Site-specific mutations are indicated underneath the sequence. Insertions of 5 and 10 bp were generated by duplication of the underlined sequences (+5/+10). Predicted BglJ–RcsB binding sites are marked by boxes with dashed lines (yecT and yqhG). The transcription start site of the yidL promoter maps to the first base of a GTG codon (underlined) that is annotated as translation start site (33).
Mapping of BglJ–RcsB binding sites
For characterization of the presumptive BglJ–RcsB binding sites and definition of a consensus binding motif, we took an iterative approach. First, we scanned the sequence of the target promoter regions for similarity to the two known BglJ–RcsB binding sites mapped at the bgl and leuO loci (4,20) using the program MEME SUITE (36). This approach yielded a putative BglJ–RcsB consensus motif, with a significant score (P-value < 10−7) in eight loci. Mutagenesis of this predicted binding motif in the promoter lacZ fusions of five loci including molR, sfsB, ygiZ, ynbA and ynjI, indeed completely abrogated their activation by BglJ–RcsB. This was demonstrated by chromosomal integration of the mutant promoter lacZ fusions and expression analyses in the absence and presence of BglJ (Figure 1). The mutations are depicted in Figure 2. Mutagenesis of a predicted motif which overlapped with the core promoter of ykiA as well as mutagenesis of predicted motifs at yidL and chiA mapping ∼150 bp upstream of the transcription start site did not affect their activation by BglJ–RcsB (not shown). Next, 50 bp DNA sequences encompassing the seven known BglJ–RcsB binding sites (including the five ones mapped here) were used to define a sequence logo using MEME SUITE (36). With this motif a putative binding site with a significant score was identified upstream of the yqhG promoter (Figure 2), but no significant hits were identified next to the promoters of ykiA, yidL and chiA. Therefore, we manually scanned the sequence upstream of these promoters for a match to the most conserved TTc–AA bases of the motif. Candidate sites for the ykiA and yidL promoters were mutated (Figure 2). These mutations resulted in the loss of activation of the promoter lacZ fusions by BglJ–RcsB (Figure 1). No BglJ–RcsB box was characterized or predicted next to the chiA promoter. Finally, the sequences of the nine BglJ–RcsB binding sites characterized in this and previous work were used to define a sequence logo shown in Figure 3 (36). This BglJ–RcsB motif was used to search the K12 genome sequence using the program FIMO (36). By this search all experimentally characterized BglJ–RcsB binding sites were confirmed (Figure 3). Further binding sites were predicted for yecT and yqhG as well as for the putative target loci rhsA and rhsB (20). The binding sites shown in Figure 3 scored among the top 14 of all sites that were predicted in the genome sequence. However, the score was lower for the binding site shown for ykiA (experimentally verified) and for yqhG (predicted). Additional motifs were predicted for other putative BglJ–RcsB targets. However, as their score was lower they require experimental validation to be shown. Interestingly, part of the BglJ–RcsB motif (including the central wwTTCykA sequence) corresponds well to the right half of the consensus sequence for the RcsA–RcsB binding site TaAGaa|tatTCctA (12), indicating that this part may be bound by the RcsB subunit. If this was indeed the case then the BglJ–RcsB motif would be oriented with RcsB facing the promoter at most promoters expect for PyidL and PyqhG.
Figure 3.
BglJ–RcsB consensus sequence. Top: Sequence logo for the BglJ–RcsB binding site. The logo was generated by MEME SUITE (36) using 120 bp sequences encompassing the mapped BglJ–RcsB binding sites of the loci shown in the middle. Middle: Sequences of the BglJ–RcsB binding sites mapped by site-specific mutagenesis and their genome position in the MG1655 reference genome sequence NC_000913. Bottom: BglJ–RcsB binding sites which were predicted in the E. coli K12 MG1655 genome by the program FIMO (36) using the BglJ–RcsB logo (top) as input. All BglJ–RcsB binding sites except ykiA and yqhG scored among the top 14 hits in the FIMO search of the E. coli K12 MG1655 genome sequence. Letters in bold indicate a match to the consensus sequence motif, with upper case letters used for highly conserved bases, and lower case letters for ambiguous conserved bases.
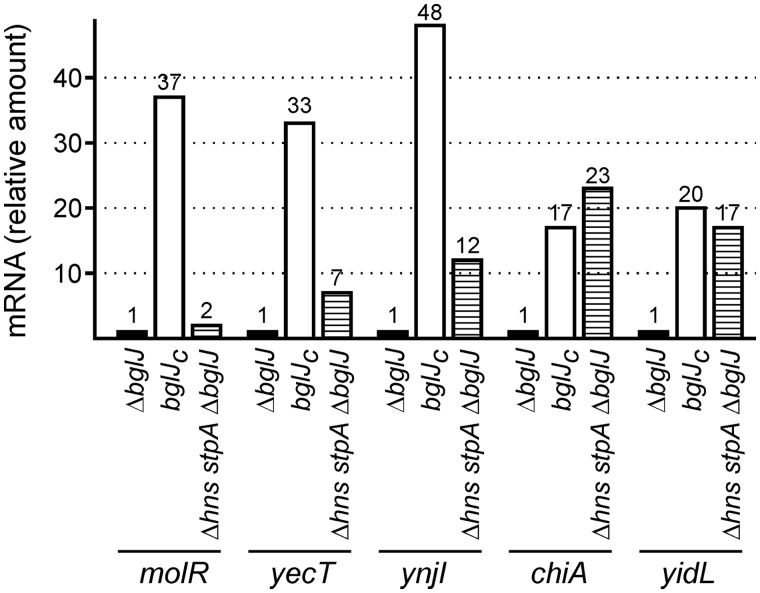
Regulation of BglJ–RcsB targets by HNS/StpA
BglJ–RcsB was first characterized as activator of the HNS repressed loci bgl and leuO (4,20). Therefore, we had assumed that BglJ–RcsB acts as HNS antagonist, activating gene expression indirectly by inhibiting repression by HNS. To address the correlation between activation of a promoter by BglJ–RcsB and repression by HNS we tested for some of the loci whether they are also repressed by HNS. For quantitative expression analyses, qRT-PCR was performed of RNA isolated from strain T75 carrying the bglJ deletion (allele ΔyjjP-yjjQ-bglJ), from the isogenic bglJC mutant (T1166) and from an isogenic Δhns stpA ΔyjjP-yjjQ-bglJ triple mutant derivative (T1048). In the latter strain (T1048), the bglJ gene was deleted, as the HNS repressed yjjQ-bglJ operon would otherwise be expressed. The normalized qRT-PCR values confirm that all tested loci are activated when BglJ is present (Figure 4). For the molR locus the expression level in the bglJ deletion strain and the Δhns stpA ΔyjjP-yjjQ-bglJ mutant was similar (Figure 4) suggesting that molR is not repressed by HNS and StpA. In contrast, expression of yecT and ynjI was moderately increased in the Δhns stpA ΔyjjP-yjjQ-bglJ and strongly increased in the bglJC background (Figure 4), indicating that these loci are to some extent directly or indirectly regulated by HNS and/or StpA, while BglJ–RcsB activates them significantly. Two further BglJ–RcsB activated loci, chiA and yidL, were expressed at similarly high levels in the Δhns stpA ΔyjjP-yjjQ-bglJ mutant as in the bglJC mutant (Figure 4). The chiA promoter is known to be repressed by HNS (32). The data imply that yidL is also HNS repressed. Taken together the data suggest, that activation by BglJ–RcsB occurs both at non-HNS repressed promoters as well as at promoters repressed by HNS and/or StpA.
Figure 4.
BglJ–RcsB activated genes and their co-regulation by HNS/StpA. For expression analysis of BglJ–RcsB activated genes, a qRT-PCR analysis was performed of RNA isolated from ΔyjjP-yjjQ-bglJ deletion strain T75 (indicated as ΔbglJ), from strain T1166 (bglJC) expressing BglJ constitutively and from the Δhns stpA ΔyjjP-yjjQ-bglJ mutant T1048 (indicated as Δhns stpA ΔbglJ). The qRT-PCR data were normalized to rpoD, and for each gene the expression level (indicated as mRNA, relative amount) was calculated relative to the data obtained for RNA isolated from strain T75 (ΔyjjP-yjjQ-bglJ), which was set as 1.
Synergistic regulation of bgl by BglJ–RcsB and CRP–cAMP
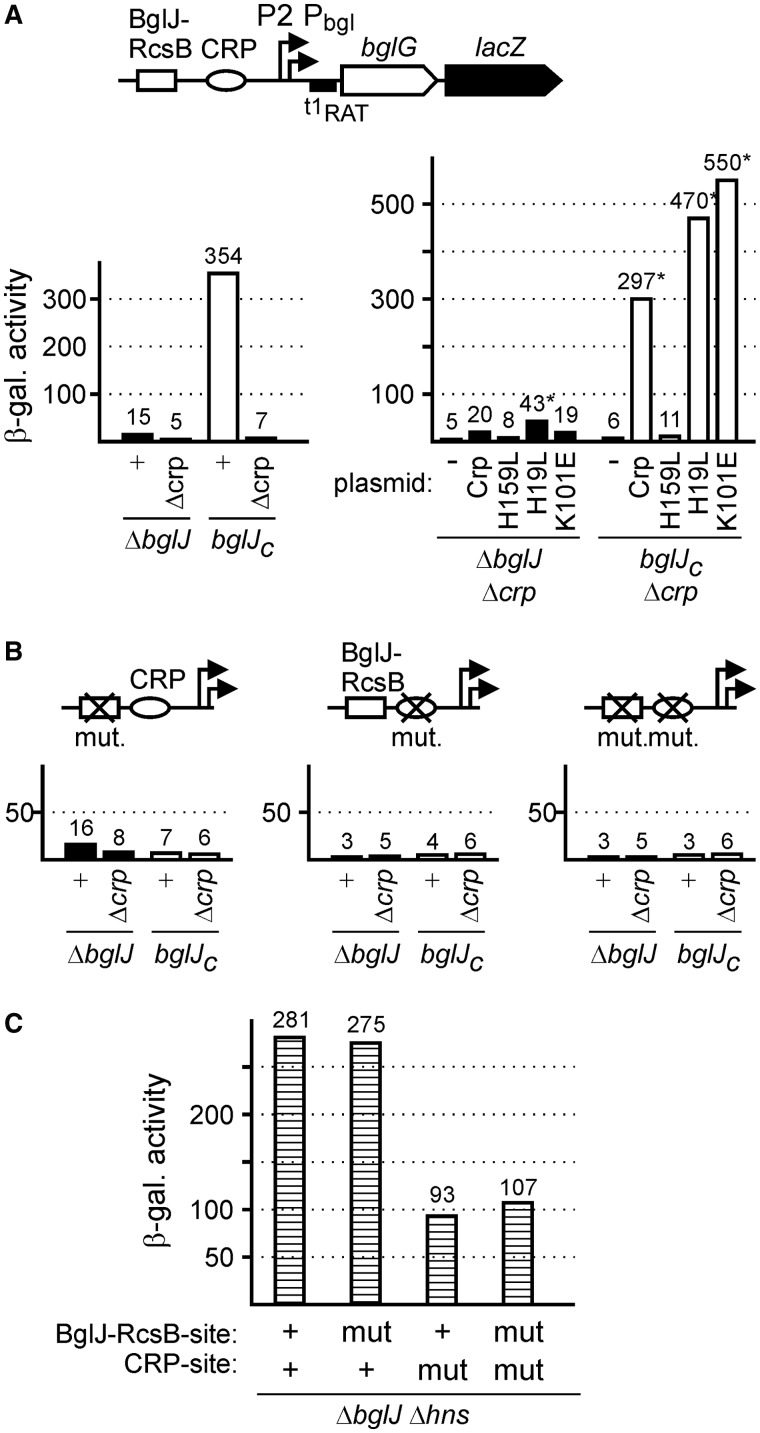
We previously characterized a sequence motif required for BglJ–RcsB-mediated activation of the bgl operon (4). Intriguingly, the center of this presumptive BglJ–RcsB binding site maps 96 bp upstream of the transcription start site of the HNS-repressed and CRP-dependent bgl promoter, Pbgl, and thus further upstream than in case of the other BglJ–RcsB activated promoters (Figure 2). To test, whether BglJ–RcsB indeed activates transcription initiation at Pbgl we performed a 5′-RACE analysis of RNA isolated from strain T1166 expressing bglJ constitutively. As controls, we analyzed RNAs isolated from strain T75 (ΔyjjP-yjjQ-bglJ), in which bgl transcription is repressed, and from a Δhns stpA ΔyjjP-yjjQ-bglJ mutant (T1048) in which the bgl promoter is active. These 5′-RACE analyses confirmed the transcription start of the known bgl promoter Pbgl in the bglJ deletion strain and the Δhns stpA ΔyjjP-yjjQ-bglJ triple mutant (Figure 2). Furthermore, it revealed a new BglJ-dependent transcription start site mapping 20 bp upstream of the start site of the known promoter. Consequently, the BglJ–RcsB binding site is located at position −76 relative to the transcription start site of the newly identified P2 promoter (Figure 2).
The previously identified bgl promoter (Pbgl) is activated by the CRP–cAMP complex with the center of the CRP-binding site located at the canonical -61.5 position of type I CRP-dependent promoters (37). However, this CRP-binding site maps at position -41.5 relative to the BglJ–RcsB activated P2 promoter, and thus at a position typical for class II CRP-dependent promoters. To test whether CRP indeed is required for BglJ–RcsB activation of P2bgl or whether this promoter is CRP-independent, we determined expression of a bgl-lacZ reporter fusion in the bglJ deletion background (allele ΔyjjP-yjjQ-bglJ), a BglJ-positive strain (bglJC) and isogenic Δcrp derivatives of these strains (Figure 5A). As expected (4), expression levels were low in the bglJ deletion background (15 units) and high (354 units) in the BglJ expressing strain (Figure 5A). However, in the isogenic crp mutants expression levels were very low (5 units) irrespective of the absence or presence of BglJ (Figure 5A). These data suggest that activation of P2bgl by BglJ–RcsB is CRP-dependent.
Figure 5.
Synergistic regulation of bgl by BglJ–RcsB and cAMP-CRP. (A) Regulation of the P2bgl and Pbgl promoters by BglJ–RcsB and CRP was analyzed by determining the expression level of a chromosomal bgl-lacZ fusion in strain T568 carrying the bglJ deletion (allele ΔyjjP-yjjQ-bglJ, indicated as ΔbglJ), its isogenic Δcrp derivative, as well as in isogenic derivatives carrying allele bglJC for constitutive expression of BglJ (left panel in A). The Δcrp mutants were complemented with plasmids pDCRP, pDCRP-H159L, pKES324 (CRP-H19L), pKES328 (CRP-K101E) and the empty vector pDctrl as control (−) (right panel in A). The bgl-lacZ reporter fusion carries all elements required for repression by HNS, and a mutation of terminator t1 (t1RAT) to render expression independent of substrate-specific regulation by the operon encoded transcriptional antiterminator BglG (38,39). The standard deviation of β-galactosidase activities marked with an asterisk (*) is 40%. (B) Expression of bgl-lacZ fusions with mutation of the BglJ–RcsB binding site, the CRP-binding site and mutations of both sites was likewise determined in the four genetic backgrounds, i.e. in the absence and presence of BglJ and CRP, respectively. (C) Expression levels directed by bgl-lacZ fusions with wild-type and mutant BglJ–RcsB and CRP-sites, respectively, as determined in hns mutant strains T729 (wild-type binding sites, indicated as +), T735 (BglJ–RcsB-site mutated), T1543 (CRP-site mutated) and T1545 (both sites mutated).
Furthermore, complementation of the Δcrp mutants with plasmid pDCRP encoding CRP under control of its native promoter (40) restored activation of bgl transcription by BglJ–RcsB (Figure 5A). However, complementation with a plasmid encoding CRP-H159L did not restore activation by BglJ–RcsB, respectively (Figure 5A). The CRP-H159L mutant protein carries a substitution in the activation region 1 (AR1) of CRP and is defective in activation of class I promoters and several class II CRP-dependent promoters (40–42). These data further support the notion that CRP is required for activation of bgl transcription by BglJ–RcsB, and they demonstrate that the AR1 of CRP is important for this activation. In addition, we tested complementation of the Δcrp mutants with CRP-H19L and CRP-K101E mutants. These mutations map in the AR2 of CRP required for activation of open complex formation by RNA polymerase at the synthetic PmelR derived −41.5 CC promoter and other class II promoters (40–42). However, in the bgl system complementation with CRP-H19L and CRP-K101E mutants restored activation by BglJ–RcsB (Figure 5A). These data suggest that AR2 of CRP is not involved in co-activation of P2bgl by BglJ–RcsB and CRP.
As a further experiment to address co-regulation of bgl transcription by BglJ–RcsB and CRP we constructed chromosomal bgl-lacZ fusions with a mutated CRP-binding site, a mutated BglJ–RcsB-site, and with both sites mutated (Figure 5B). Expression of these fusions and their activation by BglJ–RcsB was tested in the absence and presence of BglJ (using strains carrying allele ΔyjjP-yjjQ-bglJ and bglJC, respectively). In addition, isogenic crp mutant derivatives were used (Figure 5B). The expression analysis revealed that activation of bgl by BglJ–RcsB requires both an intact BglJ–RcsB binding site and an intact CRP-binding site (Figure 5B). Furthermore, expression analysis in an isogenic Δhns stpA derivative (carrying deletions of bglJ and leuO) was performed to monitor the influence of the site-specific mutations on bgl transcription independently of repression by HNS and StpA. These data demonstrate that the mutation of the BglJ–RcsB binding site does not influence the activity of the bgl promoters (Figure 5C). However, the expression levels directed by the bgl-lacZ fusions with mutant CRP-binding sites were 3-fold lower, which suggests that the CRP dependence of bgl transcription is moderate (3-fold) in the absence of HNS. This later observation is in agreement with in vitro transcription data (43). Taken together we conclude that BglJ–RcsB and CRP synergistically activate the bgl P2 promoter and relief repression by HNS and StpA.
BglJ–RcsB, a canonical transcriptional activator and an HNS antagonist
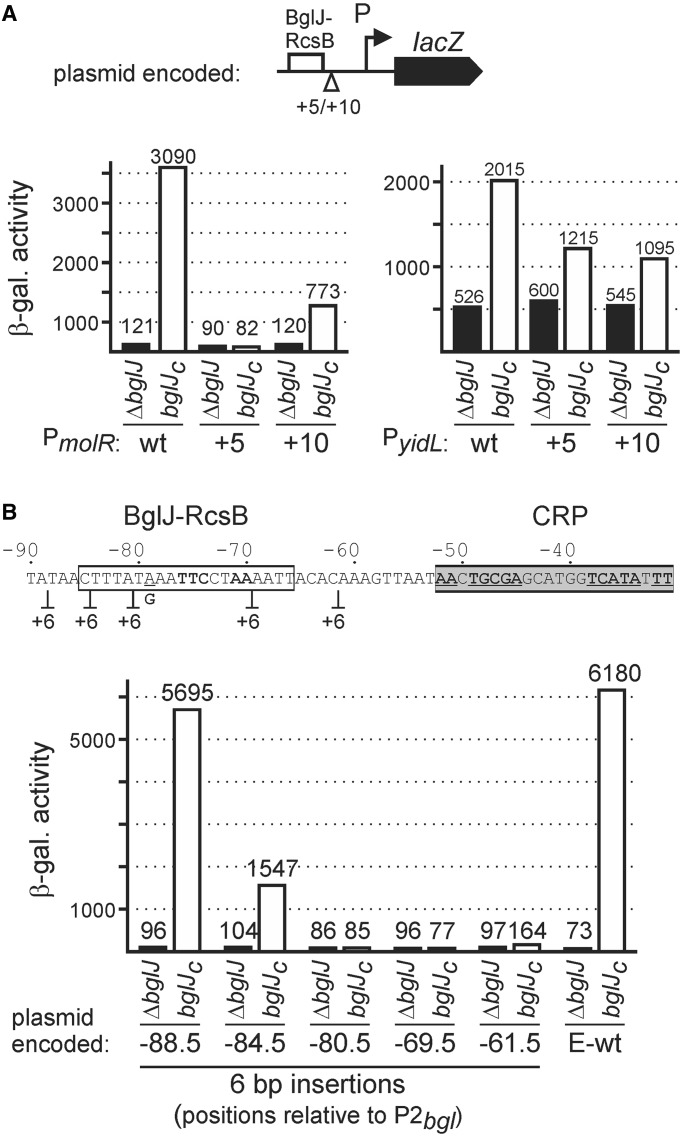
The center of the BglJ–RcsB binding sites maps 65–68 bases upstream of the transcription start site in case of ynbA, ynjI, molR, ygiZ and sfsB. At leuO the BglJ–RcsB binding site maps 57 bases upstream of the transcription start and thus roughly one helical turn closer (Figure 2), while the distance at bgl and ykiA is 76 and 79 bases, respectively, roughly corresponding to one additional helical turn (Figure 2). This may indicate that activation by BglJ–RcsB is DNA phasing dependent and that the mechanism of activation is by recruitment of RNA polymerase similar to canonical transcriptional activators. However, at yidL the center of the BglJ–RcsB binding site maps out of phase at 52 bp upstream of the transcription start site. Furthermore, some of the BglJ–RcsB target promoters are repressed by HNS, including bgl (Figure 5) (4) and yidL (Figure 4), indicating that activation by BglJ–RcsB can be indirect by inhibition of HNS mediated repression. To test whether activation by BglJ–RcsB is sensitive to DNA phasing, we chose the non-HNS repressed molR promoter and the HNS repressed yidL promoter, and constructed derivatives with 5 and 10 bp sequence duplications in between the Bgl–RcsB binding site and the core promoter (Figures 2 and 6A). In parallel, we tested a series of 6 bp insertion that map within the regulatory region of the HNS represses bgl locus (Figure 6B). Activation of the molR promoter by BglJ–RcsB was abrogated upon insertion of 5 bp, but restored to 25% of the wild-type level upon insertion of 10 bp (Figure 6A). In contrast, activation of the HNS repressed yidL promoter was similarly reduced to ∼50% of the wild-type level upon insertion of 5 and 10 bp. These data suggest that DNA phasing is important in case of activation of the molR promoter but not in case of the HNS repressed yidL promoter. In contrast, for the HNS repressed bgl promoter a 6 bp linker insertion mapping in between the BglJ–RcsB binding site and the CRP-binding site completely abrogated activation, as did insertions mapping within the BglJ–RcsB binding site (Figure 6B). Thus, DNA phasing seems to be crucial in case of the bgl promoter which is HNS repressed and synergistically activated by BglJ–RcsB and CRP.
Figure 6.
DNA phasing dependence of activation by BglJ–RcsB. (A) Activation of plasmid encoded PmolR and PyidL lacZ fusions as well as derivatives carrying 5 bp (+5) and 10 bp (+10) sequence duplications in between the BglJ–RcsB binding site and the core promoter was tested in the bglJ deletion strain T23 (ΔbglJ) and the isogenic bglJC strain T1030. (B) Activation of plasmid encoded bgl lacZ fusions carrying the bgl promoter region with a single nucleotide exchange at position −79 generating an EcoRI site (E-wt), as well as derivatives with 6 bp CAATTG linker insertions was tested in strain S541 and its isogenic bglJC derivative S3910.
DISCUSSION
BglJ is an auxiliary regulator that activates a specific set of target loci together with RcsB but independent of RcsB phosphorylation (4,20). Previously, BglJ–RcsB was identified as activator of the HNS repressed leuO and bgl loci (4,17,20). In this work, we characterized the activation of additional loci by BglJ–RcsB. Mapping of the BglJ–RcsB binding sites at seven loci and analysis of helical turn dependence at three loci suggest that BglJ–RcsB activates transcription by several mechanisms. First, at the molR promoter BglJ–RcsB may act similar to canonical activators which bind upstream of the core promoter and interact with RNA polymerase. Second, at the yidL promoter BglJ–RcsB may act as HNS antagonist and activate transcription indirectly by preventing HNS-mediated repression. Third, at the HNS-repressed bgl operon BglJ–RcsB activates transcription synergistically with CRP. This synergistic activation involves a shift of the transcription start site by 20 bp from the bgl promoter (Pbgl) to a novel P2 promoter (P2bgl). Taken together our data demonstrate a pleiotropic and versatile role of the BglJ–RcsB transcriptional activator that functions in the context of simple and more complex promoters, and that acts antagonistically as well as synergistically with additional transcription factors, such as HNS, CRP and LeuO.
The mapping of the asymmetric BglJ–RcsB consensus motif in relation to the transcription start site at several promoters (Figures 2 and 3) suggests a DNA phasing and orientation dependent positioning, with the half-site of the BglJ–RcsB motif that is presumably RcsB bound mapping closer to the promoter at most of the analyzed promoters, except yidL and yqhG (Figure 2). Furthermore, activation of molR and bgl by BglJ–RcsB turned out to be DNA phasing dependent (Figure 6) suggesting that the mechanism of transcription activation by BglJ–RcsB involves protein–protein interactions and thus resembles that of canonical activators (37). In addition, some promoters are regulated by BglJ–RcsB and other pleiotropic factors in an antagonistic or synergistic way. At the yidL promoter BglJ–RcsB and HNS control transcription antagonistically. At this locus the BglJ–RcsB binding site is inverted and maps out of phase. Activation of yidL transcription by BglJ–RcsB is independent of DNA phasing (Figure 6) and thus may be mediated by inhibition of HNS mediated repression. The leuO gene is another example for antagonistic regulation that involves BglJ–RcsB. Activation of the HNS and StpA repressed leuO gene by BglJ–RcsB is inhibited by LeuO (20). At the bgl operon, which is controlled by HNS, StpA, CRP, and LeuO, BglJ–RcsB and CRP presumably act synergistically to overcome repression by HNS (Figure 5). Synergistic activation of a transcriptional regulator together with CRP has been shown at several loci including the ara locus by AraC and CRP, malE by MalT and CRP, rha by RhaS/RhaR and CRP and asc by two CRP dimers, among others (37,44–47). However, at the bgl locus BglJ–RcsB shifts transcription initiation from a class I CRP dependent promoter to a promoter with the center of the CRP-binding site mapping at position −41.5, which is typical of class II CRP-dependent promoters (Figures 2 and 5). Interestingly though, for synergistic activation of bgl by BglJ–RcsB and CRP, the CRP AR1 but not the AR2 is important (Figure 5). This is in contrast to other class II CRP dependent promoters where CRP stimulates open complex formation by RNA polymerase (40–42). Thus, the synergistic activation of bgl by BglJ–RcsB and CRP apparently involves a novel mechanism.
The interaction of RcsB with various auxiliary transcriptional regulators may indicate that RcsB serves as a hub at which various signals are integrated by several means, such as induction of the RcsF-RcsCD phosphorelay and/or interaction with auxiliary factors (1,2). The characterization of BglJ–RcsB targets in this work adds an additional set of loci to the Rcs regulon. Remarkably, beyond leuO, bgl and chiA, most of the BglJ–RcsB targets are of unknown or predicted function, with many of them being conserved and/or encoding membrane proteins (Table 1). Furthermore, putative BglJ–RcsB targets include the complex rhsA and rhsB loci (Figure 3), among others (20). The rhs loci encode hydrophilic proteins with repetitive sequence elements (48,49), which presumably play a role in intercellular competition (50). Taken together, the spectrum of target loci indicates that the BglJ–RcsB regulon is important under conditions different from the standard laboratory conditions. Correspondingly, the yjjQ-bglJ locus has been implicated in E. coli pathogenicity (51). Yet, up to date it is not known under which physiological conditions BglJ is present in the cell. Transcription of the yjjQ-bglJ operon is repressed by HNS under standard laboratory growth conditions (15). The leuO gene encoding the only known activator of the yjjQ-bglJ operon, is likewise repressed by HNS and StpA under standard laboratory conditions (20).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [Schn 371/10-1, 371/10-2]. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
We thank Dr Thomas Stratmann and rotation students for technical support.
REFERENCES
- 1.Majdalani N, Gottesman S. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 2005;59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 2.Clarke DJ. The Rcs phosphorelay: more than just a two-component pathway. Future Microbiol. 2010;5:1173–1184. doi: 10.2217/fmb.10.83. [DOI] [PubMed] [Google Scholar]
- 3.Castanie-Cornet MP, Cam K, Bastiat B, Cros A, Bordes P, Gutierrez C. Acid stress response in Escherichia coli: mechanism of regulation of gadA transcription by RcsB and GadE. Nucleic Acids Res. 2010;38:3546–3554. doi: 10.1093/nar/gkq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venkatesh GR, Kembou Koungni FC, Paukner A, Stratmann T, Blissenbach B, Schnetz K. BglJ-RcsB heterodimers relieve repression of the Escherichia coli bgl operon by H-NS. J. Bacteriol. 2010;192:6456–6464. doi: 10.1128/JB.00807-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehti TA, Bauchart P, Kukkonen M, Dobrindt U, Korhonen TK, Westerlund-Wikström B. Phylogenetic group-associated differences in regulation of the common colonization factor Mat fimbria in Escherichia coli. Mol. Microbiol. 2013;87:1200–1222. doi: 10.1111/mmi.12161. [DOI] [PubMed] [Google Scholar]
- 6.Lehti TA, Heikkinen J, Korhonen TK, Westerlund-Wikström B. The response regulator RcsB activates expression of Mat fimbriae in meningitic Escherichia coli. J. Bacteriol. 2012;194:3475–3485. doi: 10.1128/JB.06596-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henikoff S, Wallace JC, Brown JP. Finding protein similarities with nucleotide sequence databases. Methods Enzymol. 1990;183:111–132. doi: 10.1016/0076-6879(90)83009-x. [DOI] [PubMed] [Google Scholar]
- 8.Consortium TU. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41:D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laubacher ME, Ades SE. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J. Bacteriol. 2008;190:2065–2074. doi: 10.1128/JB.01740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagiwara D, Sugiura M, Oshima T, Mori H, Aiba H, Yamashino T, Mizuno T. Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J. Bacteriol. 2003;185:5735–5746. doi: 10.1128/JB.185.19.5735-5746.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Majdalani N, Heck M, Stout V, Gottesman S. Role of RcsF in signaling to the Rcs phosphorelay pathway in Escherichia coli. J. Bacteriol. 2005;187:6770–6778. doi: 10.1128/JB.187.19.6770-6778.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wehland M, Bernhard F. The RcsAB Box. Characterization of a new operator essential for the regulation of exopolysaccharide biosynthesis in enteric bacteria. J. Biol. Chem. 2000;275:7013–7020. doi: 10.1074/jbc.275.10.7013. [DOI] [PubMed] [Google Scholar]
- 13.Sturny R, Cam K, Gutierrez C, Conter A. NhaR and RcsB independently regulate the osmCp1 promoter of Escherichia coli at overlapping regulatory sites. J. Bacteriol. 2003;185:4298–4304. doi: 10.1128/JB.185.15.4298-4304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sledjeski D, Gottesman S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc. Natl Acad. Sci. U.S.A. 1995;92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stratmann T, Madhusudan S, Schnetz K. Regulation of the yjjQ-bglJ operon, encoding LuxR-type transcription factors, and the divergent yjjP gene by H-NS and LeuO. J. Bacteriol. 2008;190:926–935. doi: 10.1128/JB.01447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sayed AK, Foster JW. A 750 bp sensory integration region directs global control of the Escherichia coli GadE acid resistance regulator. Mol. Microbiol. 2009;71:1435–1450. doi: 10.1111/j.1365-2958.2009.06614.x. [DOI] [PubMed] [Google Scholar]
- 17.Giel M, Desnoyer M, Lopilato J. A mutation in a new gene, bglJ, activates the bgl operon in Escherichia coli K-12. Genetics. 1996;143:627–635. doi: 10.1093/genetics/143.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukerji M, Mahadevan S. Characterization of the negative elements involved in silencing the bgl operon of Escherichia coli: possible roles for DNA gyrase, H-NS, and CRP-cAMP in regulation. Mol. Microbiol. 1997;24:617–627. doi: 10.1046/j.1365-2958.1997.3621725.x. [DOI] [PubMed] [Google Scholar]
- 19.Schnetz K. Silencing of Escherichia coli bgl promoter by flanking sequence elements. EMBO J. 1995;14:2545–2550. doi: 10.1002/j.1460-2075.1995.tb07252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stratmann T, Pul Ü, Wurm R, Wagner R, Schnetz K. RcsB-BglJ activates the Escherichia coli leuO gene, encoding an H-NS antagonist and pleiotropic regulator of virulence determinants. Mol. Microbiol. 2012;83:1109–1123. doi: 10.1111/j.1365-2958.2012.07993.x. [DOI] [PubMed] [Google Scholar]
- 21.Hernández-Lucas I, Calva E. The coming of age of the LeuO regulator. Mol. Microbiol. 2012;85:1026–1028. doi: 10.1111/j.1365-2958.2012.08175.x. [DOI] [PubMed] [Google Scholar]
- 22.Stoebel DM, Free A, Dorman CJ. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology. 2008;154:2533–2545. doi: 10.1099/mic.0.2008/020693-0. [DOI] [PubMed] [Google Scholar]
- 23.Chen CC, Chou MY, Huang CH, Majumder A, Wu HY. A cis-spreading nucleoprotein filament is responsible for the gene silencing activity found in the promoter relay mechanism. J. Biol. Chem. 2005;280:5101–5112. doi: 10.1074/jbc.M411840200. [DOI] [PubMed] [Google Scholar]
- 24.Bi W, Stambrook PJ. Site-directed mutagenesis by combined chain reaction. Anal. Biochem. 1998;256:137–140. doi: 10.1006/abio.1997.2516. [DOI] [PubMed] [Google Scholar]
- 25.Diederich L, Rasmussen LJ, Messer W. New cloning vectors for integration into the lambda attachment site attB of the Escherichia coli chromosome. Plasmid. 1992;28:14–24. doi: 10.1016/0147-619x(92)90032-6. [DOI] [PubMed] [Google Scholar]
- 26.Dole S, Kühn S, Schnetz K. Post-transcriptional enhancement of Escherichia coli bgl operon silencing by limitation of BglG-mediated antitermination at low transcription rates. Mol. Microbiol. 2002;43:217–226. doi: 10.1046/j.1365-2958.2002.02734.x. [DOI] [PubMed] [Google Scholar]
- 27.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson GG, Young KYK, Edlin GJ, Konigsberg W. High-frequency generalised transduction by bacteriophage T4. Nature. 1979;280:80–82. doi: 10.1038/280080a0. [DOI] [PubMed] [Google Scholar]
- 29.Miller JH. A Short Course in Bacterial Genetics. A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Plainview, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 30.Wagner EGH, Vogel J. Approaches to identify novel non-messenger RNAs in Bacteria and to investigate their biological functions: functional analysis of identified non-mRNAs. In: Hartmann RK, Bindereif A, Schön A, Westhof E, editors. Handbook of RNA Biochemistry. Weinheim: WILEY-VCH; 2005. pp. 632–642. [Google Scholar]
- 31.Madhusudan S, Paukner A, Klingen Y, Schnetz K. Independent regulation of H-NS mediated silencing of the bgl operon at two levels: upstream by BglJ and LeuO and downstream by DnaKJ. Microbiology. 2005;151:3349–3359. doi: 10.1099/mic.0.28080-0. [DOI] [PubMed] [Google Scholar]
- 32.Francetic O, Badaut C, Rimsky S, Pugsley AP. The ChiA (YheB) protein of Escherichia coli K-12 is an endochitinase whose gene is negatively controlled by the nucleoid-structuring protein H-NS. Mol. Microbiol. 2000;35:1506–1517. doi: 10.1046/j.1365-2958.2000.01817.x. [DOI] [PubMed] [Google Scholar]
- 33.Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martínez C, Fulcher C, Huerta AM, Kothari A, Krummenacker M, et al. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 2013;41:D605–D612. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daley DO, Rapp M, Granseth E, Melen K, Drew D, von Heijne G. Global topology analysis of the Escherichia coli inner membrane proteome. Science. 2005;308:1321–1323. doi: 10.1126/science.1109730. [DOI] [PubMed] [Google Scholar]
- 35.Choi YL, Nishida T, Kawamukai M, Utsumi R, Sakai H, Komano T. Cloning and sequencing of an Escherichia coli gene, nlp, highly homologous to the ner genes of bacteriophages Mu and D108. J. Bacteriol. 1989;171:5222–5225. doi: 10.1128/jb.171.9.5222-5225.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee DJ, Minchin SD, Busby SJW. Activating transcription in bacteria. Annu. Rev. Microbiol. 2012;66:125–152. doi: 10.1146/annurev-micro-092611-150012. [DOI] [PubMed] [Google Scholar]
- 38.Nagarajavel V, Madhusudan S, Dole S, Rahmouni AR, Schnetz K. Repression by binding of H-NS within the transcription unit. J. Biol. Chem. 2007;282:23622–23630. doi: 10.1074/jbc.M702753200. [DOI] [PubMed] [Google Scholar]
- 39.Radde N, Gebert J, Faigle U, Schrader R, Schnetz K. Modeling feedback loops in the H-NS mediated regulation of the Escherichia coli bgl operon. J. Theor. Biol. 2008;250:298–306. doi: 10.1016/j.jtbi.2007.09.033. [DOI] [PubMed] [Google Scholar]
- 40.Bell A, Gaston K, Williams R, Chapman K, Kolb A, Buc H, Minchin S, Williams J, Busby S. Mutations that alter the ability of the Escherichia coli cyclic AMP receptor protein to activate transcription. Nucleic Acids Res. 1990;18:7243–7250. doi: 10.1093/nar/18.24.7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.West D, Williams R, Rhodius V, Bell A, Sharma N, Zou C, Fujita N, Ishihama A, Busby S. Interactions between the Escherichia coli cyclic AMP receptor protein and RNA polymerase at Class II promoters. Mol. Microbiol. 1993;10:789–797. doi: 10.1111/j.1365-2958.1993.tb00949.x. [DOI] [PubMed] [Google Scholar]
- 42.Niu W, Kim Y, Tau G, Heyduk T, Ebright RH. Transcription activation at class II CAP-dependent promoters: two interactions between CAP and RNA polymerase. Cell. 1996;87:1123–1134. doi: 10.1016/s0092-8674(00)81806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schnetz K, Wang JC. Silencing of Escherichia coli bgl promoter: effects of template supercoiling and cell extracts on promoter activity in vitro. Nucleic Acids Res. 1996;24:2422–2429. doi: 10.1093/nar/24.12.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beatty CM, Browning DF, Busby SJW, Wolfe AJ. Cyclic AMP receptor protein-dependent activation of the Escherichia coli acsP2 promoter by a synergistic class III mechanism. J. Bacteriol. 2003;185:5148–5157. doi: 10.1128/JB.185.17.5148-5157.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schleif R. AraC protein, regulation of the L-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol. Rev. 2010;34:779–796. doi: 10.1111/j.1574-6976.2010.00226.x. [DOI] [PubMed] [Google Scholar]
- 46.Richet E. Synergistic transcription activation: a dual role for CRP in the activation of an Escherichia coli promoter depending on MalT and CRP. EMBO J. 2000;19:5222–5232. doi: 10.1093/emboj/19.19.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wickstrum JR, Santangelo TJ, Egan SM. Cyclic AMP receptor protein and RhaR synergistically activate transcription from the L-Rhamnose-responsive rhaSR promoter in Escherichia coli. J. Bacteriol. 2005;187:6708–6718. doi: 10.1128/JB.187.19.6708-6718.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao S, Sandt CH, Feulner G, Vlazny DA, Gray JA, Hill CW. Rhs elements of Escherichia coli K-12: complex composites of shared and unique components that have different evolutionary histories. J. Bacteriol. 1993;175:2799–2808. doi: 10.1128/jb.175.10.2799-2808.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sadosky AB, Davidson A, Lin RJ, Hill CW. rhs gene family of Escherichia coli K-12. J. Bacteriol. 1989;171:636–642. doi: 10.1128/jb.171.2.636-642.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koskiniemi S, Lamoureux JG, Nikolakakis KC, t'Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. Rhs proteins from diverse bacteria mediate intercellular competition. Proc. Natl Acad. Sci. U.S.A. 2013;110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li G, Ewers C, Laturnus C, Diehl I, Dai J, Antao E-M, Schnetz K, Wieler LH. Characterization of a yjjQ mutant of avian pathogenic E. coli (APEC) Microbiology. 2008;154:1082–1093. doi: 10.1099/mic.0.2007/015784-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.