Abstract
Background:
Human papilloma virus (HPV) DNA has been detected in breast carcinoma by different laboratorial techniques, suggesting that the virus could play a role in the pathogenesis of this tumor.
Materials and Methods:
It was a descriptive study. Systematic random sampling was used for selecting 55 cases of breast cancer and 51 controls of benign breast lesions from the file of Seyedshohada hospital of Isfahan since 2005-2009. A total of 106 paraffin-embedded specimens were selected and HPV DNA was analyzed by polymerase chain reaction and sequenced for different types of HPV in case of positivity for HPV DNA. Data analysis was performed by SPSS 16 software using descriptive statistic, Chi-square, and Fisher's exact tests.
Results:
Out of 55 malignant and 51 benign breast specimens, 18.2% (10) and 13.7% (7) were positive to HPV DNA, respectively (P = 0.53); 70% (7) malignant and 43% (3) benign breast specimens were positive to high-risk HPV genotypes. In malignant specimens, the most common high- and low-risk genotypes were HPV-16 (3.6%) and HPV-11 (3.6%), respectively. In benign specimens, the most common high- and low-risk genotypes were HPV-31 (3.9%) and HPV-43 (3.9%), respectively. Among malignant and benign specimens, ductal carcinoma and fibro adenoma were the most common lesions positive to different types of HPV, respectively.
Conclusion:
This study demonstrated the presence of HPV genome in both malignant and benign tumor tissues in women with breast lesions in Isfahan; therefore, further larger epidemiologic studies need to be analyzed to establish the exact role of this virus in the pathogenesis of breast cancer.
Keywords: Breast cancer, genotype, human papilloma virus
INTRODUCTION
Breast cancer is one of the major health problems worldwide, occupying first places in mortality in woman.[1] In Iran, breast cancer is the most common cancer in women[2] and its age specific incidence is 33.21 per 100000 Iranian women.[2] Breast cancer is in the sixth and third place in DALY/1000 (Disability Adjusted life years) in Iran and in Iranian women, respectively.[3] There are well-established risk factors for breast cancer, most of which relate to estrogens and growth hormones in females. Nonetheless, in 50-80% of cases, known risk factors have not been identified; this has generated the attempt to identify new factors related with this neoplasia as a viral infection.[2]
Because mouse mammary tumor virus (MMTV) is the proven cause of breast cancer in both field and experimented mice, similar viruses have long been suspected as a potential cause of human breast tumors. High-risk human papilloma viruses (HPVs), Epstein–Barr (EBV), and other virus also have been identified in human breast tumors, but there is no definitive evidence for a causal role.[4] HPV and MMTV have hormone responsive elements that appear to be associated with enhanced replication of the viruses in the presence of corticosteroid and other hormones. This biological phenomenon is particularly relevant because of the hormone dependence of breast cancer.[4]
On the other hand, any viral hypothesis as a cause of breast cancer must take into account the most striking epidemiological feature of human breast cancer, the three- to six-fold differences in mortality and up to eight-fold differences in incidence between some Asian and western populations. These differences dramatically lessen to a two- to three-fold difference within one or two generations of migration of female from low- to high-risk of breast cancer countries. A plausible explanation for these may initiate some breast cancer in most population.[4] HPV DNA has been detected in breast carcinoma by different laboratorial techniques, suggesting the virus could play a role in the pathogenesis of this tumor.[5] HPV DNA might be transported from the original site of infection to the breast tissue by the blood stream.[6]
In several studies, the results of HPV DNA analysis by polymerase chain reaction (PCR) showed that in a group of breast carcinomas 0, 6.5, 21, 24.75, 29.4, 35, 48, 61, 63, and 74% of specimens were positive to HPV-DNA.[1,5,6,7,8,9,10,11,12,13] In a group of benign breast specimens, the results of most studies showed that none of them were positive to HPV-DNA,[1,5,8,11] except in one study in which 32% normal breast tissue sample were positive as well.[13]
According to multiple reports about role of HPV in developing breast cancer, high incidence of breast cancer in Isfahan province (age specific rate 37.32 per 100/000 Iranian women population),[2] the possibility of primary prevention of breast cancer by HPV vaccine[14] (HPV vaccine has 95-100% efficacy in prevention of cervical cancer[15]) and the limited study in Iran, this study was conducted to investigate the prevalence of HPV DNA and its genotypes in patients with breast lesions in Isfahan.
MATERIALS AND METHODS
Tissue samples
It was a descriptive study including 55 cases of breast cancer and 51 controls of benign breast lesions selected from the files of the Seyedshohada hospital (Isfahan, Iran) by systematic random sampling. A total of 106 paraffin-embedded breast tissues prepared during the years 2005-2009 for routine histopathological study of breast neoplasia were obtained for the study.
DNA extraction
DNA was extracted from 10-μm sections of paraffin-embedded tissues. Five sections were cut with standard microtome from every paraffin wax block and transferred into a 1.5-ml microtube. To prevent cross contamination between the samples, the microtome blade was washed with xylene and ethanol after sectioning of each block. Six hundred microliters of 1% SDS and 0.1 M NaOH solution (pH 12.7) and 10 beads of Chelex 20 were added to each microtube. The microtubes were heated at 100°C in a water bath for 45 min. A cooling time of 5 min was allowed after heating. To withdraw the DNA solution, the top solidified wax layer was pierced by a micropipette tip. The solution beneath wax layer was withdrawn and transferred to a clean 1.5-ml microtube. The Chelex beads were not removed. Further steps of extraction and purification were performed according to the method by Shi et al.[16]
DNA purity, yield, and size
DNA purity was assessed with a spectrophotometer and calculated by ratio of the DNA optical density (OD260) and protein optical density (OD280). DNA yield was calculated from DNA optical density (OD260) for clean DNA samples. DNA size was analyzed by electrophoresis pattern of sample aliquots (5 μl) in a 1% agarose gel stained with ethidium bromide and visualized under ultraviolet light.
HPV DNA detection assay
PCR amplification of genomic DNA was performed using primers specific for human SMN and β globin genes. PCR reaction mixture contained 1 U Taq polymerase, 1X PCR buffer (50 mM KCl and 10 mM Tris-HCl at pH 8.4, AMS buffer containing 20 mM ammonium sulfate, 75 mM Tris-HCl at pH 8.8), 0.2 mM of each dNTPs, 1.5 mM MgCl2, 10 pmol of each primers, and 100-200 ng of extracted DNA. The PCR conditions were as follows: Denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for SMA primer and at 63°C for β globin primer for 1 min, and extension at 72°C for 1 min, followed by 5 min of final extension at 72°C. Purified DNA from nucleated cells of whole blood was used as a positive control and sterile distilled water was used as negative control. The PCR amplification products were run on 2% agarose gel, stained with 0.5 μg/ml ethidium bromide, and visualized under ultraviolet light. The samples yielded positive results with SMA primers and were further analyzed for presence of HPV DNA. HPV was amplified with the L1 consensus primers Gp5+/Gp6+. Primer sequences were GP5+-TTTGTTACTGTGGTAGATACTAC GP6+-GAAAAATAAACTGTAAATCATATTC.
The reaction mixture was the same as for the SMN and β globin genes, except for MgCl2 concentration, which was increased double-fold. The cycling conditions were initial denaturation at 94°C for 5 min, followed by 40 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 2 min, extension at 72°C for 2 min, and a final extension step at 72°C for 10 min. A standard HPV DNA was used as a positive control and sterile distilled water was used as a negative control. The amplicons were electrophoresed using 2% agarose gel stained with ethidium bromide and visualized under ultraviolet light.[17] HPV positive samples were subjected to genotyping. The INNO-LiPA assay performed from DNA extracted by NucliSENSE EasyMag system on the EasyMag Extraction Platform (bioMerieux, Marcy l’Etoile, France) allows for detection of 28 different HPV genotypes (6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 43, 44, 45, 51, 52, 53, 54, 56, 58, 59, 66, 68, 70, 73, 74, 82, 69, 71). The INNO-LiPA HPV Genotyping v2 is based on the reverse hybridization principle. Part of the L1 region of the HPV genome is amplified and denatured biotinylated amplicons are hybridized with specific oligonucleotide probes immobilized on the strip. After hybridization and stringent washing, streptavidin-conjugated alkaline phosphatase is added and binds to any biotinylated hybrid previously formed. Incubation with BCIP/NBT chromogen gives a purple/brown precipitate and results can be interpreted visually.[18]
Statistical data processing
Data were processed by SPSS statistical software program version 16.0. Descriptive statistic, χ2(Pearson Chi-square) and Fisher's exact test were used for analysis. Statistical significance was set at P < 0.05.
RESULTS
Spectrophotometer was used to evaluate extracted DNA purity. The mean ratio for OD 260/280 was 1.7.
The HPV DNA detected in 16% (17) breast specimens. Out of 55 malignant breast specimens (cases) and 51 benign breast specimens (controls), 18.2% (10) and 13.7% (7) were positive to HPV DNA, respectively (P = 0.53). It shows that there was no significant difference between the two groups.
Out of malignant specimens 16.7% (6) of ductal carcinoma, 20% (2) of lobular carcinoma, and 50% (2) of medullary carcinoma were positive to HPV.
Out of benign specimens 29.4% (5) of fibroadenoma, 3.4% (1) of fibrocystic change and 50% (1) of mastitis were positive to HPV.
Comparisons between malignant and benign specimens for HPV genotyping are shown in Table 1.
Table 1.
Comparison between malignant and benign breast specimens for prevalence of HPV genotypes
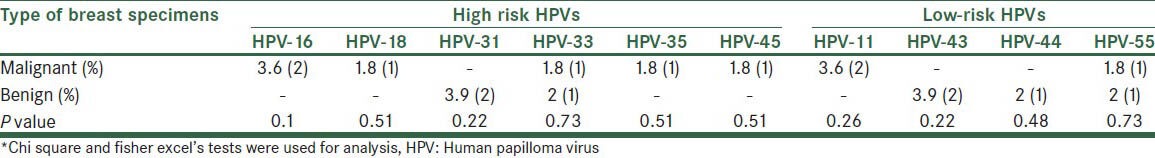
All malignant and benign specimens were negative to HPV-16 and -18 in a single specimen.
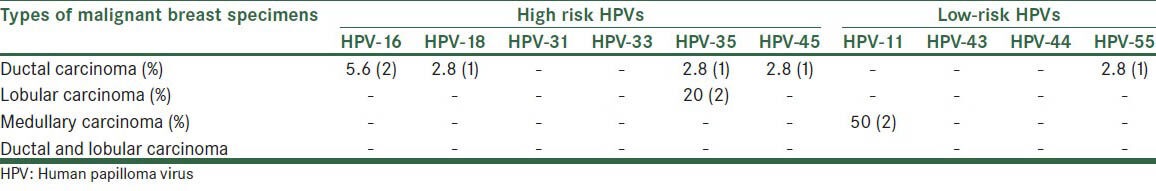
Frequency of HPV genotypes in different types of malignant breast specimens is shown in Table 2.
Table 2.
Frequency of HPV genotypes in different types of malignant breast specimens
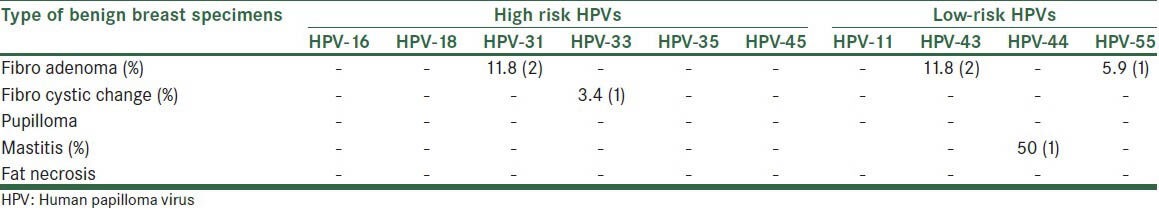
Frequency of HPV genotypes in different types of benign breast specimens is shown in Table 3.
Table 3.
Frequency of HPV genotypes in different types of benign breast specimens
Total 70% (7) malignant and 43% (3) benign breast specimens were positive to high-risk HPV genotypes (P = 0.35).
DISCUSSION
In our study, 18.2% malignant breast specimens and 13.7% benign breast specimens were positive to HPV DNA. Except in a group of Swiss women with breast cancer that had all samples negative for HPV DNA,[7] positivity to HPV DNA in malignant samples in our study was lower in comparison to the results related to other studies, Japan 21%,[9] 24.75%,[5] Mexico 29.4%,[1] 35%,[10] 48%,[11] 61%,[12] 63%,[6] and 74%.[13] But in Korean women, 6.5% breast carcinomas were HPV positive.[8] In addition, in studies conducted in Iran, Mazandaran province, Mashhad, and Isfahan, the prevalence of HPV DNA in breast specimens was higher (25.9%, 48%, and 20%, respectively).[11,19,20] On the other hand, in contrast to most studies,[1,5,8,11] except Mazandaran study (2.4%),[19] our benign breast specimens were positive to HPV DNA. The large differences in prevalence of HPV DNA may be due to variation in epidemiology of HPV.
In spite of higher prevalence of positivity to HPV DNA in malignant specimens in comparison to benign specimens, this difference was not statically significant (P = 0.53).
In malignant specimens, the most common high-risk HPV genotype was HPV-16 (3.6%) that was very lower in contrast to its prevalence of 56%,[5] 66.6%,[1] and 92%[9] in other studies.
Other high-risk HPV genotypes such as 18, 33, 35, and 45 had the same prevalence (1.8%) in malignant specimens that was lower in comparison to that in other studies (12%,[9] 20%,[1] 40%,[5] 54%,[13] and 100%[21] positive to HPV-18; 4%,[5] 13.4%[1] positive to HPV-16 and HPV-18; 4%,[9] 94.6%[13] positive to HPV-33). Another study showed that 26% and 16% breast carcinoma had infection with high-risk and low-risk HPV, respectively, and 6% of them were positive to both high- and low-risk HPV types.[11] These results may be explained by difference in HPV epidemiology in different regions.
The most common low-risk HPV genotype in malignant specimens was HPV-11. This genotype was reported in another study for breast cancer as well.[10]
Despite other studies,[1,5,8,11] benign specimens were positive to HPV DNA such as HPV-31, HPV-33, HPV-43, HPV-44, and HPV-55. On the other hand, 56% and 87.5% positivity to HPV-18[13] and HPV-33 were reported from normal breast tissues, respectively. Thus, difference in epidemiology of HPV was emphasized.
Among malignant specimens, the most common carcinoma positive to different type of high-risk HPV such as HPV-16, HPV-18, HPV-35, and HPV-45 was ducal carcinoma.
Among benign specimens, the most common lesion positive to HPV type (-31, -43, -45) was fibroadenoma. There has been no study that investigated the HPV genotypes separately in different types of benign and malignant breast lesions for comparison.
CONCLUSION
There is controversy regarding the role of HPV in the pathogenesis of breast cancer. The controversy is influenced by the technical limitations and the epidemiology of HPV in different geographical area. This study demonstrated the presence of HPV genome in both malignant and benign tumor tissues in women with breast lesions in Isfahan; further larger epidemiologic studies need to be analyzed in order to establish the exact role of this virus in the pathogenesis of breast cancer. Confirming an etiological role for HPV in breast cancer in Iranian females may help develop vaccine strategies for combating this increasingly common cancer.
Footnotes
Source of Support: Vice-chancellery for Research, Isfahan University of Medical Science (no. 188126)
Conflict of Interest: None declared.
REFERENCES
- 1.De Leon DC, Montiel DP, Nemcova J, Mykykova I, Turcios E, Villavicencio V, et al. Human puoillomavirus in breast tumors: Prevalence in a group of Mexican patients. BMC Cancer. 2009;9:26. doi: 10.1186/1471-2407-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aghajani H. 1st edition. Tehran: Tandis publication; 2011. Civil report of cancer registry 2008; pp. 54–121. [Google Scholar]
- 3.Akbari ME. 1st edition. Qom: Darrolfekr publication; 2008. Cancer in Iran; p. 14666. [Google Scholar]
- 4.Lawson JS. Do viruses cause breast cancer? Methods Mol Biol. 2009;471:421–38. doi: 10.1007/978-1-59745-416-2_21. [DOI] [PubMed] [Google Scholar]
- 5.Damin AP, Karam R, Zettler CG, Caleffi M, Alexandre CO. Evidence for an association of human papilloma virus and breast carcinomas. Breast Cancer Res Treat. 2004;84:131–7. doi: 10.1023/B:BREA.0000018411.89667.0d. [DOI] [PubMed] [Google Scholar]
- 6.Widschwendter A, Brunhuer T, Wiedemair A, Mueller-Holzner E, Marth C. Detection of human papillimaviras DNA in breast cancer of patients with cervical cancer. J Clin Virol. 2004;31:292–7. doi: 10.1016/j.jcv.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Lindel K, Forster A, Altematt HJ, Greiner G, Gruber G. Breast cancer and human papillomarirus (HAV) infection: No evidence of a viral etiology in a group of Swiss women. Breast. 2007;16:172–7. doi: 10.1016/j.breast.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Choi YL, Cho EY, Kim JH, Nam SJ, Oh YL, Song SY, et al. Detection of human papilloma virus DNA by DNA chip in breast carcinomas of Korean Women. Tumour Biol. 2007;28:327–32. doi: 10.1159/000124238. [DOI] [PubMed] [Google Scholar]
- 9.Khan NA, Castillo A, Koriyama C, Kijima Y, Umekita Y, Ohi Y, et al. Human papillomavirus detected in female breast carcinomas in Japan. Br J Cancer. 2008;99:408–14. doi: 10.1038/sj.bjc.6604502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Klimberg VS, Andrews NR, Hicks CR, Peng H, Chiriva-Internati M, et al. Human papillomavirus DNA is present in a subset of unselected breast cancers. J Hum Virol. 2001;4:329–34. [PubMed] [Google Scholar]
- 11.Seyedi Alavi GH N, Sadeghian A, Jabari H, Bahreyni M, Bagheri H. Presence of human papilloma virus sequences in breast cancer tissues and association with histopathological features. Iranian J Obstet, Gynecol Infertility. 2009;12:1–4. [Google Scholar]
- 12.Akil N, Yasmeen A, Kassab A, Ghabreau L, Darnel AD, Al Moustafa AE. High-risk human papillomavirus infections in breast cancer in Syrian women and their association with Ld-1 expression: A tissue microarray study. Br J Cancer. 2008;99:404–7. doi: 10.1038/sj.bjc.6604503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gumus M, Yumuk PF, Salepci T, Aliustaoglu M, Dane F, Ekenel M, et al. HPV DNA frequency and subset analysis in human breast cancer patients’ normal and tumoral tissue samples. J Exp Clin Cancer Res. 2006;25:515–21. [PubMed] [Google Scholar]
- 14.Heng B, Glenn WK, Ye Y, Tran B, Delprado W, Lutze-Mann L, et al. Human papilloma virus is associated with breast cancer. Br J Cancer. 2009;101:1345–50. doi: 10.1038/sj.bjc.6605282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghazal-Aswad S. Cervical cancer prevention in the human papilloma virus vaccine era. Ann N Y Acad Sci. 2008;1138:253–6. doi: 10.1196/annals.1414.030. [DOI] [PubMed] [Google Scholar]
- 16.Shi SR, Cote RJ, Wu L, Liu C, Datar R, Shi Y, et al. DNA extraction from archival formalin-fixed, paraffin embedded tissue sections based on the antigen retrieval principle: Heating under the influence of p. J Histochem Cytochem. 2002;50:1005–11. doi: 10.1177/002215540205000802. [DOI] [PubMed] [Google Scholar]
- 17.Tabanifar B, Salehi R, Asgarani E, Faghihi M, Heidarpur M, Allame T. An efficient method for DNA extraction from paraffin wax embedded tissues for PCR amplification of human and viral DNA. Iranian J Pathol. 2008;3:173–8. [Google Scholar]
- 18.Venturoli S, Leo E, Nocera M, Barbieri D, Cricca M, Costa S, et al. Comparison of abbott real time high risk HPV and hybrid capture 2 for the detection of high-risk HPV DNA in a referral population setting. J Clin Virol. 2012;53:121–4. doi: 10.1016/j.jcv.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 19.Sigaroodi A, Nadji SA, Naghshvar F, Nategh R, Emami H, Velayati AA. Human papilloma virus is associated with breast cancer in the north part of Iran. Scientific World Journal 2012. 2012 doi: 10.1100/2012/837191. 837191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohammadizade F, Khoshru S. professional thesis. Isfahan University of Medical Science; 2007. Human papilloma virus in epithelium of nipple and tumoral tissues of invasive breast carcinoma and it's association with clinicopathological parameters in Alzahra and Beheshti hospital of Isfahan University of Medical Science in 2007. [Google Scholar]
- 21.Antonsson A, Spurr TP, Chen AC, Fraancis GD, McMillan NA, Saunders NA, et al. High prevalence of human papilloma viruses in fresh frozen breast cancer samples. J Med Virol. 2011;83:2157–63. doi: 10.1002/jmv.22223. [DOI] [PubMed] [Google Scholar]


