Abstract
Background:
MicroRNAs (miRNAs) are endogenous non-coding RNAs, 19-25 nucleotides in length, involved in post-transcriptional regulation of gene expression in a considerable majority of mRNAs. Different aspects of cellular activities like cell growth, proliferation, and differentiation are regulated by miRNAs through their regulatory effects on particular RNA species. In many tumors, up- or down-regulation of different miRNAs has been reported. In acute myeloid leukemia, up-regulation of miR-92a has been reported in human in-vitro studies.
Materials and Methods:
We performed inhibition of miR-92a in an acute promyelocytic leukemia cell line (HL-60), using locked nucleic acid (LNA) Antagomir. At different time points after LNA-anti-miR92a transfection, qRT-Real-Time-polymerase chain reaction (PCR) and Annexin-V/Propidium Iodide staining were performed and the data was analyzed using the Kruskal-Wallis and Mann-Whitney tests.
Results:
The assessment of the apoptosis and necrosis indicates that miR-92a inhibition can decrease the viable HL-60 cells and this is at least partially due to induction of apoptosis.
Conclusion:
These findings suggest the inhibition of miR-92a as a novel approach for treatment of Acute Promyelocytic Leukemia (APL).
Keywords: MicroRNA, miR-92a, acute promyelocytic leukemia, locked nucleic acid
INTRODUCTION
MicroRNAs (miRNAs), 19-25 nucleotide length ribonucleic acids,[1] play a crucial role in the regulation of gene expression. Initially discovered in 1993, in Caenorhabditis elegans, miRNAs were next revealed to be involved in various biological processes, including the cell cycle, differentiation, growth and development, metabolism, aging, and apoptosis.[2,3] Many reports support their essential function in a wide range of diseases, including cardiovascular, rheumatological, infectious, inflammatory, autoimmune, and metabolic disorders.[4,5,6,7] In addition, dysregulation of miRNAs, including both down- and up-regulation of their biogenesis, mimics the phenomenon attributed to the oncogenes and tumor suppressor genes, depending on the target mRNA regulated by a particular species of miRNAs.[8,9,10,11,12,13]
Inhibition of oncogenic miRNAs (called oncomiRs) is of potential application for the treatment of cancers. Several methods have been tested for inhibition of miRNAs.[8,10,14,15,16] Locked Nucleic Acid (LNA) is a new lineage of a modified oligonucleotide that has a 2’-O 4’- C Methylene bridge in its structure, leading to stability in high temperature and resistance to enzymatic degradation.[17] The efficacy and safety of LNA-anti-miRs for inhibition of miRNAs has been shown in several studies, suggesting that they are suitable candidates for therapeutic applications.[17,18,19,20]
Acute myeloid leukemia (AML) is a disorder of hematopoietic stem cells, accompanied with obstruction in hematopoietic cell differentiation and increased clonal neoplastic proliferation. Untreated, this malignancy leads to death within a few weeks to several months.[21,22] Despite extensive studies, the biological and pathophysiological aspects of AML are not yet clearly known.[23,24] miRNAs, as influential players in cell function, can potentially shed light on the pathogenesis of this disorder. In recent times, it has been shown that there is an increased expression of miR-92a in AML.[25,26,27] This miRNA has also been shown to negatively regulate p63, leading to increased proliferation of myeloid cells.[28] miR-92a is a member of the miR-17-92 cluster located on chromosome 13q31.3. This cluster is suggested to function as oncomiR in some other malignancies.[29]
Based on these evidences, we have proposed that inhibition of miR-92a is of therapeutic effect in AML. According to the American–French–British (FAB) classification, AML is divided into eight subtypes, M0-M7.[22] In this study, HL-60 cells have been used as an in vitro model of AML-M3, also known as acute promyelocytic leukemia (APL), to investigate the effect of LNA-anti-miR-92a transfection on cell viability and apoptosis/necrosis. The results of this study may pave the way for new therapies for AML.
MATERIALS AND METHODS
Cell culture
The HL-60 cell line (Human Acute Promyelocytic Leukemia: APL) was purchased from the National Cell Bank of Iran (Pasteur Institute, Iran). The cell culture was maintained in Roswell Park Memorial Institute (RPMI) 1640 (Gibco, UK) supplemented with fetal calf serum (FCS; Gibco, UK) 15% v/v, 100 U/ml of penicillin and 100 μg/ml of streptomycin (Sigma-Aldrich, USA) in an air-saturated and humid atmosphere, consisting of 5% CO2, in 25-cm2 culture flasks (Nunc, Denmark), at 37°C. The cells were passaged twice weekly to maintain an exponential growth phase.
Cell transfection
The nucleotide sequences of miR-92a were obtained from www.mirbase.org as UAUUGCACUUGUCCCGGCCUGU (accession number MIMAT0000092). miRCURY LNA microRNA Inhibitor™ for hsa-miR-92a and microRNA inhibitor negative control (scrambled) oligonucleotides were purchased from Exiqon, Denmark. Both oligonucleotides were labeled at the 5′ end with fluorescent dye, 6-FAM.
HL-60 cell transfection was performed by using the X-treme GENE siRNA Transfection Reagent™ (Roche, Germany) according to the manufacturer instructions. Briefly, 5 × 105 cells in the exponential growth phase were cultured in six-well culture plates (Nunc, Denmark) containing 1.8 ml RPMI 1640 per well without antibiotic or FCS. The fifty picomol miRCURY LNA microRNA Inhibitor™ was mixed with 5 μL X-tremeGENE siRNA Transfection Reagent™ in 200 μl Opti-MEM I Medium™ (Gibco, UK) and incubated for 15 minutes at room temperature. The complex was then added to the cells and swirled cautiously to ensure even distribution over the entire plate surface. After eight hours of incubation, the FCS and antibiotics were added and the cells were incubated for the 24, 48, and 72 hours. Untreated cells and cells transfected with scrambled-LNA were cultured parallel to the LNA-anti-miR transfected cells. Evaluation of the transfection was detected by flow cytometry and fluorescent microscopy. LNA was conjugated with 6-FAM™ Fluoresceine (6-carboxyfluorescein) and HL-60 cells, which transfected with the LNA, and was seen by a fluorescence microscope and analyzed by a FACSCalibur flow cytometer (BD, USA).
Reverse transcriptase microRNA real time polymerase chain reaction
Reverse transcriptase (RT) microRNA real time PCR was performed to determine the efficiency of miR-92a inhibition by LNA-anti-miR. Briefly, the total cellular RNA was extracted 24, 48, and 72 hours post transfection, with a miRCURY RNA Isolation Kit™ (Exiqon, Denmark) and cDNA was synthesized with a Universal cDNA Synthesis Kit™ (Exiqon, Denmark). Real time PCR was performed by using the SYBR® Green master mix Kit™ (Exiqon, Denmark) and specific miR-92a primers were obtained from Exiqon (product No: 1204258; Exiqon, Denmark). The ABI Step One Plus (ABI, USA) instrument was used for real time PCR experiments and the ΔΔCt method was used for data calculation.
Apoptosis and necrosis assay
The Annexin-V-FLUOS Staining Kit (Roche, Germany) was used for detection of apoptosis and necrosis in HL-60 cells. Annexin-V was used to detect the phosphatidylserine on apoptotic cells. To discriminate necrotic cells Propidium Iodide (PI) staining was employed. The procedure was performed 24, 48, and 72 hours after transfection, according to the manufacturer's instruction (untreated cells were used for controls), and the cells were assessed by the FACSCalibur flow cytometer (BD, USA) with 488 nm excitation, 515 nm band pass filter for fluorescein-conjugated Annexin-V detection, and a filter > 600 nm for PI detection.
Statistical analysis
All the experiments were carried out in triplicate. The results were calculated by using the IBM SPSS (version 20) software. The Kruskal-Wallis and Mann-Whitney tests were used to assess the differences between the groups. Data were presented as Mean ± SD. Statistical significance was defined as P < 0.05.
RESULTS
miRCURY LNA microRNA inhibitor™ strongly inhibits miR-92a
For inhibition of miR-92a, the miRCURY LNA microRNA Inhibitor™ was transfected to HL-60 cells with the X-tremeGENE siRNA Transfection Reagent™. On the basis of the initial optimization experiments, transfection was performed with 50 pM of LNA-anti-miR and 5 μl of the transfection reagent. As the transfected oligonucleotides were fluorochrome-conjugated, transfection efficiencies was assessed by fluorescence microscopy and flow cytometry. At the optimized level, transfection efficiency was about 90% [Figure 1].
Figure 1.
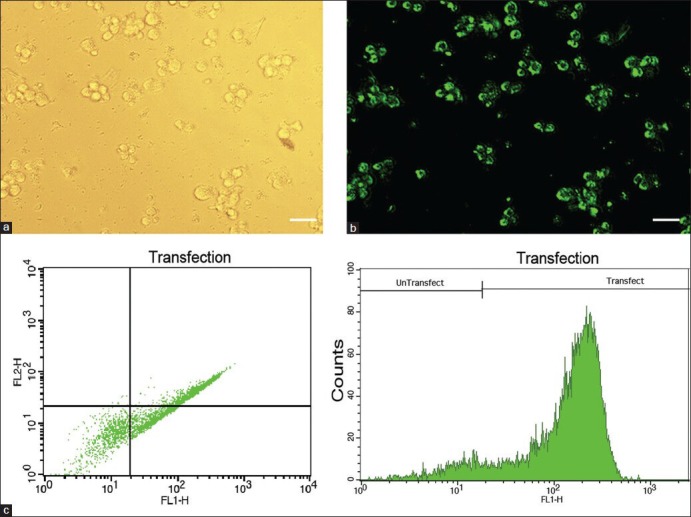
HL-60 cells have been transfected with 6-FAM™ fluorescein-conjugated LNA oligonucleotides, and then, to assess transfection efficiency, they have been observed by a fluorescent microscope and analyzed by flow cytometry. Phase contrast (a) and fluorescent (b) images of the same field of HL-60 cells show that a majority of the cells are transfected. Representative FSC-SCS and FL1-Count flow cytometry graphs are shown in (c). Scale bars: 50 μm
Expression of miR-92a was evaluated by reverse transcriptase microRNA real time PCR in HL-60 cells transfected with the miRCURY LNA microRNA Inhibitor™ (LNA-anti-miR group), transfected with the microRNA inhibitor scrambled oligonucleotides (scrambled LNA group), and untreated HL-60 cells (untreated groups), at 24, 48, and 72 hours after transfection. Although, miR-92a expression was a little bit lower in the scrambled LNA-transfected cells compared to the untreated cells, the differences were not biologically significant and the miR-92a level was in the same order in these two groups. However, in all three time points, the expression of miR-92a was considerably lower in the LNA-anti-miR group compared to the control group (P < 0.025). The expression of miR-92a was at the lowest level 24 hours after transfection and gradually increased in the next two time points [Figure 2].
Figure 2.
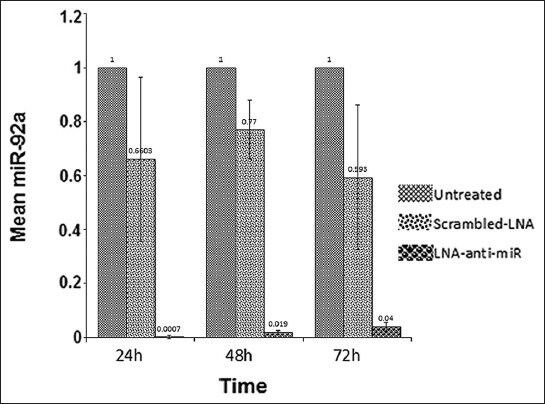
Assessment of the miR-92a level by real time PCR, 24, 48, and 72 hours after transfection. The ΔΔCt method was used for data analysis, and the untreated group was considered as a reference for each time point. Data were mean ± SD of three independent experiments
Inhibition of miR-92a increased apoptosis and necrosis in HL-60 cells
To evaluate the effect of miR-92a inhibition on apoptosis and necrosis, the cells were stained with Annexine-V and PI 24, 48 and 72 hours after transfection. The apoptotic cells were almost undetectable in the untreated cells. However, the ratio of these cells minimally increased 48 and 72 hours after scrambled LNA transfection compared to the untreated cells. MiR-92a inhibition was associated with a dramatic increase in apoptosis, as the ratio of apoptic cells in the LNA-anti-miR group was higher than that in the other groups, at the three time points (P < 0.028 for all time points; Figure 3).
Figure 3.
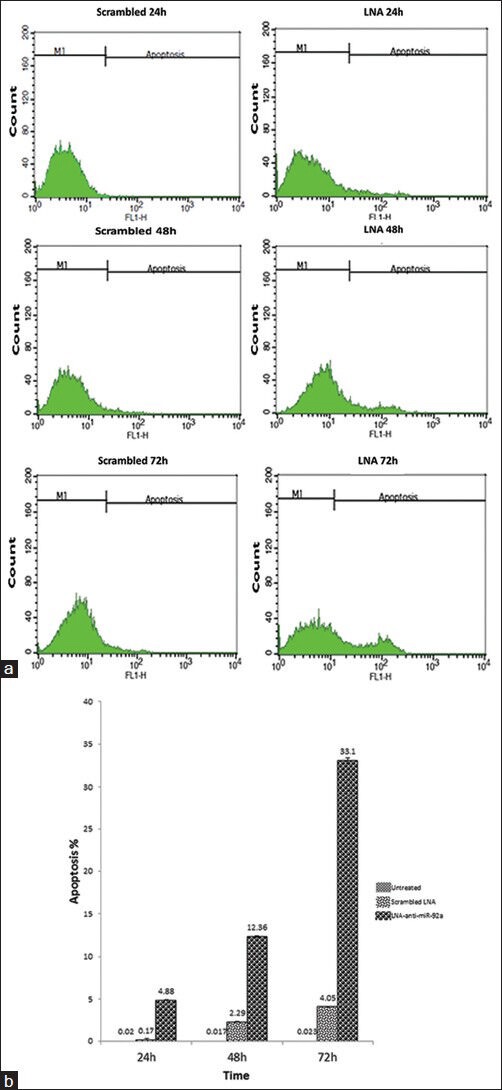
Assessment of apoptosis by Annexin V- PI staining performed 24, 48, and 72 hours after transfection. Representative cytofluorometric graphs are shown (a); data shown in the graph are mean ± SD of three independent experiments (b)
In line with the data of the apoptosis assay, necrosis in the untreated cells was undetectable, but the ratio of the necrotic cells minimally increased 48 and 72 hours after scrambled LNA transfection compared to the untreated cells. The ratio of necrotic cells in the LNA-anti-miR group was higher than that in the other groups, at the three time points, which showed that inhibition of miR-92a was associated with increment of necrosis in the HL-60 cells (P < 0.045 for all time points; Figure 4).
Figure 4.
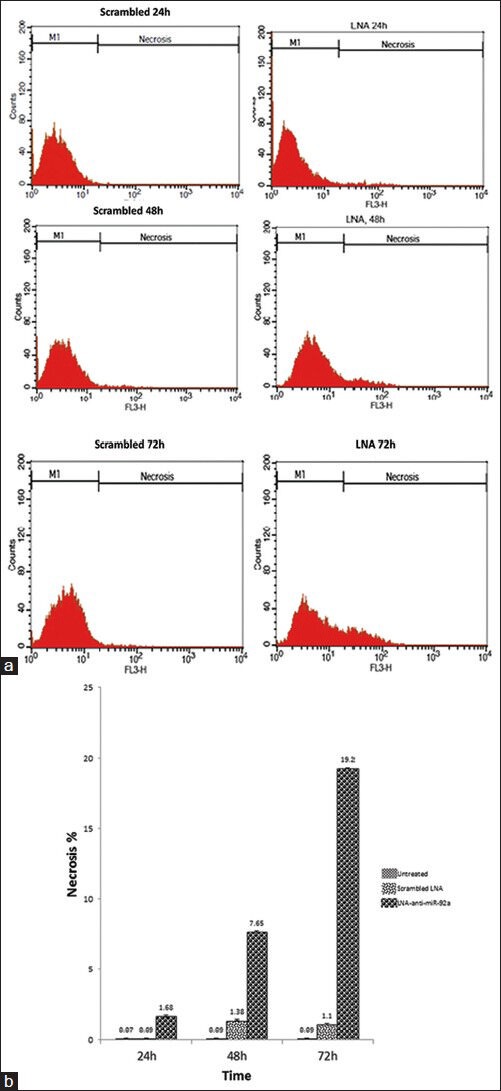
Assessment of late apoptosis/necrosis by Annexin V- PI staining performed 24, 48, and 72 hours after transfection. Representative cytofluorometric graphs are shown (a); data shown in the graph are mean ± SD of three independent experiments (b)
DISCUSSION
In this study, we have used LNA-anti-miR to inhibit miR-92a in an APL (AML-M3) cell line. Real time PCR data confirmed that this miRNA was almost entirely down-regulated after LNA-anti-miR transfection. The apoptosis/necrosis assay, showed that LNA-anti-miR-92a transfection significantly increased apoptosis and necrosis. In these assays, the effect of LNA-anti-miR-92a transfection on cell viability was much higher than in the basal transfection reagent toxicity observed with scrambled oligonucleotide transfection.
In addition, we acknowledge that our Annexin-V/PI staining method cannot discriminate between late apoptosis and necrosis. However, our data indicates that inhibition of miR-92a can increase the apoptosis and necrosis of APL cells and this is at least partially due to the induction of apoptosis.
MiR-92a seems to be involved in cell cycle regulation and cell signaling, therefore, it has a role in cell development and proliferation.[27,30] Overexpression of miR-92a may act as an oncogene or down-regulated tumor suppressor genes, or may affect the genes that control apoptosis and cell differentiation.[31] miR-92a has a function in leukemia, from AML to ALL, in hepatocellular carcinoma, and several other cancers.[25,32] There is evidence which suggests that miR-92a targets an estrogen receptor (ERβ1) mRNA and suppresses its expression in breast cancer.[33] In AML, the miR-92a increases. In addition, it is reported that this miRNA negatively regulates p63 in CD32 cells, leading to increased proliferation of murine myeloid cells.[28]
Acute myeloid leukemia is associated with a high rate of morbidity and mortality,[21] and in spite of recent progresses, the current treatment options for this disorder are suboptimal. There are a number of studies on oncomiRs as an approach to cancer treatment, and attempts have been made to treat cancer by using miRNA inhibitors.[8,9,10,15] Many studies are in the early phase and some of them are in other phases.[15] LNA is one of the Anti-miRs that is used for inhibiting oncomiRs.[17] In this study, specified inhibition of miR-92a by using LNA-anti-miR has prevented proliferation of APL cells and it seems that it is an oncomiR in APL. With regard to the treatment of cancer, destruction of cells that have uncontrolled proliferation is the aim, perhaps by using LNA-anti-miR for inhibiting miR-92a, which can treat APL. This seems to be the method for AML treatment. Chemotherapy is a routine approach for AML treatment.[21,23,24] In many cases chemotherapy does not affect AML and cannot reduce the proliferative cells. Some efforts have been initiated for treating cancer by using Anti-oncomiRs (oncomiR-inhibitors) with other drugs, for example, chemotherapy agents.[22,24] It is suggested that Anti-miR-92a can be used, accompanied by other anti-cancer drugs, for treatment of APL. All-trans retinoic acid (ATRA) is a routine drug that is used for treating APL, but it is not always successful.[22] This study suggests that Anti mir-92a can be used with or without ATRA for treating APL, which is resistant to treatment.
CONCLUSION
Our data suggests that inhibition of miR-92a with LNA-anti-miR can be potentially used for the treatment of APL. It can be used alone or in combination with the current therapies, to reduce the existing limitations in the treatment of this disorder. Further in vivo studies are definitely required to assess the feasibility of this strategy. However, efficient in vivo delivery of anti-miR oligonucleotides remains an obstacle to be tackled before we can move to clinical trials.
Footnotes
Source of Support: Isfahan University of Medical Sciences
Conflict of Interest: None declared.
REFERENCES
- 1.Griffiths-Jones SM. The micro RNA sequence database. Methods Mol Biol. 2006;342:129–38. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McManus DD, Ambros V. Circulating MicroRNAs in cardiovascular disease. Circulation. 2011;124:1908–10. doi: 10.1161/CIRCULATIONAHA.111.062117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nature cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 6.Faller M, Guo F. MicroRNA biogenesis: There's more than one way to skin a cat. Biochim Biophys Acta. 2008;1779:663–7. doi: 10.1016/j.bbagrm.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–62. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 8.Dong C, Ji M, Ji C. microRNAs and their potential target genes in leukemia pathogenesis. Cancer Biol Ther. 2009;8:200–5. doi: 10.4161/cbt.8.3.7333. [DOI] [PubMed] [Google Scholar]
- 9.Trang P, Weidhaas J, Slack F. MicroRNAs as potential cancer therapeutics. Oncogene. 2008;27:S52–S7. doi: 10.1038/onc.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho WC. MicroRNAs in cancer: From research to therapy. Biochim Biophy Acta. 2010;1805:209–17. doi: 10.1016/j.bbcan.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 11.O’Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annual review of immunology. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 12.Ruan K, Fang X, Ouyang G. MicroRNAs: Novel regulators in the hallmarks of human cancer. Cancer Lett. 2009;285:116–26. doi: 10.1016/j.canlet.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 13.George G, Mittal RD. MicroRNAs: Potential biomarkers in cancer. Indian J Clin Biochem. 2010;25:4–14. doi: 10.1007/s12291-010-0008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mishra PJ, Merlino G. MicroRNA reexpression as differentiation therapy in cancer. J Clin Invest. 2009;119:2119. doi: 10.1172/JCI40107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddy SD, Gajula RP, Pakala SB, Kumar R. MicroRNAs and cancer therapy. Cancer Biol Ther. 2010;9:479–82. doi: 10.4161/cbt.9.7.11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ceribelli A, Yao B, Dominguez-Gutierrez PR, Nahid MA, Satoh M, Chan EK. MicroRNAs in systemic rheumatic diseases. Arthritis Res Ther. 2011;13:229. doi: 10.1186/ar3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ørom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–41. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 18.Zhao X, Yang L, Hu J, Ruan J. miR-138 might reverse multidrug resistance of leukemia cells. Leuk Res. 2010;34:1078–82. doi: 10.1016/j.leukres.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Yuan Y, Zeng ZY, Liu XH, Gong DJ, Tao J, Cheng HZ, et al. MicroRNA-203 inhibits cell proliferation by repressing ΔNp63 expression in human esophageal squamous cell carcinoma. BMC Cancer. 2011;11:57. doi: 10.1186/1471-2407-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J, Liao X, Wong C. Downregulations of B-cell lymphoma 2 and myeloid cell leukemia sequence 1 by microRNA 153 induce apoptosis in a glioblastoma cell line DBTRG-05MG. Int J Cancer. 2010;126:1029–35. doi: 10.1002/ijc.24823. [DOI] [PubMed] [Google Scholar]
- 21.Shipley JL, Butera JN. Acute myelogenous leukemia. Exp Hematol. 2009;37:649–58. doi: 10.1016/j.exphem.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Richard A, McPherson MR. 21st ed. Philadelphia: Sunders; 2007. Pincus Henry's Clinical Diagnosis and Management by Laboratory Methods. [Google Scholar]
- 23.Nagarajan L. Germany: Springer Verlag; 2009. Acute Myelogenous Leukemia: Genetics, Biology and Therapy. [Google Scholar]
- 24.Estey EH. Acute myeloid leukemia: 2012 update on diagnosis, risk stratification, and management. Am J Hematol. 2012;87:89–99. doi: 10.1002/ajh.22246. [DOI] [PubMed] [Google Scholar]
- 25.Yoshizawa S, Ohyashiki J, Ohyashiki M, Umezu T, Suzuki K, Inagaki A, et al. Downregulated plasma miR-92a levels have clinical impact on multiple myeloma and related disorders. Blood Cancer J. 2012;2:e53. doi: 10.1038/bcj.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nilsson S, Möller C, Jirström K, Lee A, Busch S, Lamb R, et al. Downregulation of miR-92a Is associated with aggressive breast cancer features and increased tumour Macrophage Infiltration. PLoS One. 2012;7:e36051. doi: 10.1371/journal.pone.0036051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka M, Oikawa K, Takanashi M, Kudo M, Ohyashiki J, Ohyashiki K, et al. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS One. 2009;4:e5532. doi: 10.1371/journal.pone.0005532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manni I, Artuso S, Careccia S, Rizzo MG, Baserga R, Piaggio G, et al. The microRNA miR-92 increases proliferation of myeloid cells and by targeting p63 modulates the abundance of its isoforms. FASEB J. 2009;23:3957–66. doi: 10.1096/fj.09-131847. [DOI] [PubMed] [Google Scholar]
- 29.Van Haaften G, Agami R. Tumorigenicity of the miR-17-92 cluster distilled. Genes Dev. 2010;24:1–4. doi: 10.1101/gad.1887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uziel T, Karginov FV, Xie S, Parker JS, Wang YD, Gajjar A, et al. The miR-17 ~ 92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci. 2009;106:2812. doi: 10.1073/pnas.0809579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shigoka M, Tsuchida A, Matsudo T, Nagakawa Y, Saito H, Suzuki Y, et al. Deregulation of miR-92a expression is implicated in hepatocellular carcinoma development. Pathol Intl. 2010;60:351–7. doi: 10.1111/j.1440-1827.2010.02526.x. [DOI] [PubMed] [Google Scholar]
- 32.Hu G, Drescher KM, Chen XM. Exosomal miRNAs: Biological properties and therapeutic potential. Front Genet. 2012;3:56. doi: 10.3389/fgene.2012.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Nakhle H, Burns PA, Cummings M, Hanby AM, Hughes TA, Satheesha S, et al. Estrogen receptor β1 expression is regulated by miR-92 in breast cancer. Cancer Res. 2010;70:4778. doi: 10.1158/0008-5472.CAN-09-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]


