Abstract
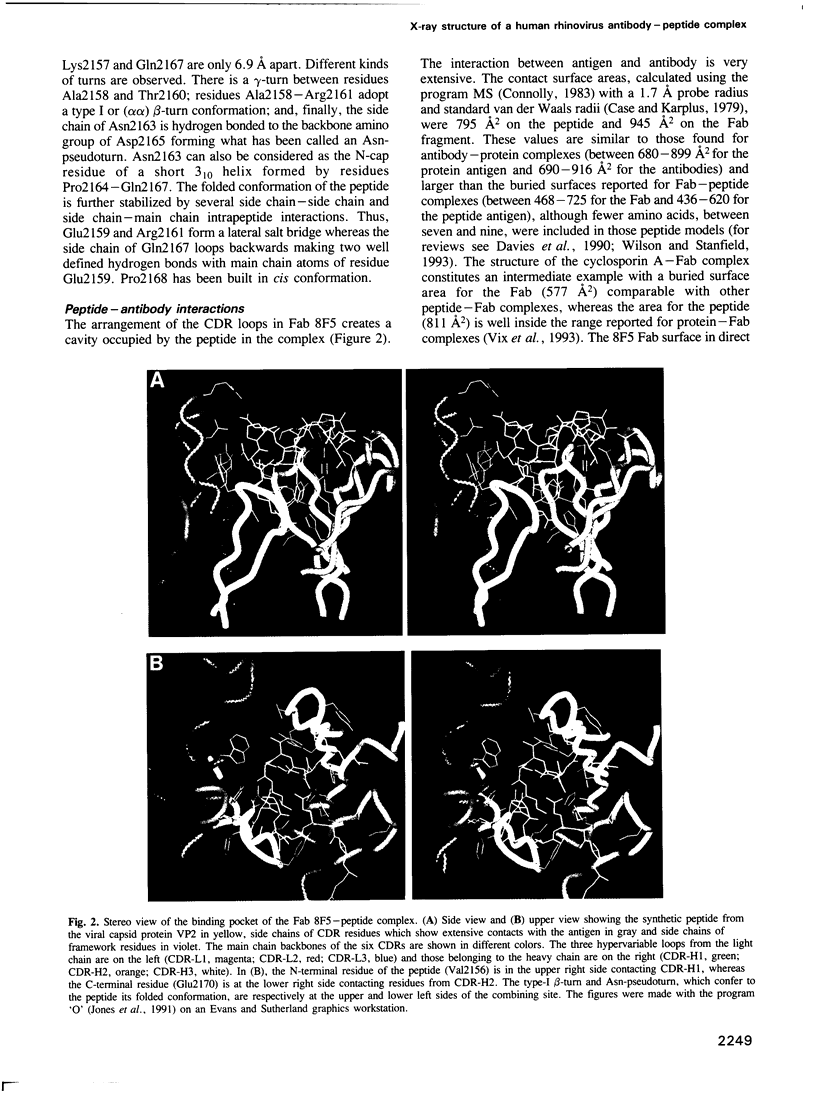
The three-dimensional structure of the complex between the Fab fragment of an anti-human rhinovirus neutralizing antibody (8F5) and a cross-reactive synthetic peptide from the viral capsid protein VP2 has been determined at 2.5 A resolution by crystallographic methods. The refinement is presently at an R factor of 0.18 and the antigen-binding site and viral peptide are well defined. The peptide antigen adopts a compact fold by two tight turns and interacts through hydrogen bonds, some with ionic character, and van der Waals contacts with antibody residues from the six hypervariable loops as well as several framework amino acids. The conformation adopted by the peptide is closely related to the corresponding region of the viral protein VP2 on the surface of human rhinovirus 1A whose three-dimensional structure is known. Implications for the cross-reactivity between peptides and the viral capsid are discussed. The peptide-antibody interactions, together with the analysis of mutant viruses that escape neutralization by 8F5 suggest two different mechanisms for viral escape. The comparison between the complexed and uncomplexed antibody structures shows important conformational rearrangements, especially in the hypervariable loops of the heavy chain. Thus, it constitutes a clear example of the 'induced fit' molecular recognition mechanism.
Full text
PDF

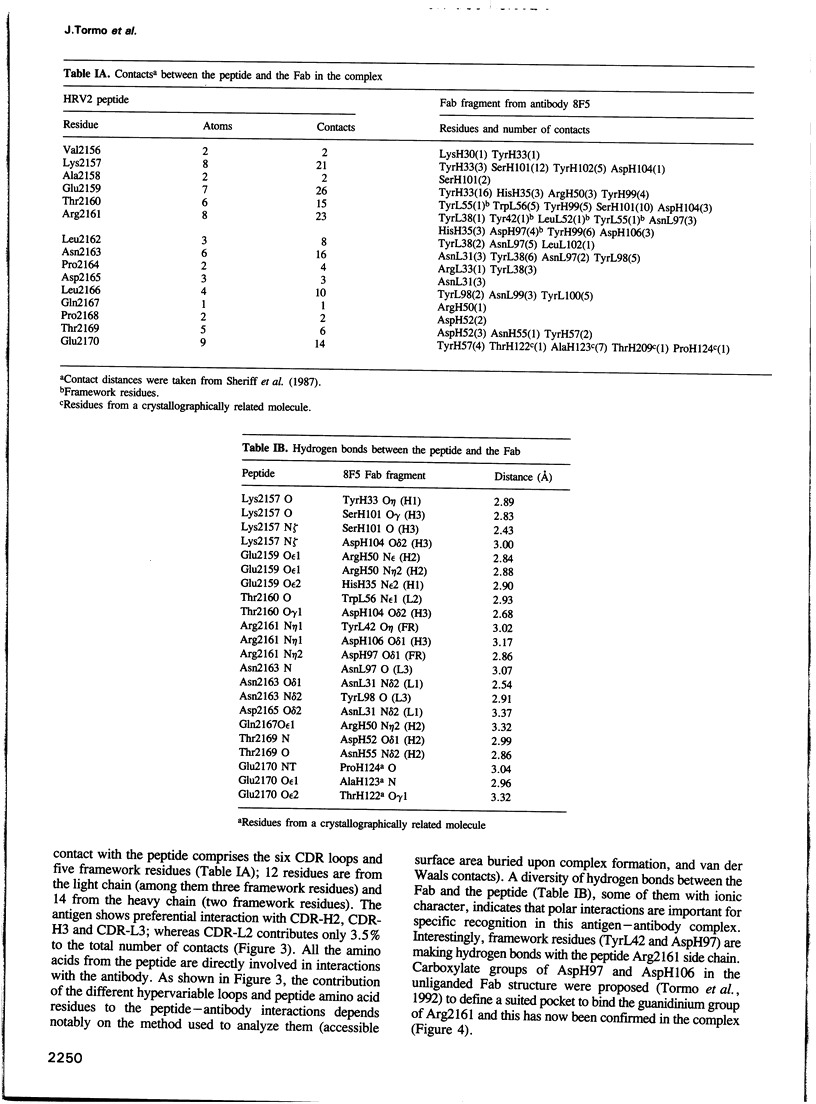
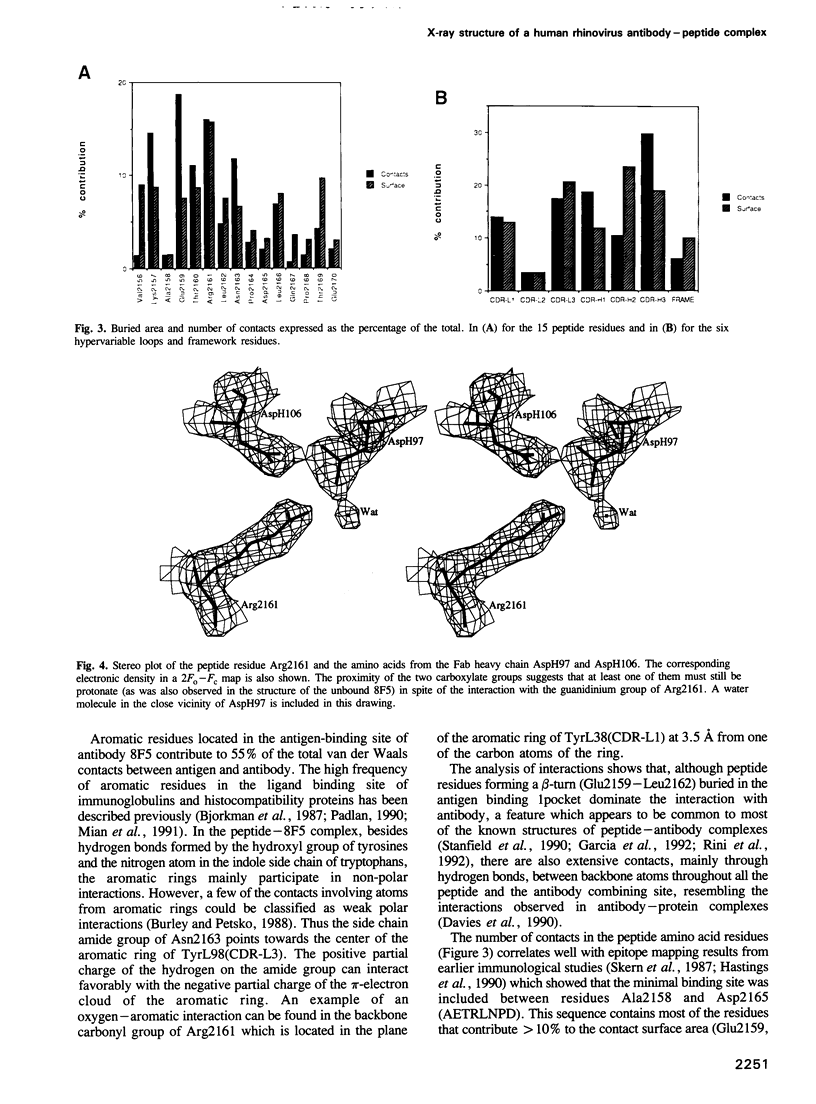
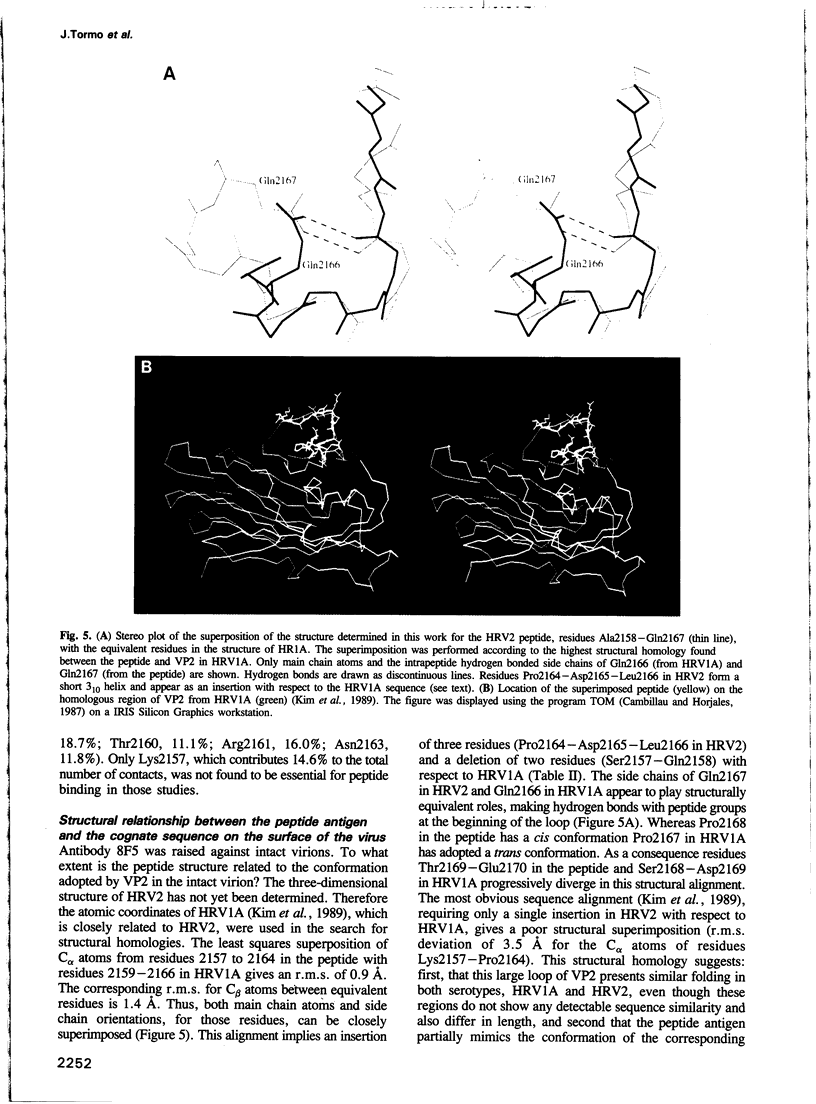
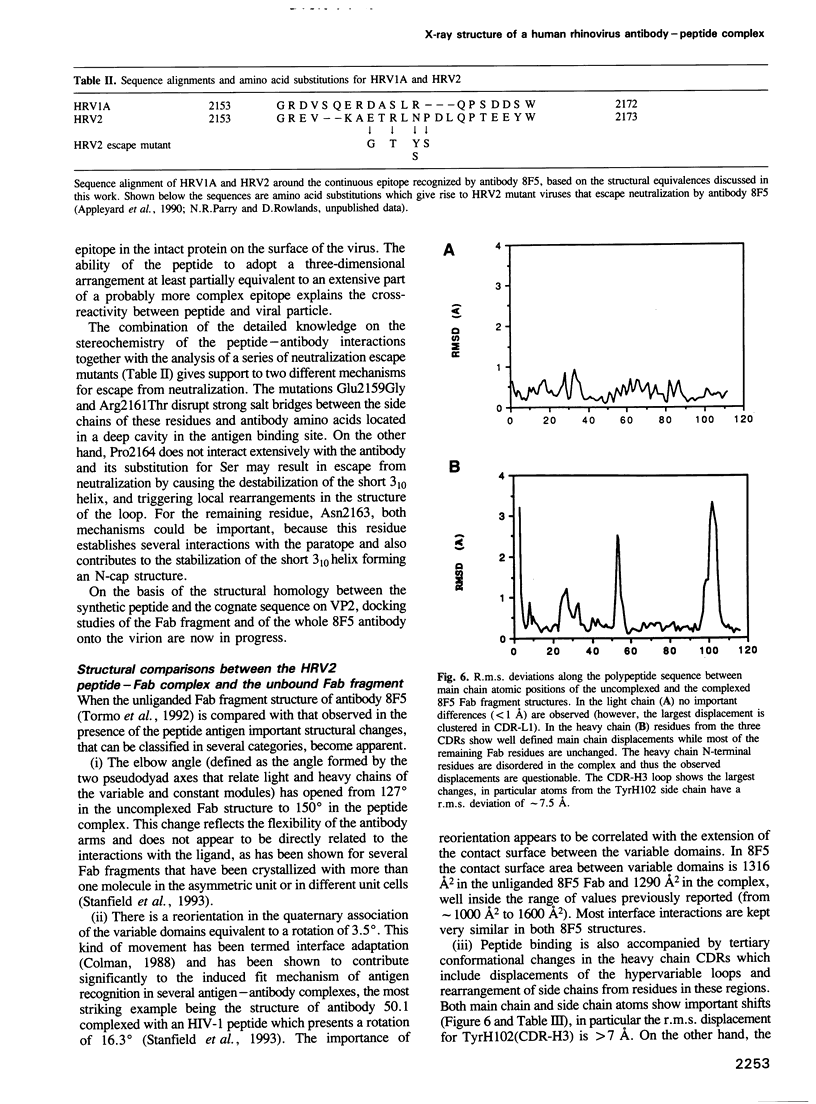
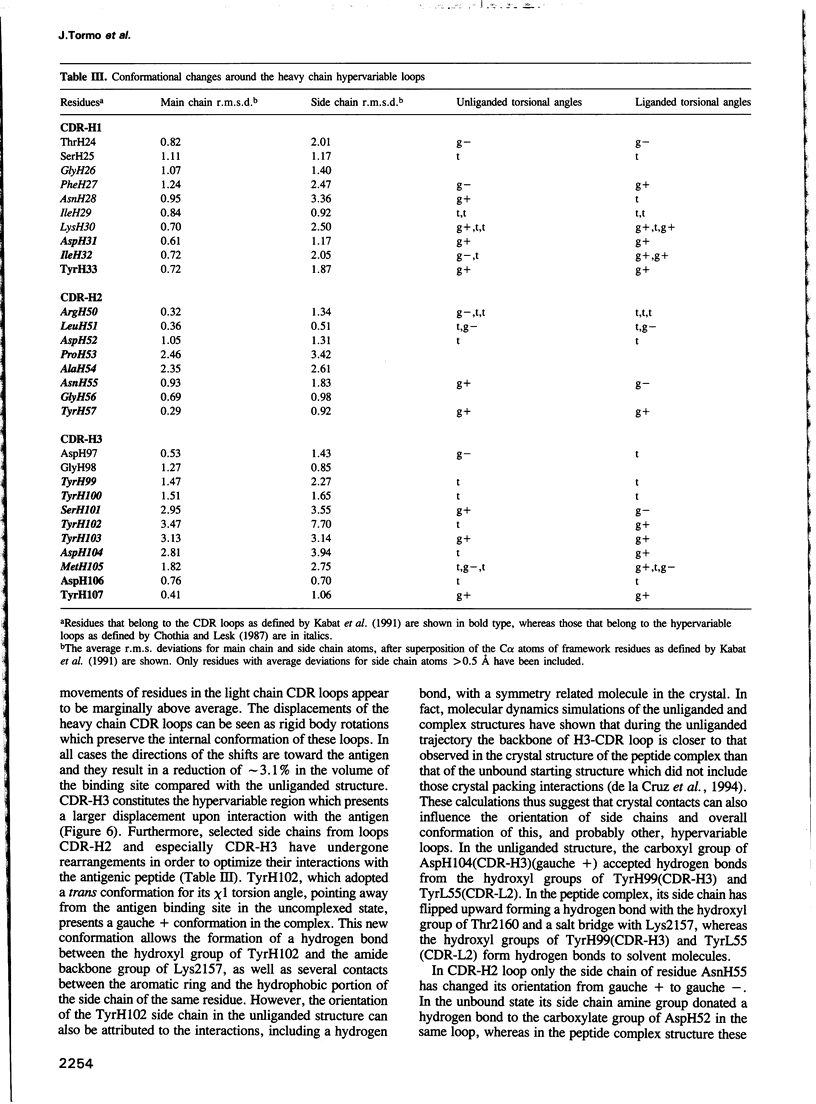


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alzari P. M., Lascombe M. B., Poljak R. J. Three-dimensional structure of antibodies. Annu Rev Immunol. 1988;6:555–580. doi: 10.1146/annurev.iy.06.040188.003011. [DOI] [PubMed] [Google Scholar]
- Amzel L. M., Poljak R. J. Three-dimensional structure of immunoglobulins. Annu Rev Biochem. 1979;48:961–997. doi: 10.1146/annurev.bi.48.070179.004525. [DOI] [PubMed] [Google Scholar]
- Appleyard G., Russell S. M., Clarke B. E., Speller S. A., Trowbridge M., Vadolas J. Neutralization epitopes of human rhinovirus type 2. J Gen Virol. 1990 Jun;71(Pt 6):1275–1282. doi: 10.1099/0022-1317-71-6-1275. [DOI] [PubMed] [Google Scholar]
- Audibert F., Jolivet M., Chedid L., Arnon R., Sela M. Successful immunization with a totally synthetic diphtheria vaccine. Proc Natl Acad Sci U S A. 1982 Aug;79(16):5042–5046. doi: 10.1073/pnas.79.16.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987 Oct 8;329(6139):506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- Burley S. K., Petsko G. A. Weakly polar interactions in proteins. Adv Protein Chem. 1988;39:125–189. doi: 10.1016/s0065-3233(08)60376-9. [DOI] [PubMed] [Google Scholar]
- Case D. A., Karplus M. Dynamics of ligand binding to heme proteins. J Mol Biol. 1979 Aug 15;132(3):343–368. doi: 10.1016/0022-2836(79)90265-1. [DOI] [PubMed] [Google Scholar]
- Chothia C., Lesk A. M. Canonical structures for the hypervariable regions of immunoglobulins. J Mol Biol. 1987 Aug 20;196(4):901–917. doi: 10.1016/0022-2836(87)90412-8. [DOI] [PubMed] [Google Scholar]
- Chothia C., Lesk A. M., Tramontano A., Levitt M., Smith-Gill S. J., Air G., Sheriff S., Padlan E. A., Davies D., Tulip W. R. Conformations of immunoglobulin hypervariable regions. Nature. 1989 Dec 21;342(6252):877–883. doi: 10.1038/342877a0. [DOI] [PubMed] [Google Scholar]
- Colman P. M. Structure of antibody-antigen complexes: implications for immune recognition. Adv Immunol. 1988;43:99–132. doi: 10.1016/s0065-2776(08)60364-8. [DOI] [PubMed] [Google Scholar]
- Davies D. R., Padlan E. A., Sheriff S. Antibody-antigen complexes. Annu Rev Biochem. 1990;59:439–473. doi: 10.1146/annurev.bi.59.070190.002255. [DOI] [PubMed] [Google Scholar]
- Francis M. J., Hastings G. Z., Sangar D. V., Clark R. P., Syred A., Clarke B. E., Rowlands D. J., Brown F. A synthetic peptide which elicits neutralizing antibody against human rhinovirus type 2. J Gen Virol. 1987 Oct;68(Pt 10):2687–2691. doi: 10.1099/0022-1317-68-10-2687. [DOI] [PubMed] [Google Scholar]
- Garcia K. C., Ronco P. M., Verroust P. J., Brünger A. T., Amzel L. M. Three-dimensional structure of an angiotensin II-Fab complex at 3 A: hormone recognition by an anti-idiotypic antibody. Science. 1992 Jul 24;257(5069):502–507. doi: 10.1126/science.1636085. [DOI] [PubMed] [Google Scholar]
- Geysen H. M., Rodda S. J., Mason T. J., Tribbick G., Schoofs P. G. Strategies for epitope analysis using peptide synthesis. J Immunol Methods. 1987 Sep 24;102(2):259–274. doi: 10.1016/0022-1759(87)90085-8. [DOI] [PubMed] [Google Scholar]
- Hastings G. Z., Speller S. A., Francis M. J. Neutralizing antibodies to human rhinovirus produced in laboratory animals and humans that recognize a linear sequence from VP2. J Gen Virol. 1990 Dec;71(Pt 12):3055–3059. doi: 10.1099/0022-1317-71-12-3055. [DOI] [PubMed] [Google Scholar]
- Hendrickson W. A. Stereochemically restrained refinement of macromolecular structures. Methods Enzymol. 1985;115:252–270. doi: 10.1016/0076-6879(85)15021-4. [DOI] [PubMed] [Google Scholar]
- Jacob C. O., Pines M., Arnon R. Neutralization of heat-labile toxin of E. coli by antibodies to synthetic peptides derived from the B subunit of cholera toxin. EMBO J. 1984 Dec 1;3(12):2889–2893. doi: 10.1002/j.1460-2075.1984.tb02226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javaherian K., Langlois A. J., LaRosa G. J., Profy A. T., Bolognesi D. P., Herlihy W. C., Putney S. D., Matthews T. J. Broadly neutralizing antibodies elicited by the hypervariable neutralizing determinant of HIV-1. Science. 1990 Dec 14;250(4987):1590–1593. doi: 10.1126/science.1703322. [DOI] [PubMed] [Google Scholar]
- Jones T. A. Diffraction methods for biological macromolecules. Interactive computer graphics: FRODO. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kim S. S., Smith T. J., Chapman M. S., Rossmann M. C., Pevear D. C., Dutko F. J., Felock P. J., Diana G. D., McKinlay M. A. Crystal structure of human rhinovirus serotype 1A (HRV1A). J Mol Biol. 1989 Nov 5;210(1):91–111. doi: 10.1016/0022-2836(89)90293-3. [DOI] [PubMed] [Google Scholar]
- Mian I. S., Bradwell A. R., Olson A. J. Structure, function and properties of antibody binding sites. J Mol Biol. 1991 Jan 5;217(1):133–151. doi: 10.1016/0022-2836(91)90617-f. [DOI] [PubMed] [Google Scholar]
- Müller G. M., Shapira M., Arnon R. Anti-influenza response achieved by immunization with a synthetic conjugate. Proc Natl Acad Sci U S A. 1982 Jan;79(2):569–573. doi: 10.1073/pnas.79.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson N. H., Kolatkar P. R., Oliveira M. A., Cheng R. H., Greve J. M., McClelland A., Baker T. S., Rossmann M. G. Structure of a human rhinovirus complexed with its receptor molecule. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):507–511. doi: 10.1073/pnas.90.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padlan E. A. On the nature of antibody combining sites: unusual structural features that may confer on these sites an enhanced capacity for binding ligands. Proteins. 1990;7(2):112–124. doi: 10.1002/prot.340070203. [DOI] [PubMed] [Google Scholar]
- Parry N. R., Barnett P. V., Ouldridge E. J., Rowlands D. J., Brown F. Neutralizing epitopes of type O foot-and-mouth disease virus. II. Mapping three conformational sites with synthetic peptide reagents. J Gen Virol. 1989 Jun;70(Pt 6):1493–1503. doi: 10.1099/0022-1317-70-6-1493. [DOI] [PubMed] [Google Scholar]
- Patarroyo M. E., Amador R., Clavijo P., Moreno A., Guzman F., Romero P., Tascon R., Franco A., Murillo L. A., Ponton G. A synthetic vaccine protects humans against challenge with asexual blood stages of Plasmodium falciparum malaria. Nature. 1988 Mar 10;332(6160):158–161. doi: 10.1038/332158a0. [DOI] [PubMed] [Google Scholar]
- Prasad B. V., Burns J. W., Marietta E., Estes M. K., Chiu W. Localization of VP4 neutralization sites in rotavirus by three-dimensional cryo-electron microscopy. Nature. 1990 Feb 1;343(6257):476–479. doi: 10.1038/343476a0. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Rini J. M., Schulze-Gahmen U., Wilson I. A. Structural evidence for induced fit as a mechanism for antibody-antigen recognition. Science. 1992 Feb 21;255(5047):959–965. doi: 10.1126/science.1546293. [DOI] [PubMed] [Google Scholar]
- Rini J. M., Stanfield R. L., Stura E. A., Salinas P. A., Profy A. T., Wilson I. A. Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6325–6329. doi: 10.1073/pnas.90.13.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M. G., Arnold E., Erickson J. W., Frankenberger E. A., Griffith J. P., Hecht H. J., Johnson J. E., Kamer G., Luo M., Mosser A. G. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985 Sep 12;317(6033):145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- Rossmann M. G., Johnson J. E. Icosahedral RNA virus structure. Annu Rev Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- Sheriff S., Hendrickson W. A., Smith J. L. Structure of myohemerythrin in the azidomet state at 1.7/1.3 A resolution. J Mol Biol. 1987 Sep 20;197(2):273–296. doi: 10.1016/0022-2836(87)90124-0. [DOI] [PubMed] [Google Scholar]
- Skern T., Neubauer C., Frasel L., Gründler P., Sommergruber W., Zorn M., Kuechler E., Blaas D. A neutralizing epitope on human rhinovirus type 2 includes amino acid residues between 153 and 164 of virus capsid protein VP2. J Gen Virol. 1987 Feb;68(Pt 2):315–323. doi: 10.1099/0022-1317-68-2-315. [DOI] [PubMed] [Google Scholar]
- Smith T. J., Olson N. H., Cheng R. H., Liu H., Chase E. S., Lee W. M., Leippe D. M., Mosser A. G., Rueckert R. R., Baker T. S. Structure of human rhinovirus complexed with Fab fragments from a neutralizing antibody. J Virol. 1993 Mar;67(3):1148–1158. doi: 10.1128/jvi.67.3.1148-1158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield R. L., Fieser T. M., Lerner R. A., Wilson I. A. Crystal structures of an antibody to a peptide and its complex with peptide antigen at 2.8 A. Science. 1990 May 11;248(4956):712–719. doi: 10.1126/science.2333521. [DOI] [PubMed] [Google Scholar]
- Stanfield R. L., Takimoto-Kamimura M., Rini J. M., Profy A. T., Wilson I. A. Major antigen-induced domain rearrangements in an antibody. Structure. 1993 Oct 15;1(2):83–93. doi: 10.1016/0969-2126(93)90024-b. [DOI] [PubMed] [Google Scholar]
- Tormo J., Fita I., Kanzler O., Blaas D. Crystallization and preliminary x-ray diffraction studies of the Fab fragment of a neutralizing monoclonal antibody directed against human rhinovirus serotype 2. J Biol Chem. 1990 Oct 5;265(28):16799–16800. [PubMed] [Google Scholar]
- Tormo J., Stadler E., Skern T., Auer H., Kanzler O., Betzel C., Blaas D., Fita I. Three-dimensional structure of the Fab fragment of a neutralizing antibody to human rhinovirus serotype 2. Protein Sci. 1992 Sep;1(9):1154–1161. doi: 10.1002/pro.5560010909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Regenmortel M. H. Structural and functional approaches to the study of protein antigenicity. Immunol Today. 1989 Aug;10(8):266–272. doi: 10.1016/0167-5699(89)90140-0. [DOI] [PubMed] [Google Scholar]
- Vix O., Rees B., Thierry J. C., Altschuh D. Crystallographic analysis of the interaction between cyclosporin A and the Fab fragment of a monoclonal antibody. Proteins. 1993 Apr;15(4):339–348. doi: 10.1002/prot.340150402. [DOI] [PubMed] [Google Scholar]
- Wang G. J., Porta C., Chen Z. G., Baker T. S., Johnson J. E. Identification of a Fab interaction footprint site on an icosahedral virus by cryoelectron microscopy and X-ray crystallography. Nature. 1992 Jan 16;355(6357):275–278. doi: 10.1038/355275a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmot C. M., Thornton J. M. Beta-turns and their distortions: a proposed new nomenclature. Protein Eng. 1990 May;3(6):479–493. doi: 10.1093/protein/3.6.479. [DOI] [PubMed] [Google Scholar]
- de la Cruz X., Mark A. E., Tormo J., Fita I., van Gunsteren W. F. Investigation of shape variations in the antibody binding site by molecular dynamics computer simulation. J Mol Biol. 1994 Mar 4;236(4):1186–1195. doi: 10.1016/0022-2836(94)90020-5. [DOI] [PubMed] [Google Scholar]