Abstract
Background:
DJ-1 (PARK7) was reported as an oncogene in a Ras-dependent manner. Recent studies have shown that DJ-1 stimulates cell proliferation, cell invasion, and cancer metastasis. However, the molecular mehchanism by which DJ-1 induces cancer cell invasion and metastasis remains unclear.
Methods:
Breast cancer cells were transfected with DJ-1 siRNA or DJ-1 overexpression to investigate the effect of DJ-1 on KLF17 expression. ID-1 luciferase promoter assay was performed to evaluate DJ-1-dependent KLF17 expression changes. In addition, Epistasis analysis of DJ-1 and KLF17 was performed to evaluate their regulatory interactions. Ras inhibitors were pretreated to determine whether DJ-1 regulates cell invasion in a Ras-dependent manner.
Results:
In the present study, we found increased DJ-1 expression in highly invasive breast cancer cells as compared with non-metastatic cells. Furthermore, DJ-1 promoted breast cancer cell invasion by downregulating E-cadherin and increasing Snail expression. Interestingly, exogenous DJ-1 overexpression markedly decreased mRNA and protein expression of KLF17, the EMT negative regulator. These data were confirmed by ID-1 promoter activity, which is directly regulated by DJ-1-dependent KLF17 transcription factor. Epistasis analysis showed that KLF17 overexpression overcomes increased cell invasion by DJ-1, suggesting that KLF17 might be one of the downstream signalling molecules of DJ-1. Acceleration of cell invasion by DJ-1 was alleviated by Ras inhibitors, suggesting that DJ-1 cooperates with Ras to increase cell invasion.
Conclusion:
Altogether, these data suggest for the first time that DJ-1 acts as an EMT-positive regulator in breast cancer cells via regulation of the KLF17/ID-1 pathway.
Keywords: DJ-1 (PARK7), breast cancer cells, invasion, epithelial–mesenchymal transition (EMT), KLF17, ID-1, Ras
DJ-1, originally identified as a novel oncogene product involved in a Ras-related signal transduction pathway (Nagakubo et al, 1997), belongs to the peptidase C56 family. The human DJ-1 gene maps to 1p36.2-36.3, a locus for many chromosome aberrations in cancers (Taira et al, 2001). It is generally known that loss-of-function DJ-1 mutations can cause early-onset Parkinson's disease. Mitochondrial membrane perturbation, oxidative stress, and proteasome dysfunction are among the several hypotheses suggested to explain the molecular basis of neuronal damage (Dawson and Dawson, 2003). DJ-1 protects several kinds of cancer cells such as pancreatic (Inberg and Linial, 2010), neuronal (Yokota et al, 2003; Zhang et al, 2010), leukaemic (Liu et al, 2008), lung (MacKeigan et al, 2003), and breast (Ismail et al, 2012) against oxidative stress-induced apoptosis. DJ-1 is reported to modulate the PTEN/Akt survival pathway inactivation (Kim et al, 2005). In addition, DJ-1 binds multiple RNA targets, implicating involvement in multiple cellular pathways (van der Brug et al, 2008).
The prognostic significance of DJ-1 has been studied in breast cancer, in which the serum level of DJ-1 is increased compared with healthy subject (Le Naour et al, 2001). In breast cancer, an increased expression of cytoplasmic DJ-1 was also observed (Kim et al, 2005). A previous study showed that DJ-1 is associated with endometriotic cell migration as well as invasion potential (Rai and Shivaji, 2011). Moreover, there was a positive association between higher DJ-1 levels and disease recurrence in stage I lung cancer (Kim et al, 2005). Bai et al (2012) also showed that DJ-1 expression is significantly correlated with lung cancer lymphatic metastasis. He et al (2012) represented that DJ-1 regulates the invasion and metastasis of pancreatic cells by activating the SRK/ERK/uPA cascade. They showed that knockdown of DJ-1 led to cytoskeleton disruption and diminished uPA activity and expression, all these effects being reversed by restoration of DJ-1 expression (He et al, 2012). Interestingly, DJ-1 was validated as a MMP substrate (Dean et al, 2007) and identified as being secreted (Butler et al, 2008), suggesting roles in both intracellular and extracellular microenvironments.
Cancer metastases, rather than primary tumours, are responsible for most of the cancer deaths (Steeg, 2006; Eccles and Welch, 2007; Steeg, 2007). Epithelial–mesenchymal transition (EMT) is a cellular program in which polarised epithelial cells undergo complex biological changes such that the epithelial cells express a mesenchymal phenotype, which induces enhanced migratory capacity, invasiveness, metastatic potential, and drug resistance (Zhou et al, 2009). A previous study showed that DJ-1 is upregulated in renal fibrosis and mediates EMT by suppressing cytoplasmic PTEN expression (Yao et al, 2011). However, the molecular mechanism by which DJ-1 inhibitis PTEN expression remains unclear.
KLF17 (Krüppel-like transcription factor 17; ZNF393) is a member of the Sp/KLF zinc finger protein family with diverse functions (Lomberk and Urrutia, 2005; van Vliet et al, 2006). Krüppel-like factor (KLF) family members are critical regulators of various cellular processes, including reprogramming of differentiated cells to stem cells (Takahashi et al, 2007), cell survival (Perkins et al, 1995), and tumour development (Foster et al, 2000; Ghaleb et al, 2005). Interestingly, KLF17 binds to the DNA consensus sequence (5′-CACCC-3′) to inhibit ID-1 transcription (van Vliet et al, 2006), resulting in the inhibition of EMT and breast cancer cell invasion (Gumireddy et al, 2009). ID-1 is a novel negative regulator of PTEN that could activate the Akt pathway and its downstream effectors such as the Wnt/TCF pathway (Lee et al, 2009), suggesting that the oncogenic function of ID-1 may be partly attributed to its PTEN inhibition in human breast carcinogenesis. ID-1 is overexpressed in highly invasive cancer cells, including prostate (Zhang et al, 2007), breast (Fong et al, 2003), cervical (Darnel et al, 2010), and bladder cancer (Ding et al, 2006). It has been suggested that the combined expression patterns of KLF17 and ID-1 can serve as a potential biomarker for lymph node metastasis in breast cancer (Gumireddy et al, 2009).
In the present study, we tried to evaluate the molecular mechanism of DJ-1 protein underlying the EMT status and in vitro cell invasion of breast cancer cells. In addition, we also studied whether Ras is involved in DJ-1-induced cell invasion.
Materials and methods
Chemicals and reagents
MTT (3-[4,5-dimethyl-2-thiazolyl]-2,5-diphenyl-2H-tetrazolium bromide) was purchased from Sigma (St. Louis, MO, USA). DMEM, RPMI, FBS and penicillin/streptomycin antibiotics were purchased from Gibco (Invitrogen, Carlsbad, CA, USA). Qiazol was purchased from Qiagen (Valencia, CA, USA), and 2 × SYBR Green PCR master mix was purchased from Takara Biotechnology (Dalian, Japan). The CytoSelect 96-well cell invasion assay kit was purchased from Cell Biolabs (San Diego, CA, USA). Rabbit polyclonal anti-DJ-1, anti-Snail, anti-KLF17, and rabbit monoclonal anti-E-cadherin antibodies were purchased from Abcam (Cambridge, UK). Mouse monoclonal anti-ID-1 antibody was purchased from Millipore (Upstate Chemicon, Temecula, CA, USA). Goat polyclonal anti-occludin, anti-fibronectin, and mouse monoclonal IgG HRP-conjugated anti-β-actin antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). HRP-conjugated goat anti-rabbit and anti-mouse secondary antibodies were purchased from Pierce (Rockford, IL, USA). ECL Western Blotting Detection Reagent was purchased from Neuronex (Daegu, South Korea). Ras inhibitors such as PD98059 and LY294002 were purchased from Sigma. All other reagents were obtained from standard commercial sources.
Cell culture
The human breast cancer cell lines MCF-7, T47D, MDA-MB-231, and MDA-MB-435 were obtained from the American Type Culture Collection and maintained in DMEM (MCF-7 and MDA-MB-231) or RPMI (T47D and MDA-MB-435) media containing 10% FBS and 1% penicillin/streptomycin solution. Cells were grown at 37 °C in a humidified atmosphere containing 5% CO2. Although the origin of the MDA-MB-435 cell line has been disputed, it has many properties similar to those of other aggressive breast cancer cell lines (Chambers, 2009).
Overexpression or knockdown of genes
MCF-7, MDA-MB-231, and MDA-MB-435 cells were transfected with mixed DJ-1 siRNA (Santa Cruz Biotechnology). Briefly, 1 × 105 cells were seeded in 60-mm plates, media being replaced on the following day with serum-free media just before transfection. Cells were then transfected with siRNA at a final concentration of 10 nM using Lipofectamine 2000 (Invitrogen) as a transfecting reagent in Opti-MEM (Invitrogen). For exogenous DJ-1 overexpression, pc3DNA-flag-DJ-1 vector (provided by Dr. Jong-In Yook at Yonsei University, Seoul, Korea) or pc3DNA control vector were transfected under the same experimental conditions. For exogenous KLF17 overexpression, pCMV6-AC-KLF17-GFP (Origene, Rockville, MD, USA) or pCMV6 control vector were transfected under the same experimental conditions. After 8 h, serum-free media containing the transfection mixture was replaced with fresh serum-containing media and incubated for the indicated periods. Cells were collected after 8–48 h of transfection and used for cell invasion assay, chromatin immunoprecipitation (ChIP) assay, real-time qPCR, and western blotting analyses.
In vitro cell invasion assay
The effect of DJ-1 expression on breast cancer cell invasion was determined using the CytoSelect 96-well cell invasion assay kit (Cell Biolabs) containing polycarbonate membrane inserts (8-μm pore size) as indicated in the instruction manual and previously described (Cabello et al, 2009). The basement membrane layer was rehydrated by adding 100 μl of warm, serum-free medium to the inner compartment and incubated for 1 h in a cell culture incubator. A cell suspension (1 × 105 per ml) in serum-free medium was cultured on the basement membrane after removing the rehydration media. In all, 150 μl of serum-containing medium was added to the feeder tray. After insertion of the basement membrane chamber, 100 μl of cell suspension was added. After 24 h of incubation, the medium in the membrane chamber was transferred to a new harvesting tray containing 150 μl of detachment solution for 30 min. The cells were dislodged completely from the underside of the membrane by gently tilting the membrane several times. Fifty microlitres of 4 × lysis buffer/CyQuant GR dye solution was added to all samples, which were then incubated for 20 min at room temperature. Fluorescence measurements were performed in a fluorescence plate reader at 480 nm/520 nm.
To evaluate the effect of Ras inhibitors on DJ-1-induced cell invasion, control vector- or DJ-1-Flag-transfected cells were seeded onto matrigel-coated transwells in matrigel invasion chambers (BD Biosciences, Franklin Lakes, NJ, USA). Then, a specific inhibitor of PI3K (LY294002, 10 μM) or BRAF/MEK inhibitor (PD98059, 25 μM) was treated to the lower well. After 48 h, non-invading cells were removed from the top well with a cotton swab while the bottom cells were fixed and stained with a 0.2% solution of crystal violet in 2% ethanol. After washing, the crystal violet stain was solubilised with a 10% acetic acid solution, and cell invasion was quantitated by measuring the absorbance at 595 nm.
Quantitative real-time PCR
The effect of DJ-1 expression on a panel of genes involved in invasion and EMT regulation in breast cancer cells was investigated using real-time quantitative RT-PCR analysis. In brief, 1 × 105 cells were plated in 60-mm dishes and grown for 24 h, followed by transfection with DJ-1 siRNA, DJ-1-Flag, or KLF17 overexpression vector for 48 h as indicated. Total RNA was extracted with Qiazol (Qiagen) according to the manufacturer's instructions. Extracted RNA (5 μg) was reverse transcribed into cDNA using a first-strand cDNA synthesis kit (Applied Biosystems, Foster City, CA, USA), and the resulting cDNA was diluted 10-fold and kept at −20 °C until use. The real-time qPCR primers were designed using the Primer Express 1.5 software (Applied Biosystems) as follows: DJ-1 forward, 5′-GTCATTTGTCCTGATGCCAGC-3′, reverse, 5′-TCAGATAAATTCTGTGCGCCC-3′ E-cadherin forward, 5′-CGACCCAACCCAAGAATCTA-3′, reverse 5′-CTCCAAGAATCCCCAGAATG-3′ Snail forward, 5′-AGCTCTCTGAGGCCAAGGATCT-3′, reverse 5′-TGTGGCTTCGGATGTGCAT-3′ occludin forward 5'-CGGTCTAGGACGCAGCAGAT-3′, reverse 5′-AAGAGGCCTGGATGACATGG-3′ fibronectin forward 5′-CAGGATCACTTACGGAGAAACAG-3′, reverse 5′-GCCAGTGACAGCAACACAGTG-3′ KLF17 forward, 5′-CTGCCTGAGCGTGGTATGAG-3′, reverse 5′-TCATCCGGGAAGGAGTGAGA-3′ ID-1 forward, 5′-CTCTACGACATGAACGGCTG-3′, reverse 5′-TGCTCACCTTGCGGTTCTG-3′ PTEN forward 5′-TCAGTGGCGGAACTTGCAA-3′, reverse 5′-CATGAACTTGTCTTCCCGTCG-3′ and GAPDH forward, 5′-AGATCATCAGCAATGCCTCCTG-3′, reverse 5′-ATGGCATGGACTGTGGTCATG-3′. The expressions of these genes were normalised to GAPDH. Real-time PCR was carried out using an ABI Prism 7500 sequence detection system (Applied Biosystems). Each 20 μl PCR reaction contained 10 μl SYBR Green PCR master mix, 4 μl diluted cDNA, and 200 nM primer set. All samples were amplified in triplicate in a 96-well plate using the following cycling conditions: 2 min at 50 °C, 10 min at 95 °C, and 40 cycles at 95 °C for 15 s followed by 1 min at 60 °C. Calculations were performed using the Δcycle threshold (ΔCt) method, normalising the average Ct value of each treatment compared with its endogenous control (GAPDH) and then calculating the 2–ΔCt for each treatment. These experiments were repeated three times.
Western blotting analyses
Cells were washed twice with cold PBS, after which 200 μl of PRO-PREP protein extraction solution (Intron, Daejon, South Korea) was added. The cell lysates were centrifuged and protein concentrations were estimated using the Coomassie protein assay reagent (Thermo Scientific, Rockford, IL, USA). Protein samples (30–40 μg) were electrophoresed on 8%–15% SDS–PAGE gels. Proteins were transferred to nitrocellulose membranes blocked in 5% skim milk in TBS (25 mM Tris base and 150 mM NaCl) for 2 h at room temperature and then incubated with primary antibody overnight at 4 °C. The membranes were then incubated with HRP-conjugated secondary antibodies at 1 : 5000 dilutions for 1 h at room temperature and then washed three times in TBST (TBS and 0.1% Tween 20). β-Actin was used as a reference protein for the normalisation of protein contents. The target proteins were detected with ECL detection reagents, and the relative intensities of the bands were analysed by the Image-J software (U.S. NIH, Bethesda, MD, USA).
ChIP assay
The effect of DJ-1 expression on ID-1 promoter activity regulation through KLF17 was investigated using ChIP assay as previously described (Gumireddy et al, 2009). Briefly, Lipofectamine 2000 control and transiently-transfected MDA-MB-231 cells were fixed in 1% paraformaldehyde at 37 °C for 10 min. After washing twice with ice-cold PBS containing protease inhibitor cocktail, cells were collected in lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris, pH 8.1), incubated for 30 min on ice, and then sonicated to shear DNA. The lysates were then centrifuged, and the supernatants were diluted 10-fold with ChIP dilution buffer (0.01% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl at pH 8.0, 150 mM NaCl, and protease inhibitors). Rabbit anti-KLF17 or normal rabbit IgG were added to the supernatants and incubated overnight at 4 °C with mild rotation. The immunocomplex was incubated with protein G-agarose, then washed with low-salt buffer, high-salt buffer, and lithium chloride wash buffer, and eluted with elution buffer (1% SDS, 0.1 M NaHCO3, and 200 mM NaCl). Reversal of crosslinking was done by heating at 65 °C overnight in the presence of NaCl. DNA was purified using PureLink PCR Purification kit (Invitrogen). The amount of immunoprecipitated DNA was analysed in triplicate using human ID-1 (sense: 5′-AGCTTGTGAGAGGCAGTGATTCT-′3; antisense: 5′-GGAACCCAGAGGTAGGCTGTT-′3) promoter primer sequences by real-time PCR with SYBR green PCR kit. Data were analysed using the 2−ΔCt method and normalised with input (HDAC1) samples.
Statistical analysis
The differences in mean values among the groups were evaluated and expressed as the means±s.d. Averages were drawn, and statistical calculations were then performed using a Student's t-test in Microsoft Excel 2010 (Redmond, WA, USA).
Results
DJ-1 expression in breast cancer cells
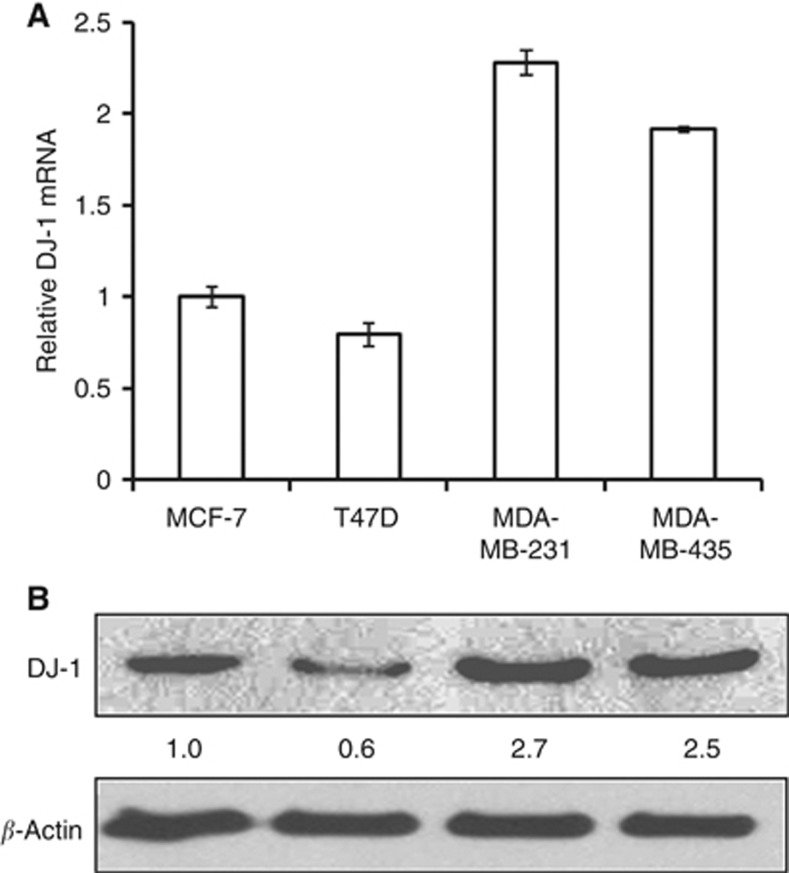
To demonstrate the relation between DJ-1 expression and breast cancer cell invasion capacity, we investigated DJ-1 mRNA and protein expression levels in non-metastatic (MCF-7 and T47D) and metastatic (MDA-MB-231 and MDA-MB-435) cancer cells. Interestingly, the expression levels of DJ-1 mRNA (Figure 1A) and protein (Figure 1B) are higher in the invasive breast cancer cells compared with non-metastatic cells.
Figure 1.
Expression of DJ-1 mRNA (A) and protein (B) in non-metastatic (MCF-7, T47D) and metastatic (MDA-MB-231, MDA-MB-435) breast cancer cells. Real-time qPCR data are from three independent experiments; each line represents s.d.
Effect of DJ-1 knockdown or overexpression on the EMT of breast cancer cells
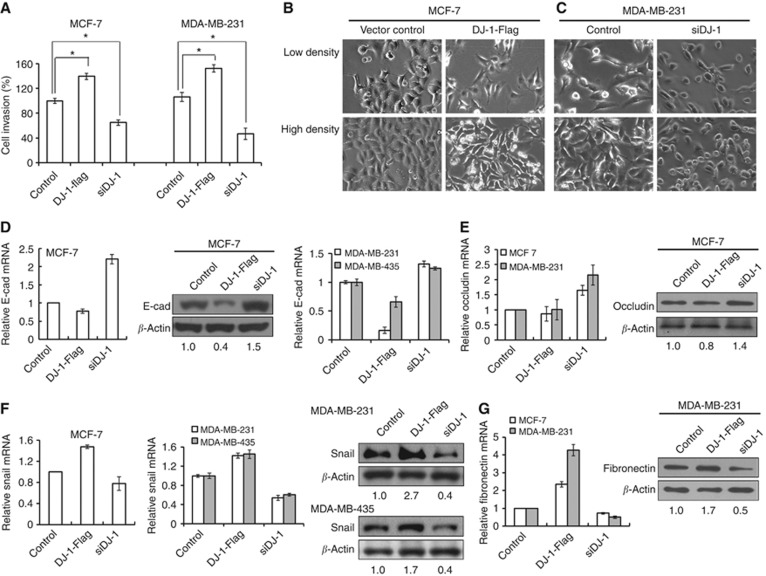
E-cadherin is one of the most important molecules in cell–cell adhesion in epithelial tissues, a hallmark of EMT being the loss of E-cadherin expression. We investigated the effect of DJ-1 on epithelial markers (E-cadherin, occludin) and mesenchymal markers (Snail, fibronectin) in breast cancer cells. DJ-1 mRNA and protein expression levels were confirmed in DJ-1-Flag- or siDJ-1-transfected cells (Supplementary Figure S1). Interestingly, exogenous DJ-1 overexpression increased the invasion capacity, and DJ-1 knockdown by siRNA treatment downregulated cell invasion of MCF-7 and MDA-MB-231 cells as compared with vector-treated cells (Figure 2A).
Figure 2.
Effect of DJ-1 overexpression and knockdown on cell invasion and EMT of breast cancer cells. (A) For cell invasion assay, DJ-1-Flag or siDJ-1 was transfected for 12 h, 48 h after which transwell invasion assay was performed. Statistical analysis was performed using the Student's t-test (*P<0.05). After transfection with DJ-1-Flag or siDJ-1 for 48 h in (B) MCF-7 and (C) MDA-MB-231 cells, respectively, cellular morphological change was evaluated by light microscope. mRNA and/or protein expression of (D, E) epithelial (E-cadherin, occludin) and (F, G) mesenchymal markers (Snail, fibronectin) were analysed by real-time qPCR or western blotting analysis after 24–48 h of transfection. These results are from three independent experiments; each line represents s.d.
To evaluate the EMT status in relation to DJ-1 expression level, morphological changes were assessed by light microscopy. As shown in Figure 2B, MCF-7 cells overexpressing DJ-1 lost their observable epithelial morphology after 48 h of DJ-1-Flag transfection. On the contrary, siDJ-1-treated MDA-MB-231 cells gained epithelial-like morphology, unlike control. To further confirm EMT by DJ-1, the expression of epithelial and mesenchymal markers was investigated by real-time qPCR and/or western blotting. The downregulation of E-cadherin mRNA and/or protein levels was observed in cells transfected with DJ-1-Flag (Figure 2D). On the contrary, siDJ-1 transfection increased E-cadherin mRNA and/or protein expression. Occludin mRNA and protein expression was also increased in siDJ-1-transfected cells. There was no remarkable change of occludin expression in DJ-1-Flag-transfected cells (Figure 2E). Conversely, Snail mRNA and/or protein expression levels were upregulated in DJ-1-Flag-transfected cells, whereas siDJ-1 decreased Snail mRNA and/or protein expression levels in breast cancer cells (Figure 2F). The fibronectin mRNA and protein expression patterns were similar to that of Snail in MCF-7 and MDA-MB-231 cells (Figure 2G).
Effect of DJ-1 and KLF17 on ID-1 gene expression
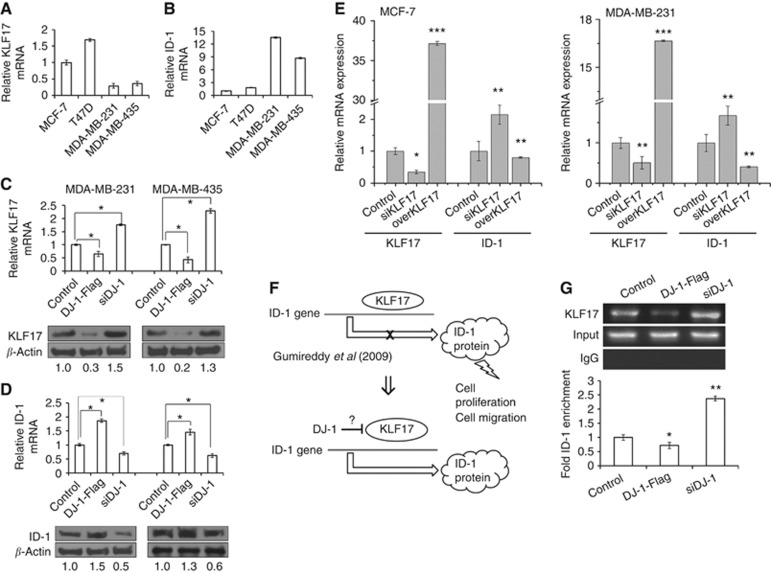
The mRNA expression level of KLF17 is higher in non-metastatic breast cancer cells as compared with invasive cells (Figure 3A), ID-1 mRNA expression showing the opposite pattern as compared with KLF17 (Figure 3B). Interestingly, DJ-1-Flag transfection decreased the expression level of KLF17 mRNA and protein, whereas DJ-1 knockdown by siRNA increased KLF17 expression (Figure 3C). On the contrary, DJ-1-Flag and siDJ-1 transfection caused ID-1 expression to increase and decrease, respectively (Figure 3D). To evaluate the effect of KLF17 on ID-1 expression in MCF-7 and MDA-MB-231 cells, siKLF17 or KLF17 overexpression vector was transfected. As shown in Figure 3E, transient KLF17 knockdown increased ID-1 mRNA expression, whereas exogenous KLF17 overexpression inhibited ID-1 gene expression (Figure 3E).
Figure 3.
The effect of DJ-1 overexpression and knockdown on KLF17 and ID-1 expression in breast cancer cells. The basal mRNA expression levels of (A) KLF17 and (B) ID-1 were analysed. MDA-MB-231 and MDA-MB-435 cells were transfected with DJ-1-Flag or siDJ-1 for 24–48 h and mRNA and protein expression levels of (C) KLF17 and (D) ID-1 were analysed using real-time qPCR and western blotting analyses. To evaluate the effect of KLF17 on ID-1 mRNA expression, pCMV6-AC-KLF17-GFP or siKLF17 was transfected for 24 h, after which (E) ID-1 expression was analysed by real-time qPCR. The effect of DJ-1 on KLF17-dependent ID-1 expression (F) was confirmed by ChIP assay. MDA-MB-231 cells were transfected with either DJ-1-Flag or siDJ-1; KLF17 binding to the ID-1 promoter at CACCC nucleototide sequence was then investigated using KLF17 antibody. (G) RT–PCR and real-time qPCR analyses were performed. Input HDAC1 and IgG were used as positive and negative controls, respectively. The statistical significances were evaluated using the Student's t-test (*P<0.05, **P<0.01, ***P<0.001). These results are from three independent experiments; each line represents s.d.
A previous study showed that KLF17 binds directly to the DNA consensus sequence (5′-CACCC-3′) of ID-1 promoter to inhibit its transcription (Gumireddy et al, 2009). Therefore, ChIP analysis was performed with KLF17 antibody to evaluate whether DJ-1 expression level affects KLF17 binding to the CACCC sequence of ID-1 promoter (Figure 3F). KLF17 mRNA expression was evaluated by RT-PCR, and input and IgG were used as positive and negative controls, respectively. As shown in Figure 3G, DJ-1-Flag transfection decreased KLF17 binding to ID-1 promoter (P<0.05), whereas siDJ-1 transfection upregulated KLF17 binding to ID-1 promoter (P<0.01). The results revealed that DJ-1 regulates KLF17 binding to the ID-1 promoter.
Epistasis analysis of DJ-1 and KLF17 on the cellular morphology and invasiveness
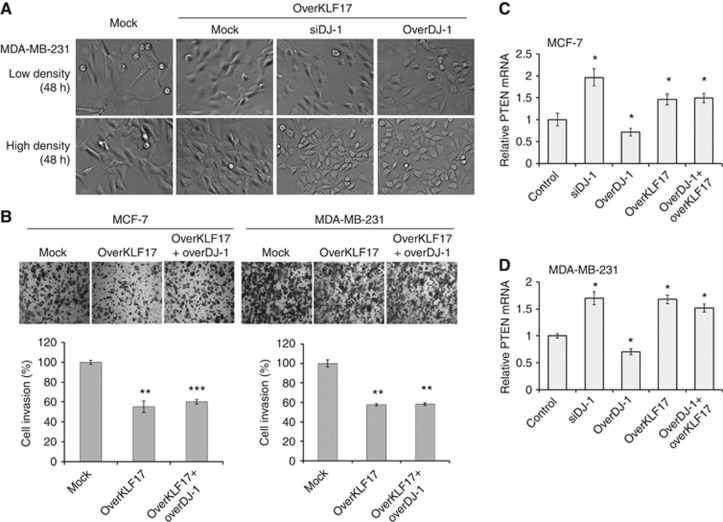
To further evaluate that DJ-1 regulates breast cancer invasion and EMT in part through KLF17, we performed double overexpression of DJ-1 and KLF17 in cancer cells. As shown in Figure 4A, MDA-MB-231 cellular morphological changes, KLF17 overexpression showed similar results independent of DJ-1 expression level. When we performed matrigel invasion assay at the same condition, KLF17 overexpression inhibits cell invasion significantly in both the cells (P<0.005) (Figure 4B). In addition, co-overexpression of KLF17 with DJ-1 results in a similar invasiveness with that of KLF17 single overexpression. To further confirm the effect of DJ-1 and KLF17 on PTEN gene expression, we investigated PTEN mRNA expression by real-time qPCR. DJ-1 knockdown or KLF17 overexpression increased PTEN mRNA expression significantly in both the cells (P<0.05) (Figures 4C and D). However, exogenous DJ-1 overexpression downregulated PTEN mRNA expression, and double overexpression with KLF17 overcomes PTEN inhibition by DJ-1. These data suggest that KLF17 might be one of the downstream signalling proteins of DJ-1.
Figure 4.
Epistatic analysis of DJ-1 and KLF17 on cellular morphology, invasion, and PTEN mRNA expression. (A) pCMV6-AC-KLF17-GFP was co-transfected with DJ-1-Flag or siDJ-1 for 48 h, after which cellular morphology was analysed with phase-contrast microscope. (B) For cell invasion assay, KLF17 overexpression vector was co-transfected with DJ-1-Flag for 12 h, 48 h after which transwell invasion assay was performed. (C, D) PTEN mRNA expression was analysed by real-time qPCR after transfection with DJ-1-Flag, siDJ-1, and/or KLF17 overexpression vector. Statistical analysis was performed using the Student's t-test (*P<0.05, **P<0.005, ***P<0.001). These results are from two or three independent experiments; each line represents s.d.
Effect of Ras inhibitor on DJ-1-enhanced cell invasion
It has been shown that DJ-1 cooperates with oncogenic Ras to promote metastasis (Moon et al, 2006). Therefore, we examined the role of Ras/Raf/MEK or PI3K/Akt in cell invasion when DJ-1 is overexpressed. DJ-1-Flag overexpression vector was transfected in MCF-7 and MDA-MB-231 bearing a PI3K and BRAF/MEK mutation, respectively, with or without specific Ras inhibitor pretreatment. LY294002 (a specific inhibitor of PI3K) or PD98059 (a specific inhibitor of MEK, which is an immediate downstream kinase target of BRAF) were used to evaluate the effect of Ras in these two cell lines.
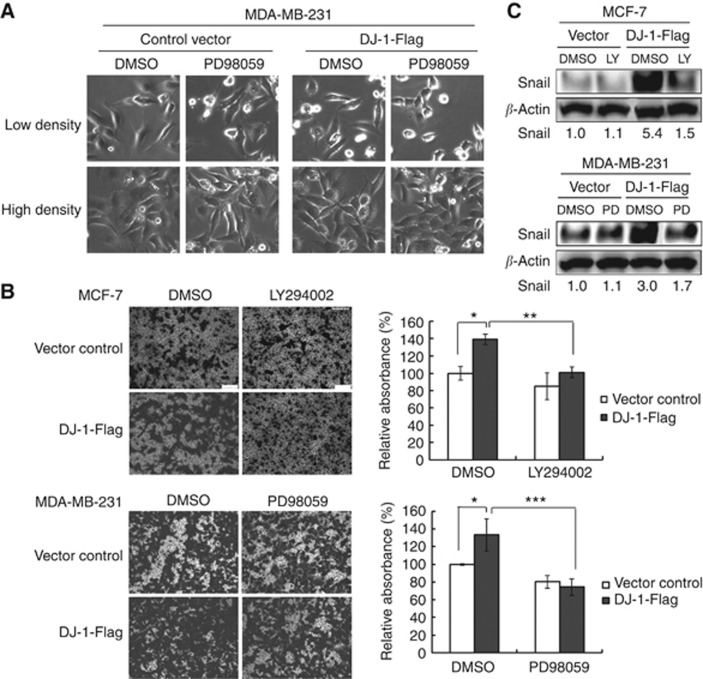
First, we evaluated the effect of PD98059 on morphological change due to exogenous DJ-1 overexpression in MDA-MB-231 cells. As shown in Figure 5A, there was no remarkable morphological change in control vector-transfected cells. In DJ-1-Flag transfected cells, however, PD98059 treatment alleviated the mesenchymal morphological change induced by DJ-1 overexpression. To further confirm the role of PI3K or MEK activation in DJ-1-promoted cell invasion, we treated the cells with LY294002 or PD98059 and measured cell invasion in DJ-1-Flag- or control vector-transfected cells. In control vector-transfected cells, LY294002 or PD98059 treatment induced no significant change in MCF-7 and MDA-MB-231 cells' invasion capacity, respectively (Figure 5B). In DJ-1-Flag-transfected cells, however, each Ras inhibitor alleviated DJ-1-promoted cell invasion significantly in both the cells (Figure 5B). We also evaluated Snail protein expression under the same experimental condition. There was no remarkable change in Snail protein expression due to Ras inhibitors in control vector-transfected cells. However, in DJ-1-Flag-transfected cells, LY294002 or PD98059 treatment decreased Snail expression remarkably (Figure 5C).
Figure 5.
The effect of Ras inhibitors on DJ-1-promoted cell invasion. (A) Morphological change was analysed in MDA-MB-231 cells after control vector or DJ-1-Flag transfection for 48 h with or without PD98059 (a specific inhibitor of MEK, which is an immediate downstream kinase target of BRAF). (B) Cell invasion was analysed in matrigel-coated transwell. After 48 h of DJ-1-Flag transfection with or without PD98059 (a specific inhibitor of BRAF-MEK) or LY294002 (a specific inhibitor of PI3K), the bottom cells were fixed and stained with a 0.2% solution of crystal violet in 2% ethanol for light microscope image analysis. After washing, the crystal violet stain was solubilised with 10% acetic acid solution, and the cell invasion was quantitated by measuring the absorbance at 595 nm. The statistical significance was evaluated using the Student's t-test (*P<0.05, **P<0.01, ***P<0.001). These results are from three independent experiments, and each line represents s.d. (C) Snail protein expression was also evaluated under the same experimental conditions.
Discussion
Lee et al (2009) showed that ID-1 negatively regulated PTEN at the transcriptional level, and this led to the Akt-mediated canonical Wnt signalling pathway, which may be partly attributed in human breast carcinogenesis. KLF17 is able to negatively regulate EMT and cell invasion by directly binding to the ID-1 promoter, and KLF17/ID-1 reciprocal expression is a critical pathway in the development of breast cancer metastasis (Gumireddy et al, 2009), indicating that regulation of this pathway may serve as a basis for the development of therapies that target early metastatic events.
Kim et al (2005) reported that DJ-1 inhibits PTEN/Akt survival pathway inactivation in breast cancer, and this has been supported by their results that DJ-1 expression correlates negatively with PTEN immunoreactivity and positively with PKB/Akt hyperphosphorylation. However, little has been known about how DJ-1 negatively regulates PTEN gene expression.
ID protein (inhibitor of differentiation or DNA binding) is a group of helix–loop–helix (HLH) proteins unable to directly bind DNA. They act as dominant-negative regulators of basic HLH (bHLH) transcription factors via heterodimerisation. As bHLH proteins are involved in tissue-specific differentiation, aberrant ID expression may interfere with cellular differentiation and growth programmes of a wide variety of cell types (Norton, 2000). Upregulation of ID-1 has been found in many types of human cancer, including breast (Lin et al, 2000). In addition, overexpression of ID-1 has been indicated as a marker for unfavourable prognosis in breast cancer (Ling et al, 2005), and the level of ID-1 protein expression is correlated with a more aggressive clinical behaviour of ovarian and prostate carcinomas (Schindl et al, 2003; Schoppmann et al, 2003).
Our data showed that DJ-1 accelerates EMT and cell invasion capacity by downregulating E-cadherin and increasing Snail expression. Exogenous DJ-1 overexpression markedly decreased the EMT negative regulator KLF17, as confirmed by investigating ID-1 expression inhibition by direct binding of KLF17 transcription factor. Nagakubo et al (1997), who first demonstrated that DJ-1 is a Ras-dependent oncogene, showed that DJ-1 transcription activity was not detected in an artificial promoter binding system. We tried to uncover whether DJ-1 binds directly to KLF17 promoter, and there seems no direct binding between DJ-1 and KLF17 promoter sequence. The exact mechanisms by which DJ-1 inhibits KLF17 expression remain unclear and will be investigated further.
To further evaluate that KLF17 is one of the downstream signalling proteins of DJ-1 mediating cellular EMT and invasion, Epistasis analysis of DJ-1 and KLF17 was performed. After double overexpression of these two genes, cellular morphology, invasion efficacy, and PTEN gene expression were analysed. Interestingly, KLF17 overexpression overcomes DJ-1-mediated EMT and invasiveness. In addition, KLF17 and DJ-1 co-overexpression recovered decreased PTEN mRNA expression by DJ-1 single overexpression. These data suggest that KLF17 might be one of the mediators of DJ-1 that accelerates EMT and cell invasion through PTEN inhibition.
Although several studies have proposed that DJ-1 is a potent modulator of the PI3K/Akt pathway in both breast cancer cells and neurons (Kim et al, 2005; Yang et al, 2005), there is little molecular basis for this assumption. Based on the present evidence that DJ-1 inhibits KLF17 expression, which is correlated negatively with PTEN immunoreactivity in primary breast cancer tissue (Kim et al, 2005), DJ-1 seems to modulate the PI3K/Akt pathway through KLF17 downregulation. A previous study finding that PTEN is required for the regulation of the PI3K/Akt pathway by DJ-1 (He et al, 2012) supports this suggestion. In addition, Nagakubo et al (1997) suggested that DJ-1 does not bind to Ras directly, suggesting that the signalling molecule(s) downstream of DJ-1 interact with Ras. Our data showed that Ras inhibitors alleviated acceleration of cell invasion by DJ-1, supporting that DJ-1 works with Ras to increase cell invasion. A previous study showed that KLF17 downregulation enhances the key role played by Ras in tumour cell invasion (Gumireddy et al, 2009). Furthermore, coexpression of ID-1 and Ras produces an abundance of rapidly growing aggressive tumours and metastasis (Swarbrick et al, 2008). Straume and Akslen (2005) also showed a significant increase in ID-1 expression in cases with BRAF mutations. These data partially explain how DJ-1 regulate cell invasion in a Ras-dependent manner.
Altogether, these data suggest for the first time that DJ-1 promotes breast cancer cell invasion and EMT in part through the downregulation of KLF17, resulting in ID-1 upregulation, thus accounting for DJ-1 inhibition of PTEN expression. Furthermore, considering the synergetic effects of ID-1 and Ras in stimulating cancer metastasis (Swarbrick et al, 2008), our data reveal ID-1 as an intermediary between DJ-1 and a Ras-related signal transduction pathway.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (2013R1A1A2008485) and Kyungpook National University Research Fund, 2013.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Bai J, Guo C, Sun W, Li M, Meng X, Yu Y, Jin Y, Tong D, Geng J, Huang Q, Qi J, Fu S. DJ-1 may contribute to metastasis of non-small cell lung cancer. Mol Biol Rep. 2012;39:2697–2703. doi: 10.1007/s11033-011-1024-7. [DOI] [PubMed] [Google Scholar]
- Butler GS, Dean RA, Tam EM, Overall CM. Pharmacoproteomics of a metalloproteinase hydroxamate inhibitor in breast cancer cells: dynamics of membrane type 1 matrix metalloproteinase-mediated membrane protein shedding. Mol Cell Biol. 2008;28:4896–4914. doi: 10.1128/MCB.01775-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello CM, Bair WB, 3rd, Lamore SD, Ley S, Bause AS, Azimian S, Wondrak GT. The cinnamon-derived Michael acceptor cinnamic aldehyde impairs melanoma cell proliferation, invasiveness, and tumor growth. Free Radic Biol Med. 2009;46:220–231. doi: 10.1016/j.freeradbiomed.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers AF. MDA-MB-435 and M14 cell lines: identical but not M14 melanoma. Cancer Res. 2009;69:5292–5293. doi: 10.1158/0008-5472.CAN-09-1528. [DOI] [PubMed] [Google Scholar]
- Darnel AD, Wang D, Ghabreau L, Yasmeen A, Sami S, Akil N, Al Moustafa AE. Correlation between the presence of high-risk human papillomaviruses and Id gene expression in Syrian women with cervical cancer. Clin Microbiol Infect. 2010;16:262–266. doi: 10.1111/j.1469-0691.2009.02774.x. [DOI] [PubMed] [Google Scholar]
- Dawson TM, Dawson VL. Molecular pathways of neurodegeneration in Parkinson's disease. Science. 2003;302:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- Dean RA, Butler GS, Hamma-Kourbali Y, Delbe J, Brigstock DR, Courty J, Overall CM. Identification of candidate angiogenic inhibitors processed by matrix metalloproteinase 2 (MMP-2) in cell-based proteomic screens: disruption of vascular endothelial growth factor (VEGF)/heparin affin regulatory peptide (pleiotrophin) and VEGF/connective tissue growth factor angiogenic inhibitory complexes by MMP-2 proteolysis. Mol Cell Biol. 2007;27:8454–8465. doi: 10.1128/MCB.00821-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Wang G, Ling MT, Wong YC, Li X, Na Y, Zhang X, Chua CW, Wang X, Xin D. Significance of Id-1 up-regulation and its association with EGFR in bladder cancer cell invasion. Int J Oncol. 2006;28:847–854. [PubMed] [Google Scholar]
- Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. 2007;369:1742–1757. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong S, Itahana Y, Sumida T, Singh J, Coppe JP, Liu Y, Richards PC, Bennington JL, Lee NM, Debs RJ, Desprez PY. Id-1 as a molecular target in therapy for breast cancer cell invasion and metastasis. Proc Natl Acad Sci USA. 2003;100:13543–13548. doi: 10.1073/pnas.2230238100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster KW, Frost AR, McKie-Bell P, Lin CY, Engler JA, Grizzle WE, Ruppert JM. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 2000;60:6488–6495. [PubMed] [Google Scholar]
- Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW. Kruppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res. 2005;15:92–96. doi: 10.1038/sj.cr.7290271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumireddy K, Li A, Gimotty PA, Klein-Szanto AJ, Showe LC, Katsaros D, Coukos G, Zhang L, Huang Q. KLF17 is a negative regulator of epithelial-mesenchymal transition and metastasis in breast cancer. Nat Cell Biol. 2009;11:1297–1304. doi: 10.1038/ncb1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zheng Z, Li J, Ben Q, Liu J, Zhang J, Ji J, Yu B, Chen X, Su L, Zhou L, Liu B, Yuan Y. DJ-1 promotes invasion and metastasis of pancreatic cancer cells by activating SRC/ERK/uPA. Carcinogenesis. 2012;33:555–562. doi: 10.1093/carcin/bgs002. [DOI] [PubMed] [Google Scholar]
- Inberg A, Linial M. Protection of pancreatic {beta}-cells from various stress conditions is mediated by DJ-1. J Biol Chem. 2010;285:25686–25698. doi: 10.1074/jbc.M110.109751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail IA, Kang HS, Lee HJ, Kwon BM, Hong SH. 2'-Benzoyloxycinnamaldehyde-mediated DJ-1 upregulation protects MCF-7 cells from mitochondrial damage. Biol Pharm Bull. 2012;35:895–902. doi: 10.1248/bpb.35.895. [DOI] [PubMed] [Google Scholar]
- Kim RH, Peters M, Jang Y, Shi W, Pintilie M, Fletcher GC, DeLuca C, Liepa J, Zhou L, Snow B, Binari RC, Manoukian AS, Bray MR, Liu FF, Tsao MS, Mak TW. DJ-1, a novel regulator of the tumor suppressor PTEN. Cancer Cell. 2005;7:263–273. doi: 10.1016/j.ccr.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Le Naour F, Misek DE, Krause MC, Deneux L, Giordano TJ, Scholl S, Hanash SM. Proteomics-based identification of RS/DJ-1 as a novel circulating tumor antigen in breast cancer. Clin Cancer Res. 2001;7:3328–3335. [PubMed] [Google Scholar]
- Lee JY, Kang MB, Jang SH, Qian T, Kim HJ, Kim CH, Kim Y, Kong G. Id-1 activates Akt-mediated Wnt signaling and p27(Kip1) phosphorylation through PTEN inhibition. Oncogene. 2009;28:824–831. doi: 10.1038/onc.2008.451. [DOI] [PubMed] [Google Scholar]
- Lin CQ, Singh J, Murata K, Itahana Y, Parrinello S, Liang SH, Gillett CE, Campisi J, Desprez PY. A role for Id-1 in the aggressive phenotype and steroid hormone response of human breast cancer cells. Cancer Res. 2000;60:1332–1340. [PubMed] [Google Scholar]
- Ling MT, Lau TC, Zhou C, Chua CW, Kwok WK, Wang Q, Wang X, Wong YC. Overexpression of Id-1 in prostate cancer cells promotes angiogenesis through the activation of vascular endothelial growth factor (VEGF) Carcinogenesis. 2005;26:1668–1676. doi: 10.1093/carcin/bgi128. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang M, Li M, Wang D, Rao Q, Wang Y, Xu Z, Wang J. Expression and role of DJ-1 in leukemia. Biochem Biophys Res Commun. 2008;375:477–483. doi: 10.1016/j.bbrc.2008.08.046. [DOI] [PubMed] [Google Scholar]
- Lomberk G, Urrutia R. The family feud: turning off Sp1 by Sp1-like KLF proteins. Biochem J. 2005;392:1–11. doi: 10.1042/BJ20051234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKeigan JP, Clements CM, Lich JD, Pope RM, Hod Y, Ting JP. Proteomic profiling drug-induced apoptosis in non-small cell lung carcinoma: identification of RS/DJ-1 and RhoGDIalpha. Cancer Res. 2003;63:6928–6934. [PubMed] [Google Scholar]
- Moon EY, Lee MR, Wang AG, Lee JH, Kim HC, Kim HM, Kim JM, Kwon BM, Yu DY. Delayed occurrence of H-ras12V-induced hepatocellular carcinoma with long-term treatment with cinnamaldehydes. Eur J Pharmacol. 2006;530:270–275. doi: 10.1016/j.ejphar.2005.11.053. [DOI] [PubMed] [Google Scholar]
- Nagakubo D, Taira T, Kitaura H, Ikeda M, Tamai K, Iguchi-Ariga SM, Ariga H. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem Biophys Res Commun. 1997;231:509–513. doi: 10.1006/bbrc.1997.6132. [DOI] [PubMed] [Google Scholar]
- Norton JD. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci. 2000;113 (Pt 22:3897–3905. doi: 10.1242/jcs.113.22.3897. [DOI] [PubMed] [Google Scholar]
- Perkins AC, Sharpe AH, Orkin SH. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- Rai P, Shivaji S. The role of DJ-1 in the pathogenesis of endometriosis. PLoS One. 2011;6:e18074. doi: 10.1371/journal.pone.0018074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindl M, Schoppmann SF, Strobel T, Heinzl H, Leisser C, Horvat R, Birner P. Level of Id-1 protein expression correlates with poor differentiation, enhanced malignant potential, and more aggressive clinical behavior of epithelial ovarian tumors. Clin Cancer Res. 2003;9:779–785. [PubMed] [Google Scholar]
- Schoppmann SF, Schindl M, Bayer G, Aumayr K, Dienes J, Horvat R, Rudas M, Gnant M, Jakesz R, Birner P. Overexpression of Id-1 is associated with poor clinical outcome in node negative breast cancer. Int J Cancer. 2003;104:677–682. doi: 10.1002/ijc.11009. [DOI] [PubMed] [Google Scholar]
- Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- Steeg PS. Cancer: micromanagement of metastasis. Nature. 2007;449:671–673. doi: 10.1038/449671a. [DOI] [PubMed] [Google Scholar]
- Straume O, Akslen LA. Strong expression of ID1 protein is associated with decreased survival, increased expression of ephrin-A1/EPHA2, and reduced thrombospondin-1 in malignant melanoma. Br J Cancer. 2005;93:933–938. doi: 10.1038/sj.bjc.6602792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbrick A, Roy E, Allen T, Bishop JM. Id1 cooperates with oncogenic Ras to induce metastatic mammary carcinoma by subversion of the cellular senescence response. Proc Natl Acad Sci USA. 2008;105:5402–5407. doi: 10.1073/pnas.0801505105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira T, Takahashi K, Kitagawa R, Iguchi-Ariga SM, Ariga H. Molecular cloning of human and mouse DJ-1 genes and identification of Sp1-dependent activation of the human DJ-1 promoter. Gene. 2001;263:285–292. doi: 10.1016/s0378-1119(00)00590-4. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- van der Brug MP, Blackinton J, Chandran J, Hao LY, Lal A, Mazan-Mamczarz K, Martindale J, Xie C, Ahmad R, Thomas KJ, Beilina A, Gibbs JR, Ding J, Myers AJ, Zhan M, Cai H, Bonini NM, Gorospe M, Cookson MR. RNA binding activity of the recessive parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc Natl Acad Sci USA. 2008;105:10244–10249. doi: 10.1073/pnas.0708518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet J, Crofts LA, Quinlan KG, Czolij R, Perkins AC, Crossley M. Human KLF17 is a new member of the Sp/KLF family of transcription factors. Genomics. 2006;87:474–482. doi: 10.1016/j.ygeno.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Yang Y, Gehrke S, Haque ME, Imai Y, Kosek J, Yang L, Beal MF, Nishimura I, Wakamatsu K, Ito S, Takahashi R, Lu B. Inactivation of Drosophila DJ-1 leads to impairments of oxidative stress response and phosphatidylinositol 3-kinase/Akt signaling. Proc Natl Acad Sci USA. 2005;102:13670–13675. doi: 10.1073/pnas.0504610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Wei H, Liu L, Bai S, Li C, Luo Y, Zeng R, Han M, Ge S, Xu G. Upregulated DJ-1 promotes renal tubular EMT by suppressing cytoplasmic PTEN expression and Akt activation. J Huazhong Univ Sci Technolog Med Sci. 2011;31:469–475. doi: 10.1007/s11596-011-0475-3. [DOI] [PubMed] [Google Scholar]
- Yokota T, Sugawara K, Ito K, Takahashi R, Ariga H, Mizusawa H. Down regulation of DJ-1 enhances cell death by oxidative stress, ER stress, and proteasome inhibition. Biochem Biophys Res Commun. 2003;312:1342–1348. doi: 10.1016/j.bbrc.2003.11.056. [DOI] [PubMed] [Google Scholar]
- Zhang SP, Du XG, Pu XP. 3-O-demethylswertipunicoside protects against oxidative toxicity in PC12 cells. Biol Pharm Bull. 2010;33:1529–1533. doi: 10.1248/bpb.33.1529. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ling MT, Wang Q, Lau CK, Leung SC, Lee TK, Cheung AL, Wong YC, Wang X. Identification of a novel inhibitor of differentiation-1 (ID-1) binding partner, caveolin-1, and its role in epithelial-mesenchymal transition and resistance to apoptosis in prostate cancer cells. J Biol Chem. 2007;282:33284–33294. doi: 10.1074/jbc.M705089200. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat Rev Drug Discov. 2009;8:806–823. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.