Abstract
In Escherichia coli, a signal recognition particle (SRP) has been identified which binds specifically to the signal sequence of presecretory proteins and which appears to be essential for efficient translocation of a subset of proteins. In this study we have investigated the function of E. coli FtsY which shares sequence similarity with the alpha-subunit of the eukaryotic SRP receptor ('docking protein') in the membrane of the endoplasmic reticulum. A strain was constructed which allows the conditional expression of FtsY. Depletion of FtsY is shown to cause the accumulation of the precursor form of beta-lactamase, OmpF and ribose binding protein in vivo, whereas the processing of various other presecretory proteins is unaffected. Furthermore, FtsY-depleted inverted cytoplasmic membrane vesicles are shown to be defective in the translocation of pre-beta-lactamase using an in vitro import assay. Subcellular localization studies revealed that FtsY is located in part at the cytoplasmic membrane with which it seems peripherally associated. These observations suggest that FtsY is the functional E. coli homolog of the mammalian SRP receptor.
Full text
PDF


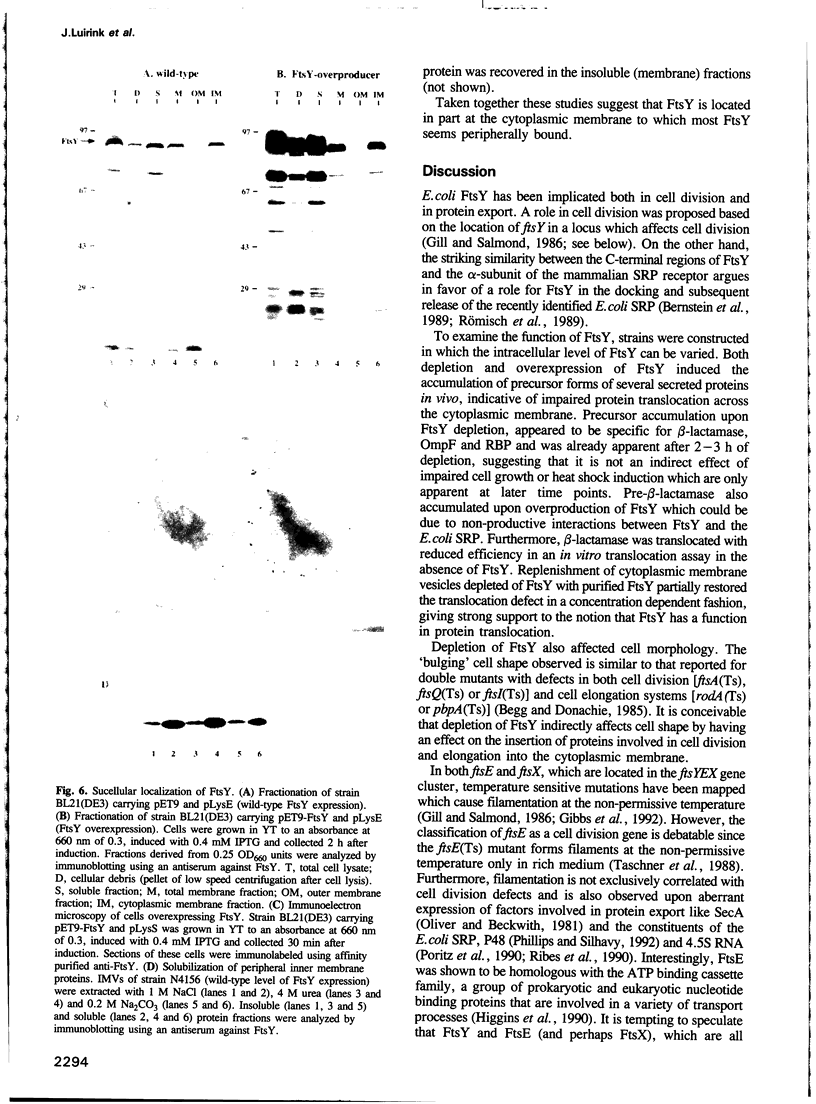


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Begg K. J., Donachie W. D. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J Bacteriol. 1985 Aug;163(2):615–622. doi: 10.1128/jb.163.2.615-622.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein H. D., Poritz M. A., Strub K., Hoben P. J., Brenner S., Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989 Aug 10;340(6233):482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- Bernstein H. D., Zopf D., Freymann D. M., Walter P. Functional substitution of the signal recognition particle 54-kDa subunit by its Escherichia coli homolog. Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):5229–5233. doi: 10.1073/pnas.90.11.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch D., Voorhout W., Tommassen J. Export and localization of N-terminally truncated derivatives of Escherichia coli K-12 outer membrane protein PhoE. J Biol Chem. 1988 Jul 15;263(20):9952–9957. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Breukink E., Demel R. A., de Korte-Kool G., de Kruijff B. SecA insertion into phospholipids is stimulated by negatively charged lipids and inhibited by ATP: a monolayer study. Biochemistry. 1992 Feb 4;31(4):1119–1124. doi: 10.1021/bi00119a021. [DOI] [PubMed] [Google Scholar]
- Cabelli R. J., Dolan K. M., Qian L. P., Oliver D. B. Characterization of membrane-associated and soluble states of SecA protein from wild-type and SecA51(TS) mutant strains of Escherichia coli. J Biol Chem. 1991 Dec 25;266(36):24420–24427. [PubMed] [Google Scholar]
- Cagnon C., Valverde V., Masson J. M. A new family of sugar-inducible expression vectors for Escherichia coli. Protein Eng. 1991 Oct;4(7):843–847. doi: 10.1093/protein/4.7.843. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I., Shales S. W. Comparison of the polypeptide composition of Escherichia coli outer membranes prepared by two methods. J Bacteriol. 1980 Oct;144(1):425–427. doi: 10.1128/jb.144.1.425-427.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vrije T., Tommassen J., De Kruijff B. Optimal posttranslational translocation of the precursor of PhoE protein across Escherichia coli membrane vesicles requires both ATP and the protonmotive force. Biochim Biophys Acta. 1987 Jun 12;900(1):63–72. doi: 10.1016/0005-2736(87)90278-1. [DOI] [PubMed] [Google Scholar]
- Ferreira L. C., Keck W., Betzner A., Schwarz U. In vivo cell division gene product interactions in Escherichia coli K-12. J Bacteriol. 1987 Dec;169(12):5776–5781. doi: 10.1128/jb.169.12.5776-5781.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Itoh T., Tomizawa J. I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
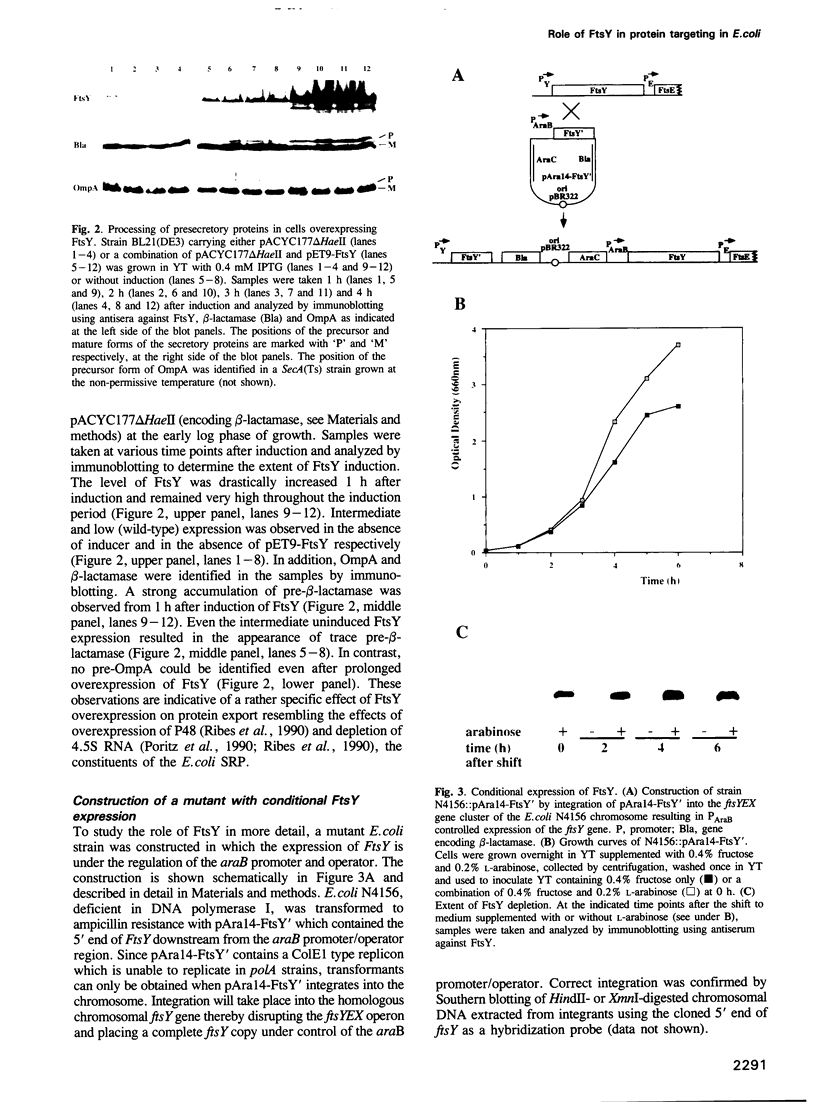
- Gibbs T. W., Gill D. R., Salmond G. P. Localised mutagenesis of the fts YEX operon: conditionally lethal missense substitutions in the FtsE cell division protein of Escherichia coli are similar to those found in the cystic fibrosis transmembrane conductance regulator protein (CFTR) of human patients. Mol Gen Genet. 1992 Jul;234(1):121–128. doi: 10.1007/BF00272353. [DOI] [PubMed] [Google Scholar]
- Gill D. R., Hatfull G. F., Salmond G. P. A new cell division operon in Escherichia coli. Mol Gen Genet. 1986 Oct;205(1):134–145. doi: 10.1007/BF02428043. [DOI] [PubMed] [Google Scholar]
- Gill D. R., Salmond G. P. The Escherichia coli cell division proteins FtsY, FtsE and FtsX are inner membrane-associated. Mol Gen Genet. 1987 Dec;210(3):504–508. doi: 10.1007/BF00327204. [DOI] [PubMed] [Google Scholar]
- Gill D. R., Salmond G. P. The identification of the Escherichia coli ftsY gene product: an unusual protein. Mol Microbiol. 1990 Apr;4(4):575–583. doi: 10.1111/j.1365-2958.1990.tb00626.x. [DOI] [PubMed] [Google Scholar]
- Hann B. C., Walter P. The signal recognition particle in S. cerevisiae. Cell. 1991 Oct 4;67(1):131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- Hartl F. U., Wiedmann M. Prokaryotic secretion: a signal recognition particle in Escherichia coli? Curr Biol. 1993 Feb;3(2):86–89. doi: 10.1016/0960-9822(93)90161-g. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Hyde S. C., Mimmack M. M., Gileadi U., Gill D. R., Gallagher M. P. Binding protein-dependent transport systems. J Bioenerg Biomembr. 1990 Aug;22(4):571–592. doi: 10.1007/BF00762962. [DOI] [PubMed] [Google Scholar]
- High S., Stirling C. J. Protein translocation across membranes: common themes in divergent organisms. Trends Cell Biol. 1993 Oct;3(10):335–339. doi: 10.1016/0962-8924(93)90103-8. [DOI] [PubMed] [Google Scholar]
- Honda K., Nakamura K., Nishiguchi M., Yamane K. Cloning and characterization of a Bacillus subtilis gene encoding a homolog of the 54-kilodalton subunit of mammalian signal recognition particle and Escherichia coli Ffh. J Bacteriol. 1993 Aug;175(15):4885–4894. doi: 10.1128/jb.175.15.4885-4894.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto C. A., Francetić O. Highly selective binding of nascent polypeptides by an Escherichia coli chaperone protein in vivo. J Bacteriol. 1993 Apr;175(8):2184–2188. doi: 10.1128/jb.175.8.2184-2188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusters R., de Vrije T., Breukink E., de Kruijff B. SecB protein stabilizes a translocation-competent state of purified prePhoE protein. J Biol Chem. 1989 Dec 15;264(35):20827–20830. [PubMed] [Google Scholar]
- Laminet A. A., Plückthun A. The precursor of beta-lactamase: purification, properties and folding kinetics. EMBO J. 1989 May;8(5):1469–1477. doi: 10.1002/j.1460-2075.1989.tb03530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B., Meijers J., Peters R., van der Hoek P., van Alphen L. Electrophoretic resolution of the "major outer membrane protein" of Escherichia coli K12 into four bands. FEBS Lett. 1975 Oct 15;58(1):254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- Luirink J., Dobberstein B. Mammalian and Escherichia coli signal recognition particles. Mol Microbiol. 1994 Jan;11(1):9–13. doi: 10.1111/j.1365-2958.1994.tb00284.x. [DOI] [PubMed] [Google Scholar]
- Luirink J., High S., Wood H., Giner A., Tollervey D., Dobberstein B. Signal-sequence recognition by an Escherichia coli ribonucleoprotein complex. Nature. 1992 Oct 22;359(6397):741–743. doi: 10.1038/359741a0. [DOI] [PubMed] [Google Scholar]
- Minsky A., Summers R. G., Knowles J. R. Secretion of beta-lactamase into the periplasm of Escherichia coli: evidence for a distinct release step associated with a conformational change. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4180–4184. doi: 10.1073/pnas.83.12.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder E., Woldringh C. L. Actively replicating nucleoids influence positioning of division sites in Escherichia coli filaments forming cells lacking DNA. J Bacteriol. 1989 Aug;171(8):4303–4314. doi: 10.1128/jb.171.8.4303-4314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S. C., Poritz M. A., Walter P. Signal recognition particle receptor is important for cell growth and protein secretion in Saccharomyces cerevisiae. Mol Biol Cell. 1992 Aug;3(8):895–911. doi: 10.1091/mbc.3.8.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D. B., Beckwith J. E. coli mutant pleiotropically defective in the export of secreted proteins. Cell. 1981 Sep;25(3):765–772. doi: 10.1016/0092-8674(81)90184-7. [DOI] [PubMed] [Google Scholar]
- Phillips G. J., Silhavy T. J. The E. coli ffh gene is necessary for viability and efficient protein export. Nature. 1992 Oct 22;359(6397):744–746. doi: 10.1038/359744a0. [DOI] [PubMed] [Google Scholar]
- Poritz M. A., Bernstein H. D., Strub K., Zopf D., Wilhelm H., Walter P. An E. coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science. 1990 Nov 23;250(4984):1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- Pugsley A. P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993 Mar;57(1):50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport T. A. Transport of proteins across the endoplasmic reticulum membrane. Science. 1992 Nov 6;258(5084):931–936. doi: 10.1126/science.1332192. [DOI] [PubMed] [Google Scholar]
- Ribes V., Römisch K., Giner A., Dobberstein B., Tollervey D. E. coli 4.5S RNA is part of a ribonucleoprotein particle that has properties related to signal recognition particle. Cell. 1990 Nov 2;63(3):591–600. doi: 10.1016/0092-8674(90)90454-m. [DOI] [PubMed] [Google Scholar]
- Römisch K., Webb J., Herz J., Prehn S., Frank R., Vingron M., Dobberstein B. Homology of 54K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature. 1989 Aug 10;340(6233):478–482. doi: 10.1038/340478a0. [DOI] [PubMed] [Google Scholar]
- Schweizer M., Hindennach I., Garten W., Henning U. Major proteins of the Escherichia coli outer cell envelope membrane. Interaction of protein II with lipopolysaccharide. Eur J Biochem. 1978 Jan 2;82(1):211–217. doi: 10.1111/j.1432-1033.1978.tb12013.x. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Taschner P. E., Huls P. G., Pas E., Woldringh C. L. Division behavior and shape changes in isogenic ftsZ, ftsQ, ftsA, pbpB, and ftsE cell division mutants of Escherichia coli during temperature shift experiments. J Bacteriol. 1988 Apr;170(4):1533–1540. doi: 10.1128/jb.170.4.1533-1540.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu T., Yuki T., Morimura S., Mori H., Yamanaka K., Niki H., Hiraga S., Ogura T. The Escherichia coli FtsH protein is a prokaryotic member of a protein family of putative ATPases involved in membrane functions, cell cycle control, and gene expression. J Bacteriol. 1993 Mar;175(5):1344–1351. doi: 10.1128/jb.175.5.1344-1351.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Putten A. J., Stegehuis F., van Bergen en Henegouwen P. M., De Graaf F. K., Oudega B. Alterations in the carboxy-terminal half of cloacin destabilize the protein and prevent its export by Escherichia coli. Mol Microbiol. 1988 Sep;2(5):553–562. doi: 10.1111/j.1365-2958.1988.tb00063.x. [DOI] [PubMed] [Google Scholar]