Abstract
Ciliates are a diverse assemblage of eukaryotes that have been the source of many discoveries including self-splicing RNAs, telomeres and trans-splicing. While analyses of ciliate morphology have given rise to robust hypotheses on relatively shallow level relationships, the deeper evolutionary history of ciliates is largely unknown. This is in part because studies to date have focused on only a single locus, small subunit ribosomal DNA (SSU-rDNA). In the present study, we use a taxon-rich strategy based on multiple loci from GenBank and recently completed transcriptomes to assess deep phylogenetic relationships among ciliates. Our phylogenomic data set includes up to 537 taxa, all of which have been sampled for SSU-rDNA and a subset of which have LSU-rDNA and up to 7 protein-coding sequences. Analyses of these data support the bifurcation of ciliates as suggested by SSU-rDNA, with one major clade defined by having somatic macronuclei that divide with intranuclear microtubules (Intramacronucleata) and the other clade containing lineages that either divide their macronuclei with microtubules external to the macronucleus or are unable to divide their macronuclei (Postciliodesmatophora). These multigene phylogenies provide a robust framework for interpreting the evolution of innovations across the ciliate tree of life.
Keywords: Phylogenomic analysis, macronucleus, Ciliophora, Postciliodesmatophora, Intramacronucleata
1. Introduction
Ciliates are characterized by the presence of cilia in at least one of their life stages and by their nuclear dimorphism (i.e. the presence of a somatic macronucleus and a germline micronucleus in each cell). Macronuclei derive from zygotic nuclei following conjugation through chromosomal rearrangements, that include fragmentation, elimination of internal sequences and amplification of the processed chromosomes; in all but one class of ciliates (Karyorelictea, see below) the resulting macronuclei divide by amitosis during asexual division (Prescott, 1994). Given their age of approximately one billion years (Parfrey et al., 2011), estimation of deep relationships within this clade are difficult.
The class Karyorelictea Corliss, 1974 had been argued to be sister to all other ciliates based on the relatively simple morphologies and the presence of nearly-diploid, non-dividing macronuclei within this clade (Raikov, 2006). Subsequent phylogenetic analyses of ciliates based only on SSU-rDNA divides ciliates into two clades, Intramacronucleata Lynn, 1996 and Postciliodesmatophora Gerassimova & Seravin,1976 (Lynn, 1996). The Intramacronucleata includes the bulk of the ciliate classes such as Oligohymenophorea (e.g. Paramecium and Tetrahymena), and are united by the feature of division of the macronucleus involving intramacronuclear microtubules (Hirt et al., 1995; Lukashenko, 2009; Lynn, 1996). In contrast, ciliates in Postciliodesmatophora either have macronuclei that cannot divide (i.e. Karyorelictea) or macronuclei that divide with microtubules external to the macronucleus (i.e. Heterotrichea).
Given the limitation of single gene trees, we use a taxon-rich strategy based on multiple loci to assess the relationships within ciliates. We expand the taxonomic sampling of SSU-rDNA to 537 species representing all major ciliate lineages and combine these data with large subunit-rDNA and up to seven protein genes from a subset of taxa. We analyzed the full data matrix as well as six submatrices to assess the impact of taxon sampling and missing data.
2. Methods
2.1. Dataset assembly
We collected small subunit ribosomal DNA (SSU-rDNA) and large subunit ribosomal DNA (LSU-rDNA) sequences for all the ciliates from GenBank using a custom Python script. The SSU-rDNA of Philasterides armatalis (FJ848877) and LSU-rDNA of Stylonychia lemnae (AF508773) were used as queries to in a Blast analysis against the GenBank nr database and one sequence ≥ 1000bp per taxon ID was kept. The taxon IDs of the ciliates from GenBank were downloaded in February 2012. Environmental and uncultured sequences were removed. As our preliminary analyses showed that the sequences of Mesodidium and Myrionecta formed a long, unstable branch as discussed elsewhere (Strüder-Kypke et al., 2006), we excluded these taxa in our final analyses. The sequences for at most two species per genus and for all the species that have available protein sequences used in the analyses (see below) were kept, resulting in 537 and 111 sequences for SSU-rDNA and LSU-rDNA, respectively. Sequences were aligned in GUIDANCE (Penn et al., 2010b) and ambiguous columns in the alignment were removed with default parameters using GUIDANCE web server (Penn et al., 2010a).
Assembly of the protein-coding gene dataset relied on a custom built pipeline that uses Python scripts to collect homologs from one of three sources: directly downloaded from GenBank, translated from EST data, or translated from transcriptome data. First, in January 2012, we downloaded all 1935 amino acid sequences from Ciliophora, excluding those from Paramecium, Tetrahymena, and Ichthyophthirius as these taxa have complete genome data. We then used Proteinortho4 (Lechner et al., 2011) to bin proteins into orthologous groups. We chose the seven proteins that had sequences available from the largest number of species (i.e. Actin, α-tubulin, β-tubulin, cytochrome oxidase subunit 1, elongation factor 1α, eukaryotic release factor 1, and histone 4). A representative of each protein was used as a query in BLASTP analysis against two Paramecium species (P. caudatum and P. tetraurelia), two Tetrahymena species (T. pyriformis andT. thermophila) and Ichthyophthirius multifiliis to capture proteins from these lineages with completed genomes. We then retrieved EST and transcriptome data (Table S1) and used Python scripts to identify homologs of the seven proteins chosen from GenBank. For each protein, we used BLASTX to compare the EST or transcriptome data to a fasta file for each of the seven proteins, with an e-value limit of 1e-15. Given difficulties in determining alleles and paralogs from non-overlapping EST/transcriptome data, we retained the longest sequence for each taxon. In order to reduce missing data, some proteins from a few key congeners were combined to represent a single taxon.
We combined inferred amino acid sequences for each protein-coding gene. These sequences were aligned using the GUIDANCE web server with default parameters and individual gene trees were examined to choose appropriate orthologs for concatenations. For example, in cases where paralogs formed a monophyletic group, the shortest branched sequence was retained. When paralogs fell into multiple locations on the tree, we aimed to maintain orthologous groups that included the greatest taxonomic representation. The elongation factor 1α of Paranophrys carnivora (AAD03258) and the cytochrome oxidase subunit 1 of Halteria grandinella (ACP43519) were excluded as they cluster within other classes, indicating the possibility of contamination or misidentification. A total of 53 actin sequences,157 α-tubulin sequences, 35 β-tubulin sequences, 35 cytochrome oxidase subunit 1 sequences, 31 elongation factor 1α sequences, 27 eukaryotic release factor 1 sequences, and 41 histone 4 sequences were used in the final analyses.
2.2. Creation of Data Matrices
Our full data matrix consisted of 9 genes (7 protein-coding genes plus SSU-rDNA and LSU-rDNA) and 537 taxa (denoted all: 9 in results/discussion). In order to assess the impact of taxon sampling and missing data, we created six data matrices by subsampling our full data matrix (Table 1). We analyzed a matrix of just SSU-rDNA sequences given that this is the locus with the broadest taxonomic sampling. The most inclusive of these matrices contained 9 genes and all taxa that had at least 2 proteins of the 9 genes (2P: 9). Similarly, the 3P: 9 matrix included all taxa with at least 3 proteins of the targeted 9 genes. To address the concern that rDNA was driving our results, we deleted it from each of the 9-gene data sets resulting in all: 7, 2P: 7, and 3P: 7 matrices. concern that rDNA was driving our results, we deleted it from each of the 9-gene data sets resulting in all: 7, 2P: 7, and 3P: 7 matrices.
Table 1.
The division of ciliates into Intramacronucleata and Postciliodesmatophora is supported by all but one analyses using subsamples of full data set.
| All: 9 | All: 7 | 2P: 9 | 2P: 7 | 3P: 9 | 3P: 7 | SSU-rDNA | |
|---|---|---|---|---|---|---|---|
| Intramacronucleata | 89 | nm | 84 | 49 | 85 | 64 | 17 |
| Armophorea | 75 | 76 | 100 | 99 | 100 | 100 | 73 |
| Litostomatea | 100 | 43 | 100 | 100 | 100 | 100 | 100 |
| Spirotrichea | nm | nm | 100 | 88 | 100 | 100 | nm |
| Colpodea | 43 | nm | -- | -- | -- | -- | nm |
| Oligohymenophorea | 98 | nm | 93 | nm | 93 | nm | 88 |
| Nassophorea | nm | -- | -- | -- | -- | -- | nm |
| Phyllopharyngea | 100 | 57 | -- | -- | -- | -- | 100 |
| Plagiopylea | 87 | nm | -- | -- | -- | -- | 54 |
| Prostomatea | nm | nm | -- | -- | -- | -- | nm |
| Postciliodesmatophora | 100 | nm | 100 | 91 | 100 | 89 | 92 |
| Heterotrichea | 100 | 34 | 100 | 100 | 100 | 98 | 65 |
| Karyorelictea | 100 | nm | -- | -- | -- | -- | 100 |
Note: nm = non-monophyletic; All: 9 = our full data matrix consisted of 9 genes (7 protein-coding genes plus SSU-rDNA and LSU-rDNA) and 537 taxa; all: 7 = data matrix consisted of 7 protein-coding genes and 537 taxa; 2P: 9 = data matrix consisted of the 9 genes and the taxa that had at least 2 proteins; 2P: 7 = data matrix consisted of 7 protein-coding genes and the taxa that had at least 2 proteins; 3P: 9 = data matrix consisted of the 9 genes and the taxa that had at least 3 proteins; 3P: 7 = data matrix consisted of 7 protein-coding genes and the taxa that had at least 3 proteins; SSU-rDNA = small subunit ribosomal DNA;
2.3. Phylogenetic Analyses
Four stramenopiles, five apicomplexans and four dinoflagellates were used as outgroups (Adl et al. 2012). Genealogies for this study were constructed using RaxML-HPC2 v7.2.8 (Stamatakis, 2006; Stamatakis et al., 2008) on CIPRES Science Gateway (Miller et al., 2010). The SSU-rDNA and LSU-rDNA partition was analyzed with GTR + gamma as this was the best fitting model available in RAxML. ProtTest3 (Darriba et al., 2011) was used to select the appropriate model of sequence evolution for the amino acid data for each protein. The mtART amino acid replacement matrix was the best for cytochrome oxidase subunit 1 and the LG amino acid replacement matrix was found to be the best-fitting model for the other proteins. 1000 rapid bootstrap replicates followed by a full maximum-likelihood search were used for all analyses except all: 9 and all: 7, for which 100 bootstrap replicates were run.
3. Results and discussion
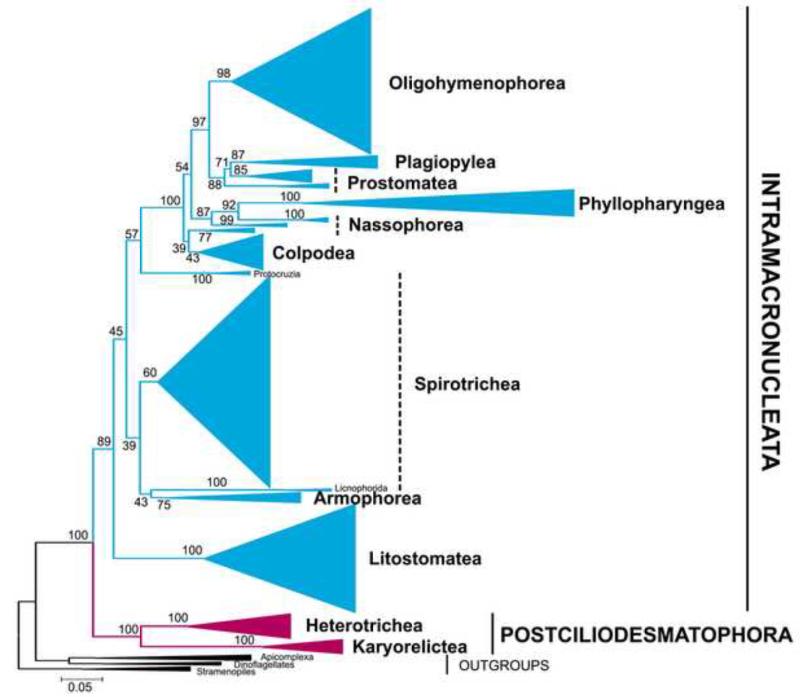
Overall, our phylogenomic analyses support the bifurcation of ciliates into two major clades defined by differences in division of the somatic macronuclei: the Intramacronucleata and Postciliodesmatophora. Both clades are recovered in the tree reconstructed in our largest dataset, including 537 ciliates and all 9 genes (i.e. 2 rDNAs + 7 protein-coding; denoted all: 9), with high support (89% BS and 100% BS respectively, Figure 1). To test whether the topology is dependent on the nearly ubiquitous SSU-rDNA gene or the linked LSU-rDNA gene, we analyzed just protein genes from fewer taxa and also recovered the same topology though with lower levels of support (Figure 2, Table 1). This topology leads to two equally parsimonious mappings on the origin of nuclear division in ciliates: 1) macronuclei capable of division are ancestral and this feature was lost in the Karyorelictea, or 2) the ability of somatic macronuclei to divide arose twice in ciliates (see also Hammerschmidt et al., 1996; Katz, 2001; Orias, 1991).
Figure 1.
Maximum likelihood (ML) tree reconstructed using 537 ciliates and all 9 genes (all: 9; SSU-rDNA, LSU-rDNA plus 7 protein genes) yields a well-resolved bifurcation of ciliates phylogenies, Intramacronucleata (in maroon) and Postciliodesmatophora (in blue). Numbers at the nodes represent ML support values. The scale bar corresponds to 0.05 expected substitutions per site.
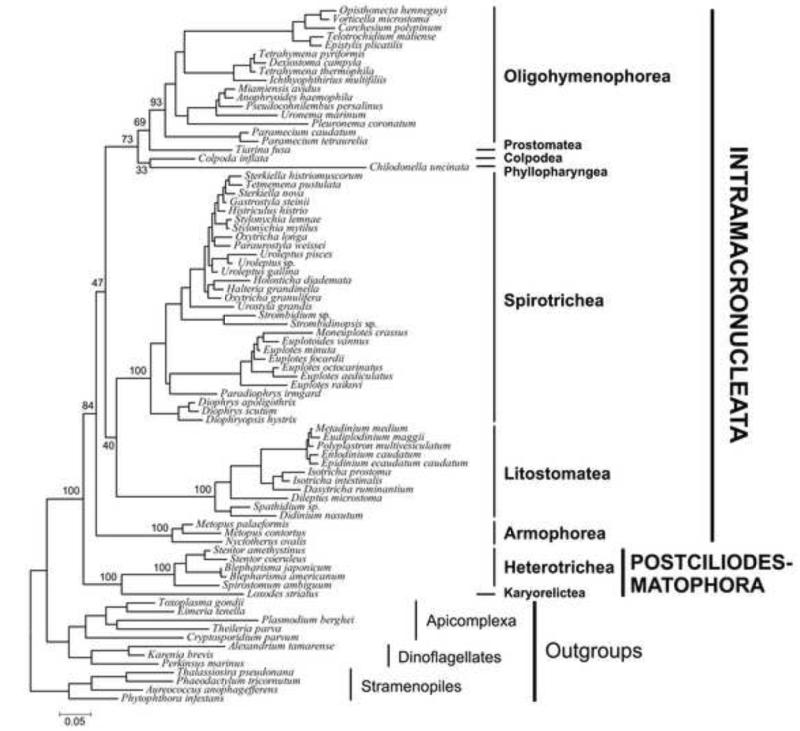
Figure 2.
Maximum likelihood (ML) tree reconstructed using 68 ciliates that had at least 2 proteins of the 9 genes (2P:7; SSU-rDNA, LSU-rDNA plus 7 protein genes; see Table 1). Numbers at the main nodes represent ML support values. The scale bar corresponds to 0.05 expected substitutions per site.
As further evidence of the robustness of our approach, many major clades predicted by SSU-rDNA trees are consistently recovered across our analyses with moderate to high support despite the relatively sparse sampling of protein-coding genes within most classes. The classes Armophorea, Litostomatea and Heterotrichea are recovered consistently (Figures 1, 2; Table 1). Other groups are not monophyletic including Nassophorea, Prostomatea and Spirotrichea, which may reflect the impact of limited taxon sampling and/or missing data. For example, the genus Protocruzia, which has unusual paradiploid and dividing macronucleus with mitotic-like division (Ruthmann and Hauser, 1974), falls separate from the rest of the Spirotrichea. As additional protein-coding sequences are made available from diverse ciliates, it will be possible to disentangle artifacts from areas requiring taxonomic revision.
The deep division among ciliates between Intramacronucleata and Postciliodesmatophora is supported in nearly all subsampled data sets (Table 1). The phylogenetic tree based on the concatenate data (i.e. 2 rDNAs + 7 protein-coding; denoted all: 9, Figure 1) shows the highest node support for the monophyly of Intramacronucleata (89% BS) and full support for Postciliodesmatophora (Table 1). These support values are higher than other analyses include for the well-sampled SSU-rDNA sequences (Table 1). We explored the impact of changing levels of missing data and found that, phylogenies of taxa that have at least two or three proteins also give a good resolution (2P: 9 and 3P: 9, Table 1). Only the phylogeny based on solely the concatenate protein sequences (all: 7, Table 1) did not show deep division between Intramacronucleata and Postciliodesmatophora, which we suspect is due to the 90% of missing data in this analysis.
Supplementary Material
Highlights.
A taxon-rich phylogenomic strategy was used to assess the evolutionary relationships within ciliates.
537 taxa combined with a moderate number of proteins were accessed from GenBank and recently released transcriptome data.
Full data matrix as well as six submatrices were analyzed to assess the impact of taxon sampling and missing data.
The multigene phylogenies support the bifurcation of ciliates into two major clades as suggested by SSU-rDNA.
Acknowledgements
This work is supported by the AREA award from the National Institutes of Health (1R15GM081865-01) to L.A.K. We would like to thank Ms. Jessica R. Grant 176 for technical help and data analyses. We also thank Dr. George B. McManus for sharing the transcriptome data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary material
Table S1: List of the EST and Transcriptome data used in the present analyses.
References
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt B, Schlegel M, Lynn D, Leipe DD, Sogin ML, Raikov IB. Insights into the evolution of nuclear dualism in the ciliates revealed by phylogenetic analysis of rRNA sequences. J. Eukaryot. Microbiol. 1996;43:225–230. doi: 10.1111/j.1550-7408.1996.tb01396.x. [DOI] [PubMed] [Google Scholar]
- Hirt RP, Dyal PL, Wilkinson M, Finlay BJ, Roberts DM, Embley TM. Phylogenetic relationships among karyorelictids and heterotrichs inferred from small subunit rRNA sequences: resolution at the base of the ciliate tree. Mol. Phylogenet. Evol. 1995;4:77–87. doi: 10.1006/mpev.1995.1008. [DOI] [PubMed] [Google Scholar]
- Katz LA. Evolution of nuclear dualism in ciliates: a reanalysis in light of recent molecular data. Int J Syst Evol Microbiol. 2001;51:1587–1592. doi: 10.1099/00207713-51-4-1587. [DOI] [PubMed] [Google Scholar]
- Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC bioinformatics. 2011;124;12 doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashenko NP. Molecular evolution of ciliates (Ciliophora) and some related groups of protozoans. Genetika. 2009;45:1013–1028. [PubMed] [Google Scholar]
- Lynn D. My journey in ciliate systematics. J. Eukaryot. Microbiol. 1996;43:253–260. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE); New Orleans, LA. 2010. pp. 1–8. [Google Scholar]
- Orias E. Evolution of amitosis of the ciliate macronucleus: gain of the capacity to divide. J. Protozool. 1991;38:217–221. doi: 10.1111/j.1550-7408.1991.tb04431.x. [DOI] [PubMed] [Google Scholar]
- Parfrey LW, Lahr DJ, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc. Natl. Acad. Sci. USA. 2011;108:13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn O, Privman E, Ashkenazy H, Landan G, Graur D, Pupko T. GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Res. 2010a;38:W23–28. doi: 10.1093/nar/gkq443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn O, Privman E, Landan G, Graur D, Pupko T. An alignment confidence score capturing robustness to guide tree uncertainty. Mol. Biol. Evol. 2010b;27:1759–1767. doi: 10.1093/molbev/msq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM. The DNA of ciliated protozoa. Microbiol. Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raikov IB. Nuclei of ciliates. In: Hausmann K, Bradbury PC, editors. Ciliates: cells as organisms. Gustav Fischer; Stuttgart: 2006. pp. 221–242. [Google Scholar]
- Ruthmann A, Hauser M. Mitosis-like macronuclear division in a ciliate. Chromosoma. 1974;45:261–272. doi: 10.1007/BF00283410. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Strüder-Kypke MC, Wright AD, Foissner W, Chatzinotas A, Lynn DH. Molecular phylogeny of litostome ciliates (Ciliophora, Litostomatea) with emphasis on free-living haptorian genera. Protist. 2006;157:261–278. doi: 10.1016/j.protis.2006.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.