Abstract
Infrared Matrix-Assisted Laser Desorption Electrospray Ionization (IR-MALDESI) Mass Spectrometry imaging of biological tissue sections using a layer of deposited ice as an energy absorbing matrix was investigated. Dynamics of plume ablation were first explored using a nanosecond exposure shadowgraphy system designed to simultaneously collect pictures of the plume with a camera and collect the FT-ICR mass spectrum corresponding to that same ablation event. Ablation of fresh tissue analyzed with and without using ice as a matrix were both compared using this technique. Effect of spot-to-spot distance, number of laser shots per pixel and tissue condition (matrix) on ion abundance was also investigated for 50 µm thick tissue sections. Finally, the statistical method called design of experiments was used to compare source parameters and determine the optimal conditions for IR-MALDESI of tissue sections using deposited ice as a matrix. With a better understanding of the fundamentals of ablation dynamics and a systematic approach to explore the experimental space, it was possible to improve ion abundance by nearly one order of magnitude.
Keywords: Mass spectrometry imaging, IR-MALDESI, tissue ablation, shadowgraphy
INTRODUCTION
Matrix assisted laser desorption/ionization (MALDI) [1, 2] and electrospray ionization (ESI)[3] have coexisted for many years as the two main ionization techniques for MS analysis of biological molecules. The idea of using ESI as a method to post ionize neutrals desorbed by laser ablation was first introduced by Shiea & Huang in 2005 when they proposed electrospray-assisted laser desorption ionization (ELDI)[4]. With ELDI, a UV laser pulse was used to non-resonantly excite and desorb dried out protein molecules from a sample plate at atmospheric pressure and post ionize the neutral molecules using ESI before being measured in the mass spectrometer. They successfully produced multiply-charged spectra of cytochrome c and protein mixtures[5] and were followed by other groups proposing similar non-resonant desorption processes [6–8].
Rather than exciting the analyte directly, Muddiman and co-workers proposed in 2006 to mix the analyte with an energy absorbing matrix to take advantage of the softness and efficiency of the MALDI resonant desorption process knowing the ion/neutral fraction was very small [9, 10]. It was also shown that charge state distribution was nearly identical for ESI and MALDESI spectra, suggesting that desorbed neutrals are ionized in an ESI-like mechanism.
In 2007, two groups demonstrated that ablation using an IR laser in combination with ESI and both proposed new acronyms for their source. Both used a wavelength of 2.94 µm, so that water, naturally occurring in biological samples, is excited at its O–H symmetric and asymmetric stretching bands. Vertes and co-workers called their method laser ablation electrospray ionization (LAESI)[11] while Murray and co-workers introduced infrared laser-assisted desorption electrospray ionization (IR LADESI)[12]. Since both methods use endogenous water as a matrix to resonantly absorb laser energy they are fundamentally equivalent to MALDESI MS; which we denote IR-MALDESI [13–17]. MALDESI combines advantages of both ESI and MALDI, including the production of multiply charged ions, high salt tolerance, ambient nature and amenability to imaging.
Mass spectrometry imaging of biological tissue sections by laser ablation technique has been introduced to the community over 15 years ago [18, 19], has since then matured and been the subject of several review articles [20–22]. Because of the significantly higher ionization yield, most work has been done using UV-MALDI, even though IR-MALDI using endogenous or exogenous water or ice as an energy absorbing matrix has also been widely explored [23–31] and its desorption/ionization processes thoroughly investigated [32–34]. Since IR-MALDESI is an ambient ionization technique that allows the direct analysis of biological samples at ambient pressure with minimal or no sample preparation, it has great potential for MS imaging of biological tissue sections[16, 17, 35–37]. In IR-MALDESI, ablation dynamics plays a critical role since the plume of material has to be ejected from the surface and reach the electrospray plume to be ionized. IR desorption/ablation can be divided into multiple intertwined stages [38, 39]. First, as the surface is excited by the laser, surface evaporation occurs. Then, upon being excited above a certain irradiance threshold, super heating and phase explosion of subsurface material occurs and more material is ejected. Those first stages occur on the nanosecond scale and are often referred to as primary material ejection. Secondary material ejection occurs at the microsecond scale and it is where most of the ablated material is ejected from the surface due to induced recoil pressure. Because of greater abundance and larger distance travelled by material ejected during secondary material ejection, it is generally accepted that those neutral molecules are those most likely to interact with the ESI plume[11].
Even though it has been demonstrated that a thin layer of deposited ice water could be used as an energy absorbing matrix for IR-MALDESI[16] of tissue sections, very little is known about what effect this layer of ice has on the ablation dynamics and how it eventually translates to greater MS signal. We are reporting here a series of experiments performed to better understand the effect of this layer of ice on the ablation process and optimize IR-MALDESI for Mass Spectrometry Imaging (MSI) of tissue sections. First, a novel flash shadowgraphy system [39–41] was implemented, which has the capability to simultaneously collect images of the plume of ablated material and record the IRMALDESI FT-ICR mass spectrum associated with that same plume. This setup was used in a series of experiments designed to better understand the effect of the ice matrix on the plume dynamics and the MS ion abundance. A second series of experiments was performed to investigate the effect of the spot-to-spot distance and the number of laser ablations per pixel on the signal ion abundance. Finally, the statistical method called design of experiments (DOE) [14, 42, 43] was used to systematically explore the experimental space and improve the sensitivity of the source by nearly an order of magnitude.
EXPERIMENTAL
Materials
HPLC grade acetonitrile, methanol and water were purchased from Burdick and Jackson (Musketon, MI, USA). Formic acid was purchased from Sigma Aldrich (St. Louis, MO, USA). Nitrogen gas (99.98 % purity) used for purging the enclosure was obtained from MWSC High Purity Gases (Raleigh, NC, USA). Chicken liver was obtained from NCSU Lake Wheeler Road field laboratory and stored at −80 °C until sectioning. Adult mouse brains were obtained from NCSU’s college of Veterinary Medicine. For all tissue samples, 50 µm tissue slices were prepared using a CM1950 Cryostat (Leica, Buffalo Grove, IL, USA) and thaw mounted on microscope slides for analysis. Optical images of tissue sections were collected using the camera mounted on a laser microdissection system (LMD 7000, Leica, Wetzlar, Germany) after complete sublimation of the ice matrix, which was achieved by leaving the sample at −10 °C in a purged, near-zero relative humidity enclosure.
Shadowgraphy
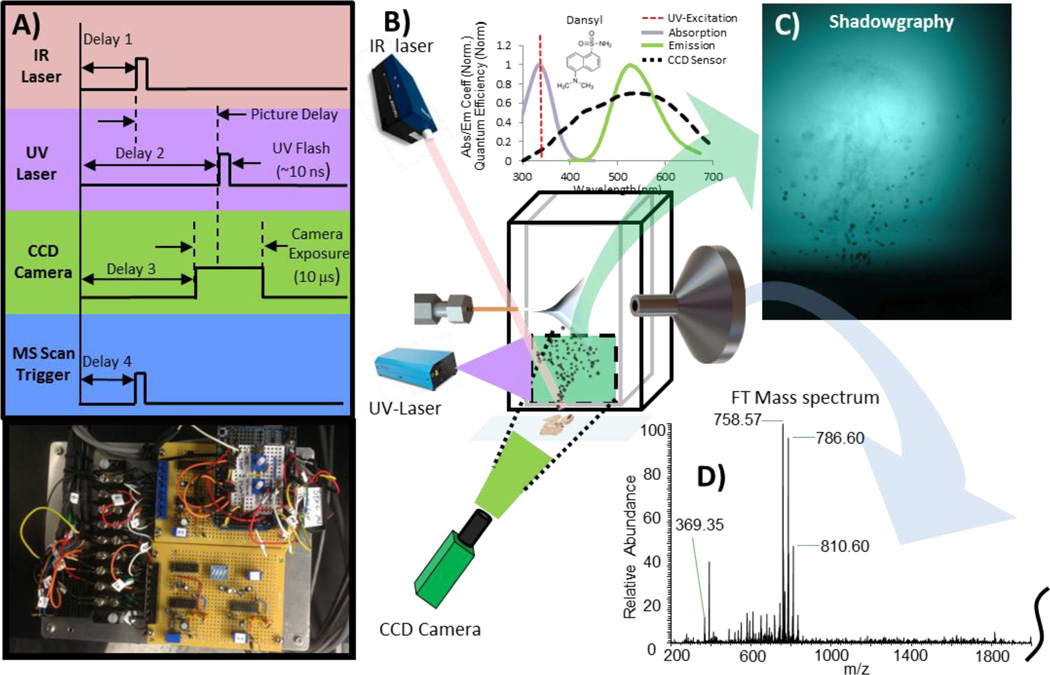
The plume visualization system uses the energy of a 5 ns pulse generated by an Explorer 349 Q-switched UV laser (Spectra Physics, Santa Clara, CA) to excite a saturated Dansyl amide solution (Sigma Aldrich, St-Louis, MO, USA) placed in a 10 mm wide quartz cuvette (VWR, Radnord, PA). The 120 µJ UV laser pulse excites the fluorophore near its maximum energy absorption peak (349 nm). The wavelength of the 10 ns fluorescent emission occurs near the sensitivity peak of the high performance QI-Click CCD camera (QImaging, Surrey, BC, Canada) and is used as background exposure light to capture phenomenon occurring at the nanosecond timescale. Mitutoyo plan apo infinity-corrected long working distance objective (2X and 10X) were alternatively used depending of the desired field of view (Mitutoyo Corp., Japan). The triggering sequence used to capture the images is presented in Figure 1. The IR laser flashlamp is continuously pulsing at 20 Hz and when the picture command is sent (push button), the next IR flashlamp output signal initiates the chain of events leading to the triggering of the IR ablation laser, UV laser pulse (exposure light), and the electronic exposure of the CCD camera sensor at the required time to capture the plume at any given time after the ablation event (Δt). This adjustable pulse delay generator was designed to generate the TTL signals and control the different delays to a microsecond resolution. It is controlled by an Arduino Uno MCU prototyping platform (Ivrea, Italy) and delays were measured using a 100 MHz Tektronix THS720A oscilloscope (Beaverton, OR, USA). An additional signal is also sent to the 7 T Thermo LTQ FT Ultra mass spectrometer (ThermoFisher Scientific, San Jose, CA, USA) so that ion injection is precisely synchronized with the ablation event, making it possible to simultaneously visualize the plume of ablated material and analyze its content by mass spectrometry. For experiments where multiple pulses per pixel are used, the triggering system can be easily adjusted to select which ablation event is to be captured by the photography system.
Figure 1.
Experimental setup of induced fluorescence (LIF) photography system synchronized with MS acquisition. A) Simplified triggering scheme used to precisely synchronize IR laser ablation, UV laser Flash, CCD camera exposure and MS scan trigger (Thermo LTQ-FT Ultra). B) Figure showing the interaction between different components for one shadowgraphy image/mass spectrum acquisition. C) Example of shadowgraphy image obtained from tissue. D) IR-MALDESI mass spectrum of the ablated material seen in shadowgraphy image.
Ice matrix application
For each experiment, sample was placed in the source enclosure on the Peltier cooled sample plate still at room temperature. Enclosure was then purged with nitrogen until relative humidity reached 0–3 % and only then the sample stage temperature was lowered to −10 °C. After allowing the sample temperature to stabilize (10–15 minutes), the enclosure door is opened to increase relative humidity and cause the deposition of the ice matrix on surface of tissue sample. Once the desired thickness of ice is reached, the enclosure is purged again and nitrogen flow adjusted to maintain a relative humidity of 10% ± 2%. Relative humidity inside the enclosure was monitored using a humidity and temperature datalogger (RHT20, Extech, Waltham, MA,USA). This ice matrix application procedure was used to prevent the condensation of liquid water on the sample surface and results in the formation of ice crystals. Indeed, if the tissue sample is cooled in a humid environment, water droplets will start to accumulate and eventually freeze over the sample rather than having small crystals of ice forming uniformly on the surface.
IR-MALDESI imaging source
The IR-MALDESI imaging source uses an OPO laser with a pulse width of 7 ns (IR Opolette, Opotek, Carlsbad, CA, USA). The wavelength of quasi-Gaussian laserbeam was set at 2.94 µm to match with the high absorption band of water (O-H stretch). Laser power was circa 0.9 mJ/pulse at the target location, measured with a laser powermeter (Nova 2, Ophir, Jerusalem, Israel). Spot size was 250–300 µm (slightly elliptical) and measured on thermal paper. Repetition rate was set at 20 Hz throughout all the experiments discussed in this article. A user interface built on the Matlab platform (Mathworks, Natick, MA) is used to select the region of interest, laser repetition rate, number of pulses per pixel, scanning pattern, and step size. When the imaging sequence is initiated, information is sent via serial communications to the stage controller and house built laser triggering mechanism. A precise handshaking communication is then initiated between laser triggering mechanism, stage controller, and mass spectrometer. One full mass spectrum is collected per pixel and internal delays are precisely adjusted so that ion injection in the instrument’s Linear Trap Quadrupole (LTQ) is synchronized with the laser ablation event. For example, if 3 pulses/pixel are used, scan triggering delay and injection time is adjusted so that ablated material resulting from those 3 ablation events are collected by the LTQ prior to be sent to the 7 T FT-ICR cell for analysis. Unless otherwise specified, mass spectra were collected with a resolving power of 100,000FWHM at m/z 400 and tube lens voltage was set at 100 V (m/z range: 200–2000). The IR-MALDESI source used for imaging experiments has been described in greater details previously [16]. Imaging data was analyzed using MSiReader [44], a free open source interface developed by our group to view and analyze mass spectrometry imaging data.
RESULTS AND DISCUSSION
Effect of ice on signal and tissue ablation
Time-lapse experiments were performed to confirm the beneficial effect of the ice layer thickness on the ion abundance. For those experiments, liver tissue was placed on the sampling stage at room temperature and the imaging experiment was initiated (3 shots/pixels and 200 µm step size). No purging enclosure was used for this proof of concept experiments. Every 2 minutes, an optical image of the sample was captured to monitor the presence of ice. The imaging region of interest was set so that all pixels were on tissue and liver was chosen since it contains several analyte molecules that are homogeneously distributed and detectable by IR-MALDESI MS. After 2 minutes, the Peltier cold stage temperature was lowered to −10 °C. As shown in Figure S1, as soon as ice started to form atop of the tissue section, a significant increase in abundance signal is also observed (> 3-fold). A procedure was implemented using a controlled atmosphere enclosure to control the ice layer formation to ensure that ice layer thickness remains constant for the duration of an imaging experiment and from one experiment to another, albeit actual thickness of layer of ice had little effect on overall signal intensity according to this data. This ice application procedure is described in great detail in the experimental section. The controlled atmosphere enclosure also greatly helped reducing the signal variation from experiment to experiment which was attributed to the fluctuation of the relative humidity in the laboratory.
Even though the favorable effect of ice as a matrix on the signal abundance could be clearly measured, very little is known about the actual effect of the layer of ice on the plume dynamics and the mechanisms leading to this higher abundance signal. A nanosecond exposure photography system using laser induced fluorescence was used to simultaneously visualize the ablation process and measure mass spectrometer signal abundance. Plume ablation dynamics and signal abundance were measured for consecutive ablation events on a 50 µm section for both fresh tissue sections analyzed at room temperature and tissue sections analyzed after adding a layer of ice as a matrix.
The entire series of time-resolved shadowgraphy images for ablation of fresh tissue sections at room temperature and with an ice matrix were collected. The first and fourth shot picture series are presented in supplementary material (Figure S2–S4) and a series of time-resolved shadowgraphy images of ablation on liquid droplet is also presented in Figure S5–S6 for comparison purposes.
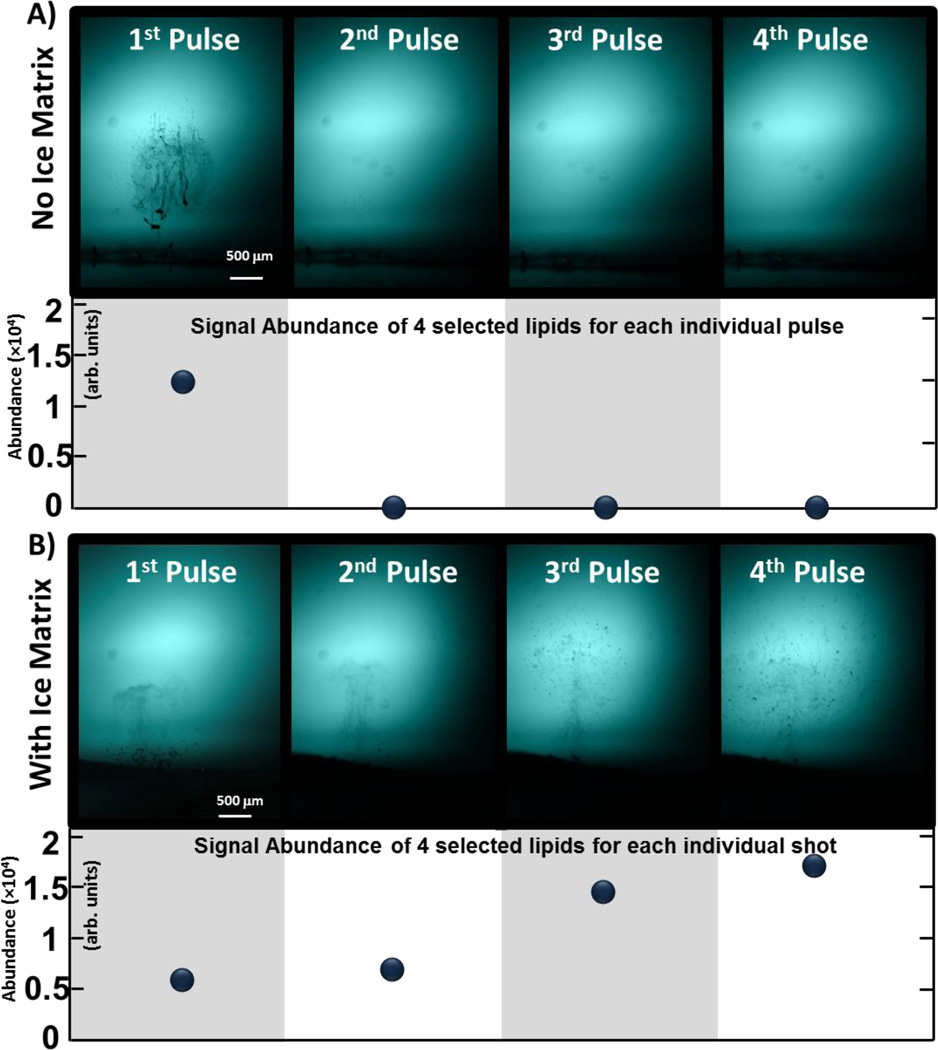
In the case of ablation of fresh tissue sections at room temperature, the energy density was sufficient to trigger plasma formation at the laser focus point in the first microsecond following laser irradiation. Primary material ejection observed was then in accordance to what was also reported by Vogel et al. [39, 45], i.e. the ejection of a relatively dense plume of small tissue fragments. It was then proposed that those fragments result from the phase explosion from the water in the tissue causing mechanical tearing of the intracellular tissue matrix. A secondary material expulsion, caused by recoil pressure, can also be observed following the smaller fragments from primary ablation. Later, secondary recoil pressure is sufficient to eject larger fragments and it can be argued that this process is accentuated in this case by the fact that thin tissue section is prone to delaminate from microscope slide and larger pieces to peel off and being expelled. Indeed, representative shadowgraphy images from consecutive laser pulses on the same pixel presented in Figure 2A clearly show that all tissue from the voxel is ablated as a result of the first laser pulse. This observation is confirmed by the MS analysis of the corresponding plumes of ablated material (n = 34 experiments) reported in the chart. When using ice as an energy absorbing matrix, the plume dynamics are far more complex, involving the ablation and interaction between tissue, ice crystals and liquid water. Shadowgraphy images show that far less tissue fragments can be observed following the first laser pulse. A plume consisting of fine liquid droplets is first ejected in primary ablation processes that resemble those observed during ablation from liquid samples. This primary ejection of material is then followed by a recoil-induced secondary expulsion of large crystals of ice. Very few tissue fragments can be observed in the plume of ablated material during this first pulse of ice matrix, which corroborate the relatively low MS signal recorded during the first laser pulses and reported in Figure 2B. Eventually, both tissue fragments and liquid droplets are ejected during primary ablation. The plume of tissue fragments are ejected first and followed by the plume of liquid droplets. In contrast to fresh tissue without ice matrix, the primary ablation of tissue fragments is not followed by the secondary ejection of larger tissue fragments and displacement of all tissue within each pixel. This observation allows one to generate signal over multiple laser pulses (up to 8–10 pulses). Despite a smaller amount of material being ablated per pulse, the abundance reached after a few pulses when using ice as a matrix is in the same order of magnitude as what is measured for the first pulse without any matrix (ice) where all material is ablated. This suggests that larger fragments ejected during secondary ablation of fresh tissue are not playing a significant role in the ionization process. It can also be argued that some signal improvement is caused by liquid extraction of neutral molecules from tissue occurring in the ablation crater between pulses and in the atmosphere as the plume of liquid droplets passes through the cluster of tissue fragments hovering above the surface after ablation. Because relatively high signal can be maintained for several consecutive pulses using ice as a matrix, it allows one to increase signal abundance by accumulating ions from multiple ablations for each pixel in the ion trap before sending them to the ICR cell for analysis. Instead than using an ice matrix to absorb laser energy, one could argue that reducing the laser fluence could be used to achieve the same end goal, i.e. obtain high signal abundance over multiple ablations. Because of the direct impact of fluence on plume ablation dynamics (e.g. plume height) and aforementioned liquid extraction potentially occurring in the voxels, such side-by-side comparison would be hard to achieve. However, the main reason to concentrate our effort on studying the ablation using ice as a matrix is the fact that thermoelectric coolers are often used to preserve tissue water content in ambient imaging techniques using mid-IR laser[30, 37]. Since it is extremely difficult to control the ambient condition to preserve water content while completely avoiding the formation of an ice layer or the sublimation of the tissue water (if a nitrogen flow is used), it is definitely an advantage to instead control the ice layer formation, understand its effect on ablation dynamics and optimize the source parameters for those conditions.
Figure 2.
Representative shadowgraphy images of consecutive ablation events (Shot 1 to Shot 4) and corresponding abundance of selected lipids for each shot (sum of m/z 369.35, 786.60, 810.60 and 758.57). Experiment was performed with fresh tissue without (top) and with an ice matrix (bottom). Without matrix, all tissue is ablated in the first pulse. With an ice matrix, abundance per pulse is greater even though smaller amount of tissue is released after every shot.
Investigation on spot-spot distance, number of pulse per pixel with and without ice matrix
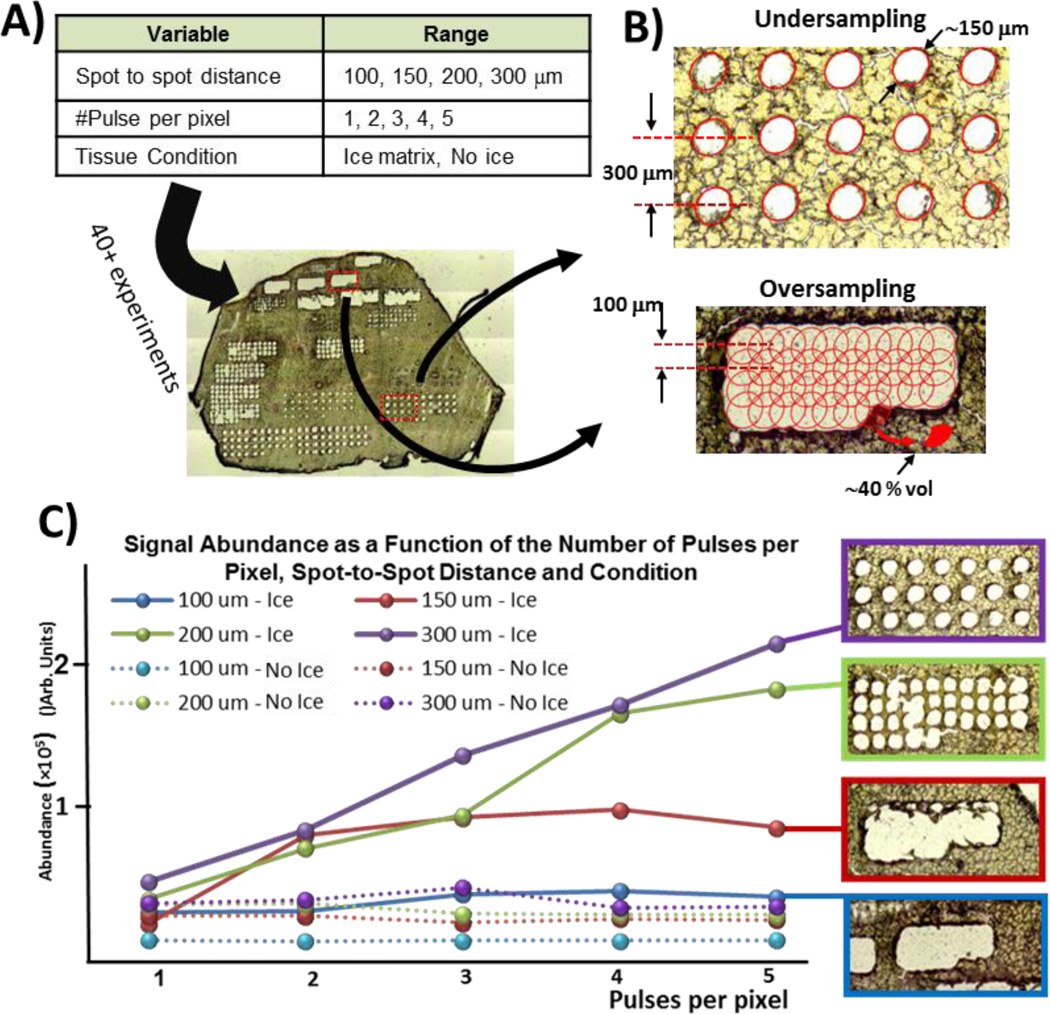
The objective of this next series of experiments was to investigate and better understand the relation between the number of shots per pixel, the spot spacing, and the tissue condition on the signal abundance. A total of 40 imaging experiments were performed to cover all possible parameter combinations (see Figure 3A). For each experiment, average abundance was computed and an optical image of the rastered ROI was acquired. Depending on the spot spacing, the imaging experiment can be performed in either undersampling or oversampling mode [46, 47]. As shown in Figure 3B, the desorption diameter or diameter for which laser fluence is above the desorption threshold for the tissue of interest was measured at 150 µm. Note that this diameter is significantly smaller than the focal diameter (250–300 µm, measured on thermal sensitive film card). When operating in oversampling mode, the step size is set smaller than ablation diameter such that only material from a fraction of the irradiated surface is ablated as shown in Figure 3B. Ablating all the way through the tissue in both undersampling and oversampling mode also ensures that amount of material used at each pixel location is constant.
Figure 3.
A) Combination of parameters tested on a liver tissue section (50 µm). For each condition, abundance of selected peaks were computed and optical image of the tissue section captured. B) Undersampling vs oversampling techniques and how overlapping pixels can lead to better spatial resolution. C) Optical images of ablated tissue and corresponding average abundance for different spot-to-spot distances and number of pulses per pixel. As expected, multi-shot imaging of tissue sections without an ice matrix did not result in greater signal since all material is ablated after first pulse (also confirmed with optical images). Greater signal can be achieved using ice as a matrix where it becomes a trade-off between spatial resolution and sensitivity.
The data shown in Figure 3C confirm that greater ion abundance can be obtained using ice as a matrix and when ions from multiple ablations are accumulated before being injected in the ICR cell for analysis. When operating in undersampling mode, abundance increases as the number of pulses per pixel increase, even for 6 or 7 pulses. All results of this series of experiments are presented in Figure S7. These results corroborate the observations made previously during shadowgraphy experiments where significant signal from individual pulses was measured even after several pulses when using ice as a matrix. Also corroborating the shadowgraphy images from fresh tissue with no ice matrix, no increase in signal could be obtained by operating in multi-injection mode since all material is ablated during the first laser pulse (also confirmed by additional optical images presented in Figure S7). As spot-to-spot distance decreases, the number of pulses necessary to ablate through all tissue diminishes and so does the maximum ion abundance that can be reached at each pixel. Importantly, these results demonstrate that multifold signal improvement can be obtained using ice as a matrix when imaging in both oversampling and undersampling modes.
Optimization of IR-MALDESI parameters for tissue imaging
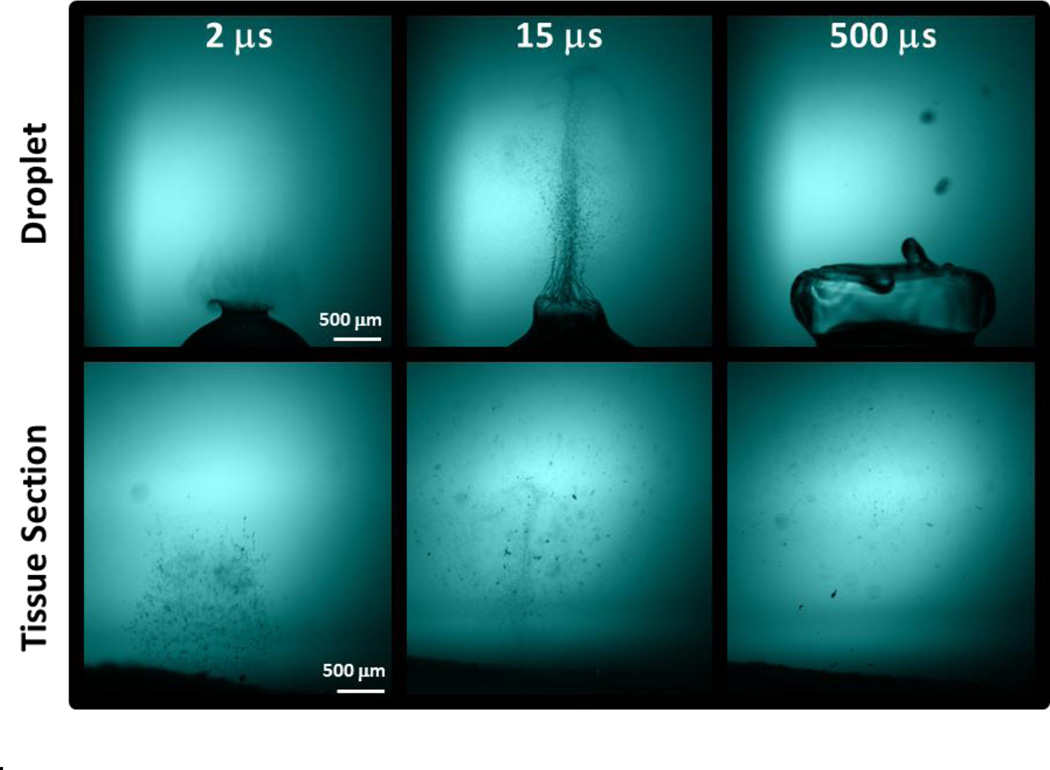
Comparison of shadowgraphy images of material ablated from both tissue and droplet samples clearly show that the plume dynamics differ greatly. As shown in the time-resolved shadowgraphy images presented in Figure 4, tissue fragments decelerate faster and do not travel as high as the liquid droplets. Indeed, a significant amount of ablated tissue fragments can still be seen hovering few millimeters above the surface 500 µs after the ablation while liquid droplets have disappeared at higher altitude. An additional series of time-resolved shadowgraphy images of the plume ablation process of tissue sections (Figure S2–S4) and water droplets (Figure S5–S6) clearly illustrate this difference in plume dynamics.
Figure 4.
Time-resolved shadowgraphy images of plume of ablated material for both a liquid droplet (top) and tissue section with ice matrix (bottom) clearly showing the difference in plume dynamics.
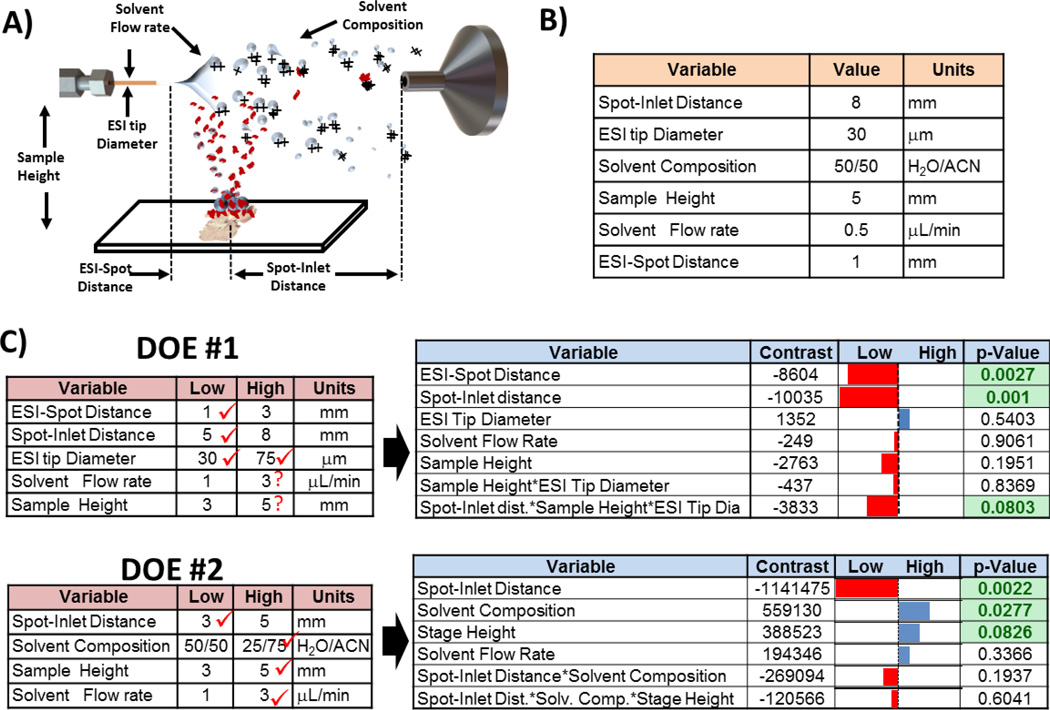
IR-MALDESI from liquid samples had previously been investigated by our group and optimal source parameters described in Figure 5A–B were found [14]. This significant difference in plume dynamics prompted us to revisit the experimental space to evaluate if optimal parameters more suitable for IR-MALDESI of tissue sections using ice as a matrix could be found. DOE was used to systematically explore the experimental space and the results of 2 full factorial screening experiments are presented in Figure 5C. For each DOE screening experiment, data was collected using different combinations of the variable investigated (2-levels: high and low). 5 datapoints were collected for each setting combinations and the sum of the following tissue ions was used as figure of merit to calculate the effect (contrast) of each variable on the signal: m/z 369.35, 622.41, 650.44, 678.47, 758,57, 806.56 and 810.60. Results of these experiments indicated that signal abundance could be significantly increased by reducing the spot-inlet distance from 8 to 3 mm and also by increasing the flow from 0.5 µL/min to 3 µL/min. It is interesting to note that this region of the experimental space (higher flow and smaller ESI distance) was explored for liquid IR-MALDESI but was found to be detrimental to signal abundance. In liquid IR-MALDESI, ions are mainly generated by the ESI-like desorption/ionization of droplets that result from the fusion between ablated droplets filled with neutrals and the charged electrosprayed solvent droplets. Since higher flow leads to the generation of larger electrospray droplets [48], it can be argued that for shorter ESI distance and greater flow, the significantly larger fused droplets do not spend enough time at atmospheric pressure to undergo an efficient ESI-like desorption/ionization, explaining the weaker signal measured in these conditions. However, when performing tissue imaging using ice as a matrix, shadowgraphy images suggest that ionization results from the interaction between tissue fragments and/or neutral molecules with the charged ESI droplets (by opposition to droplet fusion). More complete ESI droplet evaporation, different ionization pathways involving interaction of neutral or fragments with charged droplet surface and overall more efficient transmission of gas phase ions to the MS inlet (smaller ESI distance) can potentially explain the greater signal obtained under the new optimized conditions. It can also be argued that higher signal obtained in the new conditions where ablation spot is closer to ESI tip emitter (ESI-Spot distance of 1 mm) is caused by the greater interaction of ablated material with denser cluster of droplets that are closer to the ESI centerline and therefore more likely to reach the MS inlet.
Figure 5.
A) Definition of parameters evaluated in DOE experiments. B) Parameters previously found to be optimal for IR-MALDESI on liquid sample [14] C) 2 screening Design of experiments (DOE) were performed. For each of them, on the left are presented the settings that were tested and on the right are presented the corresponding results. Contrasts represent the contribution of the on the setting (or combination) and reported p-value represent the probability that such results are obtained assuming the null hypothesis.
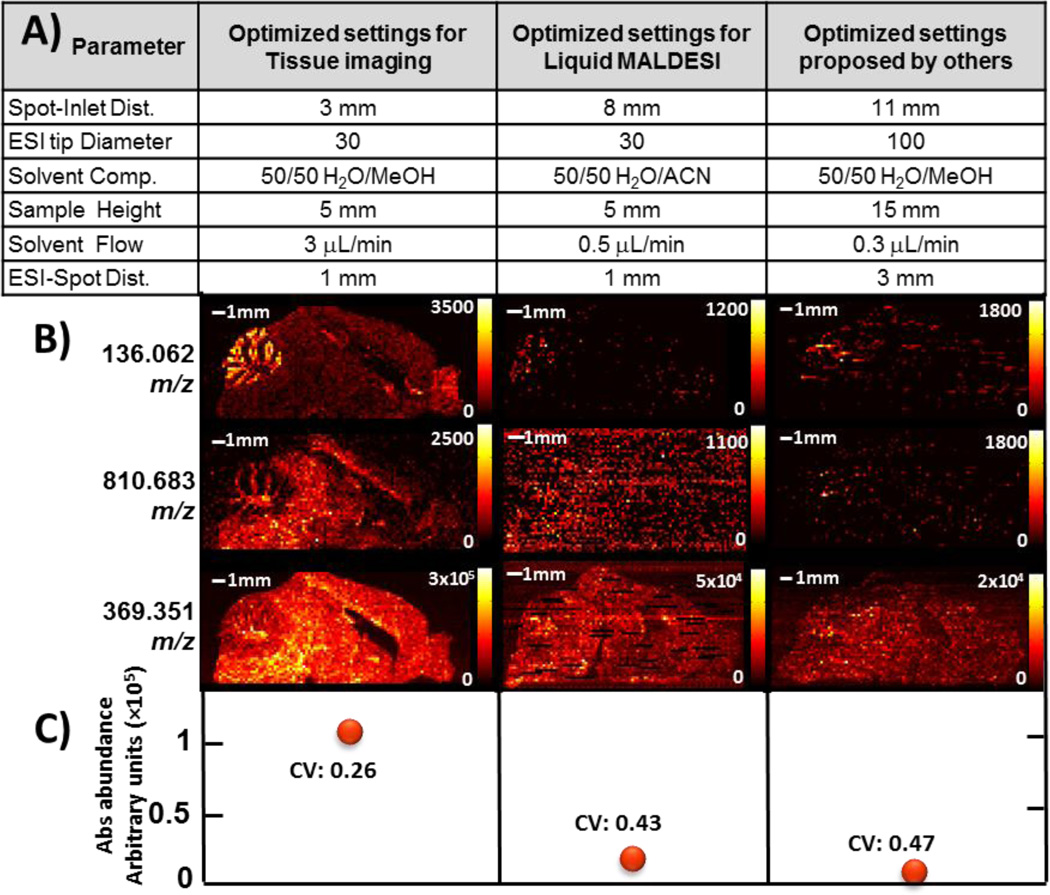
Note that additional experiments were performed to further investigate the effect of solvent composition. Imaging experiments were performed while running a linear gradient of the organic ESI solvent (5%–95%) while imaging over a homogeneous liver tissue. The experiment was repeated using ACN and MeOH as organic solvent and it was found that better and more stable results could be obtained using 50:50 methanol and water (v/v) with 0.2% formic acid [17]. The new optimized parameters for tissue imaging using ice as a matrix were compared with optimized parameters for liquid IR-MALDESI and a set of parameters found optimal for tissue imaging by others [37]. Those sets of parameters that were compared are detailed in Figure 6A. These three imaging experiments were performed on adjacent 50 µm sagittal sections of mouse brain (one using each combination of settings). Heat maps for 3 selected m/z’s are shown in Figure 6B as well as the average abundance of and coefficient of variation for all three sets of parameters that were tested. Results show that signal abundance could be improved by nearly one order of magnitude using the new proposed parameters.
Figure 6.
Comparison of the new optimized settings for tissue imaging, the settings previously found for liquid IR-MALDESI and finally parameters found optimal by others. A) Summary of tested parameters B) Comparison of those 3 sets of settings in imaging experiments performed on 50 µm mouse brain sagittal sections from same specimen. Feature on the top left represent the cerebellum. Imaging was performed with 4 pulses per pixel and 150 µm spot-to-spot distance (m/z range: 130–1300). Note that a 10 ppm m/z window size and that presented data is not interpolated. C) Average abundance and coefficient of variation (CV) of cholesterol or m/z 369.351 ± 5 ppm (M – H2O + H+)+ over a region of interest where molecule is found uniformly.
CONCLUSION
IR-MALDESI imaging of biological tissue sections using a layer of deposited ice as an energy absorbing matrix was thoroughly investigated. A novel flash shadowgraphy system that can simultaneously collect images of the plume of ablated material and acquire the FT-ICR mass spectrum resulting from that same ablation event was used to compare signal and plume dynamics with and without using ice as an energy absorbing matrix. A thorough evaluation of the effects of the number of ablations per pixel and spot spacing on the signal abundance was performed and supported with optical microscopy images. Once again, it was shown that multifold improvement could be obtained when using an ice matrix. Finally, a design of experiments was used to explore the experimental space and optimize geometry and instrument parameters for IR-MALDESI of tissue sections using ice as a matrix, resulting in an improvement in ion abundance of nearly one order of magnitude. Future experiments will focus on the effect of fluence, spot size and the use of novel remote sampling devices to improve signal in IR-MALDESI imaging.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to gratefully acknowledge the financial support received from the National Institutes of Health (R01GM087964) and North Carolina State University.
REFERENCES
- 1.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka KWH, Ido Y, Akita S, Yoshida Y, Yoshida T, Matsuo T. Protein and Polymer Analysis up to m/z 100,000 by Laser Ionization Time-of-Flight Mass Spectrometry. Rapid Commun. Mass Spectrom. 1988;2:151–153. [Google Scholar]
- 3.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 4.Shiea J, Huang MZ, Hsu HJ, Lee CY, Yuan CH, Beech I, Sunner J. Electrospray-assisted laser desorption/ionization mass spectrometry for direct ambient analysis of solids. Rapid Commun Mass Spectrom. 2005;19:3701–3704. doi: 10.1002/rcm.2243. [DOI] [PubMed] [Google Scholar]
- 5.Huang MZ, Hsu HJ, Lee LY, Jeng JY, Shiea JT. Direct protein detection from biological media through electrospray-assisted laser desorption ionization/mass spectrometry. J Proteome Res. 2006;5:1107–1116. doi: 10.1021/pr050442f. [DOI] [PubMed] [Google Scholar]
- 6.Brady JJ, Judge EJ, Levis RJ. Mass spectrometry of intact neutral macromolecules using intense non-resonant femtosecond laser vaporization with electrospray postionization. Rapid Commun Mass Spectrom. 2009;23:3151–3157. doi: 10.1002/rcm.4226. [DOI] [PubMed] [Google Scholar]
- 7.Jorabchi K, Smith LM. Single droplet separations and surface partition coefficient measurements using laser ablation mass spectrometry. Anal Chem. 2009;81:9682–9688. doi: 10.1021/ac901819r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Qiu B, Luo H. Fingerprinting of yogurt products by laser desorption spray postionization mass spectrometry. Rapid Commun Mass Sp. 2010;24:1365–1370. doi: 10.1002/rcm.4527. [DOI] [PubMed] [Google Scholar]
- 9.Sampson JS, Hawkridge AM, Muddiman DC. Direct characterization of intact polypeptides by matrix-assisted laser desorption electrospray ionization quadrupole Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun Mass Sp. 2007;21:1150–1154. doi: 10.1002/rcm.2947. [DOI] [PubMed] [Google Scholar]
- 10.Sampson JS, Hawkridge AM, Muddiman DC. Generation and detection of multiply-charged peptides and proteins by matrix-assisted laser desorption electrospray ionization (MALDESI) Fourier transform ion cyclotron resonance mass spectrometry. J Am Soc Mass Spectr. 2006;17:1712–1716. doi: 10.1016/j.jasms.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Nemes P, Vertes A. Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal Chem. 2007;79:8098–8106. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- 12.Rezenom YH, Dong J, Murray KK. Infrared laser-assisted desorption electrospray ionization mass spectrometry. The Analyst. 2008;133:226–232. doi: 10.1039/b715146b. [DOI] [PubMed] [Google Scholar]
- 13.Sampson JS, Murray KK, Muddiman DC. Intact and top-down characterization of biomolecules and direct analysis using infrared matrix-assisted laser desorption electrospray ionization coupled to FT-ICR mass spectrometry. J Am Soc Mass Spectr. 2009;20:667–673. doi: 10.1016/j.jasms.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barry JA, Muddiman DC. Global optimization of the infrared matrix-assisted laser desorption electrospray ionization (IR MALDESI) source for mass spectrometry using statistical design of experiments. Rapid Commun Mass Sp. 2011;25:3527–3536. doi: 10.1002/rcm.5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cochran KH, Barry JA, Muddiman DC, Hinks D. Direct Analysis of Textile Fabrics and Dyes Using Infrared Matrix-Assisted Laser Desorption Electrospray Ionization Mass Spectrometry. Anal Chem. 2012;85:831–836. doi: 10.1021/ac302519n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robichaud G, Barry JA, Garrard KP, Muddiman DC. Infrared Matrix-Assisted Laser Desorption Electrospray Ionization (IR-MALDESI) Imaging Source Coupled to a FTICR Mass Spectrometer. J Am Soc Mass Spectr. 2013;24:92–100. doi: 10.1007/s13361-012-0505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barry JA, Groseclose MR, Robichaud G, Castellino S, Wagner D, Muddiman DC. Localization of Lapatinib and its metabolites in liver tissue as determined by UV-MALDI and IR-MALDESI mass spectrometry imaging coupled to FT-ICR MS. Anal Bioanal Chem. 2013 Submitted. [Google Scholar]
- 18.Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat Med. 2001;7:493–496. doi: 10.1038/86573. [DOI] [PubMed] [Google Scholar]
- 19.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 20.Norris JL, Caprioli RM. Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem Rev. 2013;113:2309–2342. doi: 10.1021/cr3004295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaurand P. Imaging mass spectrometry of thin tissue sections: A decade of collective efforts. J Proteomics. 2012 doi: 10.1016/j.jprot.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 22.van Hove ERA, Smith DF, Heeren RMA. A concise review of mass spectrometry imaging. J Chromatogr A. 2010;1217:3946–3954. doi: 10.1016/j.chroma.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 23.Baltz-Knorr M, Schriver K, Haglund R. Infrared laser ablation and ionization of water clusters and biomolecules from ice. Appl Surf Sci. 2002;197:11–16. [Google Scholar]
- 24.Kraft P, Alimpiev S, Dratz E, Sunner J. Infrared, surface-assisted laser desorption ionization mass spectrometry on frozen aqueous solutions of proteins and peptides using suspensions of organic solids. J Am Soc Mass Spectr. 1998;9:912–924. doi: 10.1016/S1044-0305(98)00063-4. [DOI] [PubMed] [Google Scholar]
- 25.Berkenkamp S, Karas M, Hillenkamp F. Ice as a matrix for IR-matrix-assisted laser desorption/ionization: mass spectra from a protein single crystal. P Natl Acad Sci USA. 1996;93:7003–7007. doi: 10.1073/pnas.93.14.7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oraevsky AA, Jacques SL. Mechanism of laser ablation for aqueous media irradiated under confined-stress conditions. J Appl Phys. 1995;78:1281–1290. [Google Scholar]
- 27.Belov ME, Alimpiev SS, Mlynsky VV, Nikiforov SM, Derrick PJ. Laser ablation of organic molecules from frozen matrices. Rapid Commun Mass Sp. 1995;9:1431–1436. [Google Scholar]
- 28.Williams P. Time of flight mass spectrometry of DNA laser-ablated from frozen aqueous solutions: applications to the Human Genome Project. International Journal of Mass Spectrometry and Ion Processes. 1994;131:335–344. [Google Scholar]
- 29.Von Seggern CE, Gardner BD, Cotter RJ. Infrared atmospheric pressure MALDI ion trap mass spectrometry of frozen samples using a Peltier-cooled sample stage. Anal Chem. 2004;76:5887–5893. doi: 10.1021/ac049575v. [DOI] [PubMed] [Google Scholar]
- 30.Römpp A, Schäfer KC, Guenther S, Wang Z, Köstler M, Leisner A, Paschke C, Schramm T, Spengler B. High-resolution atmospheric pressure infrared laser desorption/ionization mass spectrometry imaging of biological tissue. Anal Bioanal Chem. 2013:1–10. doi: 10.1007/s00216-013-7180-y. [DOI] [PubMed] [Google Scholar]
- 31.Pirkl A, Soltwisch J, Draude F, Dreisewerd K. Infrared Matrix-Assisted Laser Desorption/Ionization Orthogonal-Time-of-Flight Mass Spectrometry Employing a Cooling Stage and Water Ice As a Matrix. Anal Chem. 2012;84:5669–5676. doi: 10.1021/ac300840b. [DOI] [PubMed] [Google Scholar]
- 32.Dreisewerd K, Berkenkamp S, Leisner A, Rohlfing A, Menzel C. Fundamentals of matrix-assisted laser desorption/ionization mass spectrometry with pulsed infrared lasers. Int J Mass Spectrom. 2003;226:189–209. [Google Scholar]
- 33.Menzel C, Dreisewerd K, Berkenkamp S, Hillenkamp F. Mechanisms of energy deposition in infrared matrix-assisted laser desorption/ionization mass spectrometry. Int J Mass Spectrom. 2001;207:73–96. [Google Scholar]
- 34.Venugopalan V, Nishioka N, Mikić B. Thermodynamic response of soft biological tissues to pulsed infrared-laser irradiation. Biophys J. 1996;70:2981–2993. doi: 10.1016/S0006-3495(96)79868-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nemes P, Vertes A. Ambient mass spectrometry for in vivo local analysis and in situ molecular tissue imaging. Trac-Trend Anal Chem. 2012;34:22–34. [Google Scholar]
- 36.Shrestha B, Nemes P, Nazarian J, Hathout Y, Hoffman EP, Vertes A. Direct analysis of lipids and small metabolites in mouse brain tissue by AP IR-MALDI and reactive LAESI mass spectrometry. Analyst. 2010;135:751–758. doi: 10.1039/b922854c. [DOI] [PubMed] [Google Scholar]
- 37.Nemes P, Woods AS, Vertes A. Simultaneous Imaging of Small Metabolites and Lipids in Rat Brain Tissues at Atmospheric Pressure by Laser Ablation Electrospray Ionization Mass Spectrometry. Anal Chem. 2010;82:982–988. doi: 10.1021/ac902245p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Z, Vertes A. Early plume expansion in atmospheric pressure midinfrared laser ablation of water-rich targets. Phys Rev E Stat Nonlin Soft Matter Phys. 2008;77:036316. doi: 10.1103/PhysRevE.77.036316. [DOI] [PubMed] [Google Scholar]
- 39.Apitz I, Vogel A. Material ejection in nanosecond Er: YAG laser ablation of water, liver, and skin. Appl Phys a-Mater. 2005;81:329–338. [Google Scholar]
- 40.Kokai F, Takahashi K, Shimizu K, Yudasaka M, Iijima S. Shadowgraphic and emission imaging spectroscopic studies of the laser ablation of graphite in an Ar gas atmosphere. Appl Phys a-Mater. 1999;69:S223–S227. [Google Scholar]
- 41.Thoroddsen ST, Takehara K, Etoh TG, Ohl CD. Spray and microjets produced by focusing a laser pulse into a hemispherical drop. Phys Fluids. 2009;21 [Google Scholar]
- 42.Robichaud G, Dixon RB, Potturi AS, Cassidy D, Edwards JR, Sohn A, Dow TA, Muddiman DC. Design, Modeling, Fabrication, and Evaluation of the Air Amplifier for Improved Detection of Biomolecules by Electrospray Ionization Mass Spectrometry. Int J Mass Spectrom. 2011;300:99–107. doi: 10.1016/j.ijms.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riter LS, Vitek O, Gooding KM, Hodge BD, Julian RK. Statistical design of experiments as a tool in mass spectrometry. Journal of Mass Spectrometry. 2005;40:565–579. doi: 10.1002/jms.871. [DOI] [PubMed] [Google Scholar]
- 44.Robichaud G, Garrard K, Barry J, Muddiman D. MSiReader: An Open-Source Interface to View and Analyze High Resolving Power MS Imaging Files on Matlab Platform. J Am Soc Mass Spectr. 2013;24:718–721. doi: 10.1007/s13361-013-0607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vogel A, Venugopalan V. Mechanisms of pulsed laser ablation of biological tissues. Chem Rev. 2003;103:577–644. doi: 10.1021/cr010379n. [DOI] [PubMed] [Google Scholar]
- 46.Jurchen JC, Rubakhin SS, Sweedler JV. MALDI-MS imaging of features smaller than the size of the laser beam. J Am Soc Mass Spectr. 2005;16:1654–1659. doi: 10.1016/j.jasms.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 47.Todd PJ, Schaaff TG, Chaurand P, Caprioli RM. Organic ion imaging of biological tissue with secondary ion mass spectrometry and matrix-assisted laser desorption/ionization. J Mass Spectrom. 2001;36:355–369. doi: 10.1002/jms.153. [DOI] [PubMed] [Google Scholar]
- 48.Kebarle P, Tang L. From ions in solution to ions in the gas phase-the mechanism of electrospray mass spectrometry. Anal Chem. 1993;65:972A–986A. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.