Abstract
Documenting mass spectral data is a fundamental aspect of accepted protocols. In this report we contrast MSn sequential disassembly spectra obtained from natural and synthetic glycan epitopes. The epitopes considered are clusters found on conjugate termini of lipids and N-, and O-glycans of proteins. The latter are most frequently pendant through a CID-labile HexNAc glycosidic linkage. The synthetic samples were supplied by collaborating colleagues and commercial sources and usually possessed a readily released reducing-end linker, a by-product of synthesis. All samples were comparably methylated, extracted, and MSn disassembled to compare their linkage and branching spectral details. Both sample types provide B-ion type fragments early in a disassembly pathway and their compositions are a suggestion of structure. Further steps of disassembly are necessary to confirm the details of linkage and branching. Included in this study were various Lewis and H antigens, 3- and 6-linked sialyl-lactosamine, NeuAc-2,8-NeuAc dimer, and Galα1,3Gal. Sample infusion provided high quality spectral data while disassembly to small fragments generates reproducible high signal/noise spectra for spectral matching. All samples were analyzed as sodium adducted positive ions. This study includes comparability statistics and evaluations on several mass spectrometers.
INTRODUCTION
Over the last two decades mass spectrometry (MS) has made major contributions to the structural understanding of biopolymers, usually as products of collision induced disassociation, (CID). Especially significant have been the numerous applications leading to a better understanding of protein structure. In sharp contrast, although the first biopolymer to be identified, carbohydrates have largely escaped any comparable understanding. Even with Nobel Prizes for generating stable gas phase ions [1] and for their control in electromagnetic fields [2] such advances are only starting to penetrate the complex details of carbohydrate structure. These ubiquitous biopolymers, replete with stereo and structural isomers embedded in branching arrays, have brought inordinate challenges well beyond the usual MS/MS and CID approaches. Adding to these complications are variable bond energies, where low energy collision products (glyco- and ketosidic bonds) dominate CID MS/MS spectra. These ion intervals do indicate monomer composition, but they fail to define structural continuity through linkage relationships. Interestingly, linkage differences have long been detected within spectra of small oligomers and building on these observations a full sequence understanding has remained an expectation. Unfortunately, these essential cross-ring fragments are only observed in smaller oligomers, where effective collision energies are higher. On this single constraint glycan sequencing efforts have stalled, encouraging a culture of accepting biological inference data in publications [3] and a growing use of meta data [4] for assigning the details of structure. To this specific problem the ion trap delivers a solution. Early applications in the first commercial ion traps provided the opportunity to evaluate larger oligomers by disassembly to smaller packets, exposing the ubiquitous presence of unreported structural isomers [5–7]. Fractionating ions into smaller subsets in the vacuum space of the ion trap, creates a tractable pathway parameter where energy enhancements expose linkage details in product spectra.
In applying this technology, the list of functional roles in which carbohydrates participate continues to grow, and most likely we are sampling but a small sample of the biosphere. Interestingly, as functional components are identified, small subsets of glycans are observed, referred to as epitopes. Such carbohydrate groupings are detected on different conjugate platforms and serve as receptors for protein (GBP) and lipid binding (GBL). Based on their size, (2 to 6 monosaccharides, including analogs of sulfation, phosphorylation, and acetylation), it has been estimated that these may be about 3000–4000 in number [8]. The exact number is of little significance, but the recognition of this finite number strongly suggests an achievable target for new chemical and/or enzymatic syntheses, and thereby introduces an alternative, predetermined approach to capture and define glycomic function [9–11]. An epitope’s broader range of function is likely expanded by antennal position, glycan type, (N-, and O-linkage), peptide site and finally the protein itself.
Mass spectral library matching for intact PA derivatized oligosaccharides has been reported previously showing good spectral reproducibility [12]. Spectral matching of protonated methylated epitopes has also been considered [13,14], as well as negative mode MS/MS and MS3 to a small set of native oligosaccharide epitopes [15] coupled into an LC-MS/MS strategy [16]. But in our experience sodium adduction of methylated derivatives provides superior fragmentation, particularly with respect to cross-ring cleavage products, as well as an improved evaluation of ion stereochemistry when applied to small oligomers. Thus, a library for these components and substructures was initiated, compiled and reported. These included a large set of commercially available oligomers and well characterized natural products [17]. This on-going effort is now coupled with a de novo strategy for determining complete oligosaccharide structures by MSn [18].
We demonstrate here the ability to obtain MSn spectra from well-characterized standard materials which can then be used to provide a comparative data set. Spectra from various biological samples are then compared to these standard spectra, making the viability of an MSn ion treebased comparability strategy a reality. The ability to obtain pure standard materials provides a viable strategy for molecular disassembly combined with spectral matching to make structural assignments with high fidelity employing high signal/noise spectra.
Experimental
Sample Preparation
Lacto-N-difucohexaose II (L603) was obtained from V-labs. Standard KML102 (NeuAcα2→3Galβ1→4GlcNAcβ1→3Galβ1→4GlcNAcβ1→O-Benzyl) was a kind gift from Dr. Khushi Matta. Lacto-N-difucohexaose I (GKAD-02010) and lacto-N-fucopentaose I (GKAD-02006) were obtained from Prozyme (Hayward, CA). Other synthetic glycan standards were graciously supplied by the Consortium for Functional Glycomics (CFG) (http://www.functionalglycomics.org/); structural information may be found at: http://www.functionalglycomics.org/static/consortium/resources/resourcecored2.shtml#TetraHigh. All standard materials were permethylated, with no other sample manipulation. Complete structures for the standard oligosaccharides used in this study are listed in the supplementary data.
Bovine fetuin (Sigma 3004) was treated with Sialidase S (Prozyme GK80020), prior to N-glycan release. N-linked glycans from sialidase S-treated fetuin and IgG-depleted human plasma were released with N-glycanase (Prozyme).
Glycosphingolipids (human plasma, normal human colon tissue) were extracted with 2:1 chloroform/methanol and the crude lipid extract was fractionated via silica solid phase extraction (SPE) [19]. The silica SPE cartridges were from Grace/Alltech. Dried glycosphingolipid fractions were permethylated intact.
For human plasma analysis, blood was collected in a vacutainer (Becton Dickinson) containing sodium citrate as an anticoagulant. Plasma was separated by centrifugation, and passed through a Protein A/G column (ThermoFisher/Pierce) to obtain IgG-depleted plasma and IgG fractions. N-glycans were released from each fraction enzymatically. Glycans were derivatized with 2-AA via reductive amination. Derivatized samples were separated by HPLC using a HILIC method. Collected fractions were dried and permethylated.
O-glycans were released via beta-elimination using alkaline borohydride [20]. Samples were purified via cation exchange and porous graphitized carbon solid phase extraction prior to permethylation.
Permethylation was carried out in spin columns (Harvard Apparatus) as described [21]. Sodium hydroxide beads and iodomethane were from Sigma. Purification of permethylated oligosaccharides was performed by liquid-liquid extraction with dichloromethane and 0.5M aqueous sodium chloride.
Glycan Analysis
Permethylated oligosaccharides were dissolved in 1:1 methanol/water. Except where noted, the samples were loaded onto a Triversa Nanomate (Advion) mounted onto an ion trap mass spectrometer (LTQ, ThermoFisher). Activation Q and activation time were left at the default values, 0.250 and 30 ms, respectively. Collision energy was set to 35% for all CID spectra. The scan rate was set to “Enhanced” for most spectra. Data were acquired and are displayed in profile mode. MSn peaks were selected manually. In general, the precursor ion m/z and isolation width were set to capture the full isotopic envelope. This was done to obtain complete isotope clusters of fragment ions at higher stages of MSn, and to simplify fragment ion charge state determination. For each data file, at least one scan was obtained of the isolated precursor ion with the collision energy set to 0, to record the precursor isotope envelope. The signal was averaged for a variable number of scans, with the times indicated in each spectrum. The microscan count, AGC target value, and maximum injection times were varied, depending on the signal intensity. Detailed information is included in the supplemental data, with times, scan numbers, and centroided peak lists.
Where noted, certain MSn spectra were obtained on an LTQ Orbitrap using static nanospray and the spectra were obtained using similar parameters. For spectra obtained using the Orbitrap mass analyzer resolution was set to 60000. Certain MSn spectra were also obtained on a Velos Pro (ThermoFisher) using dynamic nanospray. The collision energy was set to 30% and other parameters were left at their default values. All ions are sodium adducts.
Results and Discussion
Many of the biological interactions of glycans and their binding proteins are specific for certain well-defined structural motifs. These substructures are frequently components of larger oligosaccharide structures, be they N-linked, O-linked, or glycosphingolipids. In addition, such substructures may be found on different “scaffolds” within a class of oligosaccharides. For example, a Lewis X epitope may be found on multiple N-linked glycans of a particular sample. Targeting an analytical strategy to delineate particular epitopes can provide the analytical focus on where the biological activity is occurring. A complication of identifying these epitopes is the possibility of isomeric forms of those substructures. For example, Lewis X, Lewis A, and the H antigens are all isomeric. While differentiating structural motifs in a de novo fashion will rely on unique fragment masses, some subtle structural differences may only manifest as intensity pattern differences, with the major fragment masses being identical.
A similarity score was generated for each pair of spectra. The formula and tables for each subsequent figure are shown in the supplementary material. For our purposes, a similarity score greater than 0.850 is strongly indicative of a match using the described scoring algorithm.
Lewis A, Lewis X, H antigens, B-type, m/z 660
Lewis structures are involved in myriad intercellular interactions [22,23]. Distinguishing Lewis A from Lewis X via mass spectrometry can be particularly difficult, as they differ only in the linkage of the galactose and fucose to the N-acetylglucosamine residue. Targeted methods have been reported, using unmethylated oligosaccharides [24] and glycopeptides [25]. H antigens have the same substructure mass, but are somewhat easier to distinguish, as the fucose is attached to galactose rather than Nacetylglucosamine; these are also based on Type 1(Gal-β1,3-GlcNAc) or Type 2 (Gal-β1,4-GlcNAc) lactosamines.
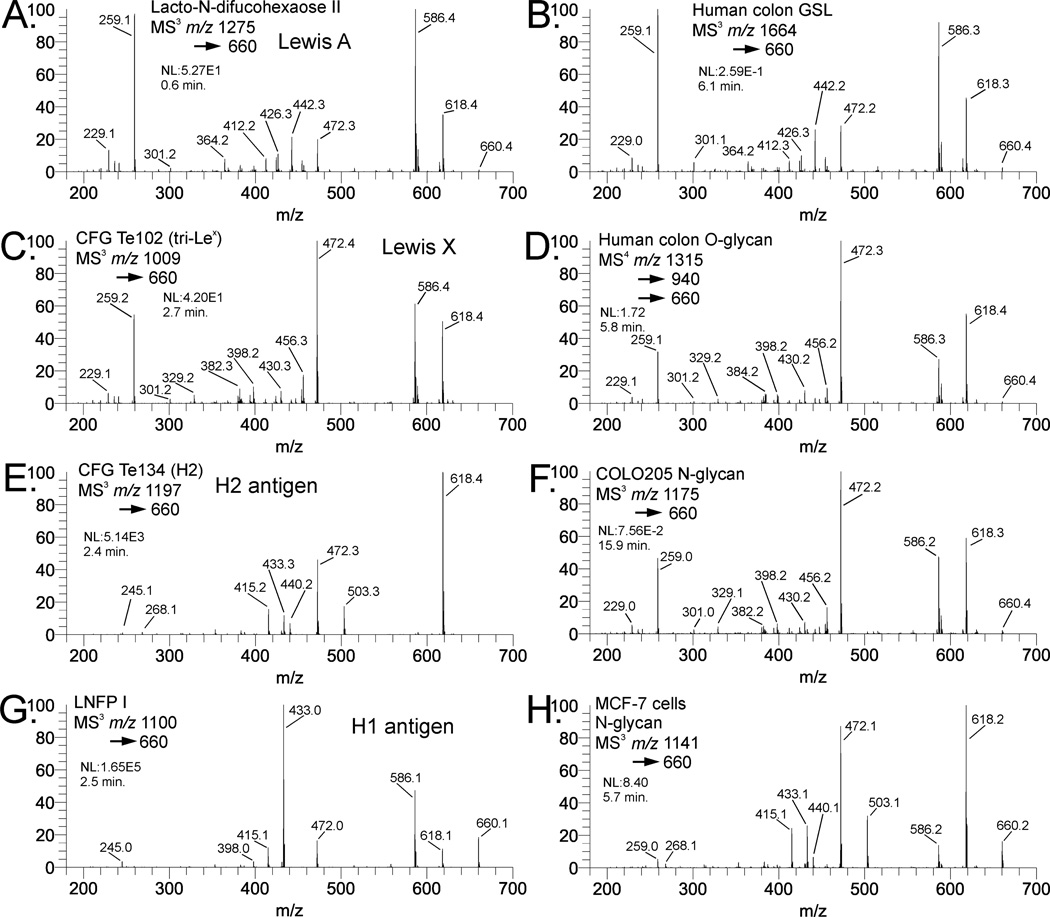
Figure 1A shows the m/z 660 fragment isolated from lacto-N-difucohexaose II, a Lewis A-containing oligosaccharide. Figure 1C shows the m/z 660 fragment isolated from the CFG standard Te102, a Lewis X containing structure. The major fragment ions are similar for both structures, with a few notable differences. The Lewis X epitope in Figure 2C shows a small but important fragment ion at m/z 329, indicative of a 3,5A cross-ring cleavage. This places the hexose at either the 4 or 6-position, and clearly distinguishes Lewis X from Lewis A. The presence of an additional cross-ring cleavage product at m/z 301, a 2,4A fragment, is consistent with a hexose at either the 3 or 4 position of the N-acetylglucosamine. The presence of both fragments in the Lewis X standard spectrum is sufficient to empirically localize galactose to the 4-position. Beyond that, this ion does not distinguish these epitopes, as it appears at similar intensity for both structures. The Lewis A standard spectrum contains a significant peak at m/z 442, representing the loss of a terminal hexose residue because neutral loss of the 3-linked substituent is highly favored over the 4-linked substituent in these and other isomeric pairs where monosaccharide residues such as Hex or deoxyHex are linked to a HexNAc.
Fig. 1.
Comparison of Lewis A, Lewis X, H1, and H2 antigens. Standard materials are: (A) Lacto-N-difucohexaose II (Lewis A), (C) CFG Te102 (Lewis X), (E) CFG Te134 (H2), and (G) Lacto-N-fucopentaose I (H1). Comparative biological materials are (B) glycosphingolipid isolated from normal human colon tissue (Lewis A), (D) O-glycan isolated from normal human colon tissue (Lewis X), and (F) fucosylated lactosamine unit from an N-glycan isolated from COLO205 cultured cells (Lewis X). In comparison to standards, (B) appears to be Lewis A, while (D) and (F) are Lewis X. The spectrum in (H) is the fucosylated lactosamine fragment from an N-glycan (composition Hex5HexNAc4dHex1) from MCF-7 cultured cells. The fragmentation pattern in (H) appears to be a composite of the H2 and Lewis X spectra, indicating a mixture of these structures.
Fig. 2.
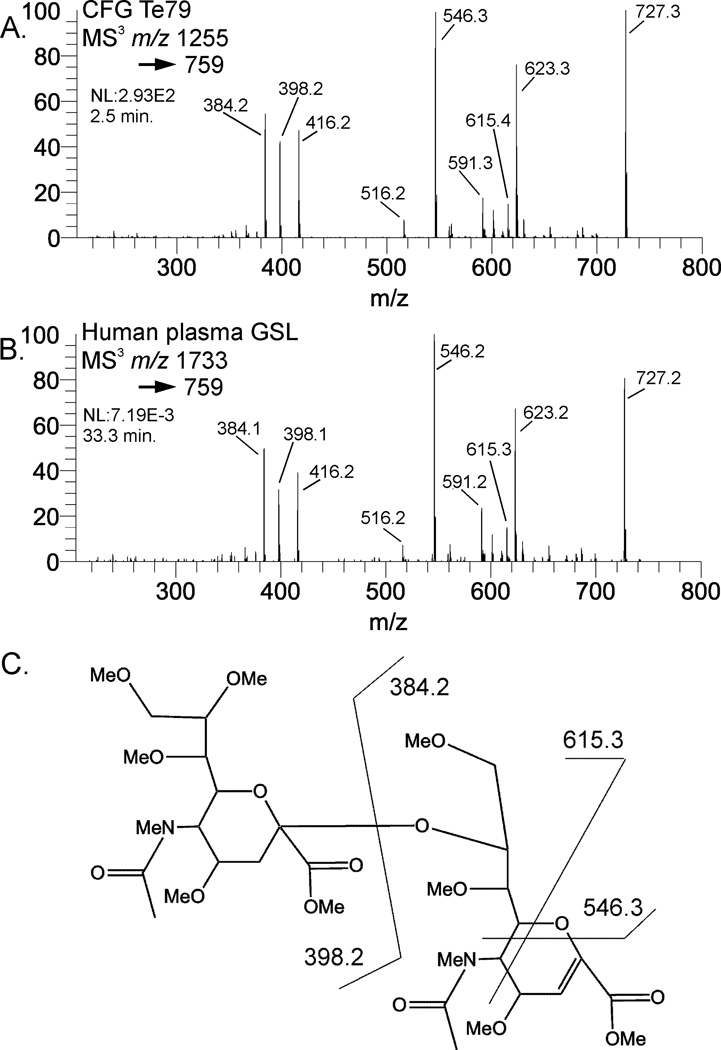
Mass spectral comparison of NeuAc-2,8-NeuAc B-type ions. 2A is MS3 of the B-type NeuAc- NeuAc isolated from the standard CFG Te79 (GD3) compared to the same mass fragment from a human plasma ganglioside (2B). The structure and fragment assignments are in 2C.
H antigens also produce an m/z 660 fragment from permethylated glycans or glycosphingolipids. As with Lewis A/X, it is necessary to isolate and dissociate this ion in order to assign the structure. Figure 1E shows the CID spectrum of the m/z 660 precursor from CFG standard Te134, which contains a Type 2 H antigen. This spectrum is clearly different from the Lewis A/X spectra, and fucose can be unambiguously localized to the galactose residue. Figure 1G shows the CID spectrum of the same ion isolated from LNFP I, an H1 antigen-containing milk glycan. The fragmentation is clearly different than that of the Lewis structures and H2. The fragment at m/z 503 in the H2 spectrum is a 3,5A cleavage placing the Fuc-Gal moiety at the 4-position of the GlcNAc; this fragment is absent from H1. The 0,4X fragment, m/z 586, can theoretically be from either H1 or H2, but is much more intense in H1. This ion also is observed in type 1 and type 2 Gal-GlcNAc B2 ions [26]. Supplemental figure S1 shows the structures and fragmentation assignments for Lewis A, Lewis X, and H antigens. In all cases, the same precursor mass appears at the MS2 (or MS3 level). Appearance of the m/z 660 ion in a mass spectrum does not imply a structure, only a fragment composition, where further CID is necessary to clearly determine the structure.
Comparison of representative biological samples indicates the utility of pattern matching for structure assignment. Figure 1B shows the CID spectrum of an m/z 660 precursor isolated from a glycosphingolipid from normal human colon tissue. This spectrum matches very well with the Lewis A standard spectrum; the similarity score compared to 1A is 0.983, while for 1C is 0.771. Figure 1D shows the MS3 spectrum of the m/z 660 fragment from an O-glycan isolated from normal human colon tissue. Figure 1F shows the similar fragment from an N-glycan isolated from COLO205 cultured cells. Both spectra 1D and 1F match exceptionally well with the Lewis X standard (similarity scores 0.950 and 0.987, respectively), and are easily distinguishable from Lewis A (0.544 and 0.679, respectively). Interestingly, these spectra were isolated at different stages of MSn (1D is MS4 and 1F is MS3). In principle, the particular MSn stage of the spectrum is not important, so long as the proper structural fragment is isolated and the spectral quality is sufficient.
Mixtures of isomeric structures are also possible with complex biological samples. Figure 1H shows a fucosylated lactosamine spectrum obtained from an N-glycan isolated from MCF-7 cultured cells (precursor m/z 11412+, composition Hex5HexNAc4dHex1). This spectrum is a composite of the standard spectra of H2 and Lewis X, indicating a mixture of the two epitopes. While the similarity scores are 0.756 for Lewis X and 0.946 for H2, visual inspection of the spectrum indicates the mixture contains a small amount of Lewis X. The fragments at m/z 259 and 586 are present in Lewis X but not H2, while the m/z 268, 415, 433, and 503 ions are present in H2 but not Lewis X. While the similarity score provides a quantitative measure of spectrum similarity, it does not absolve the analyst of visually inspecting spectra. We are working on more sophisticated computational techniques to deal with mixed spectra.
Neu5Ac-Neu5Ac dimer, B-type, m/z 759
Oligo- and polysialic acids can occur in multiple types of glycoconjugates [27]. The ganglioside GD3 contains a sialic acid dimer purported to play several biological roles, including intracellular signaling [28], and is also highly expressed in neural stem cells [29]. Frequently, gangliosides are detected and identified via thin layer chromatography (TLC), sometimes coupled with mass spectrometry [30,31]. GD3 has been analyzed via TLC coupled with negative mode CID using a QToF instrument [32]. One advantage of permethylation is that cross-ring cleavages are frequently observed, which provide empirical data indicating linkage position. Permethylation and CID of the B-type sialic acid dimer isolated from CFG standard 79 is shown in Figure 2A. The fragment mass assignments are shown in Figure 2C. This same sialic acid dimer was isolated from a human plasma glycosphingolipid (GD3) (Figure 2B). In the case of the Neu5Ac dimer, cross-ring cleavage fragments are detectable, but only localize the linkage to the 7, 8, or 9 positions, making a de novo linkage assignment difficult. The availability of a comparative standard provides the added benefit of being able to assess the actual fragmentation of the known, authentic structure. The high degree of similarity between the spectra (similarity score 0.983), even among peaks that are not clearly identifiable indicates the value of spectral library matching for making structural assignments.
Sialylated Lewis X, B-type tetrasaccharide (m/z 1021) and B/Y-type trisaccharide (m/z 646)
Figure S2 shows a comparison of sialyl Lewis X substructure fragments isolated from a standard, CFG Te140, and a human plasma N-linked glycan. Typically, at the MS/MS stage, both m/z 646 and 1021 fragments are detectable; sometimes the m/z 1021 fragment ion is very weak or not detectable. CID of the m/z 1021 ion produces the spectra shown in Figure S2A, S2B, and S2C, isolated from CFG Te140, a human plasma N-glycan, and a normal human colon tissue O-glycan, respectively. These spectra are relatively simple due to the lability of the sialic acid linkage favoring formation of the m/z 646 fragment. Further CID of the m/z 646 ion provides a much more informative spectrum (Figures S2D, S2E, and S2F). All three samples exhibit very similar spectra, both in terms of fragment masses and the overall intensity pattern, despite the differences in sample and precursor ion structures. Supplemental figure S3shows putative fragment assignments for this structure. As with Lewis X, the 3,5A-type cleavage across the N-acetyl-glucosamine residue positions the galactose residue at the 4-position rather than the 3-position. This provides additional de novo evidence of the SLex structure rather than SLea.
Further comparison could be made between these same mass fragments generated from a sialylated Lewis A standard. Reliable mass spectrometric distinction of SLex and SLea would have significant potential utility [33]. Unfortunately, we are not aware of any suitable standard that is available. The SLea tetrasaccharide is commercially available, but is unsuitable for this analysis, as it would not produce the correct B-type fragment. One could use the tetrasaccharide to prepare a glycoside (other than methyl) prior to permethylation. In our prior experience, while such structures produce the same mass fragments, they produce different intensity patterns than is obtained from cleavage of glycosidic bonds. The reasons behind this are not well understood, however, a suitable standard typically must have at least two monosaccharide residues between the epitope of interest and any aglycone functional group, to be used in this way.
Lewis Y, Lewis B, B-type tetrasaccharide (m/z 834) and B/Y trisaccharide (m/z 646)
Lewis Y is a tumor-associated carbohydrate antigen [34–36]. Studies have suggested it plays a role in cell cycle perturbation [36], and it has been the target of vaccine development [37,38]. Lewis B is also a difucosylated Hex-HexNAc, but built upon a type 1 lactosamine. Figure S4A and S4D show the MS3 and MS4 spectra of the tetrasaccharide B ion m/z 834 and trisaccharide B/Y ion m/z 646, respectively, for a Lewis B standard (LNDFH I). Figures S4B and S4E show the same spectra isolated from a Lewis Y standard (CFG Te118). At the MS3 level, the spectra show subtle intensity pattern differences, but they do not clearly distinguish these two structures. At the trisaccharide level, however, the differences are much more pronounced and easily serve to distinguish the two structures. Figures S4C and S4D show the same spectra obtained from an N-glycan antenna (precursor m/z 16913+, composition Hex10HexNAc9dHex4) isolated from MCF-7 cultured cells. The MS3 spectra are virtually identical to those of the Lewis Y standard (similarity score 0.952) despite the nearly three orders of magnitude greater intensity for the standard spectra; the MS4 spectra shown are also identical (similarity score 0.996), despite four orders of magnitude difference in intensity.
Gal-α1,3-Gal, B-type trisaccharide (m/z 690) and B-type disaccharide (m/z 445)
The Gal-α1,3-Gal epitope has particular importance to the pharmaceutical industry, as it is highly antigenic in humans [39]. Recombinant biopharmaceutical materials may be expressed in systems that synthesize this structure, including Chinese hamster ovary cells [40]. Methods have been devised for detecting this motif [41], and rely on antibody binding, chromatographic (or electrophoretic) retention, or mass spectrometric detection of a hexose extension to a lactosamine unit. The latter technique gives strong inferential information, but does not provide direct empirical evidence for the 3-linkage. For example, galactosyl extension to a lactosamine can also be in a 4-linkage [42,43]. Mass spectrometry of the permethylated structure provides cross-ring cleavages which clearly distinguish the 3-linkage from a 4-linkage, or determine whether a mixture is present [26]. The Gal-α1,3-Gal epitope has been determined in glycosphingolipids using permethylation and MSn, including assessing and quantifying isomeric mixtures [44]. N-linked glycans containing a galactosylated lactosamine will generate a distinct fragment at m/z 690 in MS/MS, consistent with B-type Gal-Gal-GlcNAc [42,43], that can be easily detected, isolated, and further fragmented. In many cases, such spectra might be a mixture, with a small amount of Gal-GlcNAc-Man fragment also present, though these are easily distinguishable by isolating and fragmenting the m/z 445 ion to provide empirical evidence of a 3-linkage within the Gal-Gal moiety.
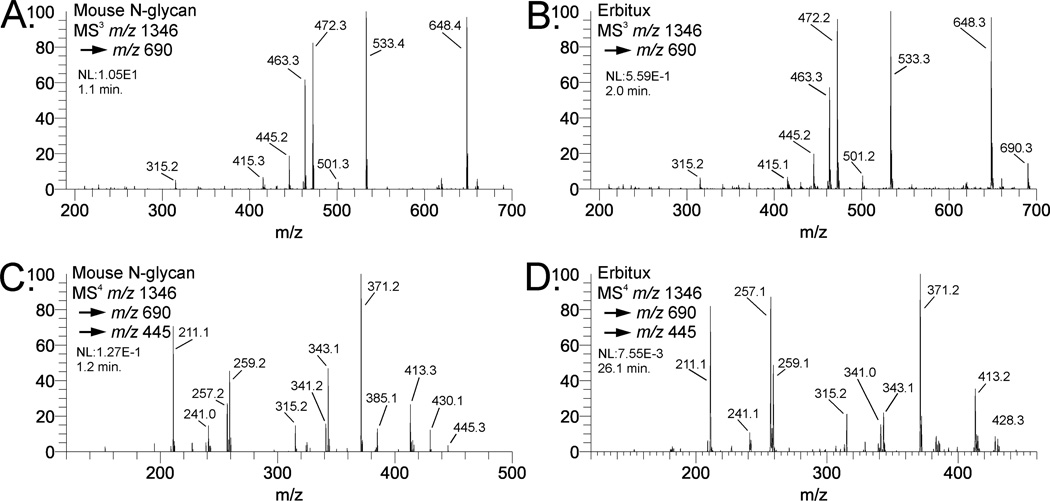
Figure 3 shows a selection of MSn spectra from two different samples containing the Gal-α1,3- Gal moiety. Figure 3A shows an MS3 spectrum of the m/z 690 fragment isolated from the (Hex)4 (HexNAc)2 (Deoxyhexose)1 + (Man)3(GlcNAc)2 N-linked glycan isolated from mouse tissue. This composition is biantennary with core fucosylation and each antenna has a Hex-Hex-HexNAc composition. Figure 3B shows the same fragment (m/z 690) isolated from the same N-glycan composition from the biopharmaceutical product Cetuximab (Erbitux), which contains the Gal-α1,3-Gal moiety [45]. The spectra are identical with respect to fragment masses and intensity patterns, and do not have significant amounts of Gal-GlcNAc-Man. To further determine the Gal-Gal linkage, the MS4 spectrum of the m/z 445 ion from the mouse N-glycan (figure 3D) is compared to the spectrum for the Cetuximab N-glycan; both clearly indicate a 3-linkage [26]. The structures and fragment assignments are 13 shown in Figure 3E and 3F. The diagnostic cross-ring cleavages are clearly more observable at the MS4 stage (disaccharide) than at MS3 (trisaccharide).
Fig. 3.
Comparison of mass spectra of Gal-(α1,3)-Gal-(β1,4)-GlcNac B-type trisaccharides and Gal-(α1,3)-Gal B-type disaccharides. Spectra in (A) and (C) are N-glycans isolated from a mouse. Spectra in (C) and (D) are N-glycans isolated from Erbitux. Structures and fragmentation are shown for the Gal-Gal-GlcNAc B-type trisaccharide (E) and the Gal-1,3-Gal B-type disaccharide (F).
Sialylated Lactosamines, B/Y Gal-GlcNAc, m/z 472
Determination of sialic acid linkage to lactosamine units presents an analytical challenge for mass spectrometry-based approaches. Glycosidic bonds to sialic acids are typically labile, although their bond dissociation energies have been noted to differ [46]. While permethylation renders the linkage stable enough to ionize, when subjected to collision-induced dissociation (CID), these bonds still break readily and it is difficult to obtain cross-ring cleavage fragments containing intact sialic acid glycosidic bonds. Typically, cross-ring cleavage fragments are the most informative for determining linkage positions [5–7,26]. In the absence of these cleavages, other strategies must be adopted. Isolation of the lithiated B/Y-type galactose residue from permethylated sialylated lactosamine antennae has been performed by MSn [47]. The 3 or 6 position of the hydroxyl group on galactose induces different fragmentation patterns when subjected to CID. While this strategy provides very good discrimination of 3- vs. 6-linked sialic acid, some samples may be below the sensitivity limit needed to obtain a good quality MSn spectrum by this approach.
On the other hand, we have observed that the sodiated B/Y-type lactosamine ion, m/z 472, can be more readily isolated from N- or O-linked glycans, providing distinct and reproducible intensity patterns even though most of the major fragment masses are identical. Supplemental figure S6 shows putative fragmentation assignments, which are similar for both 3- and 6-linked fragments. Figure S5 shows a representative data set of CID spectra of m/z 472 ions from various sialylated lactosamine motifs by MSn from synthetic standards, as well as N-linked and O-linked glycans. Figure S5A shows the MS4 spectrum of m/z 472 from the CFG standard Te175, containing a NeuAcα2-3Gal linkage. Figure S5B shows the CID spectrum of m/z 472 from CFG standard 176, containing a 6-linked sialylated lactosamine. Although these materials contain an azido-ethyl functional group for click chemistry reactions, this does not preclude permethylation or MSn analysis. Comparison of the indicated spectra reflects a high similarity of fragment masses, with a distinct difference in their ion abundance patterns. In particular, the m/z 245 ion in Figure S5A is less intense than m/z 227 or m/z 268. Furthermore, m/z 430, while representing a usually uninformative loss of an acetyl group, is much less intense than any of the aforementioned fragments; it is also significantly less intense than m/z 440. In Figure S5B, by contrast, the intensity pattern of these fragments is distinctly different. The m/z 227 ion is much less intense than the m/z 245 or 268 ions, and m/z 430 ion is relatively much more intense in S5B vs. S5A, vs. the m/z 440/442 ions.
Given the indicated intensity pattern differences, with similar masses, reproducibility and consistency are a concern. Therefore, comparisons were initiated with other materials to validate the spectrum matching strategy. Figure S5C shows the standard synthetic oligosaccharide KML102 which contains a 3-linked sialylated lactosamine. Comparing this spectrum to Figure S5A shows the same distinct intensity pattern: 245<<227<268>315; 430<440. Figure S5D shows the same fragment mass isolated from a sialidase S-treated fetuin N-glycan. Sialidase S specifically removes 3-linked sialic acid [48], so the remaining oligosaccharides should have only 6-linked sialic acid on the N-linked sialylated lactosamine antennae. Comparison of this spectrum (S5D) with Figure S5B shows a similar intensity pattern: 227<<245<268<315; 430>440/442.
Taking these four spectra as standards (S5A–S5D), we then obtained a set of m/z 472 ions from various biological samples. Figure S5E shows an m/z 472 spectrum of an O-linked glycan released from KG1A cultured cells. The intensity pattern matches very closely with S5A (0.962) and S5C (0.976). Figures S5F and S5H display the m/z 472 fragment spectra from two different human plasma N-glycans. Figure S5F is from IgG-depleted human plasma; S5H is from the IgG fraction from the same plasma sample. Both were fractionated by protein A/G, enzymatically released, derivatized with 2-AA, collected as HPLC fractions, permethylated, and infused. The spectra are virtually identical and match the standard spectra in Figure S5B and S5D, indicating a 6-linked NeuAc.
Library matching strategies also lead to issues of transferability between instruments, and reproducibility over time. This can only be assessed empirically through repeated analysis on the same, as well as different, instruments. Figure S5G shows an m/z 472 spectrum obtained from a human IgG N-linked glycan, reduced and permethylated, obtained December, 2006 on an LTQ Orbitrap, using the ITMS (linear ion trap) for mass analysis. Figure S5J shows the same sample and fragmentation pathway obtained on the same instrument, on the same day, using the FTMS (Orbitrap mass analyzer). Figure S5I shows another human IgG N-glycan sample preparation, also reduced and permethylated, obtained on a Velos Pro instrument, from December, 2009. All three spectra share the same fragmentation pathway. Figures S5G and S5I, the ITMS Orbitrap and Velos Pro spectra, respectively, are nearly identical to the other 6-linked NeuAc spectra, with similarity scores greater than 0.900, despite being different sample preparations obtained three years apart. Only the FTMS spectrum is somewhat different, having similarity scores between 0.200 and 0.300, although the overall intensity pattern (227<<245<268<315; 430>440/442) is similar. Given these data, transferability between instruments of the same type of mass analyzer appears feasible, whereas different mass analyzer designs might require separate library building.
Conclusions
The use of antibodies and lectins is now well established for performing global analysis of glycan epitopes, however, mass spectrometry offers several potential advantages. The data from mass spectral analysis provides additional information about the precursor ion and its composition which can be useful or necessary, such as in biopharmaceutical characterization applications. In addition, mass spectrometry can provide evidence of isomeric mixtures, especially in biological samples that may contain multiple positional isomers [42,49,50]. While de novo structural assignments rely on fragment masses as the primary information, ion intensity patterns also provide unique information that is useful when subtle structural differences produce fragments of the same mass. Structural problems of this type require comparison of spectra to standard materials with known structure and high purity to provide relevant benchmarks. A requirement for choosing suitable standard materials is that they must be larger than the substructure of interest to provide for fragment generation inside the ion trap mass spectrometer. Ideally, standards are larger oligosaccharides containing the epitope of interest. Synthetic glycoconjugates with an aglycone, such as the azido linkers found on the CFG standards, are also useful.
Inevitable to glycomic methodological discussions are questions of sensitivity and required sample amounts. While typical sequential mass spectrometric analyses may require somewhat greater sample amounts than fluorescence chromatography or online LC-MS, the level of information acquired vs. the level of structural detail needed to answer a particular question also should be considered. It has been frequently acknowledged that glycans offer a tremendous challenge due to their structural complexity and the potential for multiple isomeric forms, especially among larger oligosaccharides [51]. While a multitude of methods are available which require very little sample, these also provide comparatively less structural information. While sample quantities may limit the depth of analysis, the informational limitations of the chosen methodologies should not be ignored and structures that are assumed from mass composition only should also be acknowledged as such. The possibility of isomerism in large complex oligosaccharides remains a frequently ignored or oversimplified analytical problem, owing to the inability of many common methods to approach this issue. In short, comparison of methods primarily in terms of sensitivity disregards the relative amounts of information obtainable using the chosen strategies.
Supplementary Material
Structures and important fragmentation of Lewis X, Lewis A, H2, and H1 antigen.
Comparison of B-type Sialyl-Lewis X tetrasaccharide ion and B/Y-type trisaccharide ion spectra. Spectra (A), (B), and (C) show the tetrasaccharide spectra of CFG Te140, human plasma N-glycan, and normal human colon tissue O-glycan, respectively. Spectra (D), (E), and (F) show the trisaccharide spectra of CFG Te140, human plasma N-glycan, and normal human colon tissue, respectively. At the tetrasaccharide level, the spectra are dominated by NeuAc loss; at the trisaccharide level (m/z 646), the fragmentation is much more informative.
Structure and fragmentation of Sialyl Lewis X fragment ion m/z 646.
Comparison of spectra of B-type Lewis B and Lewis Y tetrasaccharide and B/Y-type trisaccharide ions. Standard spectra were obtained from Lacto-N-difucohexaose I (Lewis B) and CFG Te118 (Lewis Y). Spectra (C) and (F) represent the analogous spectra isolated from an N-glycan (Hex10HexNAc9dHex4, triply-charged m/z 16913+) from MCF-7 cultured cells.
Gal(β,1-4)GlcNAc B/Y type disaccharides isolated by MSn from sialylated lactosamines. Standard materials represented are (A) CFG Te175 (3-linked sialyl-lactosamine), (B) CFG Te176 (6-linked sialyl-lactosamine), (C) Chemically synthesized KML102 (3-linked sialyl-lactosamine), and (D) Sialidase S-treated bovine fetuin disialylated N-glycan (6-linked sialyl-lactosamine). B/Y lactosamines from biological materials include (E) O-glycan from KG1a cultured cells, (F) 2-AA-labeled IgG-depleted human plasma N-glycan, (H) 2-AA-labeled human plasma IgG N-glycan. Spectra in (G), (I), and (J) display the same B/Y lactosamine isolated from Sigma human IgG glycan (G2FS) obtained via (G) Orbitrap ITMS (December, 2006), (I) Thermo Velos Pro (December, 2009), and (J) Orbitrap FTMS (December, 2006), respectively. Other spectra were obtained between Fall 2012 through Spring 2013 on LTQ.
Structure and fragmentation of sialylated lactosamine fragment ion m/z 472. The 3- and 6-NeuAc structures differ in the position of the hydroxyl group; the ion fragments for each have the same mass.
Acknowledgements
The authors thank James M. Paulson (Scripps Research Institute) and the Consortium for Functional Glycomics for the synthetic standards, Robert Sackstein and Cristina I. Silvescu (Brigham and Women’s Hospital) for the human colon tissue and COLO205 cultured cells, Joseph T.Y. Lau (Roswell Park Cancer Institute) for the mouse sample, Khushi L. Matta for synthetic standard KML102, and Dipak K. Banerjee (University of Puerto Rico) for the MCF-7 cultured cells. This work was supported, in part, by a Program of Excellence in Glycosciences grant (P01 HL107146, PI Robert Sackstein).
Abbreviations
- ECD
electron capture dissociation
- IRMPD
infrared multiphoton dissociation
- EDD
electron detachment dissociation
References
- 1.Cook KD. ASMS members John Fenn and Koichi Tanaka share Nobel: the world learns our "secret". American Society for Mass Spectrometry. J Am Soc Mass Spectrom. 2002;13:1359. doi: 10.1016/s1044-0305(02)00803-6. [DOI] [PubMed] [Google Scholar]
- 2.Paul W. Electromagnetic Traps for Charged and Neutral Particles. Uspekhi Fizicheskikh Nauk. 1990;160:109–127. [Google Scholar]
- 3.Packer NH, Karlsson NG. There are no facts, only interpretations. J Proteome Res. 2006;5:1291–1292. doi: 10.1021/pr0627286. [DOI] [PubMed] [Google Scholar]
- 4.Campbell MP, Nguyen-Khuong T, Hayes CA, Flowers SA, Alagesan K, Kolarich D, Packer NH, Karlsson NG. Validation of the curation pipeline of UniCarb-DB: Building a global glycan reference MS/MS repository. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbapap.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 5.Weiskopf AS, Vouros P, Harvey DJ. Characterization of oligosaccharide composition and structure by quadrupole ion trap mass spectrometry. Rapid Commun Mass Spectrom. 1997;11:1493–1504. doi: 10.1002/(SICI)1097-0231(199709)11:14<1493::AID-RCM40>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 6.Sheeley DM, Reinhold VN. Structural characterization of carbohydrate sequence, linkage, and branching in a quadrupole Ion trap mass spectrometer: neutral oligosaccharides and N-linked glycans. Anal Chem. 1998;70:3053–3059. doi: 10.1021/ac9713058. [DOI] [PubMed] [Google Scholar]
- 7.Reinhold VN, Sheeley DM. Detailed characterization of carbohydrate linkage and sequence in an ion trap mass spectrometer: glycosphingolipids. Anal Biochem. 1998;259:28–33. doi: 10.1006/abio.1998.2619. [DOI] [PubMed] [Google Scholar]
- 8.Cummings RD. The repertoire of glycan determinants in the human glycome. Mol Biosyst. 2009;5:1087–1104. doi: 10.1039/b907931a. [DOI] [PubMed] [Google Scholar]
- 9.Yagi H, Saito T, Yanagisawa M, Yu RK, Kato K. Lewis X-carrying N-glycans regulate the proliferation of mouse embryonic neural stem cells via the Notch signaling pathway. J Biol Chem. 2012;287:24356–24364. doi: 10.1074/jbc.M112.365643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canepa GE, Mesias AC, Yu H, Chen X, Buscaglia CA. Structural features affecting trafficking, processing, and secretion of Trypanosoma cruzi mucins. J Biol Chem. 2012;287:26365–26376. doi: 10.1074/jbc.M112.354696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kannicht C, Ramstrom M, Kohla G, Tiemeyer M, Casademunt E, Walter O, Sandberg H. Characterisation of the post-translational modifications of a novel, human cell line-derived recombinant human factor VIII. Thromb Res. 2012 doi: 10.1016/j.thromres.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 12.Takegawa Y, Ito S, Yoshioka S, Deguchi K, Nakagawa H, Monde K, Nishimura S. Structural assignment of isomeric 2-aminopyridine-derivatized oligosaccharides using MSn spectral matching. Rapid Commun Mass Spectrom. 2004;18:385–391. doi: 10.1002/rcm.1341. [DOI] [PubMed] [Google Scholar]
- 13.Viseux N, de Hoffmann E, Domon B. Structural analysis of permethylated oligosaccharides by electrospray tandem mass spectrometry. Anal Chem. 1997;69:3193–3198. doi: 10.1021/ac961285u. [DOI] [PubMed] [Google Scholar]
- 14.Viseux N, de Hoffmann E, Domon B. Structural assignment of permethylated oligosaccharide subunits using sequential tandem mass spectrometry. Anal Chem. 1998;70:4951–4959. doi: 10.1021/ac980443+. [DOI] [PubMed] [Google Scholar]
- 15.Everest-Dass AV, Kolarich D, Campbell MP, Packer NH. Tandem mass spectra of glycan substructures enable the multistage mass spectrometric identification of determinants on oligosaccharides. Rapid Commun Mass Spectrom. 2013;27:931–939. doi: 10.1002/rcm.6527. [DOI] [PubMed] [Google Scholar]
- 16.Everest-Dass AV, Abrahams JL, Kolarich D, Packer NH, Campbell MP. Structural Feature Ions for Distinguishing N- and O-Linked Glycan Isomers by LC-ESI-IT MS/MS. J Am Soc Mass Spectrom. 2013;24:895–906. doi: 10.1007/s13361-013-0610-4. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Singh S, Reinhold VN. Congruent strategies for carbohydrate sequencing. 2. FragLib: an MS(n) spectral library. Anal Chem. 2005;77:6263–6270. doi: 10.1021/ac050725r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lapadula AJ, Hatcher PJ, Hanneman AJ, Ashline DJ, Zhang H, Reinhold VN. Congruent strategies for carbohydrate sequencing. 3. OSCAR: an algorithm for assigning oligosaccharide topology from MS(n) data. Anal Chem. 2005;77:6271–6279. doi: 10.1021/ac050726j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garner B, Priestman DA, Stocker R, Harvey DJ, Butters TD, Platt FM. Increased glycosphingolipid levels in serum and aortae of apolipoprotein E gene knockout mice. J Lipid Res. 2002;43:205–214. [PubMed] [Google Scholar]
- 20.Carlson DM. Oligosaccharides isolated from pig submaxillary mucin. J Biol Chem. 1966;241:2984–2946. [PubMed] [Google Scholar]
- 21.Kang P, Mechref Y, Klouckova I, Novotny MV. Solid-phase permethylation of glycans for mass spectrometric analysis. Rapid Commun Mass Spectrom. 2005;19:3421–3428. doi: 10.1002/rcm.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coombs PJ, Graham SA, Drickamer K, Taylor ME. Selective binding of the scavenger receptor C-type lectin to Lewisx trisaccharide and related glycan ligands. J Biol Chem. 2005;280:22993–22999. doi: 10.1074/jbc.M504197200. [DOI] [PubMed] [Google Scholar]
- 23.Larkin M, Ahern TJ, Stoll MS, Shaffer M, Sako D, O'Brien J, Yuen CT, Lawson AM, Childs RA, Barone KM, et al. Spectrum of sialylated and nonsialylated fucooligosaccharides bound by the endothelial-leukocyte adhesion molecule E-selectin. Dependence of the carbohydrate binding activity on E-selectin density. J Biol Chem. 1992;267:13661–13668. [PubMed] [Google Scholar]
- 24.Hashii N, Kawasaki N, Itoh S, Harazono A, Matsuishi Y, Hayakawa T, Kawanishi T. Specific detection of Lewis x-carbohydrates in biological samples using liquid chromatography/multiple-stage tandem mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:3315–3321. doi: 10.1002/rcm.2190. [DOI] [PubMed] [Google Scholar]
- 25.Hashii N, Kawasaki N, Itoh S, Nakajima Y, Harazono A, Kawanishi T, Yamaguchi T. Identification of glycoproteins carrying a target glycan-motif by liquid chromatography/multiple-stage mass spectrometry: identification of Lewis x-conjugated glycoproteins in mouse kidney. J Proteome Res. 2009;8:3415–3429. doi: 10.1021/pr9000527. [DOI] [PubMed] [Google Scholar]
- 26.Ashline D, Singh S, Hanneman A, Reinhold V. Congruent strategies for carbohydrate sequencing. 1. Mining structural details by MSn. Anal Chem. 2005;77:6250–6262. doi: 10.1021/ac050724z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janas T. Membrane oligo- and polysialic acids. Biochim Biophys Acta. 2011;1808:2923–2932. doi: 10.1016/j.bbamem.2011.08.036. [DOI] [PubMed] [Google Scholar]
- 28.Higuchi Y, Miura T, Kajimoto T, Ohta Y. Effects of disialoganglioside GD3 on the mitochondrial membrane potential. FEBS Lett. 2005;579:3009–3013. doi: 10.1016/j.febslet.2005.04.054. [DOI] [PubMed] [Google Scholar]
- 29.Yanagisawa M, Yoshimura S, Yu RK. Expression of GD2 and GD3 gangliosides in human embryonic neural stem cells. ASN Neuro. 2011;3 doi: 10.1042/AN20110006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schiopu C, Vukelic Z, Capitan F, Kalanj-Bognar S, Sisu E, Zamfir AD. Chip-nanoelectrospray quadrupole time-of-flight tandem mass spectrometry of meningioma gangliosides: A preliminary study. Electrophoresis. 2012;33:1778–1786. doi: 10.1002/elps.201200044. [DOI] [PubMed] [Google Scholar]
- 31.Levery SB. Glycosphingolipid structural analysis and glycosphingolipidomics. Methods Enzymol. 2005;405:300–369. doi: 10.1016/S0076-6879(05)05012-3. [DOI] [PubMed] [Google Scholar]
- 32.Zamfir AD, Serb A, Vukeli Z, Flangea C, Schiopu C, Fabris D, Kalanj-Bognar S, Capitan F, Sisu E. Assessment of the molecular expression and structure of gangliosides in brain metastasis of lung adenocarcinoma by an advanced approach based on fully automated chip-nanoelectrospray mass spectrometry. J Am Soc Mass Spectrom. 2011;22:2145–2159. doi: 10.1007/s13361-011-0250-5. [DOI] [PubMed] [Google Scholar]
- 33.Ragupathi G, Damani P, Srivastava G, Srivastava O, Sucheck SJ, Ichikawa Y, Livingston PO. Synthesis of sialyl Lewis(a) (sLe (a), CA19-9) and construction of an immunogenic sLe(a) vaccine. Cancer Immunol Immunother. 2009;58:1397–1405. doi: 10.1007/s00262-008-0654-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madjd Z, Parsons T, Watson NF, Spendlove I, Ellis I, Durrant LG. High expression of Lewis y/b antigens is associated with decreased survival in lymph node negative breast carcinomas. Breast Cancer Res. 2005;7:R780–R787. doi: 10.1186/bcr1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nudelman E, Levery SB, Kaizu T, Hakomori S. Novel fucolipids of human adenocarcinoma: characterization of the major Ley antigen of human adenocarcinoma as trifucosylnonaosyl Ley glycolipid (III3FucV3FucVI2FucnLc6) J Biol Chem. 1986;261:11247–11253. [PubMed] [Google Scholar]
- 36.Liu D, Liu J, Lin B, Liu S, Hou R, Hao Y, Liu Q, Zhang S, Iwamori M. Lewis y Regulate Cell Cycle Related Factors in Ovarian Carcinoma Cell RMG-I in Vitro via ERK and Akt Signaling Pathways. Int J Mol Sci. 2012;13:828–839. doi: 10.3390/ijms13010828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuster M, Umana P, Ferrara C, Brunker P, Gerdes C, Waxenecker G, Wiederkum S, Schwager C, Loibner H, Himmler G, Mudde GC. Improved effector functions of a therapeutic monoclonal Lewis Y-specific antibody by glycoform engineering. Cancer Res. 2005;65:7934–7941. doi: 10.1158/0008-5472.CAN-04-4212. [DOI] [PubMed] [Google Scholar]
- 38.Heimburg-Molinaro J, Lum M, Vijay G, Jain M, Almogren A, Rittenhouse-Olson K. Cancer vaccines and carbohydrate epitopes. Vaccine. 2011;29:8802–8826. doi: 10.1016/j.vaccine.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macher BA, Galili U. The Gal alpha 1,3Gal beta 1,4GlcNAc-R (alpha-Gal) epitope: A carbohydrate of unique evolution and clinical relevance. Biochimica Et Biophysica Acta-General Subjects. 2008;1780:75–88. doi: 10.1016/j.bbagen.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bosques CJ, Collins BE, Meador JW, 3rd, Sarvaiya H, Murphy JL, Dellorusso G, Bulik DA, Hsu IH, Washburn N, Sipsey SF, Myette JR, Raman R, Shriver Z, Sasisekharan R, Venkataraman G. Chinese hamster ovary cells can produce galactose-alpha-1,3-galactose antigens on proteins. Nat Biotechnol. 2010;28:1153–1156. doi: 10.1038/nbt1110-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szabo Z, Guttman A, Bones J, Shand RL, Meh D, Karger BL. Ultrasensitive Capillary Electrophoretic Analysis of Potentially Immunogenic Carbohydrate Residues in Biologics: Galactose-alpha-1,3-Galactose Containing Oligosaccharides. Molecular Pharmaceutics. 2012;9:1612–1619. doi: 10.1021/mp200612n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashline DJ, Lapadula AJ, Liu YH, Lin M, Grace M, Pramanik B, Reinhold VN. Carbohydrate structural isomers analyzed by sequential mass spectrometry. Anal Chem. 2007;79:3830–3842. doi: 10.1021/ac062383a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reinhold V, Zhang H, Hanneman A, Ashline D. Toward a platform for comprehensive glycan sequencing. Mol Cell Proteomics. 2013;12:866–873. doi: 10.1074/mcp.R112.026823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Zhou D, Xia C, Wang PG, Levery SB. Sensitive quantitation of isoglobotriaosylceramide in the presence of isobaric components using electrospray ionizationion trap mass spectrometry. Glycobiology. 2008;18:166–176. doi: 10.1093/glycob/cwm127. [DOI] [PubMed] [Google Scholar]
- 45.Qian J, Liu T, Yang L, Daus A, Crowley R, Zhou Q. Structural characterization of N-linked oligosaccharides on monoclonal antibody cetuximab by the combination of orthogonal matrix-assisted laser desorption/ionization hybrid quadrupole-quadrupole time-of-flight tandem mass spectrometry and sequential enzymatic digestion. Anal Biochem. 2007;364:8–18. doi: 10.1016/j.ab.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 46.Seymour JL, Costello CE, Zaia J. The influence of sialylation on glycan negative ion dissociation and energetics. J Am Soc Mass Spectrom. 2006;17:844–854. doi: 10.1016/j.jasms.2006.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anthony RM, Nimmerjahn F, Ashline DJ, Reinhold VN, Paulson JC, Ravetch JV. Recapitulation of IVIG anti-inflammatory activity with a recombinant IgG Fc. Science. 2008;320:373–376. doi: 10.1126/science.1154315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dugan AS, Eash S, Atwood WJ. An N-linked glycoprotein with alpha(2,3)-linked sialic acid is a receptor for BK virus. J Virol. 2005;79:14442–14445. doi: 10.1128/JVI.79.22.14442-14445.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hanneman AJ, Rosa JC, Ashline D, Reinhold VN. Isomer and glycomer complexities of core GlcNAcs in Caenorhabditis elegans. Glycobiology. 2006;16:874–890. doi: 10.1093/glycob/cwl011. [DOI] [PubMed] [Google Scholar]
- 50.Prien JM, Ashline DJ, Lapadula AJ, Zhang H, Reinhold VN. The high mannose glycans from bovine ribonuclease B isomer characterization by ion trap MS. J Am Soc Mass Spectrom. 2009;20:539–556. doi: 10.1016/j.jasms.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laine RA. A calculation of all possible oligosaccharide isomers both branched and linear yields 1.05 × 10(12) structures for a reducing hexasaccharide: the Isomer Barrier to development of single-method saccharide sequencing or synthesis systems. Glycobiology. 1994;4:759–767. doi: 10.1093/glycob/4.6.759. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structures and important fragmentation of Lewis X, Lewis A, H2, and H1 antigen.
Comparison of B-type Sialyl-Lewis X tetrasaccharide ion and B/Y-type trisaccharide ion spectra. Spectra (A), (B), and (C) show the tetrasaccharide spectra of CFG Te140, human plasma N-glycan, and normal human colon tissue O-glycan, respectively. Spectra (D), (E), and (F) show the trisaccharide spectra of CFG Te140, human plasma N-glycan, and normal human colon tissue, respectively. At the tetrasaccharide level, the spectra are dominated by NeuAc loss; at the trisaccharide level (m/z 646), the fragmentation is much more informative.
Structure and fragmentation of Sialyl Lewis X fragment ion m/z 646.
Comparison of spectra of B-type Lewis B and Lewis Y tetrasaccharide and B/Y-type trisaccharide ions. Standard spectra were obtained from Lacto-N-difucohexaose I (Lewis B) and CFG Te118 (Lewis Y). Spectra (C) and (F) represent the analogous spectra isolated from an N-glycan (Hex10HexNAc9dHex4, triply-charged m/z 16913+) from MCF-7 cultured cells.
Gal(β,1-4)GlcNAc B/Y type disaccharides isolated by MSn from sialylated lactosamines. Standard materials represented are (A) CFG Te175 (3-linked sialyl-lactosamine), (B) CFG Te176 (6-linked sialyl-lactosamine), (C) Chemically synthesized KML102 (3-linked sialyl-lactosamine), and (D) Sialidase S-treated bovine fetuin disialylated N-glycan (6-linked sialyl-lactosamine). B/Y lactosamines from biological materials include (E) O-glycan from KG1a cultured cells, (F) 2-AA-labeled IgG-depleted human plasma N-glycan, (H) 2-AA-labeled human plasma IgG N-glycan. Spectra in (G), (I), and (J) display the same B/Y lactosamine isolated from Sigma human IgG glycan (G2FS) obtained via (G) Orbitrap ITMS (December, 2006), (I) Thermo Velos Pro (December, 2009), and (J) Orbitrap FTMS (December, 2006), respectively. Other spectra were obtained between Fall 2012 through Spring 2013 on LTQ.
Structure and fragmentation of sialylated lactosamine fragment ion m/z 472. The 3- and 6-NeuAc structures differ in the position of the hydroxyl group; the ion fragments for each have the same mass.