Abstract
Objective
To characterize the epidemiology of MRSA transmission and infections in a level IIIC neonatal intensive care unit (NICU) and identify barriers to MRSA control.
Setting and Design
Retrospective cohort study in a university-affiliated NICU with an MRSA control program including weekly nares cultures of all neonates and admission nares cultures for neonates transferred from other hospitals or admitted from home.
Methods
Medical records were reviewed to identify neonates with NICU-acquired MRSA colonization or infection between April 2007 and December 2011. Compliance with hand hygiene and a MRSA decolonization protocol were monitored. Relatedness of MRSA strains were assessed using pulsed-field gel electrophoresis (PFGE).
Results
Of 3,536 neonates, 74 (2.0%) had a culture grow MRSA, including 62 neonates with NICU-acquired MRSA. Nineteen of 74 neonates (26%) had an MRSA infection, including 8 who became infected before they were identified as MRSA colonized, and 11 of 66 colonized neonates (17%) developed a subsequent infection. Of the 37 neonates that underwent decolonization, 6 (16%) developed a subsequent infection, and 7 of 14 (50%) that remained in the NICU for ≥21 days became recolonized with MRSA. Using PFGE, there were 14 different strain types identified with USA300 being the most common (31%).
Conclusions
Current strategies to prevent infections including active identification and decolonization of MRSA colonized neonates are inadequate as infants develop infections before being identified as colonized or after attempted decolonization. Future prevention efforts would benefit from improving detection of MRSA colonization, optimizing decolonization regimens, and identifying and interrupting reservoirs of transmission.
Background
The Centers for Disease Control and Prevention (CDC) estimate that approximately 1.7 million healthcare associated infections (HAIs) occur in U.S. hospitals every year, including more than 33, 000 in the neonatal intensive care unit (NICU).1 Despite appropriate therapy, neonatal infections can have long-term sequelae, including poor neurodevelopmental and growth outcomes.2,3 These infections contribute to the ballooning costs to care for preterm infants.
After coagulase-negative staphylococci, Staphylococcus aureus is the second most common pathogen causing HAIs in neonates.4 Antibiotic resistant S. aureus strains, specifically methicillin-resistant S. aureus (MRSA), have emerged and become prevalent in NICUs.4 In the U.S, the incidence of late-onset MRSA infections in NICUs increased more than 300% from 1995 to 2004.5 Because MRSA colonization predisposes neonates to MRSA infection, preventing MRSA transmission is an important component of programs to reduce morbidity and mortality from MRSA in NICUs. Current strategies to prevent MRSA transmission in NICUs include identifying colonized neonates and placing them on contact precautions, cohorting, healthcare worker hand hygiene, environmental cleaning, and in some cases decolonization of colonized neonates and/or healthcare workers.6 These measures have been successful in controlling MRSA outbreaks. Additional data, however, are needed to guide NICU MRSA control programs during non-outbreak settings. Our objectives were to characterize the epidemiology of MRSA transmission and infections in a NICU with an aggressive MRSA infection prevention program, to identify barriers to MRSA control and suggest future areas of investigation to prevent MRSA disease in this vulnerable population.
Materials and Methods
Setting and design
The Johns Hopkins Hospital (JHH) is a tertiary-care academic medical center with an embedded 175-bed Children's Center that houses a 45-bed, level IIIC NICU. The NICU had 12 private rooms and 3 open bays until the unit moved in April 2011 to a new location with all private rooms. We retrospectively identified a cohort of neonates admitted to the NICU between April 15 2007 and December 31 2011. We conducted an observational study, to assess the burden of MRSA transmission and infections in the setting of an aggressive MRSA control program. The institutional review board approved this study with a waiver of informed consent to review retrospective data.
Infection control and prevention program
Following a cluster of MRSA infections in 2007, enhanced MRSA control measures were implemented in April 2007, including nares swabs performed weekly by nurses to identify MRSA colonized neonates and at the time of NICU admission for neonates transferred from other hospitals or admitted from home; hand hygiene education; hand hygiene monitoring and feedback of compliance (started third quarter of 2008); contact precautions; private rooms if available or cohorting; decolonization of MRSA colonized neonates; and periodic screening and decolonization of healthcare workers (HCWs).7 Decolonization of neonates consisted of mupirocin applied to the nares twice a day for 5 days and two baths with 2% chlorhexidine gluconate-impregnated clothes, administered 48 hours apart for infants >36 weeks gestational age or > 4 weeks chronological age. HCW decolonization consisted of mupirocin applied to the nares twice daily for 5 days.
Data collection and outcome ascertainment
We searched a computerized surveillance system (Theradoc, Hospira, Lakeforest, IL) to identify patients with surveillance cultures and cultures sent during clinical care that grew MRSA during the study period. The two primary study outcomes were NICU-acquired MRSA colonization and NICU-onset MRSA infections. All infants that were born at JHH and had a culture grow MRSA were classified as NICU-acquired MRSA. Neonates that were born at another hospital were classified as NICU-acquired MRSA if they had surveillance or clinical culture obtained ≥3 days after admission to the NICU grow MRSA and had no known prior cultures grow MRSA. MRSA infections were ascertained by a trained infection preventionist (S.W.) who reviewed medical records of patients in whom MRSA grew in a culture sent at the discretion of the patient's treating clinician. National Healthcare Safety Network's (NHSN's) surveillance definitions for HAIs were applied to distinguish infection versus colonization.8
Laboratory Methods
All surveillance swabs collected between April 2007 and December 2011 were inoculated on selective and differential media to detect MRSA as previously described.9,10 Suspicious colonies were confirmed as S. aureus by Gram stain and slide coagulase testing. We performed pulsed field gel electrophoresis (PFGE) on available stored isolates as previously described and considered isolates to be related if their patterns had ≤ 3 band differences.11,12
Statistical Analysis
MRSA prevalence was calculated as the number of neonates with a culture that grew MRSA as a proportion of all admitted NICU patients. Trends in compliance with hand hygiene and the decolonization protocol were assessed using linear regression. Data were maintained in Microsoft Access 2007 (Microsoft Corp., Redmond, WA, USA) and analyzed using Stata version 11.0 (StataCorp., College Station, TX, USA) and MS Excel 2007(Microsoft Corp., Redmond, WA, USA).
Results
During the study period, 3,536 patients were admitted to the NICU accounting for 66,695 patient days. Patients were 55% male, 49% African-American, and 702 patients (20%) were either transferred from another hospital or admitted from home. Median length of stay in the NICU was 8 days (range 1-185). Seventy-four neonates (2.0%) had a culture grow MRSA. Of the neonates with MRSA, 65 (88%) were initially detected by surveillance cultures and 9 were initially detected by a culture sent during clinical care. Eight of 9 neonates detected by clinical culture met the NHSN criteria for infection, and the one that did not meet NHSN criteria was considered colonized (see Figure 1). Sixty-two neonates (84%) acquired MRSA in the NICU while 12 (16%) were identified on admission. The mean quarterly incidence of unit-acquired MRSA was 1.0 per 1000 patient days (95% CI 0.28, 2.45) (see Figure 2). Mean quarterly incidence of NICU-onset MRSA infection was 0.3 per 1000 patient-days (95% CI 0.0, 0.8). MRSA transmission and infections continued during the study period despite increases in hand hygiene compliance (p<0.001) and a statistically non-significant increase in compliance with a decolonization protocol (p=0.11).
Figure 1.
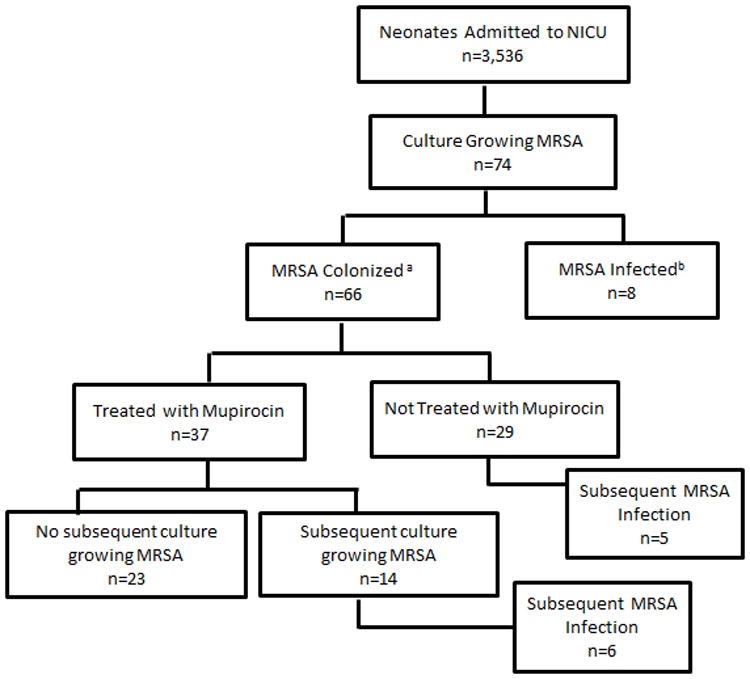
A flowchart detailing the identification of neonates with methicillin-resistant Staphylococcus aureus (MRSA) colonization and infection in a neonatal intensive care unit with an active surveillance and decolonization program. aNeonate did not meet NHSN criteria for infection at time of first culture growing MRSA; b Neonate met NHSN criteria for infection at time of first culture growing MRSA.
Figure 2.
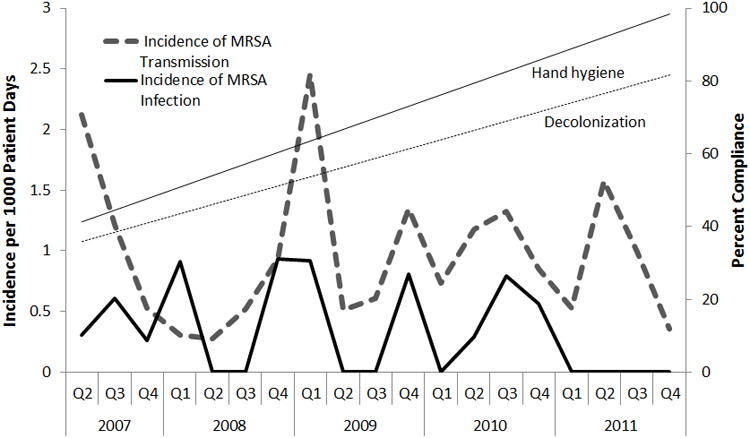
Quarterly incidence of MRSA transmission and infection from 2007 to 2011 in a setting of active surveillance and decolonization. Straight lines represent the trend in compliance with hand hygiene (solid line; Prob>F=0.0001; R2=0.75) and an MRSA decolonization protocol (broken line; Prob>F=0.11; R2=0.14) as estimated by linear regression.
The relatedness of MRSA strains was investigated using PFGE. Sixty-six (89%) of the 74 MRSA colonized or infected neonates had isolates available for strain typing. Fourteen unrelated strains were identified from neonates. The most frequently isolated strain was PFGE-type USA300 (31% of isolates) followed by PFGE-type USA100 (25%) and PFGE-type USA800 (19%). Within the most commonly identified PFGE-types, there were multiple subtypes with ≤ 3 band differences including 4 USA300s, 6 USA100s, and 6 USA800s. Figure 3 demonstrates the distribution of MRSA strains acquired over time. Of the 62 unit-acquired MRSA cases, 58 (94%) had isolates available for typing and 12 unrelated strains were found. The most frequently isolated strain was PFGE-type USA300 (34% of isolates) followed by PFGE-type USA100 (27%) and PFGE-type USA800 (20%).
Figure 3.
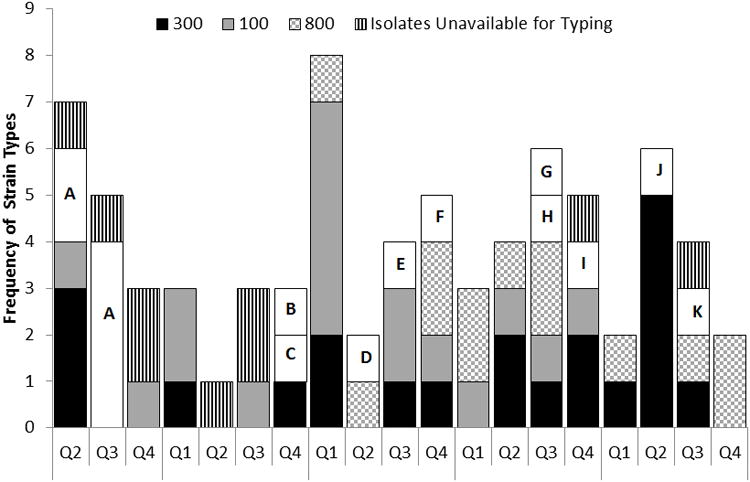
Distribution of MRSA strains isolated from patients. Each letter represents a unique strain.
Given continued MRSA transmission, all 204 NICU HCWs were screened in 2011 by using nares specimens. Seven HCWs (3.4%) were MRSA colonized. HCW MRSA strains included PFGE-types USA800 (N=3), USA300 (N=2), USA100 (N=1) and one unique strain. All 7 HCWs were decolonized. Because there was evidence of continued MRSA transmission, the five previously colonized HCWs who remained in the NICU 4-6 months later had repeat nares cultures. Four (80%) had recurrent colonization (including two with USA800) and they were again decolonized.
Of the 74 neonates that had a culture grow MRSA, 19 neonates (26%) had an MRSA infection: one was present on admission and 18 were acquired in the unit. Five neonates had multiple MRSA infections. Infections included four bacteremias, 14 lower respiratory infections, one upper respiratory infection, five skin and soft tissue infections, and one conjunctivitis. Three of the 19 infected neonates (16%) died but only one death was due to an MRSA infection. Eight neonates became infected before they were identified as MRSA colonized (see Figure 1), including 6 that had between two and six negative nares cultures prior to MRSA infection. Of 66 colonized neonates, 11 (17%) developed a subsequent infection in the NICU. The median time from identification of MRSA colonization to infection was five days (range 0-29).
Of 66 MRSA colonized neonates, 37 (66%) underwent decolonization. Twenty-three of 37 (62 %) decolonized neonates had no further MRSA positive cultures prior to unit discharge. Seven of 14 (50%) neonates that underwent decolonization and remained in the NICU for 21 days or more became recolonized with MRSA. Six of 37 neonates (16%) developed a subsequent infection despite attempted decolonization, including five neonates that had become recolonized and one neonate that became infected despite a negative nares culture. Of 29 patients in whom decolonization was not attempted, 13 (45%) were discharged from the unit within two days of the positive surveillance culture, and five (17%) developed a subsequent infection during their NICU stay.
Discussion
Control of MRSA remains a continuous challenge in NICUs. Despite aggressive infection control measures, including routine surveillance cultures, private rooms or cohorting and contact precautions, decolonization of infants and healthcare workers, and increasing hand hygiene compliance, we found on-going MRSA transmission and infection in our NICU over a four-year study period. Using data from an active surveillance program, this is the first study to describe the molecular epidemiology of endemic MRSA transmission in a USA NICU and identify clusters of related strains occurring amidst a background of unrelated MRSA strains.9,13 Our data highlight limitations to MRSA decolonization as an infection prevention strategy in a city with high endemicity of community-acquired MRSA and in this high-risk population. MRSA infections frequently occur before neonates are identified as colonized and subsequent infections occur in colonized neonates despite attempted decolonization.
Our findings confirm previous studies showing that multiple strains of MRSA can be identified circulating in NICUs during a non-outbreak setting. Carey et al characterized the molecular epidemiology of MRSA strains isolated from MRSA infected and colonized neonates in a NICU over a period of 8 years.14 Although the authors did not have data from routine weekly surveillance cultures to assess periods without MRSA infections, they similarly found that colonization and infection with multiple strain types occurred throughout the study period. Gregory et al reported on a long-standing MRSA control program including routine MRSA surveillance cultures, but they did not have isolates available for strain typing and their conclusions about transmission were based on antibiotic susceptibility patterns.15 In a NICU in Taiwan with highly endemic MRSA (41% of neonates colonized), Huang et al demonstrated molecular diversity of acquired MRSA strains after implementation of infection control measures.16 During our comprehensive assessment of the molecular epidemiology of endemic MRSA transmission in a NICU, we found 14 distinct strains including on-going transmission of USA800, USA300, and USA100 and 10 strains that were each identified only once. All three studies illustrate that endemic transmission persists despite current strategies including healthcare worker hand hygiene, environmental disinfection, contact precautions and cohorting, and, in some cases, identifying asymptomatic MRSA carriers and attempting decolonization.6
Uncovering reservoirs for on-going MRSA transmission in NICUs has proved challenging. Our findings agree with previous reports of healthcare workers as a source of MRSA transmission.17,18 Recurrent colonization of HCWs with USA800 may have contributed to acquisition of this strain by 13 neonates in the NICU. However, the distribution of strain types identified in colonized HCWs does not fully mirror those strains found in colonized or infected neonates in the NICU, suggesting additional bacterial reservoirs. Previous studies have identified mothers and fathers as potential reservoirs for MRSA transmission to neonates in the NICU.19,20 Healthy newborns frequently acquire S. aureus colonization in the first few months of life from their mothers.21,22 New technologies including genome-based analyses may help to further clarify MRSA transmission dynamics.23 In a changing NICU environment where parents are encouraged to have direct physical contact with their child, the role of parents in MRSA transmission in NICUs requires further study.
There is high correlation between strains that colonize neonates and cause subsequent infections; therefore, some NICUs attempt either targeted or universal decolonization as a MRSA infection prevention strategy. 7,16,24-27However, our data highlight the limitations of targeted decolonization as an MRSA prevention strategy. First, despite weekly surveillance cultures, 42% of infections occurred before the neonates were identified as MRSA colonized, leaving no opportunity to attempt decolonization. This finding confirms observations in previous studies that a large proportion of NICU MRSA infections occur before neonates are identified as colonized by active surveillance.15,28 Second, the interval between colonization and infection in many neonates was short (median 5 days), suggesting a narrow window of opportunity for intervention in colonized neonates to reduce risk of subsequent infection.28,29 Third, 38% of neonates who received decolonization treatment became recolonized during their NICU stay and 16% developed an MRSA infection, so the efficacy of decolonization to eradicate MRSA colonization and prevent MRSA infections may be limited. Some authors have described universal treatment of all neonates with mupirocin to successfully reduce MRSA infections in neonates.26,27 A universal approach in some settings had led to emergence of mupirocin resistance and the long-term impact of indiscriminately treating all neonates, including those not colonized with S. aureus, is unknown.30 A prospective randomized trial is needed to formally evaluate the efficacy and safety of mupirocin with or without chlorhexidine for eradicating MRSA colonization and preventing MRSA infections in neonates. This may need to include those who surround the infant, beyond healthcare providers and include family members or care givers.
A number of limitations should be considered when interpreting these data. Only nares cultures were collected routinely and not all neonates were cultured on admission, so we may have misclassified MRSA colonized neonates. Sampling multiple sites may have identified some neonates as colonized prior to becoming infected. Compliance with the use of CHG clothes in our NICU, as a part of the decolonization protocol, was not captured in these data, so the impact of this practice cannot be measured here. Furthermore, treatment options for skin, rectal or throat colonization, such as topical chlorhexidine, oral chlorhexidine rinses, and systemic antibiotics, are not routinely used in this population due to lack of safety and efficacy data.31 This was a single center observational study in an academic tertiary care NICU, and the findings may not be generalizable to other settings. We had limited data on healthcare worker MRSA colonization prevalence, so this assessment likely underestimated MRSA strains colonizing healthcare workers during the study period. Finally, we did not explore host-specific factors such as indwelling endotracheal tubes and organism specific factors including mupirocin resistance that may have impacted the efficacy of our intervention.
MRSA has become endemic in many NICUs and continues to cause invasive disease and even death. Neonates may acquire MRSA from on-going low-level healthcare workers transmission or from undiscovered MRSA reservoirs, possibly parents, and thus wider decolonization may be needed in this setting. Current strategies to prevent infections including active identification and decolonization of MRSA colonized neonates may be inadequate as many infants develop infections before being identified as colonized or after attempted decolonization. Future studies should evaluate and compare strategies to prevent MRSA disease in this population, including improved detection of MRSA colonization, optimized decolonization regimens, or identifying and interrupting reservoirs of transmission.
Acknowledgments
We thank Sue Culp, Mike DiJulia, Mary O'Neill, Sonali Advani, Melanie Gavin, the JHH Microbiology laboratory staff, the JHH NICU nursing staff, and The JHH Department of Hospital Epidemiology and Infection Control for their support of this study.
Financial support: This work was supported by the National Institute of Allergy and Infectious Disease, National Institutes of Health (1 K23 AI081752-04 to AM).
A.M.M. and T.M.P received grant support from Sage Products, Inc. K.C. serves on scientific advisory boards of NanoMR and Quidel and received grant support from Curetis and Nanosphere. T. M.P. receives grant support from MedImmune and Merck, was on an advisory board for Pfizer, and has received travel expenses from BioMerieux.
Footnotes
Potential conflicts of interest: All other authors report no disclosures.
Data were presented in part at the Annual Meeting of the Infectious Diseases Society of America, October 2012, San Diego, CA.
Bibliography
- 1.Klevens RM, Edwards JR, Richards CL, Jr, et al. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep. 2007;122(2):160–166. doi: 10.1177/003335490712200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stoll BJ, Hansen NI, Adams-Chapman I, et al. Neurodevelopmental and growth impairment among extremely low-birth-weight infants with neonatal infection. JAMA. 2004;292(19):2357–2365. doi: 10.1001/jama.292.19.2357. [DOI] [PubMed] [Google Scholar]
- 3.Schlapbach LJ, Aebischer M, Adams M, et al. Impact of Sepsis on Neurodevelopmental Outcome in a Swiss National Cohort of Extremely Premature Infants. Pediatrics. 2011;128(2):e348–e357. doi: 10.1542/peds.2010-3338. [DOI] [PubMed] [Google Scholar]
- 4.Hocevar SN, Edwards JR, Horan TC, Morrell GC, Iwamoto M, Lessa FC. Device-associated infections among neonatal intensive care unit patients: incidence and associated pathogens reported to the National Healthcare Safety Network, 2006-2008. Infect Control Hosp Epidemiol. 2012;33(12):1200–1206. doi: 10.1086/668425. [DOI] [PubMed] [Google Scholar]
- 5.Lessa FC, Edwards JR, Fridkin SK, Tenover FC, Horan TC, Gorwitz RJ. Trends in incidence of late-onset methicillin-resistant Staphylococcus aureus infection in neonatal intensive care units: data from the National Nosocomial Infections Surveillance System, 1995-2004. Pediatr Infect Dis J. 2009;28(7):577–581. doi: 10.1097/INF.0b013e31819988bf. [DOI] [PubMed] [Google Scholar]
- 6.Gerber SI, Jones RC, Scott MV, et al. Management of outbreaks of methicillin-resistant Staphylococcus aureus infection in the neonatal intensive care unit: a consensus statement. Infect Control Hosp Epidemiol. 2006;27(2):139–145. doi: 10.1086/501216. [DOI] [PubMed] [Google Scholar]
- 7.Milstone AM, Budd A, Shepard JW, et al. Role of decolonization in a comprehensive strategy to reduce methicillin-resistant Staphylococcus aureus infections in the neonatal intensive care unit: an observational cohort study. Infect Control Hosp Epidemiol. 2010;31(5):558–560. doi: 10.1086/652449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.CDC. NHSN Patient Safety Component. CDC/NHSN Surveillance Definitions for Specific Types of Infections. 2011 Jun; [Google Scholar]
- 9.Milstone AM, Carroll KC, Ross T, Shangraw KA, Perl TM. Community-associated methicillin-resistant Staphylococcus aureus strains in pediatric intensive care unit. Emerg Infect Dis. 2010;16(4):647–655. doi: 10.3201/eid1604.090107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Popoola VO, Carroll KC, Ross T, Reich NG, Perl TM, Milstone AM. Impact of Colonization Pressure and Strain Type on MRSA Transmission in Children. Clin Infect Dis. 2013 Aug 13; doi: 10.1093/cid/cit542. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. JClin Microbiol. 2003;41(11):5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milstone AM, Goldner BW, Ross T, Shepard JW, Carroll KC, Perl TM. Methicillin-resistant Staphylococcus aureus colonization and risk of subsequent infection in critically ill children: importance of preventing nosocomial methicillin-resistant Staphylococcus aureus transmission. Clin Infect Dis. 2011;53(9):853–859. doi: 10.1093/cid/cir547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klevens RM, Morrison MA, Nadle J, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298(15):1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 14.Carey AJ, Della-Latta P, Huard R, et al. Changes in the molecular epidemiological characteristics of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit. Infect Control Hosp Epidemiol. 2010;31(6):613–619. doi: 10.1086/652526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregory ML, Eichenwald EC, Puopolo KM. Seven-year experience with a surveillance program to reduce methicillin-resistant Staphylococcus aureus colonization in a neonatal intensive care unit. Pediatrics. 2009;123(5):e790–796. doi: 10.1542/peds.2008-1526. [DOI] [PubMed] [Google Scholar]
- 16.Huang YC, Lien RI, Su LH, Chou YH, Lin TY. Successful control of methicillin-resistant Staphylococcus aureus in endemic neonatal intensive care units--a 7-year campaign. PLoS One. 2011;6(8):e23001. doi: 10.1371/journal.pone.0023001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saiman L, Cronquist A, Wu F, et al. An outbreak of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit. Infect Control Hosp Epidemiol. 2003;24(5):317–321. doi: 10.1086/502217. [DOI] [PubMed] [Google Scholar]
- 18.Hall IM, Barrass I, Leach S, Pittet D, Hugonnet S. Transmission dynamics of methicillin-resistant Staphylococcus aureus in a medical intensive care unit. J R Soc Interface. 2012;9(75):2639–2652. doi: 10.1098/rsif.2012.0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morel AS, Wu F, Della-Latta P, Cronquist A, Rubenstein D, Saiman L. Nosocomial transmission of methicillin-resistant Staphylococcus aureus from a mother to her preterm quadruplet infants. Am J Infect Control. 2002;30(3):170–173. doi: 10.1067/mic.2002.119819. [DOI] [PubMed] [Google Scholar]
- 20.Al-Tawfiq JA. Father-to-infant transmission of community-acquired methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit. Infect Control Hosp Epidemiol. 2006;27(6):636–637. doi: 10.1086/505097. [DOI] [PubMed] [Google Scholar]
- 21.Mitsuda T, Arai K, Fujita S, Yokota S. Epidemiological analysis of strains of methicillin-resistant Staphylococcus aureus (MRSA) infection in the nursery; prognosis of MRSA carrier infants. J Hosp Infect. 1995;31(2):123–134. doi: 10.1016/0195-6701(95)90167-1. [DOI] [PubMed] [Google Scholar]
- 22.Jimenez-Truque N, Tedeschi S, Saye EJ, et al. Relationship between maternal and neonatal Staphylococcus aureus colonization. Pediatrics. 2012;129(5):e1252–1259. doi: 10.1542/peds.2011-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nubel U, Nachtnebel M, Falkenhorst G, et al. MRSA transmission on a neonatal intensive care unit: epidemiological and genome-based phylogenetic analyses. PLoS One. 2013;8(1):e54898. doi: 10.1371/journal.pone.0054898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang YC, Su LH, Wu TL, Lin TY. Molecular surveillance of clinical methicillin-resistant Staphylococcus aureus isolates in neonatal intensive care units. Infect Control Hosp Epidemiol. 2005;26(2):157–160. doi: 10.1086/502520. [DOI] [PubMed] [Google Scholar]
- 25.Milstone AM, Song X, Coffin S, Elward A. Identification and eradication of methicillin-resistant Staphylococcus aureus colonization in the neonatal intensive care unit: results of a national survey. Infect Control Hosp Epidemiol. 2010;31(7):766–768. doi: 10.1086/653615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delaney HM, Wang E, Melish M. Comprehensive strategy including prophylactic mupirocin to reduce Staphylococcus aureus colonization and infection in high-risk neonates. J Perinatol. 2012 doi: 10.1038/jp.2012.102. [DOI] [PubMed] [Google Scholar]
- 27.Hitomi S, Kubota M, Mori N, et al. Control of a methicillin-resistant Staphylococcus aureus outbreak in a neonatal intensive care unit by unselective use of nasal mupirocin ointment. J Hosp Infect. 2000;46(2):123–129. doi: 10.1053/jhin.2000.0786. [DOI] [PubMed] [Google Scholar]
- 28.Huang YC, Chou YH, Su LH, Lien RI, Lin TY. Methicillin-resistant Staphylococcus aureus colonization and its association with infection among infants hospitalized in neonatal intensive care units. Pediatrics. 2006;118(2):469–474. doi: 10.1542/peds.2006-0254. [DOI] [PubMed] [Google Scholar]
- 29.Maraqa NF, Aigbivbalu L, Masnita-Iusan C, et al. Prevalence of and risk factors for methicillin-resistant Staphylococcus aureus colonization and infection among infants at a level III neonatal intensive care unit. Am J Infect Control. 2011;39(1):35–41. doi: 10.1016/j.ajic.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 30.Miller MA, Dascal A, Portnoy J, Mendelson J. Development of mupirocin resistance among methicillin-resistant Staphylococcus aureus after widespread use of nasal mupirocin ointment. Infect Control Hosp Epidemiol. 1996;17(12):811–813. doi: 10.1086/647242. [DOI] [PubMed] [Google Scholar]
- 31.Chapman AK, Aucott SW, Milstone AM. Safety of chlorhexidine gluconate used for skin antisepsis in the preterm infant. J Perinatol. 2012;32(1):4–9. doi: 10.1038/jp.2011.148. [DOI] [PubMed] [Google Scholar]


