Abstract
Robust immune responses are essential for eliminating pathogens, but must be metered to avoid prolonged immune activation and potential host damage. Upon recognition of microbial DNA, the cytosolic DNA sensor cyclic GMP-AMP (cGAMP) synthetase, or cGAS, produces the second messenger cGAMP to initiate the STING pathway and subsequent interferon (IFN) production. We report that the direct interaction between cGAS and the Beclin-1 autophagy protein not only suppresses cGAMP synthesis to halt IFN production upon double stranded (ds)DNA stimulation or herpes simplex virus-1 infection, but also enhances autophagy-mediated degradation of cytosolic pathogen DNAs to prevent excessive cGAS activation and persistent immune stimulation. Specifically, this interaction releases Rubicon, a negative autophagy regulator, from the Beclin-1 complex, activating phosphatidylinositol 3-kinase class III activity and thereby inducing autophagy to remove cytosolic pathogen DNAs. Thus, the cGAS-Beclin-1 interaction shapes innate immune responses by regulating both cGAMP production and autophagy, resulting in well-balanced anti-microbial immune responses.
INTRODUCTION
Early detection of invading microbes by the host depends on a limited number of specific intracellular pattern recognition receptors (PRRs) that can detect microbes’ conserved patterns and activate signal transduction cascades, thereby triggering type I IFN-mediated anti-microbial defense mechanisms. While several DNA sensors, such as DAI, IFI16 and DDX41 (Takaoka et al., 2007; Unterholzner et al., 2010; Zhang et al., 2011), have been reported to induce IFN, recent studies have reported that the cGAS acts as an intracellular PRR to sense cytosolic pathogen DNAs and subsequently generates the second messenger cGAMP (Sun et al., 2013; Wu et al., 2013). cGAMP possesses unique 2’–5’ phosphodiester linkages that can bind the downstream signaling molecule STING, leading to the production of type I IFNs (Ablasser et al., 2013; Gao et al., 2013). Indeed, cGAS-deficient mice failed to produce IFNs and other cytokines in response to DNA transfection or DNA virus infection and were more susceptible to lethal herpes simplex virus (HSV-1) infection than wild type mice (Li et al., 2013). Furthermore, extensive structural studies have rapidly added insights of how cGAS functions as a DNA-sensing enzyme (Civril et al., 2013; Kranzusch et al., 2013; Zhang et al., 2013). cGAS contains a highly positively charged and poorly conserved N-terminal domain and a central nucleotidyl transferase (NTase) domain that partially overlaps with a C-terminal male abnormal 21 (Mab21) domain. cGAS possesses a remarkable structural similarity to the antiviral cytosolic double-stranded RNA sensor 2’–5’ oligoadenylate synthase 1 (OAS1), but contains a unique zinc thumb that recognizes B-form double-stranded DNA (Civril et al., 2013; Hartmann et al., 2003; Kranzusch et al., 2013). These suggest potential shared molecular mechanism between OAS1-mediated dsRNA and cGAS-mediated dsDNA innate immune sensing machinery.
Autophagy is an important homeostatic mechanism involving the formation of double-membrane vesicles called autophagosomes, which sequester damaged organelles, protein aggregates, and invading intracellular pathogens in the cytoplasm for degradation (Levine et al., 2011). Conserved from yeast to humans, autophagy takes place through a series of steps that include vesicle initiation, nucleation, and elongation, followed by vesicle fusion with lysosomes for degradation of the cargo (Rodgers et al., 2014). Extensive studies have demonstrated that Beclin-1 plays pivotal roles in autophagy induction and autophagosome maturation by forming various complexes with its positive (Atg14 and UVRAG) or negative (Bcl-2 and Rubicon) regulators (Liang et al., 2006; Matsunaga et al., 2009; Zhong et al., 2009). A recent study has revealed an unexpected link between DNA sensing and autophagy (Watson et al., 2012). During macrophage infection with Mycobacterium tuberculosis (Mtb), the recognition of bacterial DNA by the STING-dependent cytosolic pathway elicits ubiquitin-mediated autophagy, which then delivers bacilli to autophagosomes for degradation. Furthermore, besides its binding-dependent activation of STING pathway to initiate host defense, cGAMP also induces the activation of ULK1 autophagy kinase that subsequently phosphorylates STING and blocks its function, initiating negative-feedback control of the STING pathway and thus preventing the persistent transcription of innate immune genes (Konno et al., 2013). These data suggest that while a robust cGAS-STING-autophagy pathway is essential for eliminating intracellular microbial infection, this response must be well controlled to prevent excessive production of inflammatory cytokines. However, the molecular link between the intracellular DNA sensing pathway and the autophagy pathway remains elusive despite its critical function in the development of a balanced immune defense against pathogens. Here, we report crosstalk between the intracellular DNA sensing pathway and autophagy machinery by demonstrating the direct interaction between the cGAS DNA sensor and the Beclin-1 autophagy protein, which not only suppresses cGAS NTase activity to halt cGAMP production but also enhances the autophagy-mediated degradation of cytosolic microbial DNAs. These results indicate that the interaction between cGAS and Beclin-1 regulates cGAMP production and autophagy pathway, ultimately allowing the host to develop appropriate immunity against pathogens.
RESULTS
cGAS interacts with Beclin-1
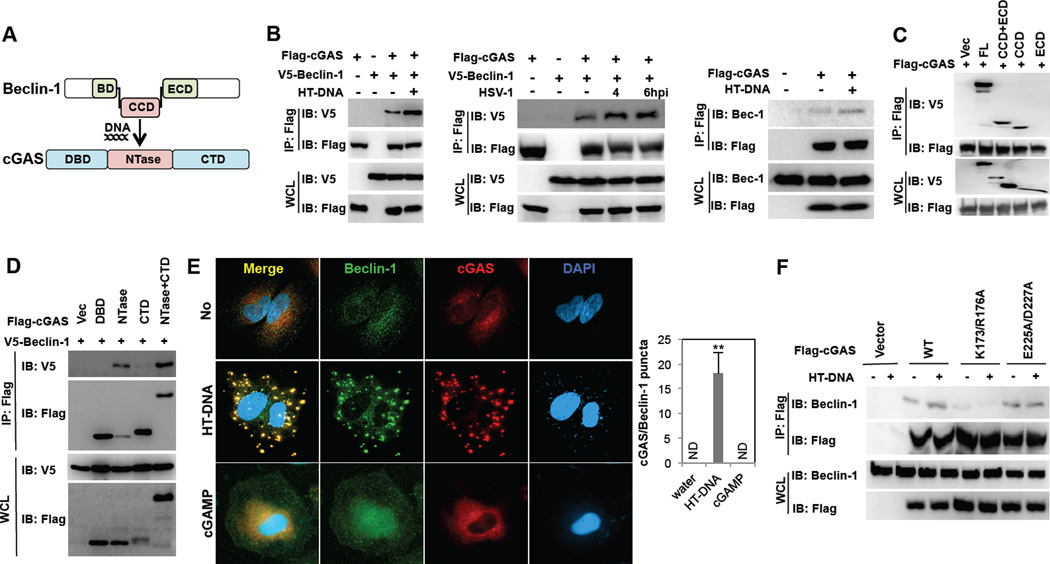
cGAS contains an N-terminal DNA binding domain (DBD), a central NTase domain and a C-terminal domain (CTD) (Kranzusch et al., 2013) (Figure 1A). Seven trials of yeast two-hybrid screens with full length cGAS, the N-terminal region (aa1–160), and the central and C-terminal region (aa160–522) of cGAS using a cDNA library from human lymphocytes collectively revealed the interaction between cGAS and Beclin-1 autophagy protein. Co-immunoprecipitation (co-IP) showed that Flag-cGAS readily interacted with V5-Beclin-1 upon transient overexpression in 293T cells and this interaction detectably increased upon herring testis (HT) DNA transfection or HSV-1 infection (Figure 1B). In addition, Flag-cGAS interaction with endogenous Beclin-1 detectably increased upon HT-DNA stimulation (Figure 1B). The central region of Beclin-1 contains the Bcl-2 binding domain (BD) for cellular and viral Bcl-2 interaction, the coiled-coil domain (CCD) for Atg14L and UVRAG interaction, and the evolutionarily conserved domain (ECD) for PI3KC3 (Vps34) interaction (Liang et al., 2006; Matsunaga et al., 2009; Zhong et al., 2009) (Figure 1A). Since the positive clones from yeast two hybrid screens contained the central CCD and ECD of Beclin-1, we tested whether the CCD and/or ECD were required for cGAS interaction. A binding assay showed that the central CCD of Beclin-1 and the central NTase domain of cGAS were responsible for their interaction (Figure 1CD). Further detailed mapping with four fragments of the CCD (CCD1–4) indicated that the CCD1–2 fragment of Beclin-1 was capable of binding cGAS as efficiently as the full-length CCD, while the CCD3–4 displayed little or no cGAS binding activity (Figure S1A). Upon gel filtration chromatography, Beclin-1 and cGAS were eluted in fractions 16–22 and 26–32, respectively, under unstimulated conditions (Figure S1B). However, under HT-DNA stimulation conditions, the elution profiles of Beclin-1 shifted right in fractions 18–30 and the elution profiles of cGAS shifted left in fractions 22–32, so that both proteins were eluted together in fractions 22–28 (Figure S1B). Furthermore, Beclin-1 and cGAS showed diffuse localization in the cytoplasm without stimulation, whereas they showed predominantly cytoplasmic punctate staining with extensive co-localization upon stimulation with HT-DNA or Immune Stimulatory DNA (ISD) or HSV-1 infection (Figure 1E and Figure S2A). Interestingly, the E225A/D227A enzymatically dead cGAS mutant showed Beclin-1 binding activity, Beclin-1 colocalization, and cytoplasmic punctate formation similar to the WT cGAS, whereas the K173A/R176A DNA-binding cGAS mutant displayed much weaker Beclin-1 binding activity than the WT and showed neither cytoplasmic punctate formation nor Beclin-1 colocalization (Figure 1F and Figure S2B). Finally, neither the cGAMP stimulation nor the STING expression was necessary for the cGAS-Beclin-1 interaction and the cytoplasmic punctate formation (Figure 1E and Figure S1CDE). These results indicate that cGAS efficiently binds Beclin-1 upon dsDNA stimulation, which is dependent on the DNA binding activity of cGAS.
Figure 1. cGAS interacts with Beclin-1.
(A) Domain structures of cGAS and Beclin-1. BD, Bcl-2 binding domain; CCD, coiled-coil domain; ECD, evolutionarily conserved domain; DBD, DNA binding domain; CTD, C-terminal domain. (B) cGAS interacts with exogenously expressing (left two panels) and endogenously expressing Beclin-1 (right panel) preferentially upon HT-DNA stimulation in 293T cells. At 48 h post-transfection with Flag-cGAS and V5-Beclin-1 (left and middle) or Flag-cGAS only (right), 293T cells were stimulated with HT-DNA (2µg/ml) or HSV-1 (MOI=5) for 4h, and whole cell lysates (WCLs) were subjected to immunoprecipitation (IP) and immunoblotting (IB) with indicated antibodies. (C) The Beclin-1 CCD binds to cGAS. WCLs of 293T cells transfected with Flag-cGAS and V5-Beclin-1 full-length or deletion mutants were used for IP and IB with indicated antibodies. (D) The cGAS NTase domain interacts with Beclin-1. WCLs of 293T cells transfected with V5-Beclin-1 and Flag-cGAS mutants were used for IP and IB with indicated antibodies. (E) Beclin-1 colocalizes with cGAS upon HT-DNA transfection. At 36h post-transfection with Flag-cGAS, Hela cells were stimulated with HT-DNA (2µg/ml) or cGAMP (5µM) for 4 h, fixed, stained with anti-Flag (red) and anti-Beclin-1 (green) antibodies and subjected to confocal analysis. DAPI stains the nucleus. The right panel shows the colocalization index (# of punctate/cell) between cGAS and Beclin-1. (F) At 48 h post-transfection with Flag-cGAS WT, K173A/R176A DNA-binding mutant, or D225A/D227A enzymatic dead mutant, 293T cells were stimulated with HT-DNA (2µg/ml) for 4h, and WCLs were subjected to IP and IB with indicated antibodies. (**p<0.005) See also Figure S1 and S2.
Beclin-1 negatively regulates cGAS-mediated IFN responses
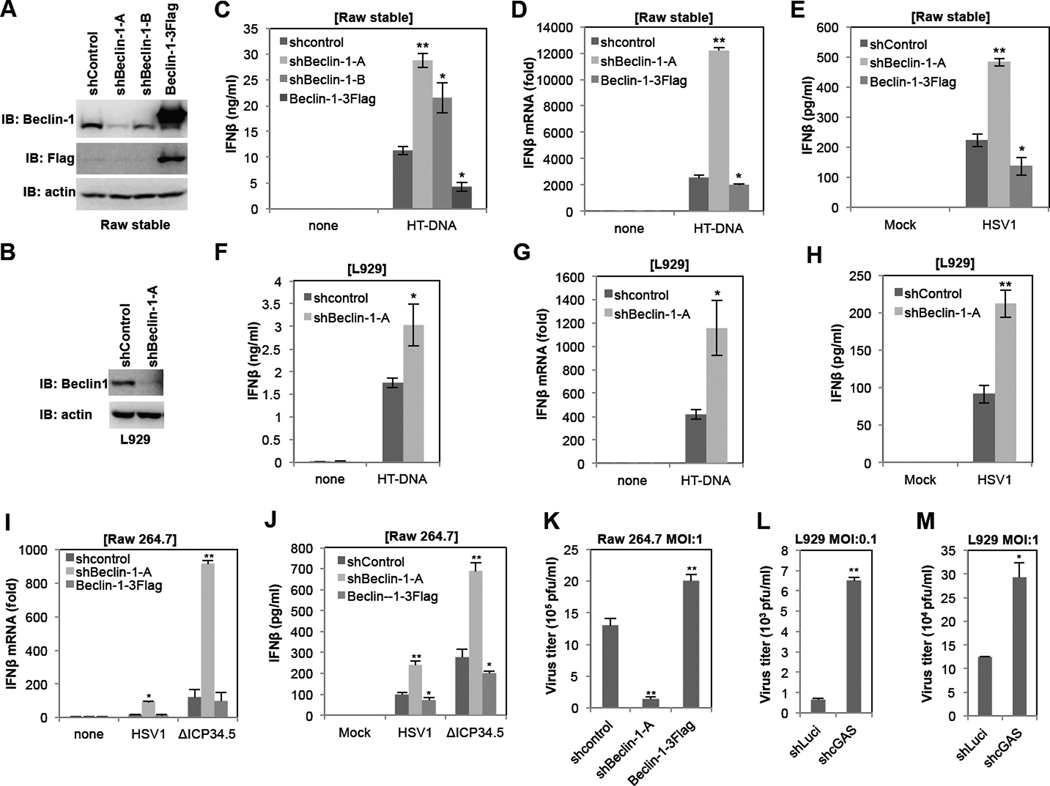
To test the effect of Beclin-1 expression on cGAS signaling activity, Raw264.7 and L929 cells were stably infected with lentiviruses containing Beclin-1-specific shRNAs or Flag-Beclin-1, resulting in the specific decrease or increase of Beclin-1 expression, respectively (Figure 2AB). These cells were then stimulated with HT-DNA or infected with the HSV-1 WT or the ΔICP34.5 Beclin-1 binding mutant, and the levels of IFNβ mRNA production and secretion and HSV-1 replication were measured (Figure 2C–K). We included the HSV-1 ΔICP34.5 mutant virus because the ICP34.5 protein inhibits autophagosome formation by interacting with Beclin-1 (Leib et al., 2009; Orvedahl et al., 2007). This assay showed a direct inverse correlation between Beclin-1 expression and cGAS signaling activity: depletion of Beclin-1 led to an increase of IFNβ mRNA and secretion and a decrease of HSV-1 replication, whereas overexpression of Beclin-1 led to a decrease of IFNβ mRNA and secretion and an increase of HSV-1 replication (Figure 2C–K). On the other hand, depletion of cGAS led to an increase of HSV-1 replication (Figure 2LM). It should be noted that while infection of either the HSV-1 WT or the ΔICP34.5 mutant virus induced IFN responses in Raw 264.7 cells, the ΔICP34.5 mutant virus was a more potent IFN inducer than the wild-type HSV-1 (Figure 2IJ). Finally, while depletion of Beclin-1 had a moderate effect on cGAMP-induced IFNβ production, it robustly enhanced poly I:C- or Sendai virus-mediated IFNβ production (Figure S3A–D), suggesting that the Beclin-1-mediated autophagy pathway tightly deregulates RNA-mediated IFN responses.
Figure 2. Beclin-1 negatively regulates cGAS-mediated IFN responses.
(A and B) Lentivirus-shRNA-mediated Beclin-1 knockdown and overexpression in Raw 264.7 cells (A) or L929 cells (B). Raw 264.7 cells and L929 cells were infected with lentivirus-shRNA-control (shcontrol), two different lentivirus-shRNA-Beclin-1 (shBeclin-1-A or -B) or lentivirus-Beclin-1–3Flag and selected by puromycin for two weeks. Knockdown and overexpression of Beclin-1 were verified by IB with indicated antibodies. (C–E) Raw 264.7 stable cell lines were stimulated with HT-DNA (2µg/ml) or HSV-1 (MOI=5). IFNβ mRNA levels were measured by qPCR at 6h post-stimulation and IFNβ production was measured by ELISA at 12h post-stimulation. (F–H) L929 cells were stimulated with HT-DNA (2µg/ml) or HSV-1 (MOI=5). IFNβ mRNA levels were measured by qPCR at 6h post-stimulation and IFNβ production was measured by ELISA at 12h post-stimulation. (I and J) Raw-shcontrol, Raw-shBeclin-1 and Raw-Beclin-1–3Flag cell lines were infected with wild-type HSV-1 or ΔICP34.5 mutant at MOI=5. IFNβ mRNA levels were measured by qPCR at 6h post-infection (I) and IFNβ production was measured by ELISA at 12h post-infection (J). (K) Raw-shcontrol, Raw-shBeclin-1 and Raw-Beclin-1–3Flag cell lines were infected with wild-type HSV-1 at MOI=1. Culture mediums were collected at 24h postinfection and subjected to standard plaque assay. (L and M) L929-shLuci and L929-shcGAS cell lines were infected with wild-type HSV-1 at MOI=0.1 (L) or MOI=1 (M). Culture mediums were collected at 24h postinfection and subjected to standard plaque assay. (*p<0.05, **p<0.005) See also Figure S3.
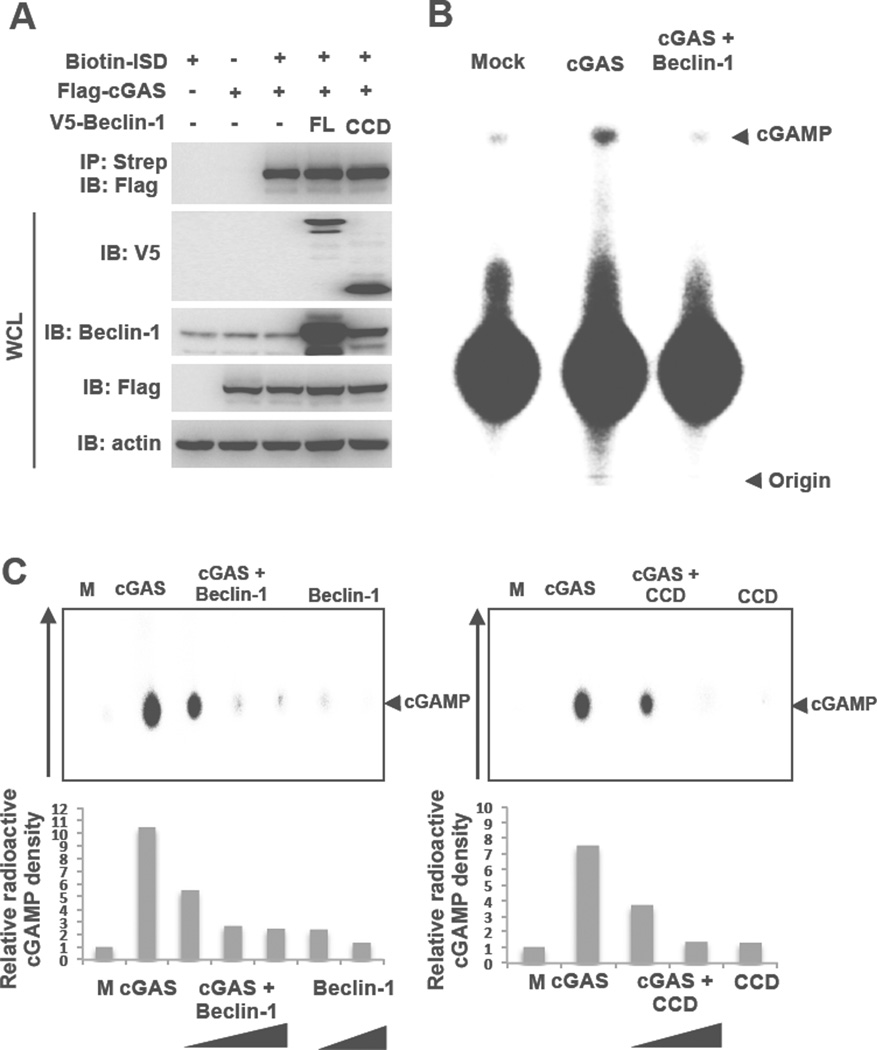
We then tested whether Beclin-1 affected either the DNA-binding ability or NTase enzymatic activity of cGAS. 293T cells were transfected with biotin-labeled ISD, Flag-cGAS, and V5-Beclin-1 (full-length or CCD alone), followed by biotin-labeled DNA pulldown with Streptavidin-conjugated beads and immunoblotting with anti-Flag antibody. This showed that either V5-Beclin-1 full-length or CCD alone showed little or no effect on cGAS DNA-binding activity (Figure 3A). Furthermore, neither depletion nor overexpression Beclin-1 affected the colocalization of cGAS with ISD in Raw 264.7 cells (Figure S3E). To measure the NTase activity of cGAS in the presence or absence of Beclin-1, we purified Flag-cGAS and Flag-Beclin-1 full-length proteins from 293T cells as well as MBP-cGAS, cGAS, and His-Beclin-1 CCD alone from E. coli. It should be noted that both the mammalian cell-purified Flag-cGAS protein and the bacterially purified MBP-cGAS fusion protein displayed basal levels of NTase activity in the absence of DNA stimulation. Using these purified proteins, we reconstituted cGAMP production by cGAS in the presence or absence of Beclin-1 full-length or CCD and analyzed the products using thin layer chromatography. Full-length Beclin-1 and CCD both effectively suppressed the NTase activity of cGAS, leading to the significant reduction of cGAMP production (Figure 3BC and Figure S3F). These results indicate that Beclin-1 interaction suppresses the NTase activity of cGAS, decreasing cGAMP synthesis and thereby decreasing IFNβ production.
Figure 3. Beclin-1 blocks cGAS enzymatic activity.
(A) Beclin-1 does not affect the DNA-binding activity of cGAS. Biotin-labeled ISD, Flag-cGAS and V5-Beclin-1 full-length or CCD domain (aa 150–224) only were transfected into 293T cells with indicated combinations. WCLs were harvested and subjected to Streptavidin-agarose pull down and IB with indicated antibodies. (B and C) Beclin-1 blocks cGAS enzymatic activity. In vitro enzymatic assays were performed in the presence of P32-α- GTP and cold GTP/ATP with full-length cGAS and Beclin-1 purified from mammalian cells (B) or MBP-cGAS (141–507aa) and full-length Beclin-1 or CCD domain (aa 150–224) purified from E. coli (C). cGAMP production was analyzed by TLC and autoradiograph. The arrow shows running direction and the arrow heads indicate the spotted origin (bottom) and the migrated cGAMP (top). See also Figure S3.
cGAS is required for dsDNA stimulation-induced autophagy
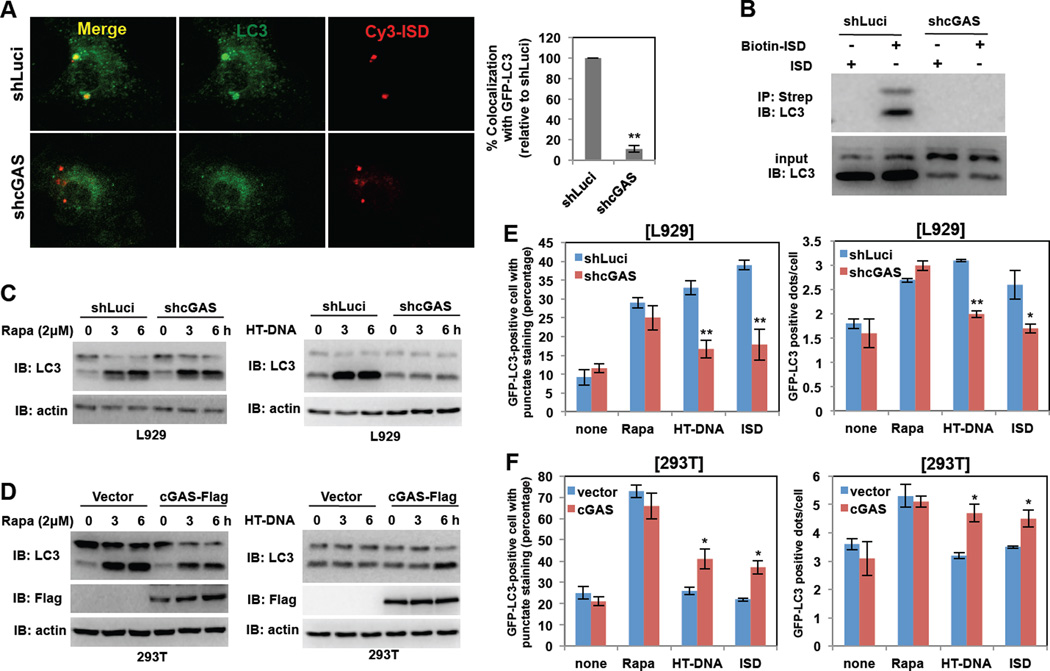
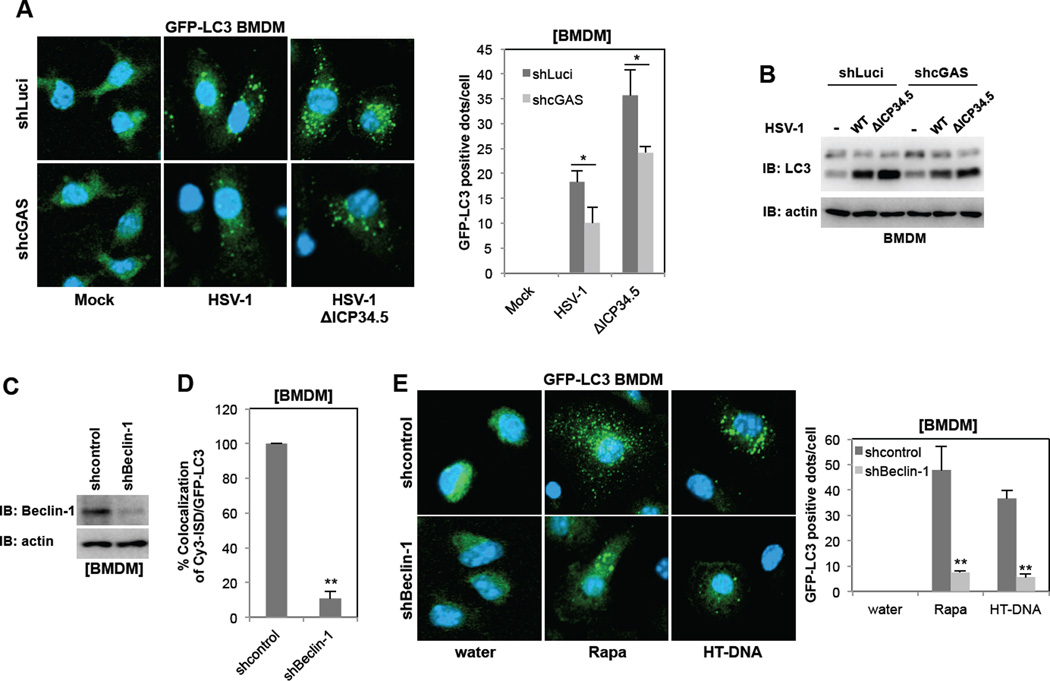
A recent study has demonstrated that cytosolic DNAs trigger the STING-dependent delivery of microbes to autophagosomes, which ultimately eliminates intracellular pathogens, indicating there is a potential link between DNA sensing and autophagy (Watson et al., 2012). Using LC3 as an autophagosome marker, we first compared GFP-LC3 staining patterns: upon autophagic stimulation, GFP-LC3 shifts from diffuse cytoplasmic staining to punctate cytoplasmic labeling, specifically of preautophagosomal and autophagosomal membranes (Mizushima et al., 2004). Stimulation of bone marrow derived macrophages (BMDMs) from GFP-LC3 transgenic mice with Cy3-labeled ISD led to the robust formation of cytoplasmic GFP-LC3 punctate structures, showing nearly complete colocalization of these structures with Cy3 or Cy5-labelded ISD or LAMP1 late endosomal marker (Figure 4A and Figure S4AB). Human THP-1 macrophage cells also showed punctate formation and colocalization of Cy3-ISD and endogenouse LC3 upon ISD treatment (Figure S4B). This was specific to dsDNA stimulation since the stimulation of BMDMs with Cy3-labeled ssDNA did not lead to punctate formation or colocalization (Figure S4C). Furthermore, depletion of the cGAS expression in BMDMs reduced cytoplasmic GFP-LC3 punctate formation and eliminated colocalization with Cy3-labeled ISD (Figure 4A). Finally, mouse BMDMs were infected with control luciferase-specific shRNA lentivirus or cGAS-specific shRNA lentivirus (Figure S4D), followed by transfection of biotin-labeled ISD and Streptavidin-mediated pulldown. This resulted in efficient recovery of an ISD-LC3 complex in control shRNA lentivirus-treated BMDMs, but not in cGAS-depleted BMDMs (Figure 4B). These results indicate that cytosolic ISD induces autophagy and also recruits LC3-positive autophagic vesicles in a cGAS-dependent manner.
Figure 4. cGAS is required for dsDNA stimulation-induced autophagy.
(A) Depletion of cGAS expression blocks the colocalization of Cy3-ISD and GFP-LC3 in BMDMs. At 48h post-infection with lentivirus-shRNA-Luciferase (shLuci) or lentivirus-shRNA-cGAS (shcGAS), GFP-LC3 BMDMs were stimulated with Cy3-ISD for 4h, fixed and analyzed by confocal microscope (left panel). Colocalization efficiency (%) of Cy3-ISD with GFP-LC3 in shcGAS-BMDMs was compared with that of shLuci-BMDMs (100%). (B) Depletion of cGAS expression blocks the association between ISD and LC3. At 48h postinfection with shLuci or shcGAS, BMDMs were stimulated with Biotin-ISD for 4h, and WCLs were harvested and subjected to Streptavidin-agarose pull down and IB with LC3 antibody. (C) Depletion of the cGAS expression blocks HT-DNA-mediated (right panel) but not rapamycin-mediated (left panel) LC3 processing. L929-shLuci and L929-shcGAS cells were stimulated with rapamycin (2µM) or HT-DNA (2µg/ml). WCLs were harvested at indicated time points and analyzed by IB with indicated antibodies. (D) Overexpression of cGAS promotes HT-DNA-mediated (right panel) but not rapamycin-mediated (left panel) LC3 processing. 293T-vector and 293T-cGAS-Flag cells were stimulated with rapamycin (2µM) or HT-DNA (2µg/ml). WCLs were harvested at indicated time points and analyzed by IB with indicated antibodies. (E) Depletion of cGAS expression impairs HT-DNA- or ISD-mediated, but not rapamycin-mediated LC3 punctate formation. At 36h post-transfection with GFP-LC3, L929-shLuci and L929-shcGAS cells were stimulated with rapamycin, HT-DNA (2µg/ml) or ISD (2µg/ml) for 4h and fixed to examine GFP-positive LC3 punctate under confocal microscope. (F) Overexpression of cGAS promotes HT-DNA- or ISD-mediated but not rapamycin-mediated LC-3 punctate formation. At 36 h post-transfection with GFP-LC3, 293T-vector and 293T-cGAS-Flag cells were stimulated with rapamycin, HT-DNA or ISD for 4 h and fixed to examine GFP-positive LC3 punctate under confocal microscope. (*p<0.05, **p<0.005) See also Figure S4 and S5.
To further test whether cGAS-mediated DNA sensing affects autophagy, L929 cells were stably infected with lentivirus containing cGAS-specific shRNA and 293T cells were infected with lentivirus containing Flag-cGAS, resulting in the specific decrease and increase of cGAS expression, respectively (Figure S4D). Immunoblotting was then performed with an antibody against LC3 to measure autophagic activity. During autophagy, an LC3 precursor (LC3-I) undergoes lipidation to yield the processed form (LC3-II). A large portion of LC3-I was converted to LC3-II following rapamycin treatment in both L929 and 293T cells, regardless of a decrease or increase in cGAS expression (Figure 4CD and Figure S4EF). By striking contrast, there was little processed LC3-II detected in L929 cells with cGAS depletion upon HT-DNA treatment, whereas increased LC3-II conversion was detected in 293T cells with cGAS overexpression under the same conditions (Figure 4CD and Figure S4EF). Upon rapamycin treatment, the numbers of GFP-LC3 punctate-positive cells and the numbers of GFP-LC3 positive dots per cells were not detectably affected by the levels of cGAS expression (Figure 4EF). Upon HT-DNA or ISD treatment, however, the numbers of GFP-LC3 punctate-positive cells and the numbers of GFP-LC3 positive dots per cells decreased in cells with cGAS depletion and increased in cells with cGAS overexpression (Figure 4EF). While infection of either the HSV-1 WT or the ΔICP34.5 mutant induced autophagy (LC3 punctate formation and processing) in BMDMs of WT or GFP-LC3 transgenic mice, the ΔICP34.5 mutant virus led to higher levels of autophagy than the WT virus (Figure 5AB). In contrast, decreased cGAS expression resulted in the reduction of autophagy (Figure 5AB), suggesting that the DNA sensor cGAS contributes to HSV-1 infection-induced autophagy. Finally, depletion of Beclin-1 expression suppressed dsDNA-induced or rapamycin-induced LC3 punctate formation in BMDMs of WT or GFP-LC3 transgenic mice (Figure 5C–E), further indicating a critical role for Beclin-1 in autophagy.
Figure 5. The role of cGAS in HSV-1-induced autophagy and the role of Beclin-1 in dsDNA-induced autophagy.
(A) GFP-LC3-shLuci-BMDMs and GFP-LC3-shcGAS-BMDMs were infected with wild-type HSV-1 or ΔICP34.5 mutant at MOI=5 for 3h, fixed, and subjected to confocal microscopy analysis. (B) shLuci-BMDMs and shcGAS-BMDMs were infected with wild-type HSV-1 or ΔICP34.5 mutant at MOI=5 for 3h, lysed, and subjected to IB with indicated antibodies. (C) Lentivirus-shRNA-mediated Beclin-1 knockdown in BMDMs. (D) GFP-LC3-shcontrol-BMDMs and GFP-LC3-shBeclin-1-BMDMs were transfected with Cy3-ISD for 6h, fixed, and subjected to confocal microscopy analysis for the colocalization efficiency (%) of Cy3-ISD and GFP-LC3. (E) GFP-LC3-shcontrol-BMDMs and GFP-LC3-shBeclin-1-BMDMs were treated with rapamycin or HT-DNA for 4h, fixed, and subjected to confocal microscopy analysis. (*p<0.05, **p<0.005)
To investigate the roles of cGAS and STING in dsDNA-induced autophagy, 293T-vector, 293T-cGAS, 293T-cGAS/STING, WT BMDMs, cGAS-depleted BMDMs and STING−/− BMDMs were stimulated with HT-DNA or cGAMP. GFP-LC3 punctate formation and LC3 processing were subsequently analyzed. HT-DNA stimulation induced a low level of LC3 processing in 293T-cGAS cells and an increased level of LC3 processing in 293T-cGAS/STING cells (Figure S5A). Interestingly, HT-DNA stimulation, but not cGAMP stimulation, still induced GFP-LC3 punctate formation and LC3 processing in STING−/− BMDMs, although at low levels (Figure S5B–D). In contrast, cGAMP stimulation, but not HT-DNA stimulation, induced GFP-LC3 punctate formation and LC3 processing in cGAS-depleted BMDMs (Figure S5B–D). Colocalization efficiency of GFP-LC3 punctate structures with Cy3-labeled ISD was lower in STING−/− BMDMs and much lower in cGAS-depleted BMDMs than in WT BMDMs (Figure S5E). These results collectively indicate that cytosolic DNAs stimulate cGAS, leading to the robust induction of autophagy and triggering the delivery of DNA to autophagosomes. Furthermore, cytosolic DNA-induced autophagy pathway appears to be complex since cGAMP-induced autophagy is primarily dependent on STING, whereas the cGAS-Beclin-1-induced autophagy is less dependent on STING.
cGAS promotes PI3KC3 kinase activity
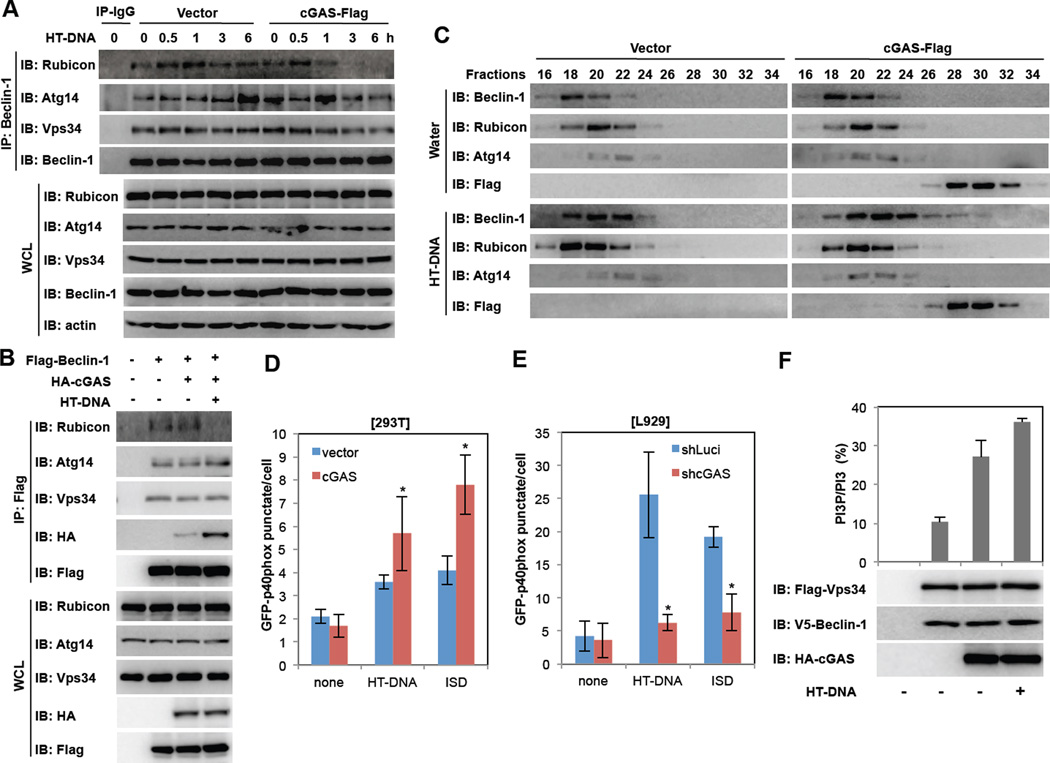
The Beclin-1-PI3KC3 (Vps34) core complex plays a crucial role in the induction of the autophagic process by generating PtdIns-3-P-rich membranes, which act as platforms for autophagy protein recruitment and autophagosome nucleation. Specifically, Atg14L and Rubicon associate with the Beclin-1-PI3KC3 (Vps34) core complex where Atg14L positively regulates autophagy at various steps, whereas Rubicon negatively regulates both autophagy and endocytosis at the membrane fusion step by suppressing PI3KC3 lipid kinase activity (Matsunaga et al., 2009; Sun et al., 2011; Zhong et al., 2009). To examine how cGAS can induce autophagy, 293T-vector or 293T-cGAS-Flag cells were stimulated with HT-DNA for various periods, followed by the purification of Beclin-1 complexes. This showed that while Rubicon continuously associated with Beclin-1 complexes in 293T-vector cells upon HT-DNA stimulation, it progressively dissociated from Beclin-1 complex in 293T-cGAS-Flag cells under the same conditions (Figure 6A). Furthermore, upon co-expression of Flag-Beclin-1 and HA-cGAS, levels of endogenous Rubicon associated with Flag-Beclin-1 markedly decreased following HT-DNA but not cGAMP stimulation, whereas endogenous Atg14L and PI3KC3 (Vps34) associated with Flag-Beclin-1 was not affected (Figure 6B and Figure S5F). On the other hand, the interaction between Flag-Beclin-1 and HA-cGAS considerably increased upon HT-DNA stimulation similarly to that shown in Figure 1B (Figure 6B). Gel filtration chromatography also showed that upon HT-DNA stimulation, the elution profiles of Beclin-1 shifted right in fractions 18–30 and the elution profiles of cGAS shifted left in fractions 20–32, showing both proteins were co-eluted in fractions 22–30 (Figure 6C). It was specific for HT-DNA stimulation since this shift of Beclin-1 elution profile was not observed upon rapamycin stimulation conditions (Figure S6A). Since both cGAS and Rubicon efficiently bound the first two CCDs (aa 144–207) (Figure S1A and S6B), increasing the cGAS-Beclin-1 interaction led to decreasing the Rubicon-Beclin-1 interaction in a dose-dependent manner (Figure S6C). These results indicate that cGAS competes with Rubicon for Beclin-1 binding, leading to the dissociation of Rubicon from the Beclin-1 complex.
Figure 6. cGAS promotes PI3KC3 kinase activity.
(A) Negative autophagy regulator Rubicon dissociates from Beclin-1 complex upon HT-DNA stimulation. 293T-vector or 293T-cGAS-Flag cells were stimulated with HT-DNA (2µg/ml) and WCLs were harvested at indicated time points for IP and IB with indicated antibodies. (B) cGAS suppresses Beclin-1 interaction with Rubicon. 293T cells were transfected with Flag-Beclin-1 and HA-cGAS with or without HT-DNA stimulation (2µg/ml) and WCLs were subjected to IP and IB with indicated antibodies. (C) Gel filtration of Beclin-1 complexes upon HT-DNA stimulation. 293T-vector or 293T-cGAS-Flag cells were stimulated with HT-DNA (2µg/ml) for 6h and WCLs were harvested, fractionated, and analyzed by IB with indicated antibodies. (D and E) cGAS is required for dsDNA stimulation-induced PI3P production. At 2 days post-transfection with GFP-p40phox (PX), 293T-vector and 293T-cGAS-Flag cells (D) or L929-shLuci and L929-shcGAS cells (E) cells were stimulated with HT-DNA (2µg/ml) or ISD (2µg/ml) for 4h. The intensity and distribution of PI3P GFP-p40phox (PX) punctate were analyzed by confocal microscope. (F) cGAS increases PI3KC3 (Vps34) kinase activity in vitro. Flag-PI3KC3 and V5-Beclin-1 were transfected into 293T cells with or without HA-cGAS or HT-DNA (2µg/ml) stimulation. Flag-PI3KC3 complexes were immunoprecipitated by anti-Flag M2 antibody and subjected to an in vitro kinase assay. (*p<0.05) See also Figure S6.
Finally, we examined PI3KC3 enzymatic activity by measuring intracellular phosphatidylinositol 3-phosphate (PtdIns-3-P) levels and distributions. For this, we used p40(phox) PX-eGFP fusion protein as a noninvasive probe since the PX domain of p40(phox) specifically binds PtdIns-3-P (Vieira et al., 2001). This showed that stimulation with HT-DNA or ISD led to a significant increase in p40(phox) PX-eGFP punctate structures, representing the presence of PtdIns-3-P-rich vesicles as a PI3KC3 enzymatic activity in 293T and L929 cells (Figure 6DE). The intensity and number of p40(phox) PX-eGFP-stained vesicles considerably increased upon cGAS expression, whereas they drastically decreased upon cGAS depletion (Figure 6DE). Specifically, HT-DNA-induced PI3KC3 enzymatic activity depended strictly on cGAS expression and partially on STING expression, whereas cGAMP-induced PI3KC3 enzymatic activity depended on STING but not on cGAS (Figure S6D). Also, depletion of Beclin-1 resulted in a decrease of PI3KC3 enzymatic activity in BMDMs, but depletion of Rubicon led to an increase of PI3KC3 enzymatic activity and thereby a reduction of IFNβ production (Figure S6E–H). To further confirm the cGAS-mediated increase of PI3KC3 enzymatic activity, we expressed Beclin-1 alone or together with cGAS and then measured PI3KC3 activity using an in vitro lipid kinase and PtdIns-3-P production assay. This also showed that the presence of cGAS efficiently enhanced PI3KC3 activity and HT-DNA stimulation further augmented the PI3P production. (Figure 6F). These results suggest that the interaction of cGAS with Beclin-1 leads to the dissociation of the negative autophagy factor Rubicon from the Beclin-1 complex, allowing the activation of PI3KC3 kinase activity and thereby increasing autophagy.
DISCUSSION
While a robust anti-microbial response is essential for controlling pathogen infection, this response must be metered and ultimately stopped when it reaches a late stage to avoid excessive production of inflammatory cytokines, which could otherwise lead to deleterious effects on the host. This suggests that halting PRR signaling is as important as initiating it. Indeed, recent studies have revealed that while the second messenger cGAMP binds STING to induce type I IFN production (Wu et al., 2013), it also activates the ULK1 autophagy kinase that subsequently phosphorylates and blocks STING (Konno et al., 2013). We report here that the direct interaction between the cGAS DNA sensor and the Beclin-1 autophagy protein not only suppresses the cGAS NTase activity to stop cGAMP production but also releases the Rubicon-mediated inhibition of autophagy, thereby delivering cytosolic microbial DNA to autophagosomes for degradation. These indicate that cGAS-STING pathway has at least two independent, feedback-negative regulatory mechanisms such as the Beclin-1-mediated suppression of cGAS activity and the cGAMP-mediated inhibition of STING activity, ultimately maintaining host immune system in check.
Previous studies have shown that cGAMP contains a distinct phosphodiester 2'–3’ linkage and binds to the adaptor protein STING, leading to a ligand-induced conformational change and activation of STING (Ablasser et al., 2013; Gao et al., 2013). However, we found that STING expression was not essential for the cGAS-Beclin-1 interaction, as Beclin-1 colocalized with cGAS in stimulation-induced punctate compartments in STING−/− cells. Furthermore, the interaction between cGAS and Beclin-1 was triggered by dsDNA stimulation or DNA virus infection, but not by cGAMP. Indeed, the cGAS DNA binding mutant (K173A/R176A) failed to bind to Beclin-1, whereas the enzymatically dead mutant (E225A/D227A) still bound to Beclin-1 as strongly as wild-type cGAS did. These data suggest that the DNA recognition activity, but not cGAMP production activity, of cGAS is required for its interaction with Beclin-1 and that the cross regulation between cGAS and Beclin-1 is separate from, or lies upstream of, the cGAMP-STING signaling event. Furthermore, while cytosolic DNAs trigger autophagy to eliminate intracellular pathogens, this pathway appears to be complex since cGAMP-induced autophagy is primarily dependent on STING, whereas the cGAS-Beclin-1-induced autophagy is less dependent on STING. Finally, besides cGAS, IFI16, DDX41 and other intracellular molecules sense cytosolic DNAs and mount various anti-viral responses (Bhat and Fitzgerald, 2013). For instance, DNA viruses, such as HSV-1, trigger autophagy during infection and recognition of viral DNA by the cellular autophagy sensor is a critical step for initiation of autophagy. However, it has not been demonstrated whether IFI16, DDX41 or other cytosolic DNA sensors also target Beclin-1 and/or induce autophagy. Further detailed and mechanistic analysis is necessary to delineate the mechanism of cytosolic DNA-triggered autophagy.
Rubicon interacts with the central CCD of Beclin-1, leading to the inhibition of PI3KC3 activity and thereby blocking the maturation step of autophagy (Matsunaga et al., 2009; Zhong et al., 2009). This suggests that cGAS competitively binds the central CCD of Beclin-1, ultimately releasing Rubicon from the Beclin-1 autophagy complex. Subsequently, this leads to PI3KC3 activation and autophagy induction to remove cytosolic pathogen DNAs. This suggests that at the late stage of the host immune response, cGAS may shuttle between the STING-mediated IFN pathway and the Beclin-1-mediated autophagy pathway to elicit IFN production, while inducing autophagy-mediated DNA degradation to avoid persistent immune stimulation. Thus, the cGAS cytosolic DNA sensor orchestrates IFN and autophagy pathways to ultimately optimize the timing and efficiency of host anti-microbicidal activity in response to various environmental stimuli and infections.
EXPERIMENTAL PROCEDURES
Reagents and Plasmid construction
Herring testis (HT) DNA, rapamycin, IgG, BSA, anti-Flag M2-agarose and all chemicals were purchased from Sigma unless otherwise indicated. ISD was prepared from equimolar amounts of the sense and antisense DNA oligonucleotides (sense strand sequsence: 5’-TACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACA-3’). The oligonucleotides were heated at 95 °C for 5 min and cooled to room temperature. ATP and GTP were obtained from Roche. cGAMP were purchased from Biolog. Poly I:C were purchased from Invivogen. Sendai virus was purchased form Charles River Laboratories. All constructs for transient and stable expression in mammalian cells were derived from the pEBG-GST mammalian fusion vector and the pER-IRES-Puro expression vector. DNA fragments corresponding to the coding sequences of the human cGAS and Beclin-1 genes were amplified by polymerase chain reaction (PCR) and subcloned into pEF-IRES-puro and selected for stable transfectants. GST-tagged Beclin-1 CCD and its mutants were cloned into a pEBG derivative encoding an N-terminal GST epitope tag between the BamHI and NotI sites. Flag-tagged truncated mutant constructs of cGAS were created by subcloning the PCR products of cDNA fragments containing each domain of the target genes into pER-IRES-Puro. V5-tagged truncated mutant constructs of Beclin-1 were created by subcloning the PCR products of cDNA fragments containing each domain of the target genes into pER-IRES-Puro. Flag-hVps34, Flag-Rubicon, Flag-Beclin-1, and V5-Beclin-1 were described previously (Liang et al., 2013). All constructs were sequenced using an ABI PRISM 377 automatic DNA sequencer to verify 100% correspondence with the original sequence.
Cell culture
Cells including HEK293T, Raw 264.7, L929 and primary bone marrow-derived macrophages (BMDMs) of WT, GFP-LC3 or STING−/− mice were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco-BRL) containing 4 mm glutamine and 10% FBS. THP-1 cells were grown in RPMI 1640/glutamin supplemented with 10% FBS and treated with 20 nM PMA (Sigma-Aldrich) for 24 h to induce differentiation into macrophage-like cells, followed by washing three times with PBS. Transient transfections were performed with Lipofectamine 2000 (Invitrogen), or calcium phosphate (Clontech), according to the manufacturer’s instructions. Raw 264.7 and L929 stable cell lines were generated using a standard selection protocol with 2–5 µg/ml of puromycin.
Lentivirus mediated shRNA knockdown
Hairpin-forming oligonucleotides were designed and cloned into pLKO.1 lentiviral vector (Addgene) and these lentiviral vectors were employed to knockdown the expressions of Beclin-1 or cGAS in Raw 264.7, L929 or BMDM cells by using the following targeting sequences (only sense strand sequence is shown): mouse-cGAS: 5’- GGATTGAGCTACAAGAATA-3’; mouse-Beclin-1-A: 5’-CCCTATGGAAATCATTCCTAT-3’; mouse-Beclin-1-B: 5’-CGGACAGTTTGGCACAATCAA-3’.
Protein purification
To purify human cGAS, 10cm culture flasks (~30 plates) were transfected with 10µg pIRES-hcGAS-3×Flag per plate. DNA was mixed with 20µl PEI (Polyethylenimine) transfection reagent (Polysciences, Inc.) and incubated for 10 min. After 48 hours, cells were scraped and washed with PBS. Cell pellet was obtained by centrifugation and was lysed in 1% NP40 buffer [50mM Tris (pH8.0), 150mM NaCl, 1% NP-40, EDTA-free protease inhibitor] and sonicated with 20% amplitude for 20 sec. Cell lysate was collected by centrifugation and filtered using 0.45µm filter. Cell lysate was cleared twice by 150µl and 100µl of Sepharose beads rotating at 4°C for 2hr. The cleared cell lysates were incubated with FLAG-M2 beads for 4hrs. cGAS-bound beads were washed 5 times with cell lysis buffer and eluted using 3×Flag peptides (Sigma). The eluted protein was dialysed and was concentrated using Amicon® Ultra centrifugal filters (Milipore). The concentrated protein was flash frozen in liquid nitrogen and stored at −80 °C for the cGAS enzyme assay.
The truncated form of murine MBP-cGAS fusion protein was expressed in BL21 (DE3, RIPL strain (Ku et al., 2012). When E. coli were grown at 37°C until reaching OD600=0.6, the temperature was then shifted to 18°C and grown overnight by adding 1mM IPTG. Cells were collected by centrifugation and lysed with 1% NP40 buffer as described above. Clarified lysates were mixed with Ni-NTA agarose (Invitrogen) and washed with lysis buffer prior to elution of protein using 150mM imidazole. MBP tagged cGAS protein was digested with AcTEV (Life technologies) for overnight at 4°C, cGAS protein was separated from MBP by two times incubation with Ni-NTA agarose and the eluted cGAS was dialyzed with 40mM Tris-Cl (pH 7.5) and 100mM NaCl. The concentrated protein was stored at −80 °C for the in vitro cGAS enzyme assay.
In vitro cGAS activity assay
For in vitro cGAS enzyme reaction, 1µM MBP-cGAS was mixed with 250µM GTP and 10µCi P32 alpha phosphate radiolabed ATP in buffer [20mM Tris-Cl (pH 7.5), 150mM NaCl, 5mM MgCl2, 1mM DTT]. After a 2hr incubation at 37°C, the reaction was stopped and 2µl of reaction solution was spotted onto high-performance TLC plates (HPTLC silica gel 60 F254, 20×10 cm, cat # 1.50628.001). TLC conditions have been previously published (Gao et al., 2013). The solvent used was NH4HCO3:C2H5OH:H20 = 0.2M:70%:30%) and the reaction molecules were separated at 25°C for 1 hr. Occasionally, the reaction was treated with 5 units of Alkaline phosphatase (Roche) for 30 minutes to stop the reaction and 1µl of reaction solution was spotted onto TLC plates with the solvent 1:1.5 [v/v] 1M (NH4)2SO4 and 1.5M KH2PO4. To visualize the reaction, the TLC plates were air-dried and subjected to Fuji Phosphor Imager.
Immunoblot analysis and immunoprecipitation
Cell lysates were collected in 1% NP-40 buffer and quantified by Bradford protein assay (Thermo Scientific). Proteins were separated by SDS-PAGE and transferred to PVDF membrane (Bio-Rad) by semi-dry transfer at 25V for 30 minutes. All membranes were blocked in 5% milk in PBST and probed overnight with indicated antibodies in 5% BSA at 4°C. Primary antibodies included: mouse Flag (Sigma), rabbit V5 (Bethyl Laboratories), rabbit HA (Covance), mouse GST (Santa Cruz), mouse and rabbit Beclin-1 (Cell Signaling), rabbit Atg14 (Cell Signaling), rabbit Vps34 (Cell Signaling), rabbit Rubicon (Bethyl Laboratories), mouse LC3 (Cosmo Bio), and actin (Santa Cruz). Appropriate HRP-conjugated secondary antibodies were incubated on membranes in 5% milk and bands were developed with ECL reagent (Thermo Scientific) and imaged on a Fuji LAS-4000 imager.
For immunoprecipitation, cells were harvested and then lysed in 1% NP-40 buffer supplemented with a complete protease inhibitor cocktail (Roche). After pre-cleared with protein A/G agarose beads for 1 h at 4°C, whole cell lysates were used for immunoprecipitation with the indicated antibodies. Generally, 1–2 µg of commercial antibody was added to 1 ml of cell lysates and incubated at 4°C for 4 to 12 h. After the addition of protein A/G agarose beads for 1 h, immunoprecipitates were extensively washed with lysis buffer and eluted with SDS loading buffer by boiling for 5 min.
Enzyme-linked immunosorbent assay
Raw 264.7 and L929 cells were treated as indicated and processed for analysis by sandwich ELISA. Cell culture supernatants were analyzed for IFNβ production using a mouse IFNβ ELISA kit (PBL Biomedical laboratories) according to the manufacturer’s protocols (Liang et al., 2011a; Liang et al., 2011b).
RNA extraction and quantitative RT-PCR
Total RNA was isolated from cells with the RNeasy Mini Kit (Qiagen) and treated with RNase-free DNase as the manufacturer’s protocol. Complementary DNA was reverse transcribed from 2 µg of total RNA using the iScript cDNA synthesis kit (Bio-Rad) and qPCR was performed with iQ SYBR Green Supermix (Bio-Rad). Primer sequences for qPCR were as follows: mouse IFNβ, sense: TCCGAGCAGAGATCT TCAGGAA, anti-sense: TGCAACCACCACTCATTCTGAG; mouse HPRT, sense: CAGTCCCAGCGTCGTGATTAG, anti-sense: AAACACTTTTTCCAAATCCTCGG; mouse cGAS, sense: ACCGGACAAGCTAAAGAAGGTGCT, anti-sense: GCAGCAGGCGTTCCACAACTTTAT.
Confocal fluorescence microscopy
For immunostaining, HeLa, RAW 264.7 and BMDM cells were seeded on 12-well culture dishes that contained 18 mm diameter round glass coverslips (105 cells per well). After transfection or stimulations, cells were fixed with 4% paraformaldehyde in PBS at 4°C for 10 min and permeabilized with 0.25% Triton X-100 in PBS for 15 min before being treated with 5% BSA for 1 hr at 25°C. Cultures were then stained with primary antibodies, including rabbit anti-Flag (Sigma), mouse anti-Beclin1 (Cell Signaling), mouse anti-LAMP1 (Abcam), and mouse anti-LC3 (Cosmo Bio) overnight at 4°C. After washing to remove excess primary antibodies, the cultures were incubated for 1h at room temperature with the following fluorescently labeled secondary antibodies: anti-rabbit IgG-FITC or TRITC, and anti-mouse IgG-FITC or TRITC (Molecular Probes). Cells were imaged with laser-scanning confocal microscopy (Nikon Eclipse C1).
PI3KC3 kinase assay
PI3KC3 (hVps34) kinase assay was performed in vitro as previously described (Liang et al., 2013). Briefly, Flag-tagged hVps34 and V5-tagged Beclin-1 were co-expressed in HEK293T cells in the presence or in the absence of HA-tagged cGAS. Flag-tagged hVps34 complexes were immunoprecipitated with anti-Flag M2 beads (Sigma), washed successively with lysis buffer, washing buffer (100 mM Tris-HCl at pH 7.4, 500 mM NaCl) and reaction buffer (10 mM Tris-HCl pH 8, 100 mM NaCl, 1 mM EDTA, and 100 mM MnCl2) and incubated with 2 nmol PI substrate and 1.25 nmol ATP in 25 µl reaction mixture at room temperature for 1 h. The PI(Liang et al.)P product was measured using Class 3 PI3-Kinase Kit according to the manufacturer’s protocols (Echelon Biosciences Inc.).
Gel filtration chromatography
Cells were harvested at 6 h after HT-DNA transfection, washed with cold PBS, resuspended in buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 20 mM b-glycerophosphate, 1 mM sodium orthovanadate, 10% glycerol, 0.5 mM EGTA, 0.5 % Triton X-100 or 0.2% NP-40, 1 mM PMSF and 10ug/ml leupeptin), and lysed by three rounds of freezing and thawing followed by 10 s of sonication in a microultrasonic cell disrupter. Cell lysates were clarified by centrifugation at 13000 rpm for 30 min at 4°C followed by passage through a 0.22 µM filter. The supernatants were fractionated on a Superose 6 gel column with a HPLC system (BioRad) (Liang et al., 2012). Fractions were analyzed by Western blot.
Statistical analysis
All data were analyzed using a 2-tailed student’s t-test with a minimum n=3. P-values of less than 0.05 were considered significant.
Supplementary Material
Highlights.
DNA sensor cGAS binds Beclin-1 autophagy protein upon HSV-1 infection or dsDNA exposure
Beclin-1 binding deregulates cGAS activity to block cGAMP and subsequent IFN production
cGAS binding to Beclin-1 releases Rubicon, inducing autophagy of cytosolic microbial DNA
ACKNOWLEDGEMENTS
This work was partly supported by 1F32AI096698 (MR); CA82057, CA31363, CA115284, CA180779, DE023926, AI105809, AI073099, HL110609, Hastings Foundation, and Fletcher Jones Foundation (JUJ); AI083025 (JUJ, KPH, and TJH); Howard Hughes Medical Institute (TJH); GRL Program (K20815000001) from National Research Foundation of Korea (BHO. and JUJ.). We thank Rina Amatya for manuscript preparation and Drs. M. Noboru, G. Barber, C. Liang, W. Yuan and P. Feng for transgenic mice and reagents. Finally, we thank all of JJ’s lab members for their discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2'-5'-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat N, Fitzgerald KA. Recognition of Cytosolic DNA by cGAS and other STINGdependent sensors. European journal of immunology. 2013 doi: 10.1002/eji.201344127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner KP. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, et al. Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann R, Justesen J, Sarkar SN, Sen GC, Yee VC. Crystal structure of the 2'-specific and double-stranded RNA-activated interferon-induced antiviral protein 2'-5'- oligoadenylate synthetase. Mol Cell. 2003;12:1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- Konno H, Konno K, Barber GN. Cyclic Dinucleotides Trigger ULK1 (ATG1) Phosphorylation of STING to Prevent Sustained Innate Immune Signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzusch PJ, Lee AS, Berger JM, Doudna JA. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Rep. 2013;3:1362–1368. doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku B, Lee KH, Park WS, Yang CS, Ge J, Lee SG, Cha SS, Shao F, Heo WD, Jung JU, et al. VipD of Legionella pneumophila targets activated Rab5 and Rab22 to interfere with endosomal trafficking in macrophages. PLoS Pathog. 2012;8:e1003082. doi: 10.1371/journal.ppat.1003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leib DA, Alexander DE, Cox D, Yin J, Ferguson TA. Interaction of ICP34.5 with Beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ Tcell responses. J Virol. 2009;83:12164–12171. doi: 10.1128/JVI.01676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAScGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Liang Q, Chang B, Brulois KF, Castro K, Min CK, Rodgers MA, Shi M, Ge J, Feng P, Oh BH, et al. Kaposi's Sarcoma-Associated Herpesvirus K7 Modulates Rubicon- Mediated Inhibition of Autophagosome Maturation. J Virol. 2013;87:12499–12503. doi: 10.1128/JVI.01898-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Deng H, Li X, Wu X, Tang Q, Chang TH, Peng H, Rauscher FJ, 3rd, Ozato K, Zhu F. Tripartite motif-containing protein 28 is a small ubiquitin-related modifier E3 ligase and negative regulator of IFN regulatory factor 7. Journal of immunology. 2011a;187:4754–4763. doi: 10.4049/jimmunol.1101704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Deng H, Sun CW, Townes TM, Zhu F. Negative regulation of IRF7 activation by activating transcription factor 4 suggests a cross-regulation between the IFN responses and the cellular integrated stress responses. Journal of immunology. 2011b;186:1001–1010. doi: 10.4049/jimmunol.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Fu B, Wu F, Li X, Yuan Y, Zhu F. ORF45 of Kaposi's sarcomaassociated herpesvirus inhibits phosphorylation of interferon regulatory factor 7 by IKKepsilon and TBK1 as an alternative substrate. J Virol. 2012;86:10162–10172. doi: 10.1128/JVI.05224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell host & microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Rodgers MA, Bowman JW, Liang Q, Jung JU. Regulation where autophagy intersects the inflammasome. Antioxidants & redox signaling. 2014;20:495–506. doi: 10.1089/ars.2013.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zhang J, Fan W, Wong KN, Ding X, Chen S, Zhong Q. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition, and autophagy suppression. J Biol Chem. 2011;286:185–191. doi: 10.1074/jbc.M110.126425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira OV, Botelho RJ, Rameh L, Brachmann SM, Matsuo T, Davidson HW, Schreiber A, Backer JM, Cantley LC, Grinstein S. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J Cell Biol. 2001;155:19–25. doi: 10.1083/jcb.200107069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1- phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.