SUMMARY
Pdx1 is a homeobox-containing transcription factor that plays a key role in pancreatic development and adult β-cell function. In this study, we traced the fate of adult β-cells after Pdx1 deletion. As expected, β-cell-specific removal of Pdx1 resulted in severe hyperglycemia within days. Surprisingly, a large fraction of Pdx1-deleted cells rapidly acquired ultrastructural and physiological features of α-cells, indicating that a robust cellular reprogramming had occurred. Reprogrammed cells exhibited a global transcriptional shift which included de-repression of the α-cell transcription factor MafB, resulting in a transcriptional profile that closely resembled that of α-cells. These findings indicate that Pdx1 acts as a master regulator of β-cell fate by simultaneously activating genes essential for β-cell identity and repressing those associated with α-cell identity. We discuss the significance of these findings in the context of the emerging notion that loss of β-cell identity contributes to the pathogenesis of type 2 diabetes.
Keywords: β-cells, reprogramming, Pdx1, diabetes, MafB
INTRODUCTION
The mammalian pancreas is composed of an exocrine compartment, which is responsible for secreting digestive enzymes into the intestine, and an endocrine compartment, which is responsible for the production and regulated secretion of hormones into the bloodstream. Pancreatic endocrine cells cluster together within the islets of Langerhans, which contain five different types of cells, each producing a distinct hormone product. The most essential of these is the β-cell, which is the only mammalian cell type capable of making insulin for regulating blood glucose uptake in peripheral tissues. Diabetes is a prevalent lifelong, chronic illness caused by either the lack of (i.e. type 1) or dysfunction of (i.e. type 2) β-cells. In addition, the production of the hormone glucagon from islet α-cells – a hormone that normally balances insulin action – is dysregulated in type 2 diabetes (T2DM), the preponderant form of the disease.
All pancreatic compartments are derived from a set of progenitor cells that express the pancreas and duodenum homeobox1 (Pdx1) gene during pancreas development (Gu et al., 2002). Pdx1 is the first transcription factor produced in the developing pancreas, and humans or mice lacking this factor suffer from pancreatic agenesis due to the inability to produce duct, exocrine, or endocrine cell types (Jonsson et al., 1994; Offield et al., 1996; Stoffers et al., 1997). Notably, Pdx1 expression becomes confined to β-cells over the course of development. Conditional removal of Pdx1 from forming β-cells using insulin-driven Cre lines results in hyperglycemia, compromised insulin+ (Ins+) cells, and increased glucagon+ (Glu+) cells (Ahlgren et al., 1998; Gannon et al., 2008). However, the cellular and molecular mechanisms underlying this requirement for Pdx1 in β-cells are unknown. Notably, the increase in Glu+ cell numbers upon embryonic Pdx1 deletion in the latter study (Gannon et al., 2008) was concluded to result from enhanced proliferation.
Several lines of evidence suggest that α-cells and β-cells share a close developmental relationship. First, early pancreas development is characterized by a wave of differentiation that gives rise to cells that express both glucagon and insulin, although these cells do not go on to populate the islet (Herrera, 2000). Second, cell ablation studies in which more than 99% of β-cells were killed demonstrated that α-cells can be converted into Ins+ cells (Thorel et al., 2010). Third, β-cell-specific deletion of DNA methyltransferase1 (Dnmt1) results in their conversion to Glu+ cells through an Nkx2.2-dependent de-repression of the α-cell determination factor Arx (Dhawan et al., 2011; Papizan et al., 2011). Fourth, forced Pax4 expression in α-cells promotes conversion into β-like-cells (Collombat et al., 2009). Finally, forced expression of Pdx1 in embryonic endocrine progenitor cells results in conversion of peri-natal α-cells into β-like-cells through an intermediate stage characterized by insulin/glucagon co-expression (Yang et al., 2011). Importantly, however, such changes in cell phenotype – i.e. conversion from a Glu+ cell to an Ins+ cell – cannot on their own serve as evidence of “reprogramming,” since a genuine stable cellular interconversion involves a transformation far more complex than a change in expression of one or even a few cell-type-specific markers. Presently, the precise cellular state that the β-cells adopt under these various conditions remains poorly defined.
Recently, Talchai et al. (2012) reported that mice with a conditional β-cell-specific deletion of the FoxO1 transcription factor exhibit a loss of β-cell identity, with affected cells adopting either an Ngn3+ hormone− progenitor-like or α-like state. Moreover, they proposed that the pathogenesis of human T2DM involved both β-cell de-differentiation to NGN3-like progenitor cells and trans-differentiation events. In the current study, we conditionally and specifically deleted Pdx1 in mature β-cells and followed their fate with a lineage tracer. As predicted from the earlier experiments using insulin-Cre, Pdx1 deletion resulted in a loss of β-cell identity. Surprisingly, however, within days these cells acquired ultrastructural, physiological, and global transcriptional signatures of β-to-α-cell reprogramming, without enhanced Ngn3 expression. The islet α-cell-enriched MafB transcription factor was among the most significantly up-regulated genes associated with this conversion, whereas induction of other key α-cell transcription factors (i.e. Arx and Brn4) was not observed. We found that Pdx1 normally binds within the MafB and glucagon promoters in β-cells and obtained evidence that MafB de-repression in Pdx1-depleted cells was responsible for gene activation. Significantly, these results highlight the importance of β-cell Pdx1 in actively inhibiting α-cell identity and provide novel mechanistic insight into repressive mechanisms involved in regulating islet β-cell identity and function, information that is relevant to the loss of Ins+ cell mass in T2DM and efforts to generate β-cells for therapeutic treatment.
RESULTS
Pdx1 maintains β-cell identity
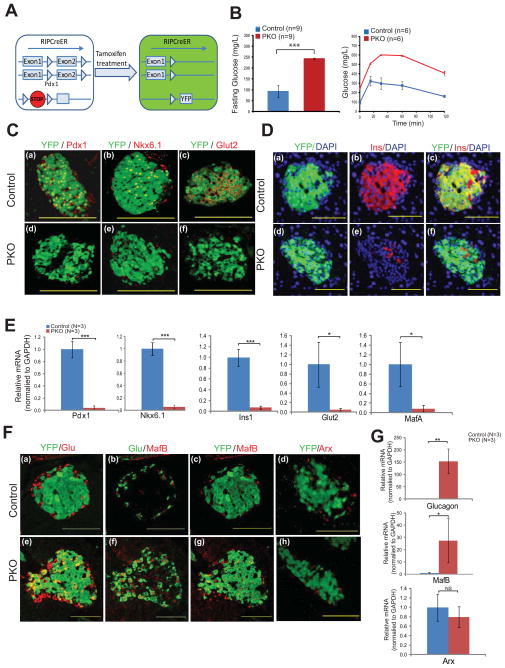
Several mechanisms could account for the previous observation that Pdx1 loss in β-cells leads to diabetes (Ahlgren et al., 1998; Gannon et al., 2008). These include (i) β-cell death, (ii) loss of β-cell identity factors resulting in dysfunctional β-like cells, or (iii) transdifferentiation to another cell type. To distinguish between these possibilities, we deleted Pdx1 in adult β-cells and tracked their fate using a RosaYFP lineage label. This was achieved by generating RIP-CreER; Pdx1fl/fl; RosaYFP mice (PKO mice). Within the pancreas, the RIP-CreER strain mediates recombination exclusively in β-cells (Dor et al., 2004 and data not shown) and administering tamoxifen (TAM) to 1 month-old mice resulted in the simultaneous deletion of Pdx1 and expression of the YFP lineage label specifically in β-cells (Fig. 1A; Fig. S1A). Thirty days after Pdx1 deletion, PKO mice displayed overt diabetes, as indicated by basal hyperglycemia and an abnormal response to glucose challenge (Fig. 1B). Importantly, these changes in glucose tolerance were not due to haploinsufficiency for Pdx1, as TAM-treated RIP-CreER; Pdx1f//+; RosaYFP mice exhibited a normal basal glucose level and normal glucose clearance rates (Fig. S1B). We confirmed efficient deletion by immunostaining for Pdx1 protein, which demonstrated a loss of nuclear staining in over 90% of islet cells (Fig. 1Ca, d). Importantly, the few islet cells that retained Pdx1 were YFP-negative, indicating that YFP staining serves as a robust surrogate for cells that have lost Pdx1. Notably, YFP+ Pdx1-deficient cells were still present in abundance in PKO islets; hence, Pdx1 is not absolutely required for adult β-cell survival (Fig. 1C). Such cells no longer expressed β-cell-specific markers such as Ins, Nkx6.1, and Glut2 (Fig. 1C, D). These expression changes were confirmed at the RNA level in sorted YFP+ cells from PKO and control islets (Fig. 1E). Thus, as expected, Pdx1 deficiency is associated with a loss of β-cell identity.
Figure 1. Adult islet β-cells lose their identity and acquire islet α-like features following Pdx1 deletion.
(A) Schematic showing the strategy for Pdx1 deletion and lineage tracing. Mice with a “floxed” allele of Pdx1 and a Cre-sensitive YFP reporter were crossed to RIP-CreER mice. Treatment of animals with tamoxifen (TAM) resulted in simultaneous deletion of exon 2 from both copies of Pdx1 and permanent heritable labeling of the cell with YFP. Mice having one “floxed” and one wild-type allele of Pdx1 were used as controls for all experiments.
(B) Fasting blood glucose levels and glucose tolerance tests performed 4 weeks after TAM administration to 4 week-old mice with two “floxed” copies of Pdx1 (“PKO” mice) or controls.
(C) Immunofluorescent images taken 4 weeks after TAM administration showing loss of Pdx1, Nkx6.1, and Glut 2 in PKO cells compared to control. Note the persistence of Pdx1 in some YFP− cells from PKO islets (but the complete absence of staining in YFP+ cells), indicating that YFP is a reliable reporter for Pdx1-deleted cells.
(D) Immunofluorescent images taken 4 weeks after TAM administration showing loss of insulin (Ins) in PKO cells compared to control. Note the lack of insulin staining in YFP+ cells from PKO islets.
(E) Relative abundance of transcripts encoding β-cell genes (Pdx1, Nkx6.1, Ins1, Glut2, MafA), as measured by qPCR following islet isolation and single-cell sorting of YFP+ cells from control and PKO islets (n=3 animals each group).
(F) Immunofluorescent images taken 4 weeks after TAM administration showing expression of glucagon (Glu) and MafB, but not Arx, in YFP+ cells in PKO islets. In control islets, YFP staining does not overlap with Glu, MafB, or Arx staining.
(G) Relative abundance of transcripts encoding α-cell genes (MafB, Glucagon, Arx) as measured by qPCR following islet isolation and single-cell sorting of YFP+ cells from control and PKO islets (n=3 animals each group).
Bar graph data are presented as mean +/− standard deviation (SD) in this and subsequent figures. *p<0.05; **p<0.01; ***p<0.001 by 2-tailed Student’s t test.
We next tested the hypothesis that β-cells adopt another endocrine cell fate upon Pdx1 loss. Hence, we performed co-immunostaining for YFP and a panel of endocrine hormones in PKO mice. Co-staining of YFP with pancreatic polypeptide (PP) or somatostatin did not reveal overlap (Fig. S1C). However, substantial portions of the lineage-labeled Pdx1-deficient cells expressed both YFP and glucagon (Fig. 1Fa, d; Fig. S2A, S2C). We then examined PKO islets for the expression of MafB and Arx, two transcription factors that are critical for the development of murine α-cells (Bramswig and Kaestner, 2011). MafB, whose expression is normally limited to α-cells in adult islets (Artner et al. (2006); Fig. 1Fb, c), was clearly evident in the YFP+ cells of PKO animals (Fig. 1Ff, g). Notably, the expression of Arx, a transcription factor required for α-cell development (Collombat et al., 2003), did not change in newly formed Glu+ MafB+ YFP+ cells (Fig. 1Fd, h). This pattern was confirmed at the RNA level on sorted cells from PKO and control islets (Fig. 1G). Moreover, we were unable to detect any effect after Pdx1 deletion on expression of Ngn3, a key regulator of endocrine progenitor cell formation during development that was recently shown to activated in islet β-cells in rodent T2DM models (Talchai et al. (2012); Fig. S3). These results show that MafB and glucagon, but not Arx or Ngn3, are induced in β-cells that have lost Pdx1.
Changes in cell identity occur rapidly following Pdx1 deletion
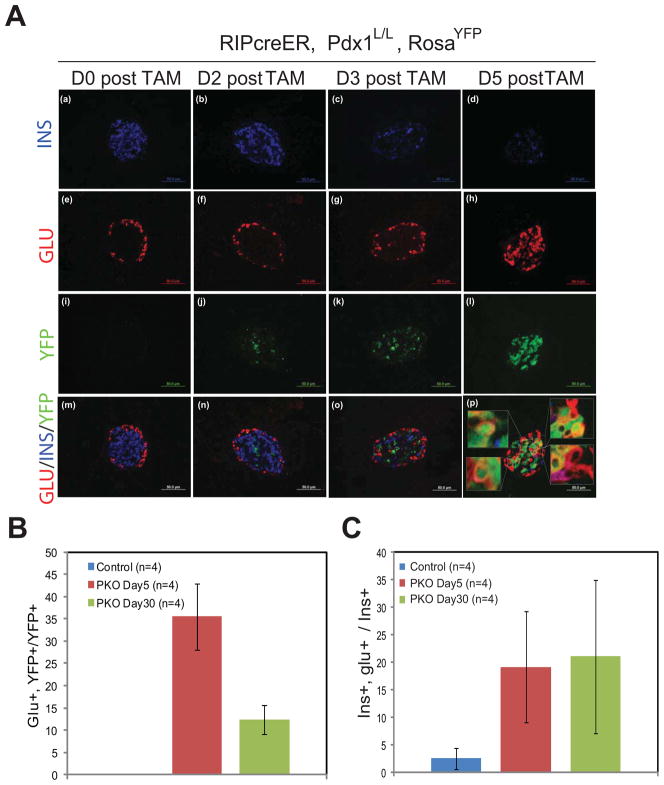
A time-course analysis was performed to address the dynamics of these phenotypic changes. PKO and control animals were injected with two consecutive daily doses of TAM and processed at 0, 2, 3, or 5 days afterwards. Low levels of YFP expression were detected as early as 2d post-TAM, reflecting a short lag between genomic recombination and the accumulation of YFP protein (Fig. 2Ai–j). YFP staining subsequently increased, such that the majority of islet cells were YFP+ by 5d post-TAM (Fig. 2Ak–l). In parallel, Glu staining became prevalent in the center of islets (Fig. 2Ae–h; Fig. S2A), while Ins staining was rapidly lost (Fig. 2Aa–d; Fig. S2B) and remained persistently absent (Fig. S2C). A loss in mouse insulin I and II mRNA levels was also clearly evident in PKO islets as early as D5 post-TAM, with more dramatic decrease observed at D30 post-TAM (Figure S2D).
Figure 2. Dynamics of islet insulin and glucagon expression following Pdx1 deletion.
(A) Representative images showing co-staining for YFP, glucagon, and insulin at either 0, 2, 3, or 5 days following TAM administration to PKO animals. Insets in (p) show cells that co-express YFP and glucagon.
(B) Quantification of Glu/YFP “double positive” cells as a fraction of the total number of YFP+ cells in control or PKO animals either 5 days after TAM treatment or 30 days after TAM treatment. No Glu+YFP+ cells were seen in control animals (n=4 each group).
(C) Quantification of Ins/Glu “double positive” cells as a fraction of the total number of Ins+ cells in control or PKO animals either 5 days after TAM treatment or 30 days after TAM treatment (n=4 each group).
Approximately 35% of YFP+ cells co-stained for Glu 5d after TAM, a percentage that decreased to under 15% by 30d post-TAM (Fig. 2B). However, this change in Glu+ YFP+ cell number was not due to marked changes in proliferation or apoptosis (Fig. S1D, E). Although the majority of islet cells in the 5d period following TAM treatment expressed either Ins or Glu, as many as 20% of the Ins+ cells were also Glu+ (Fig. 2C; Fig. S2B). These results suggest that conversion from an Ins+ to a Glu+ cell occurs rapidly following removal of Pdx1 and that co-hormone producing cells represent an intermediate cellular stage.
Pdx1-deficient cells acquire ultrastructural features of α-cells
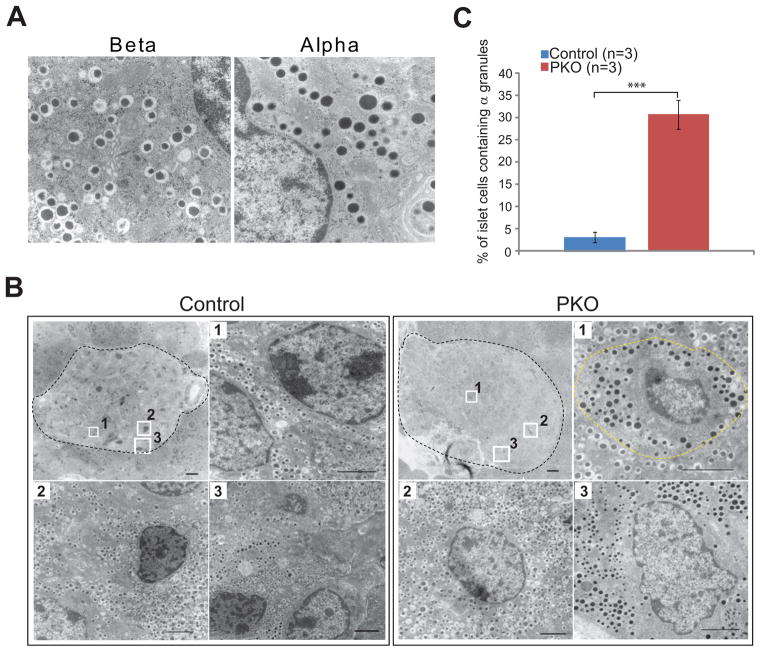
Islet α-cells and β-cells reside in distinct locations in rodents, with β-cells occupying the islet center and α-cells residing in an outer ring. Moreover, α- and β-cells package their hormone products into ultrastructurally distinct secretory granules that can be distinguished by transmission electron microscopy (TEM). Specifically, mature “β granules” are typified by electron dense insulin crystals surrounded by a clear “halo,” whereas glucagon-containing “α granules” lack any such halo (Fig. 3A).
Figure 3. PKO islets exhibit ultrastructural changes.
(A) Electron micrographs showing typical β granules (left) and α granules (right).
(B) Low and high magnification electron micrographs of control and PKO islets (outlined in black). In a control islet (left), cells in the interior (cells #1 and #2) contain β granules while cells at the margin (cell #3) contain α granules. In a PKO islet (right), cells containing α granules can be found in the islet center (cell #1; outlined in yellow).
(C) Quantification of cells containing α granules as a percentage of total islet cells, comparing control and PKO animals. (n=3 animals each group, greater than 6 islets counted per animal).
Scale bars: 100 μM. High magnification: 20 μM.
To determine whether there was a change in cellular ultrastructure following Pdx1 deletion, we isolated islets from one-month-old control and PKO and scored cells according to granule type. In control islets, approximately 3% of cells contained α granules; as expected, these cells were found exclusively at the islet periphery (Fig. 3B, C). By contrast, α granule-containing cells were readily detected in the center of PKO islets, a location where they would not normally be found (e.g. Fig. 3B, PKO cell #1). Quantification revealed that approximately 30% of islet cells in PKO animals contained α granules, a ten-fold increase over control (Fig. 3C). Thus, Pdx1 deletion in β-cells results in their conversion into Glu+ cells that have morphological features of α-cells.
Pdx1-deficient cells acquire physiological features of α-cells
PKO mice were subjected to a number of physiological tests to determine the functional consequences of Pdx1 loss. Under normal circumstances, β-cells take up glucose through the Glut2 transporter and sense glucose levels through glucokinase (hexokinase 4), resulting in depolarization via an ATP-dependent potassium channel, calcium influx, and exocytosis of insulin granules. Glycine or mixed amino acids (AAM) cause glucagon release from α-cells by similar processes (Li et al., 2012).
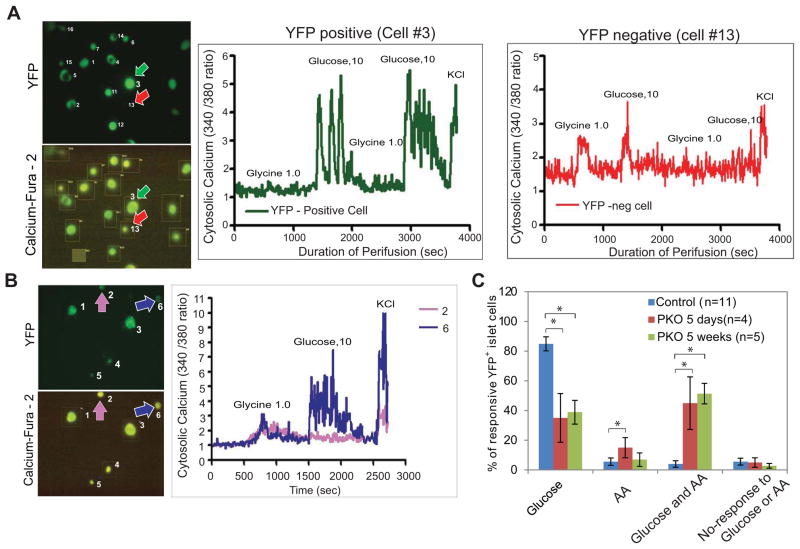
To examine the physiologic features of lineage-traced β-cells which had lost Pdx1, we measured changes in calcium flux at the single cell level using Fura-2, a sensitive indicator of intracellular calcium (Li et al., 2010; Li et al., 2012). First, we compared the calcium flux response of individual cells isolated from 2 month-old control mice to a glucagon secretagogue (either 1mM glycine or 4mM mixed amino acids) or an insulin secretagogue (10mM glucose). For this analysis, we examined two classes of cells: YFP+ β-cells and YFP-negative non-β-cells. As predicted, a change in intracellular calcium was principally observed in YFP+ cells treated with glucose (Fig. 4A; cell #3) and YFP-negative cells treated with glycine (Fig. 4A, cell #13), as the glycine receptor is not expressed on β-cells (Li et al., 2012). Next, we tested the calcium response of YFP+ islet cells from 2 month-old control and TAM-treated PKO animals. In addition to cells that responded to glucose but not glycine (β-cells) or only to glycine (α-cells), we also found a third category of cells in PKO islets that responded to both glycine and glucose (Fig. 4B, cell #6).
Figure 4. Pdx1 deletion results in cells with physiological features of α-cells.
(A) Tracing of calcium flow in islet cells from control animals measured by dual wavelength fluorescence microscopy. A YFP+ cell (cell #3), representative of a β-cell, shows responsiveness to 10mM glucose but not 1 mM glycine, while a YFP− cell (cell #13), representative of an α-cell, shows responsiveness to glycine but not glucose. Images on the left show YFP- and Fura-2-derived fluorescence of cells measured for the tracings (arrows).
(B) Tracing of calcium flow in YFP+ islet cells from PKO animals. PKO islets contain YFP+ cells that exhibit responsiveness to glycine (cell #2) and cells that exhibit responsiveness to both glycine and glucose (cell #6).
(C) Quantification of results from (A) and (B). The response of cells from control islets, islets from PKO animals 5 days post-TAM, or islets from PKO animals 5 weeks post-TAM was determined and plotted according to their glucose, glycine, dual responsiveness, or lack of responsiveness as a percentage of the total number cells counted. For each experimental animal, measurements were taken from at least 10 cells.
See also Figure S4.
We then quantified the calcium responsiveness in individual cells. In control islets, stimulation occurred in more than 85% of cells after glucose treatment and in only 5% of cells after treatment with mixed amino acids (AA), consistent with the expected abundance of α-cells and β-cells in normal islets (Fig. 4C). In PKO islets, by contrast, there was a significant increase within 5d of Pdx1 deletion in the percentage of cells that responded to AA but not glucose (Fig. 4C). In addition, approximately 40% of cells exhibited calcium flux in response to both glucose and glycine, a behavior that was rarely observed in control islets (Fig. 4C). Importantly, PKO islets lost the ability to secrete insulin and gained the ability to secrete glucagon, either spontaneously or in response to amino acids (Fig. S4). These results indicate that the new Glu+ cells in PKO mice respond to external secretion stimuli in a manner that is a hybrid between the normal α- and β-cell responses, resulting in α-like cells that secrete glucagon in a non-physiological manner.
Global α-cell-like transcriptional reprogramming occurs in Pdx1-deleted islet β-cells
Although our ultrastructural and functional studies suggested that PKO β-cells turn into cells with many of the features of α-cells, important differences relative to endogenous cells were also found upon initial characterization. First, only ~35% of the Pdx1-deleted YFP+ cells stained for Glu, with the bulk of the remainder lacking staining for any hormone. Second, Glu+ YFP+ cells produced just one of the two key transcription factors strongly associated with α– cell development and function – MafB – and not Arx. To get a more complete picture of the transcriptional changes associated with Pdx1 loss in β-cells, we conducted an mRNA microarray comparing normal islet β-cells and α-cells to the reprogrammed cells from PKO mice. To enrich for genes directly affected by Pdx1 loss, we chose the early time-point for analysis of PKO mice (5d after TAM administration). Control mRNA profiling was performed on FACS sorted islet YFP+ α-cells and β-cells obtained from 2 month-old glucagon-Cre; RosaYFP and RIP-CreER; Pdx1fl/+, RosaYFP mice, respectively (Fig. S5).
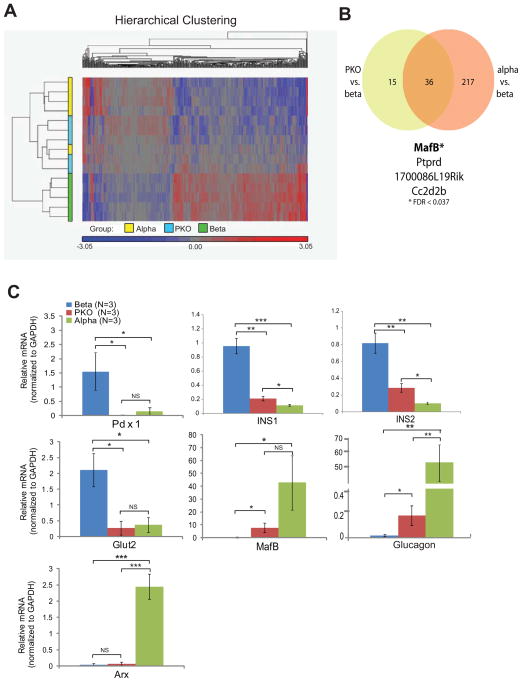
Analysis of 28,350 transcripts revealed 416 genes that were differentially expressed across the three groups, with a false discovery rate (FDR) of less than 0.1 (Supp. Tables 1 and 2). When hierarchical clustering was applied, β-cells had a distinct transcriptional profile, while α- and YFP+ PKO cells showed significant overlap (Fig. 5A). By filtering gene sets to focus on groups that exhibited a greater than 2-fold change in expression (and FDR<0.1), 253 genes were found to be differentially expressed between β-cells and α-cells (Supp. Table 3), while 51 genes were found to be differentially expressed between β-cells and PKO cells (Supp. Table 4). Notably, a total of 36 genes were differentially expressed in both of these datasets (Supp. Table 5); of these, MafB showed the greatest statistical expression difference between PKO and β-cells (Fig. 5B). We confirmed this change in the expression of MafB and other selected targets at the transcriptional level by qPCR. Transcripts levels for Pdx1, Ins1, and Glut2 were all significantly reduced in PKO cells (Fig. 5C). Conversely, MafB and glucagon transcripts were present at increased levels in PKO cells, although not to the levels present in α-cells (Fig. 5C). Arx transcript levels were unchanged in 5d PKO islets, consistent with our prior observations from islets 30 days after Pdx1 deletion. These results suggest that the mRNA profile of PKO cells closely resembles that of α-cells despite the absence of Arx.
Figure 5. Pdx1 deletion leads to an islet α-cell transcriptional reprogramming.
(A) Heat map showing relative expression of 416 genes that were differentially expressed in a microarray comparison of α-cells, β-cells, and PKO YFP+ cells isolated 5d after Pdx1 deletion. (See Experimental Procedures for microarray methodology). By hierarchical clustering, β-cells (green samples) constituted a distinct group, whereas α-cells (yellow samples) and PKO cells (blue samples) were more closely related.
(B) Venn comparison showing the subset of differentially expressed genes whose expression differed by more than 2-fold between groups in the microarray. Using these criteria, α-cells and β-cells had 253 differentially expressed genes and β-cells and PKO cells had 51 differentially expressed genes, with an overlap of 36 genes. The four most significantly different are shown.
(C) Confirmation of differences in selected targets across the three sample groups measured by qPCR.
Pdx1-mediated repression of MafB is required for maintenance of β-cell identity
MafB is required for the production of both α- and β-cells during pancreas organogenesis, although expression is silenced in β-cells soon after birth (Artner et al., 2007; Artner et al., 2010; Nishimura et al., 2006). MafB binds to and directly activates hormone gene expression in these cells (Artner et al., 2006). In contrast, sustained production of closely related MafA in islet β-cells is essential for production of functional cells (Artner et al., 2007; Artner et al., 2010; Artner et al., 2006; Zhang et al., 2005).
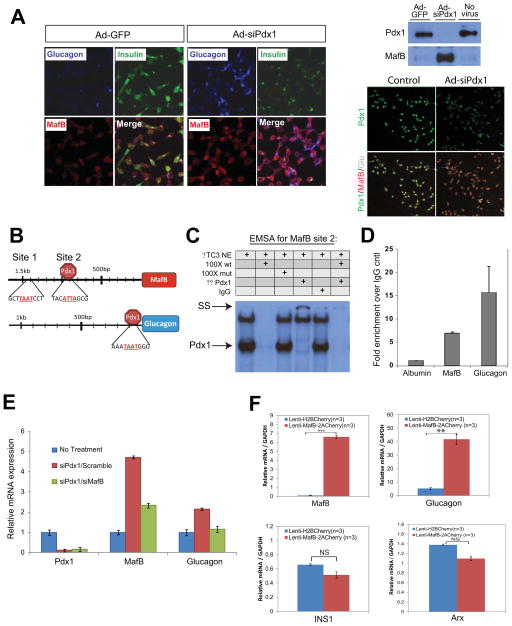
The dramatic induction of MafB in Glu+ PKO cells led us to hypothesize that this factor may be a driving force behind the shift towards an α-like identity. To investigate this possibility, a recombinant adenovirus-producing siRNA against Pdx1 was used to determine if depletion also induced MafB expression in the rat insulinoma Ins-1 cell line. Both MafB and Glu expression were rapidly up-regulated and insulin down-regulated upon Pdx1 knockdown (Fig. 6A). Expression of a dominant-negative acting Pdx1 in Ins-1 cells has been shown to selectively induce Glu expression but not Arx or Brn4 (Wang et al., 2001), a result also observed here (data not shown). Because these properties closely resembled those of PKO cells, we next examined the influence of Pdx1 on MafB transcription and the significance of MafB protein induction to Glu expression.
Figure 6. MafB and glucagon transcription is stimulated in β-cells by eliminating Pdx1 promoter binding, with the increased MafB protein levels activating glucagon.
(A) Immunostaining of Min6 cells infected with adenoviruses expressing GFP or Pdx1 siRNA illustrating how efficient reduction in Pdx1 protein (assessed by immunofluorescence and western blotting) greatly intensifies MafB and glucagon cellular staining while decreasing insulin staining.
(B) Schematic view of the two potential Pdx1 binding sites in the on MafB promoter and bona fide glucagon G1 element (Wang et al., 2001). Core homeodomain protein binding sequences are designated in red, Pdx1 only bound to MafB Site 2 in gel shifts, and both endogenous Site 2 and G1 regions by ChIP.
(C) Results from EMSA using nuclear extracts (NE) from βTC3 cells showing Pdx1 binding to MafB Site 2. Competition with wild type and mutant Site 2 as well as Pdx1 antibody (α-Pdx1) addition analysis was performed. The location of the Pdx1:Site 2 and the super-shifted (SS) complex is indicated.
(D) Pdx1 binds to the endogenous MafB Site 2 and Glucagon G1 regions in Min6 cells. ChIP assays and corresponding qPCR reactions were performed in triplicate; control reactions were performed analyzing the IgG immunoprecipitation and albumin promoter binding. n≥3.
(E) Simultaneous knockdown of Pdx1 and MafB reveals that MafB is required for glucagon expression in Ins-1 cells. qPCR data from Ins-1 cells following individual or combined knockdown of Pdx1 and MafB by siRNA. n≥3.
(F) Overexpression of MafB reveals that MafB is sufficient for the induction of endogenous glucagon expression in Ins-1 β-cells, as shown by qPCR analysis of transcript levels from either control (Lenti-H2BCherry) or after MafB overexpression (Lenti-MafB-2ACherry).
See also Table S6.
ChIP-seq analysis performed in Min6 β-cell lines identified Pdx1 binding around −800 and −1500 base pairs (bp) upstream of the MafB coding region (data not shown). Gel mobility shift assays were next performed with βTC3 nuclear extract to determine whether Pdx1 bound to bioinformatically-identified sites at −1325/−1316bp (i.e. Site 1) and −942/−933 bp (Site 2) within these regions (Fig. 6B). Pdx1 binding was only observed at Site 2, as determined by antibody addition and wild type and mutant competitor analysis (Fig. 6C and data not shown). Moreover, chromatin immunoprecipitation studies demonstrated that Pdx1 specifically bound to this region of the endogenous MafB gene in Min6 cells (Fig. 6D). In addition, this experiment illustrated the ability of Pdx1 to bind promoter proximal glucagon sequences spanning the G1 element associated with Pdx1-mediated repression (Fig. 6D; (Wang et al., 2001)). These results strongly suggest that Pdx1 directly represses MafB expression in islet β-cells.
To investigate the importance of MafB in glucagon activation following Pdx1 depletion in β-cells, we developed siRNA reagents to simultaneously eliminate Pdx1 and MafB proteins from Ins1 cells. Under these circumstances, the stimulation of glucagon mRNA and protein expression that normally accompanied Pdx1 elimination in β-cells was prevented (Fig. 6E and data not shown), strongly suggesting that MafB de-repression was responsible for glucagon induction. Furthermore, lentiviral delivery of MafB cDNA to Ins-1 cells resulted in increased production of glucagon transcripts, but not Ins1 or Arx (Fig. 6F). Taken together, these data demonstrate that MafB de-repression is responsible for the induction of glucagon, and likely other essential functional α-cell genes in PKO islets.
DISCUSSION
Cell identity is maintained by the expression of self-reinforcing transcriptional networks. Understanding the nature of these networks is a central question in developmental biology, but delineating the molecular mechanisms underlying cellular identity also has broader implications. In the field of regenerative medicine, for example, a detailed picture of the molecular mechanisms that underlie cellular differentiation states could facilitate cellular reprogramming for therapy. Likewise, determining how nuclear factors act to enforce one differentiation program at the expense of another could have implications for understanding degenerative diseases, in which loss of cell identity may be associated with a loss of tissue function.
Our work provides new insight into Pdx1 function in adult β-cells. Using conditional gene ablation and lineage tracing, we have shown that deletion of the Pdx1 transcriptional regulator in β-cells results in a rapid change in cellular differentiation state. Within days of Pdx1 deletion, β-cells lose multiple markers of β-cell identity, resulting in overt diabetes in PKO animals. Importantly, β-cells did not die following Pdx1 loss, but rather took on one of two phenotypes: an α-like phenotype, characterized by the expression of glucagon and MafB, or a loss of β-cell identity characterized by the absence of any pancreatic hormone. The abundance of Pdx1−YFP+ cells in PKO islets, and the failure to detect an increase in cleaved-caspase3 staining in Pdx1-deleted β-cells, suggests that Pdx1 is not absolutely required for survival. Nevertheless, the possibility remains that Pdx1 loss is associated with an unstable intermediate state that could lead to cellular loss over time, a prospect consistent with the finding that the proportion of Glu+YFP+ cells decreases from 35% to less than 15% between d5 and d30 following Pdx1 deletion (Fig. 2B). Within the pancreas, RIP-CreER mediates recombination solely in the β-cells. However, RIP-Cre-mediated recombination has previously been described in extra-pancreatic cells in the brain (Wicksteed et al., 2010). Thus, it remains possible that Pdx1 deletion in cells outside of the pancreas could modestly contribute in a non-cell-autonomous manner to the observed phenotypes.
We are not the first to report cellular “reprogramming” between the α- and β-cell lineages. Glu+ cells become Ins+ following forced expression of Pdx1 or Pax4 (Collombat et al., 2009; Yang et al., 2011). Moreover, Ins+ cells become Glu+ following deletion of Dnmt1 or Foxo1 (Dhawan et al., 2011; Talchai et al., 2012) and in the setting of extreme β-cell loss (Thorel et al., 2010). Moreover, Ahlgren et al. (1998) and Gannon et al. (2008) found that developmental deletion of Pdx1 by insulin-Cre increased the Glu+ to Ins+ cell ratio. And most recently, Bramswig et al. (Bramswig and Kaestner, 2011) demonstrated that changes in histone methyltransferase activity could influence β- to α-transdifferentiation. Hence, our finding that β-cells acquire α-cell features following Pdx1 deletion is not unprecedented.
Importantly, however, these prior studies did not define the extent to which such changes in cell phenotype represent a global change in cell identity. As a consequence, changes in cell phenotype observed following these manipulations might have merely reflected β-cells that had lost their identity and become Glu+ rather than a “global” β-to-α-cell reprogramming event. To address this possibility, we performed a set of rigorous experiments to characterize the lineage-traced cells in detail. We found that lineage-traced Pdx1-deleted β-cells gained ultrastructural features of α-cells (α granules), acquired many physiological features of α-cells (Glu secretion and calcium flux in response to amino acids), and a transcriptional profile that was highly similar to that of endogenous α-cells. Based on these studies, we believe that Pdx1 loss leads to the reprogramming of β-cells into cells that closely resemble α-cells. Furthermore, we propose that Pdx1 acts as a transcriptional repressor at the MafB locus and that de-repression of MafB is an important driver of this β- to α-cell reprogramming event.
Importantly, the reprogrammed cells we observe following Pdx1 deletion differ in one significant respect from bona fide α-cells: they lack expression of the Arx transcription factor, a marker of mature α-cells. This result is surprising in the context of reports that Dnmt1-mediated maintenance of β-cell identity is associated with methylation of the Arx locus (Dhawan et al., 2011) and ectopic expression of Arx during embryonic development causes pancreatic β- to α-cell conversion (Collombat et al., 2007). Our data suggest that adult islet α-cells, in contrast to their embryonic counterparts, can arise from β-cells in an Arx-independent manner, an idea supported by the recent demonstration that Arx is not required for the maintenance of islet α-cell identity in adult animals (Wilcox et al., 2013). Appreciating the differences in both the expression levels of these two α-cell identity products and the physiological secretion properties of the GFP+ cells from TAM-treated PKO mice and endogenous α-cells, collaborative interactions between MafB and Arx may be critical in controlling distinct, as well as similar, parts of the α-cell program (e.g. Pdx1 for MafB and Nkx2.2 for Arx; (Papizan et al., 2011)). As such, one would predict that MafB is of greater value to islet α-cells, considering its fundamental importance to glucagon transcription and the observations of Wilcox e al. (Wilcox et al., 2013). In the future, it will be important to further delineate the contributions of these two transcription factors to the “transcriptional wiring” of the α-cell.
Our results are reminiscent of the recent finding that FoxO1 deletion in rodent β-cells results in hyperglycemia that is associated with a loss of β-cell identity and conversion into other endocrine cell types, particularly Glu+ cells (Talchai et al., 2012). An implication of that work, and the current study, is the possibility that loss of cell identity contributes at least partially to β-cell failure in T2DM, a scenario that contrasts with the prevailing view that β-cell failure is largely a consequence of an apoptosis-driven reduction in β-cell mass (Butler et al., 2007). In support of the Talchai et al. (2012) findings, PDX1 and a small subset of other key islet-enriched transcription factors were found at dramatically reduced levels in human T2DM islet β-cells (Guo et al., 2013). Moreover, a recent study (Spijker et al., 2013) has shown that isolated human β cells possess an intrinsic ability to transdifferentiate into α cells. Our study is thus part of a growing body of evidence which indicates that β cell dysfunction, and not merely loss of β-cell mass, contributes to T2DM pathogenesis.
We found no evidence that an endocrine progenitor cell state was induced following Pdx1 deletion (e.g. Ngn3, Fig. S3). Significantly, human T2DM islet samples did not have activated levels of the progenitor cell regulators found in prior rodent studies (i.e. NEUROGENIN 3, NANOG, POU5F1, and MYCL1 (Guo et al., 2013) illustrating mechanistic differences between species in their responsiveness to T2DM stress conditions. Nevertheless, our observation that Pdx1 deletion leads to a phenotype similar to the one seen in T2DM islets – characterized by the loss of β-cell enriched transcription factors and a decrease or absence in insulin expression – is consistent with the idea that changes in the differentiated β-cell state play a role in β-cell failure. Of note, induction of MafB expression has been observed in the β-cells of glucose-intolerant mice fed a high fat diet (Lu et al., 2012). Our findings hint at one mechanism that could contribute to the α-cell dysregulation and hyperglucagonemia that frequently accompany T2DM – the formation of glucagon-secreting α-like cells that are not physiologically regulated (Fig. S4). Hence, it will be interesting to determine if human T2DM islet α-cells not only lack MafB (Guo et al., 2013), but also Arx.
A decrease in Pdx1 expression has previously been observed in rodents with chronic hyperglycemia (Harmon et al., 1999; Jonas et al., 1999) and more recently in human T2DM patients (Guo et al., 2013), lending further support to the notion that Pdx1 loss contributes to this disease. Nevertheless, it is important to stress that human T2DM pathogenesis is likely to be multifactorial, involving both β-cell loss and changes in β-cell identity and function following reduction in the expression of β-cell-enriched transcription factors like Pdx1 or Nkx6.1 (Taylor et al., 2013). In the future, identifying ways to “re-induce” the self-reinforcing transcriptional networks that maintain β-cell identity, including Pdx1, may be a viable treatment strategy for treating T2DM.
Experimental Procedures
Animals
We used a Cre/LoxP system to specifically delete Pdx1 in the pancreatic β-cell lineage. Pdx1fl/fl mice (Ahlgren et al., 1998) were bred to the RIP-CreER strain, which permits tamoxifen (TAM)-dependent recombination in β-cells (Dor et al., 2004), to create RIP-CreER; Pdx1fl/fl mice. As some RIP-Cre strains have been reported to exhibit recombination in cells other than pancreatic β-cells (Wicksteed et al., 2010), we sought to confirm the specificity of labeling within the pancreas; to this end, we counted more than 1000 islet cells in 1-month post-tamoxifen-treated control animals (RIP-CreER; Pdx1L/+; RosaYFP), and did not find any glucagon+/YFP+ cells. This result demonstrates that within the pancreas, the RIP-CreER strain is β-cell-specific. RIP-CreER animals were further bred to RosaYFP mice (JAX) to generate RIP-CreER; Pdx1fl/fl; RosaYFP mice (PKO mice), permitting lineage tracing of Pdx1-deleted cells. To activate recombination, 5 daily doses of TAM (6 mg each) in corn oil were given by oral gavage to 30 day old animals. Under these conditions, the labeling efficiency was 91% (expressed as % of insulin+ cells that become YFP+ in RIP-CreER; RosaYFP control animals). As controls, we used animals with one intact Pdx1 allele (i.e. RIP-CreER; Pdx1fl/+, RosaYFP mice). Results are representative of 6–7 animals in either control or mutant groups unless otherwise described. P-values were calculated by Student’s t-test. For glucose tolerance testing, control and PKO underwent an overnight fast. The next morning, 2 grams/kg body weight of 20% D-glucose was given by IP injection, after which blood glucose level was measured by glucometer at 15, 30, 60, and 120 minutes.
Islet studies
Calcium flow parameters of islet cells was measured by dual wavelength fluorescence microscopy using a Zeiss AxioVision system as indicator of cell type-specific response to either glycine, mixed amino acids (AAM), or glucose. Dispersed single islet cells were first incubated with Fura-2AM for 30 min and then perfused with different stimuli. Cytosolic calcium flux was calculated based on the 340nm/380nm emission ratio, which served as an indicator of cell stimulation. For hormone secretion measurements, 35 islets from either control or PKO mice were incubated with various stimuli (0.4 mM AAM, or 4 mM AAM + 10 mM Glucose) for 1 hour and tested for either insulin or glucagon secretion by HFRF assay (Cisbio, Inc.).
Histological Analysis and Immunostaining
For immunostaining of cultured Ins-1 cells, cells were washed with PBS, fixed in a 3.2% paraformaldehyde/PBS, permeabilized with 0.2% Triton X-100/PBS, and then stained overnight with primary antibodies to insulin, glucagon and MafB. For immunostaining histological sections, pancreas tissues were fixed in 4% paraformaldehyde overnight at 4°C followed by paraffin embedding, and sectioning at 5 μm thickness, followed by immunostaining as previously described (Zong et al., 2009). Primary antibody sources and application were as following: Goat anti-GFP (Abcam) at 1:500; Chicken anti-GFP (Abcam) at 1:500; Guinea pig anti-insulin (Abcam) at 1:500; Rabbit anti-glucagon (Millipore) at 1:500; Rabbit anti-Pdx1 (Abcam) at 1:500; Rabbit anti-somatostatin (DAKO) at 1:500; Rabbit anti-MafB (Bethyl Laboratories) at 1:2000; Goat anti-PP (Everest) at 1:500; Rabbit anti-ghrelin (Beta Cell Biology Consortium) at 1:500; Rabbit anti-Arx(Gift from Dr. Kanako Miyabayashi) at (1:1000); Rabbit anti-caspase-3 (Cell Signaling) at 1:1000.
For electron microscopy, pancreas samples were fixed in 2.5% glutaraldehyde, 2.0% paraformaldehyde, 0.1M sodium cacodylate buffer (pH 7.4) overnight at 4°C. After subsequent buffer washes, the samples were post-fixed in 2.0% osmium tetroxide for 1 hour at room temperature and rinsed in distilled H2O prior to en bloc staining with 2% uranyl acetate. After dehydration through a graded ethanol series, the tissue was infiltrated and embedded in EMbed-812 (Electron Microscopy Sciences, Fort Washington, PA). Thin sections were stained with uranyl acetate and lead citrate and examined with a JEOL 1010 electron microscope fitted with a Hamamatsu digital camera and AMT Advantage image capture software.
Microarray and Quantitative RT-PCR Analysis
For the collection of RNA from β-cells or lineage labeled Pdx1 knockout cells, PKO mice (RIPcreER; PDX1fl/fl, RosaYFP) and control mice (RIPcreER; PDX1fl/+, RosaYFP) were given TAM and sacrificed 5d later. Islets were isolated and dispersed into a single cell suspension by standard techniques before being sorted for YFP+ cells. For collection of RNA from α-cells, glucagon-Cre mice (Herrera, 2000) were crossed to RosaYFP mice. Single-cell suspensions were made from islets derived from bigenic animals of the appropriate genotype. Total RNA was extracted from YFP+ cells using an RNeasy total RNA isolation Kit (Qiagen) according to manufacturer’s protocol. 500ng of total RNA were used for the synthesis of cDNA, followed by amplification and biotin labeling. A total of 1.5 μg of biotinylated cRNAs was hybridized to the Mouse Gene 1.0ST microarray. After staining with streptavidin-phycoerythrin and biotinylated anti-streptavidin, samples were scanned and data collected by GCS3000 laser scanner. Statistical analyses were performed using Ingenuity Pathway Analysis database and under the R language environment. A heat map of expression was generated by Treeview (Eisen et al., 1998). Quantitative RT-PCR analysis was carried out using SYBR green gene expression assays (QIAGEN) according to the manufacturer’s instructions.
Adenoviral Pdx1 knockdown, mobility shift assays, chromatin immunoprecipitation, and MafB overexpression
Ins-1 cells/well (7.5×105) were seeded into 6-well plates and siRNAs against MafB or scrambled control (Dharmacon) introduced into cells using Lipofectamine 2000 (Invitrogen). Adenovirus expressing GFP or the Pdx1 siRNA were mixed with serum free DMEM to a concentration of 5×107 ifu/mL and infected for 48h before harvesting for immunofluorescence, protein, and RNA analysis. Nuclear extract from βTC3 cells was prepared as described previously (Schreiber et al., 1989), and binding analysis was performed with double-stranded mouse oligonucleotides (MafB Site 1 −1335 GCTTGGATCAGCTTAATCCTTACAAAACGT −1306, Site 2 −952 CCCCACCGCATACATTAGCGCAGACAGAGC −923, and Site 2 mutant (MafB site 1 mt: −952 GCTCTGTCTGCGCGCCGGTATGCGGTGGGG −923, mutated bases are underlined). Competition analysis was performed with at 100x molar excess of unlabeled competitor to labeled probe. Antibody super-shift analyses were performed by pre-incubating nuclear extract protein with Pdx1 antibody prior to adding probe. Samples were electrophoresed on 6% non-denaturing polyacrylamide gels at 150 V for 2 h in 1x TBE buffer (Tris-borate-EDTA). Gels were then dried and visualized by autoradiography. Chromatin immunoprecipitation (ChIP) assays were performed on three 10cm plates of 60% confluent Min6 cells as described previously (Raum et al., 2006). Quantitative real-time PCR was performed on immunoprecipitated DNA using SYBR Green master mix and a Roche LightCycler 480II. Primers used for amplification were as follows: MafB site1: Forward: −938 TTAGCGCAGACAGAGCTACCGAAA −915, Reverse: −742 ATACTCTTTACACTCCCACCCTCG −765; Glucagon G1 element: Forward: −148 CGTAAAAAGCAGATGAGCAAAGTG −125, Reverse: +46 GAACAGGTGTAGACAGAGGGAGTCC +70; Albumin distal TAAT-containing region: Forward: −3342 TGGGAAAACTGGGAAAACCATC −3319, Reverse: −3164 CACTCTCACACATACACTCCTGCTG −3188.
For ectopic expression studies, a MafB cDNA was cloned in-frame into a lenti-H2BCherry vector via a viral 2A peptide (Gao et al., 2013), resulting in simultaneous expression of MafB and an H2BCherry fluorescent marker. Lentivirus was generated and used to infect INS-1 cells as described previously (Zong et al., 2009). RNA extraction and RT-PCR analysis were performed as described previously (Gao et al., 2013). Lenti-H2BCherry infected-cells, which express H2BCherry protein without MafB, were used as a control.
Supplementary Material
Research Highlights.
Islet β-cells survive after Pdx1 deletion and undergo β- to α-cell reprogramming
Reprogramming involves transcriptional, ultrastructural, and physiological changes
Pdx1 binds to, and represses, the MafB promoter
De-repression of MafB following Pdx1 deletion contributes to reprogramming
Acknowledgments
We thank D. Melton and P. Herrera for providing RIP-CreER and glucagon-Cre mice, respectively, and K. Miyabayashi for the gift of Arx antibodies. We are grateful to M. Sander for sharing unpublished data and C. May and C. Wright for helpful discussions. This work was supported by grants from NIH/NIDDK (DK083355 and DK083111 to B.Z.S., DK050203 to R.S., DK089529 to C.L., and Vanderbilt Molecular Endocrinology Training Program grant 5T32 DK07563 to B.M.), the Penn Center for Molecular Studies in Digestive and Liver Diseases (P30-DK050306), the Penn Institute for Diabetes Obesity and Metabolism, the Penn Diabetes Research Center (DK19525), the Pew Charitable Trusts, and the Abramson Family Cancer Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 1998;12:1763–1768. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artner I, Blanchi B, Raum JC, Guo M, Kaneko T, Cordes S, Sieweke M, Stein R. MafB is required for islet beta cell maturation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3853–3858. doi: 10.1073/pnas.0700013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artner I, Hang Y, Mazur M, Yamamoto T, Guo M, Lindner J, Magnuson MA, Stein R. MafA and MafB regulate genes critical to beta-cells in a unique temporal manner. Diabetes. 2010;59:2530–2539. doi: 10.2337/db10-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artner I, Le Lay J, Hang Y, Elghazi L, Schisler JC, Henderson E, Sosa-Pineda B, Stein R. MafB: an activator of the glucagon gene expressed in developing islet alpha-and beta-cells. Diabetes. 2006;55:297–304. doi: 10.2337/diabetes.55.02.06.db05-0946. [DOI] [PubMed] [Google Scholar]
- Bramswig NC, Kaestner KH. Transcriptional regulation of alpha-cell differentiation. Diabetes Obes Metab. 2011;13(Suppl 1):13–20. doi: 10.1111/j.1463-1326.2011.01440.x. [DOI] [PubMed] [Google Scholar]
- Butler PC, Meier JJ, Butler AE, Bhushan A. The replication of beta cells in normal physiology, in disease and for therapy. Nat Clin Pract Endocrinol Metab. 2007;3:758–768. doi: 10.1038/ncpendmet0647. [DOI] [PubMed] [Google Scholar]
- Collombat P, Hecksher-Sorensen J, Krull J, Berger J, Riedel D, Herrera PL, Serup P, Mansouri A. Embryonic endocrine pancreas and mature beta cells acquire alpha and PP cell phenotypes upon Arx misexpression. The Journal of clinical investigation. 2007;117:961–970. doi: 10.1172/JCI29115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Mansouri A, Hecksher-Sorensen J, Serup P, Krull J, Gradwohl G, Gruss P. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev. 2003;17:2591–2603. doi: 10.1101/gad.269003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Xu X, Ravassard P, Sosa-Pineda B, Dussaud S, Billestrup N, Madsen OD, Serup P, Heimberg H, Mansouri A. The ectopic expression of Pax4 in the mouse pancreas converts progenitor cells into alpha and subsequently beta cells. Cell. 2009;138:449–462. doi: 10.1016/j.cell.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan S, Georgia S, Tschen SI, Fan G, Bhushan A. Pancreatic beta cell identity is maintained by DNA methylation-mediated repression of Arx. Dev Cell. 2011;20:419–429. doi: 10.1016/j.devcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon M, Ables ET, Crawford L, Lowe D, Offield MF, Magnuson MA, Wright CV. pdx-1 function is specifically required in embryonic beta cells to generate appropriate numbers of endocrine cell types and maintain glucose homeostasis. Dev Biol. 2008;314:406–417. doi: 10.1016/j.ydbio.2007.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T, Zhou D, Yang C, Singh T, Penzo-Mendez A, Maddipati R, Tzatsos A, Bardeesy N, Avruch J, Stanger BZ. Hippo signaling regulates differentiation and maintenance in the exocrine pancreas. Gastroenterology. 2013;144:1543–1553. 1553, e1541. doi: 10.1053/j.gastro.2013.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Guo S, Dai C, Guo M, Taylor B, Harmon JS, Sander M, Robertson RP, Powers AC, Stein R. Inactivation of specific beta cell transcription factors in type 2 diabetes. The Journal of clinical investigation. 2013 doi: 10.1172/JCI65390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon JS, Gleason CE, Tanaka Y, Oseid EA, Hunter-Berger KK, Robertson RP. In vivo prevention of hyperglycemia also prevents glucotoxic effects on PDX-1 and insulin gene expression. Diabetes. 1999;48:1995–2000. doi: 10.2337/diabetes.48.10.1995. [DOI] [PubMed] [Google Scholar]
- Herrera PL. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development. 2000;127:2317–2322. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- Jonas JC, Sharma A, Hasenkamp W, Ilkova H, Patane G, Laybutt R, Bonner-Weir S, Weir GC. Chronic hyperglycemia triggers loss of pancreatic beta cell differentiation in an animal model of diabetes. The Journal of biological chemistry. 1999;274:14112–14121. doi: 10.1074/jbc.274.20.14112. [DOI] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Li C, Chen P, Palladino A, Narayan S, Russell LK, Sayed S, Xiong G, Chen J, Stokes D, Butt YM, et al. Mechanism of hyperinsulinism in short-chain 3-hydroxyacyl-CoA dehydrogenase deficiency involves activation of glutamate dehydrogenase. The Journal of biological chemistry. 2010;285:31806–31818. doi: 10.1074/jbc.M110.123638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Liu C, Nissim I, Chen J, Chen P, Doliba N, Zhang T, Daikhin Y, Stokes D, Yudkoff M, et al. Regulation of Glucagon Secretion in Normal and Diabetic Human Islets by Gamma-Hydroxybutyrate and Glycine. The Journal of biological chemistry. 2012 doi: 10.1074/jbc.M112.385682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Hamze Z, Bonnavion R, Herath N, Pouponnot C, Assade F, Fontaniere S, Bertolino P, Cordier-Bussat M, Zhang CX. Reexpression of oncoprotein MafB in proliferative beta-cells and Men1 insulinomas in mouse. Oncogene. 2012;31:3647–3654. doi: 10.1038/onc.2011.538. [DOI] [PubMed] [Google Scholar]
- Nishimura W, Kondo T, Salameh T, El Khattabi I, Dodge R, Bonner-Weir S, Sharma A. A switch from MafB to MafA expression accompanies differentiation to pancreatic beta-cells. Dev Biol. 2006;293:526–539. doi: 10.1016/j.ydbio.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Papizan JB, Singer RA, Tschen SI, Dhawan S, Friel JM, Hipkens SB, Magnuson MA, Bhushan A, Sussel L. Nkx2.2 repressor complex regulates islet beta-cell specification and prevents beta-to-alpha-cell reprogramming. Genes Dev. 2011;25:2291–2305. doi: 10.1101/gad.173039.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raum JC, Gerrish K, Artner I, Henderson E, Guo M, Sussel L, Schisler JC, Newgard CB, Stein R. FoxA2, Nkx2.2, and PDX-1 regulate islet beta-cell-specific mafA expression through conserved sequences located between base pairs −8118 and −7750 upstream from the transcription start site. Molecular and cellular biology. 2006;26:5735–5743. doi: 10.1128/MCB.00249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spijker HS, Ravelli RB, Mommaas-Kienhuis AM, van Apeldoorn AA, Engelse MA, Zaldumbide A, Bonner-Weir S, Rabelink TJ, Hoeben RC, Clevers H, et al. Conversion of mature human beta-cells into glucagon-producing alpha-cells. Diabetes. 2013;62:2471–2480. doi: 10.2337/db12-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffers DA, Zinkin NT, Stanojevic V, Clarke WL, Habener JF. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nature genetics. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell. 2012;150:1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL, Liu FF, Sander M. Nkx6.1 is essential for maintaining the functional state of pancreatic Beta cells. Cell reports. 2013;4:1262–1275. doi: 10.1016/j.celrep.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorel F, Nepote V, Avril I, Kohno K, Desgraz R, Chera S, Herrera PL. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 2010;464:1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Maechler P, Ritz-Laser B, Hagenfeldt KA, Ishihara H, Philippe J, Wollheim CB. Pdx1 level defines pancreatic gene expression pattern and cell lineage differentiation. The Journal of biological chemistry. 2001;276:25279–25286. doi: 10.1074/jbc.M101233200. [DOI] [PubMed] [Google Scholar]
- Wicksteed B, Brissova M, Yan W, Opland DM, Plank JL, Reinert RB, Dickson LM, Tamarina NA, Philipson LH, Shostak A, Bernal-Mizrachi E, Elghazi L, Roe MW, Labosky PA, Myers MG, Jr, Gannon M, Powers AC, Dempsey PJ. Conditional gene targeting in mouse pancreatic ss-Cells: analysis of ectopic Cre transgene expression in the brain. Diabetes. 2010;59:3090–3098. doi: 10.2337/db10-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox CL, Terry NA, Walp ER, Lee RA, May CL. Pancreatic alpha-Cell Specific Deletion of Mouse Arx Leads to alpha-Cell Identity Loss. PloS one. 2013;8:e66214. doi: 10.1371/journal.pone.0066214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YP, Thorel F, Boyer DF, Herrera PL, Wright CV. Context-specific alpha- to-beta-cell reprogramming by forced Pdx1 expression. Genes Dev. 2011;25:1680–1685. doi: 10.1101/gad.16875711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Moriguchi T, Kajihara M, Esaki R, Harada A, Shimohata H, Oishi H, Hamada M, Morito N, Hasegawa K, et al. MafA is a key regulator of glucose-stimulated insulin secretion. Molecular and cellular biology. 2005;25:4969–4976. doi: 10.1128/MCB.25.12.4969-4976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Y, Panikkar A, Xu J, Antoniou A, Raynaud P, Lemaigre F, Stanger BZ. Notch signaling controls liver development by regulating biliary differentiation. Development. 2009;136:1727–1739. doi: 10.1242/dev.029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.