Abstract
Pitx2 is the last effector of the left-right (LR) cascade known to date and plays a crucial role in the patterning of LR asymmetry. In Xenopus embryos, the expression of Pitx2 gene in the left lateral plate mesoderm (LPM) is directly regulated by Xnr1 signaling, which is mediated by Smads and FoxH1. Previous studies suggest that the suppression of Pitx2 gene in the left LPM is a potential cause of cardiac/laterality defects in Oculo-Facio-Cardio-Dental (OFCD) syndrome, which is known to be caused by mutations in BCL6 co-repressor (BCOR) gene. Recently, our work has revealed that the BCL6/BCOR complex blocks Notch-dependent transcriptional activity to protect the expression of Pitx2 in the left LPM from the inhibitory activity of Notch signaling. These studies indicated that uncontrolled Notch activity in the left LPM caused by dysfunction of BCOR may result in cardiac/laterality defects of OFCD syndrome. However, this Notch-dependent inhibitory mechanism of Pitx2 gene transcription still remains unknown. Here we report that transcriptional repressor ESR1, which acts downstream of Notch signaling, inhibits the expression of Pitx2 gene by binding to a left side-specific enhancer (ASE) region in Pitx2 gene and recruiting histone deacetylase 1 (HDAC1) to this region. Once HDAC1 is tethered, histone acetyltransferase p300 is no longer recruited to the Xnr1-dependent transcriptional complex on the ASE region, leading to the suppression of Pitx2 gene in the left LPM. The study presented here uncovers the regulatory mechanism of Pitx2 gene transcription which may contribute to an understanding of pathogenesis of OFCD syndrome.
Keywords: Notch signaling, Pitx2, left-right patterning, Xenopus, OFCD syndrome
Introduction
Anatomical LR asymmetry of the internal organs, such as the orientation of the cardiovascular system, visceral organs and the number of lung lobes, is conserved in vertebrates (Blum et al., 2009; Levin, 2005; Palmer, 2004). Although the mechanisms involved in breaking LR symmetry during very early vertebrate development may not be conserved, the left-side specific expression of genes in the LPM such as Xnr1(a Xenopus ortholog of mouse Nodal), Lefty as well as Pitx2 have been observed in all vertebrates studied to date and these factors are essential for the patterning of LR asymmetry (Boorman and Shimeld, 2002; Hamada et al., 2002; Kato, 2011; Raya and Belmonte, 2006; Speder et al., 2007).
Pitx2 is a homeobox transcription factor that plays an important role for establishing LR asymmetry during development. In fact, knockout of Pitx2 in mice results in severe cardiac/laterality defects including transposition of the great arteries, double-outlet right ventricle, septal defect, right cardiac isomerism and right lung isomerism (Gage et al., 1999; Lin et al., 1999; Lu et al., 1999). Furthermore, ectopic expression of Pitx2 in the right side of chick, zebrafish and frog embryos affected the direction of heart looping and gut coiling (Essner et al., 2000; Logan et al., 1998; Ryan et al., 1998). During LR patterning, Pitx2 acts downstream of TGFβ superfamily Xnr1 in the left LPM of Xenopus embryos (Campione et al., 1999; Schweickert et al., 2000) and this molecular cascade in the left LPM is conserved in all vertebrates examined to date (Long et al., 2003; Meno et al., 1998; Piedra et al., 1998; Yoshioka et al., 1998). The Xnr1 signaling pathway, which induces the expression of Pitx2 in the left LPM, is mediated by the transcriptional complex including Smad2/3, Smad4 and FoxH1. Transcription factor FoxH1 directly binds to the ASE region in Pitx2 gene and initiates transcription of Pitx2 gene (Shiratori et al., 2001). Interestingly, previous studies indicated that the suppression of Pitx2 in the left LPM induced by dysfunction of BCOR may be a cause of laterality defects in the heart and other viscera of patients with OFCD syndrome (Hilton et al., 2007; Lin et al., 2000).
OFCD syndrome is an X-linked disorder characterized by ocular, dental, cardiac/laterality and skeletal anomalies as well as mental retardation (Aalfs et al., 1996; Gorlin et al., 1996; Hayward, 1980; Hilton et al., 2007; Lin et al., 2000; Marashi and Gorlin, 1990, 1992; Wilkie et al., 1993). Frequent ocular defects include congenital cataracts and microphthalmia. Facial anomalies include septate nasal tip, high nasal bridge, midface hypoplasia as well as palatal anomalies. Congenital cardiac abnormalities comprise septal defects and mitral valve defects. Dental irregularities include canine radiculomegaly, delayed and persistent dentition as well as hypodontia. Skeletal anomalies contain syndactyly and hammer-type flexion deformities. Defective lateralization includes dextrocardia, asplenia and intestinal malrotation. Genetic studies have shown that mutations in BCOR gene at chromosomal location Xp11.4 cause OFCD syndrome (Ng et al., 2004). These mutations in BCOR gene result in premature termination of the protein with deletion of the C-terminal domain (Horn et al., 2005; Ng et al., 2004; Oberoi et al., 2005). BCOR was originally identified as a co-repressor of transcriptional repressor BCL6 (Huynh et al., 2000). BCOR has been reported to interact with histone deacetylase (HDAC), demethylase and H2A ubiquitin ligase (Gearhart et al., 2006; Huynh et al., 2000; Sanchez et al., 2007; Tsukada et al., 2006), suggesting that BCOR may mediate epigenetic silencing to repress transcription of target genes. Therefore, uncontrolled factors whose transcription is supposed to be blocked by BCOR likely cause the phenotypes of OFCD syndrome. Recently, our work has shown that uncontrolled Notch activity inhibits the expression of Pitx2 in the left LPM of Xenopus embryos but only when the function of the BCL6/BCOR complex is incompetent (Sakano et al., 2010). This abnormal activity of Notch signaling is mediated by transcriptional repressor ESR1, a direct downstream factor of Notch signaling (Lamar and Kintner, 2005). These studies indicated that abnormally-activated Notch signaling in the left LPM induced by dysfunction of BCOR suppresses the expression of Pitx2, resulting in cardiac/laterality defects of patients with OFCD syndrome. However, it is not known how this Notch activity blocks transcription of Pitx2 gene.
To understand this inhibitory mechanism by Notch signaling, we employed the frog Xenopus laevis as the animal model system in this study. Xenopus produces a large number of embryos with each fecundation. Xenopus also exhibits a very short developmental time during early development, external development as well as close homology with human genes. The anatomical structure of Xenopus is similar to human; particularly both are tetrapods. Furthermore, this model easily enables the study of gene function in vivo by gain-of-function and loss-of-function experiments with microinjection of synthetic RNAs or antisense oligonucleotides. These favorable features are very suitable to study the pathogenesis of human congenital diseases. Here we report that the expression of Pitx2 gene is inhibited by binding of ESR1 to the ASE region in Pix2 gene. After ESR1 recruits histone deacetylase 1 (HDAC1) to the ASE region, histone acetyltransferase p300 cannot be recruited to the Xnr1-dependent transcriptional complex on the ASE region, resulting in the suppression of Pitx2 gene in the left LPM. Our study reveals the inhibitory mechanism of Pitx2 gene transcription induced by uncontrolled Notch signaling which is caused by dysfunction of the BCL6/BCOR complex.
Results
Notch-ESR signal targets the Xnr1-dependent transcriptional complex in the left LPM
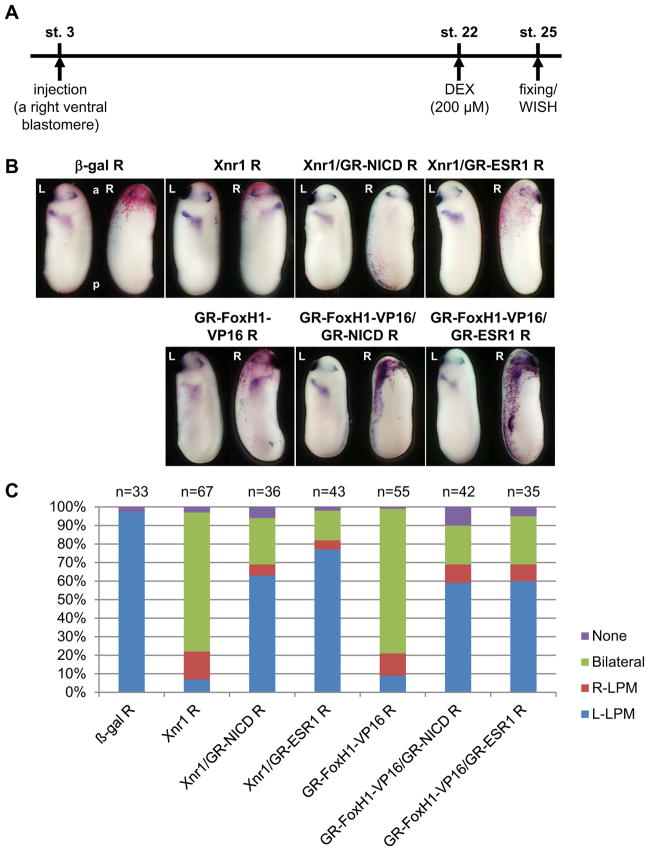
To understand how uncontrolled Notch activity suppresses the expression of Pitx2 in the left LPM, we first determined the target of Notch-dependent inhibitory activity in Xnr1 signaling that induces the expression of Pitx2 in the left LPM (Campione et al., 1999). We tested whether Notch activity suppresses the expression of Pitx2 in the LPM induced by ectopic expression of either Xnr1 or FoxH1-Vp16 which is an active form of FoxH1 (Watanabe and Whitman, 1999). For this purpose, we used hormone-inducible constructs including GR-FoxH1-VP16, GR-NICD as well as GR-ESR1, and these are activated at stage 22 by dexamethasone (DEX) when the expression of Xnr1 in the left LPM has already started (Fig. 1A). Hormone-inducible constructs were used because Notch signaling induces the bilateral expression of Xnr1 around the gastrocoel roof plate (GRP) at the neurula stage, which is essential for the expression of Xnr1 in the left LPM at the early tailbud stage (Sakano et al., 2010; Schweickert et al., 2010). It was necessary to avoid abnormal effects of Notch signaling at the neurula stage, prior to the tailbud stage. Xnr1 or GR-FoxH1-VP16 RNA was ectopically expressed in the right LPM (Fig. 1B, C). In either case, Pitx2 was bilaterally expressed or expressed at the right side in about 90 % of injected embryos (Fig. 1C). When GR-NICD, a constitutively active form of Notch1 receptor, or GR-ESR1 RNA (Sakano et al., 2010) was co-injected with Xnr1 or GR-FoxH1-VP16, the number of embryos with bilaterally or right expressing Pitx2 in either Xnr1 or GR-FoxH1-VP16 injected embryos was reduced to about 30 % (Fig. 1C). This result indicates that the potential target of Notch-ESR1 signal is the Xnr1-dependent transcriptional activity, which activates the expression of Pitx2 gene in the LPM, such as the function of FoxH1, co-activators and chromatin modifying factors.
Fig. 1. The Xnr1-dependent transcriptional complex is a potential target of Notch signaling.
A: The experimental strategy. B: One pg Xnr1 or 50 pg GR-FoxH1-VP16 RNA were injected into a right ventral blastomere of 4-cell-stage embryos with or without 2 ng GR-NICD or 1 ng GR-ESR1 RNA. DEX was added into the culture medium at stage 22. The expression of Pitx2 at stage 25 was tested by whole mount in situ hybridization. C: The quantitative assessment of the injections in (B). At least three independent experiments were performed. “n” indicates the number of injected embryos. R: injection into the right side of embryo.
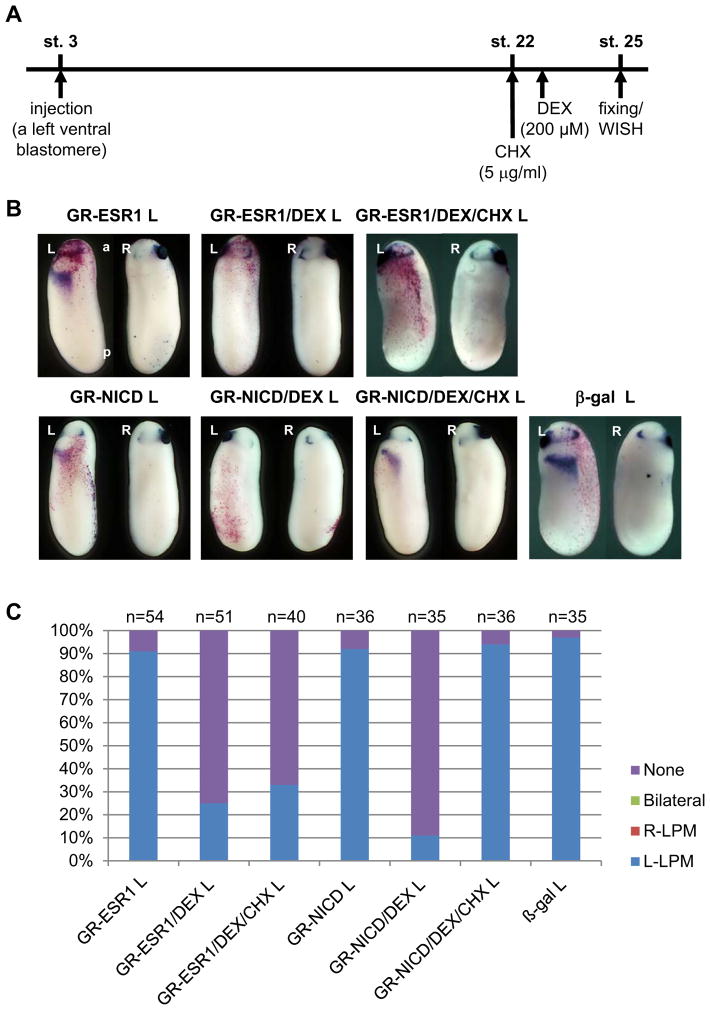
ESR1 is a transcriptional repressor with the basic-helix-loop-helix (bHLH) DNA-binding domain and a frog ortholog of mammalian Hes5 (Davis and Turner, 2001; Lamar and Kintner, 2005). Therefore, it is possible that ESR1 binds to the ASE region in Pitx2 gene, on which the Xnr1-dependent transcriptional complex is to initiate transcription (Shiratori et al., 2001), and inhibits the expression of Pitx2 in the left LPM. To confirm this possibility, we examined whether ESR1 inhibits the expression of Pitx2 without new protein synthesis. GR-ESR1 RNA was expressed in the left LPM and new protein synthesis was blocked by cycloheximide (CHX) at stage 22. After that, GR-ESR1 was activated by adding DEX (Fig. 2A). The expression of Pitx2 in the left LPM was suppressed in about 75 % of GR-ESR1 injected embryos with DEX or about 70 % of GR-ESR1 injected embryos with DEX and CHX (Fig. 2B, C). This result demonstrated that new protein synthesis was not required for ESR1 to suppress the expression of Pitx2. As a control, a similar experiment using GR-NICD was performed to test the effect of CHX. The expression of Pitx2 in the left LPM was suppressed in about 90 % of GR-NICD injected embryos with DEX or about 5 % of GR-NICD injected embryos with DEX and CHX (Fig. 2C). This showed that the CHX treatment blocked the synthesis of ESR1 protein, resulting in the rescue of Pitx2 gene expression in the LPM. This result reveals that ESR1 inhibits the expression of Pitx2 in the left LPM without induction of downstream factors.
Fig. 2. ESR1 directly blocked the expression of Pitx2 gene in the left LPM.
A: The experimental strategy. B: One ng GR-NICD or 1 ng GR-ESR1 RNA was injected into a left ventral blastomere of 4-cell-stage embryos. DEX was added into the culture medium at stage 22 and CHX was added 30 minutes before the addition of DEX. C: The quantitative assessment of the injections in (B). At least three independent experiments were performed. “n” indicates the number of injected embryos. L: injection into the left side of embryo.
Taken together these results suggest that ESR1 binds to the ASE region in Pitx2 gene and blocks the function of the Xnr1-dependent Smad/FoxH1 complex on the ASE region.
ESR1 blocks the recruitment of histone acetyltransferase p300 to the ASE region in Pitx2 gene
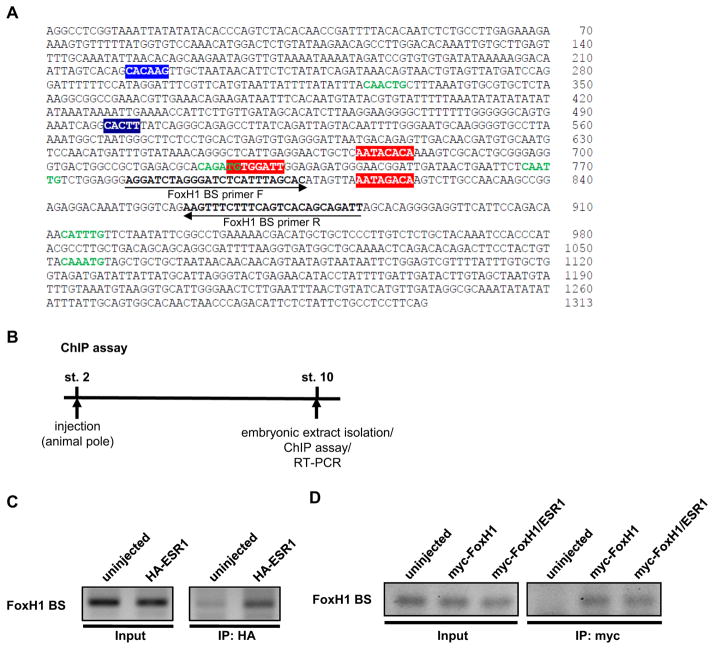
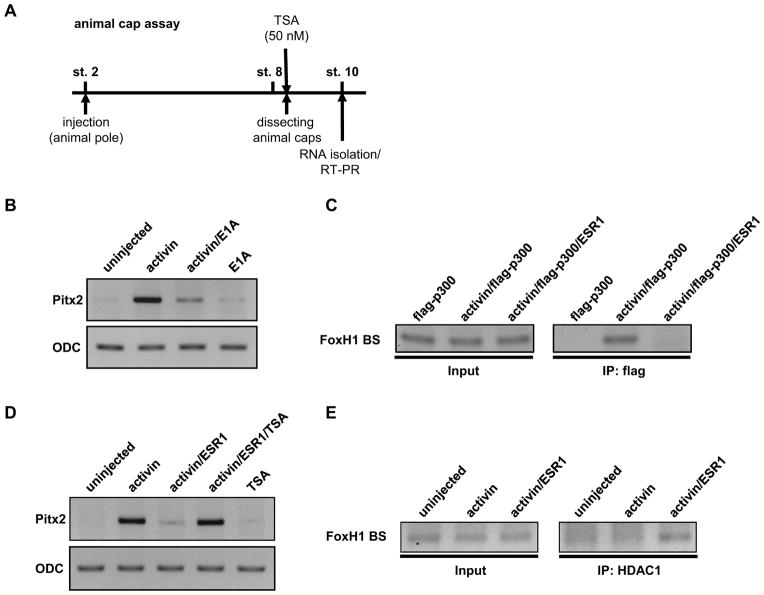
To determine how Notch-ESR1 signal suppresses the expression of Pitx2, two hypotheses were examined. The first hypothesis is that ESR1 blocks the interaction between FoxH1 and the ASE region by masking the FoxH1-binding sites on the ASE region in Pitx2 gene. The ASE region contains three FoxH1-binding sites (red boxes in Fig. 3A), one N-box (a blue box in Fig. 3A), to which the Hes factors preferentially bind, and five E-boxes (green letters in Fig. 3A), to which most bHLH transcriptional factors bind (Hirata et al., 2000; Kageyama et al., 2007; Sasai et al., 1992; Shiratori et al., 2001). First, the interaction between ESR1 and the ASE region was examined by chromatin-immunoprecipitation (ChIP) assay (Fig. 3B). HA-ESR1 was over-expressed in Xenopus embryos and extract from these embryos was used for ChIP assay. HA-ESR1 was co-immunoprecipitated with the DNA fragment including the FoxH1-binding site in the ASE region (Fig. 3C). Next, we tested whether ESR1 interferes with the interaction between FoxH1 and the ASE region. The interaction between FoxH1 and the ASE region was detected in the embryos expressing myc-FoxH1 and this interaction was not blocked by the expression of HA-ESR1 (Fig. 3D). Although the different amount of myc-FoxH1 RNA (100 pg–2 ng) was co-injected with HA-ESR1, HA-ESR1 could not block the interaction between FoxH1 and the ASE region (data not shown). These results showed that the first hypothesis seems to be incorrect. The second hypothesis is that ESR1 prevents the recruitment of histone acetyltransferase p300 to the Xnr1-dependent transcriptional complex. This hypothesis was developed based on two observations: 1) over-expression of ESR1 could inhibit the activity of FoxH1-VP16, which does not need activated Smad2 to initiate transcription of Pitx2 gene, (Fig. 1, 2), and 2) p300 interacts with Smad2 (Janknecht et al., 1998) or the VP16 domain (Wang et al., 2000) to initiate transcription. First, we tested whether p300 function is required for the Xnr1-dependent transcriptional activity in the expression of Pitx2 gene. The expression of Pitx2 was induced by activin in animal cap explants representing the left LPM, which activates the Smad2-FoxH1 pathway similar to Xnr1 and induces the expression of Pitx2 in animal cap explants (Schweickert et al., 2000; Watanabe and Whitman, 1999) (Fig. 4A). As expected, co-injection of E1A RNA, which is an inhibitor of p300 (Howe et al., 1990; Kato et al., 1999), decreased the expression of Pitx2, indicating that p300 is necessary to induce the expression of Pitx2 (Fig. 4B). Next, the interaction between p300 and the ASE region was examined by ChIP assay (Fig. 3B). In the Xenopus embryos flag-p300 was co-expressed with activin, which stimulates the generation of endogenous Smads/FoxH1 complex, and the extract from these embryos was used for ChIP assay with α-flag antibody. The interaction between p300 and the ASE region was detected (Fig. 4C). These data show that p300 is involved in the initiation of Pitx2 gene transcription. Next, we tested whether ESR1 can block the recruitment of p300 to the ASE region. ESR1 and p300 were co-expressed in Xenopus embryos and the interaction between p300 and the ASE region was examined by ChIP assay. Co-expression of ESR1 prevented the interaction between p300 and the ASE region (Fig. 4C). These results demonstrate that ESR1 binds to the ASE region in Pitx2 gene and blocks the recruitment of p300 to the Xnr1-dependent transcriptional complex.
Fig. 3. ESR1 and FoxH1 independently bind to the ASE region in Pitx2 gene.
A: Nucleotide sequence of the ASE region in Xenopus Pitx2 gene. Three FoxH1-binding sites (red boxes), one N-box (blue box) and five E-boxes (green letters) are indicated within the ASE region. B: The experimental strategy of ChIP assay. C: One ng HA-ESR1 RNA was injected into 2-cell-stage embryos, and embryonic extracts were isolated at stage 10 for ChIP analysis. ChIP assays were performed using α-HA antibody. D: One ng myc-FoxH1 RNA was injected into 2-cell-stage embryos with or without 1 ng flag-ESR1 RNA, and nuclear extracts were isolated at stage 10 for ChIP analysis. ChIP assays were performed using α-myc antibody.
Fig. 4. ESR1 recruits HDAC1 to the ASE region to exclude p300 from the Xnr1-dependent transcriptional complex.
A: The experimental strategy of animal cap assay. Zygotic transcription starts at stage 8. B: Twenty pg activin RNA was injected into 2-cell-stage embryos with or without 100 pg E1A RNA and animal caps were dissected at the stage 8. Caps were cultured until sibling embryos reached stage 10, and Pitx2 gene expression was evaluated by semi-quantitative RT-PCR analysis. C: Two ng flag-p300 RNA and/or 20 pg activin RNA was injected into 2-cell-stage embryos with or without 2 ng HA-ESR1 RNA, and embryonic extracts were isolated at stage 10 for ChIP analysis. ChIP assays were performed using α-flag antibody. D: Twenty pg activin RNA was injected into 2-cell-stage embryos with or without 2 ng HA-ESR1 RNA, and animal caps were dissected at the stage 8. Caps were cultured with or without 50 nM TSA until sibling embryos reached stage 10, and Pitx2 gene expression was evaluated by semi-quantitative RT-PCR analysis. E: Twenty pg activin RNA was injected into 2-cell-stage embryos with or without 2 ng HA-ESR1 RNA, and embryonic extracts were isolated at stage 10 for ChIP analysis. ChIP assays were performed using α-HDAC1 antibody.
The next question was how ESR1 blocks the recruitment of p300 to the ASE region. Since mammalian Hes proteins interacts with Groucho/TLE proteins (Fisher et al., 1996; Grbavec et al., 1998; Grbavec and Stifani, 1996) and recruits histone deacetylases (HDACs) to shut down transcription (Chen et al., 1999), ESR1 may also recruit HDACs to the ASE region and repress the expression of Pitx2 gene. Therefore, we tested whether Trichostatin A (TSA), an inhibitor of the class I and II HDACs (Khan et al., 2008), blocks the inhibitory function of ESR1 in the expression of Pitx2 (Fig. 4A). In animal cap explants, co-injection of ESR1 with activin reduced the expression of Pitx2. When TSA was added to the culture medium, the expression level of Pitx2 in the ESR1 injected explants was restored to a level similar to one induced by activin alone (Fig. 4D). This suggests that the function of class I and II HDACs is necessary for the suppression of Pitx2 gene by ESR1. In order to test whether ESR1 recruits HDACs to the ASE region, the interaction between HDAC1 and the ASE region in the presence of ESR1 was examined by ChIP assay. Although HDAC1 was not precipitated in the activin stimulation, HDAC1 was recruited to the ASE region when ESR1 was expressed (Fig. 4E). This result shows that ESR1 recruits HDAC1 to the ASE region in Pitx2 gene.
Taken together, our data demonstrate that ESR1 induced by abnormal activation of Notch signaling binds to the ASE region in Pitx2 gene and prevents the recruitment of p300 to Xnr1-dependent transcriptional complex on the ASE region by recruiting HDAC1 (Fig. 5). HDAC1 may alter local chromatin structure to shut down transcription of Pitx2 gene.
Fig. 5. The model of the inhibitory mechanism by Notch-ESR1 signal in transcription of Pitx2 gene.
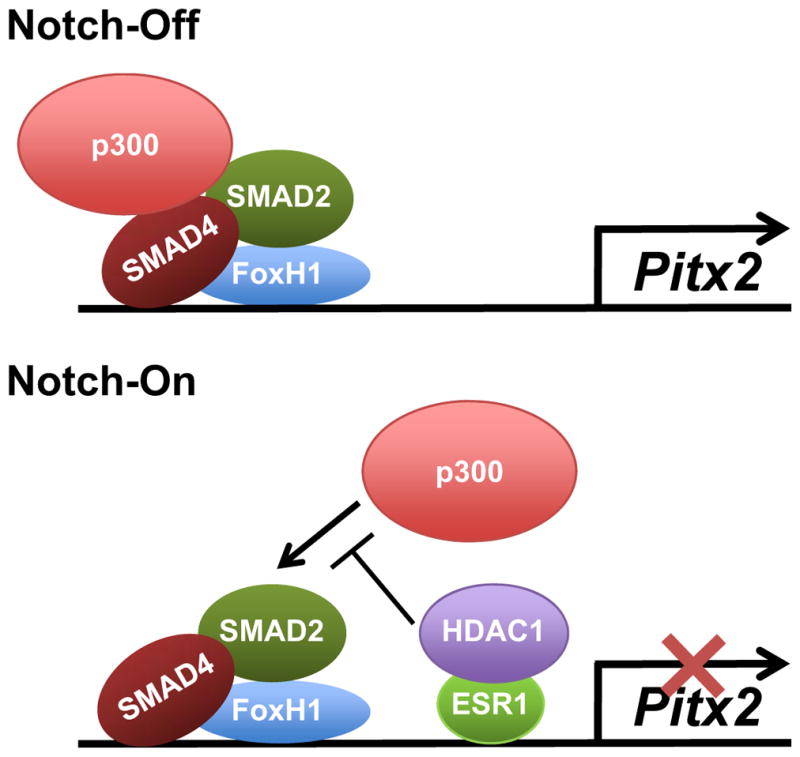
When the BCL6/BCOR complex malfunctions in the left LPM, Notch signaling excessively initiates the expression of ESR1 gene. ESR1 binds to the ASE region in Pitx2 gene and recruits HDAC1 to this region. HDAC1 may alter local chromatin structure, and prevents the recruitment of p300 to the Xnr1-dependent transcriptional complex on the ASE region. Therefore, Pitx2 expression is suppressed in the left LPM, resulting in laterality defects.
Discussion
The analysis presented here provides the inhibitory mechanism of Pitx2 gene transcription by uncontrolled Notch signaling in the left LPM of Xenopus embryos when the function of BCL6/BCOR complex is blocked. Transcriptional repressor ESR1 induced by abnormally-activated Notch signaling binds to the ASE region in Pitx2 gene and recruits HDAC1. Then, HDAC1 blocks the expression of Pitx2 gene in the left LPM by preventing the recruitment of p300 to the Xnr1-dependent transcriptional complex.
X-linked OFCD syndrome is caused by mutations in BCOR gene including nonsense, frameshift, deletion and splicing mutations (Ng et al., 2004). This syndrome is characterized by ocular, dental, cardiac/laterality and skeletal anomalies as well as mental retardation (Aalfs et al., 1996; Gorlin et al., 1996; Hilton et al., 2009; Lin et al., 2000). Importantly, previous studies showed that dysfunction of BCOR causes laterality defects of the heart and other viscera and induces the suppression of Pitx2 gene in the left LPM (Hilton et al., 2007; Lin et al., 2000), indicating that the suppression of Pitx2 gene may trigger cardiac/laterality defects of patients with OFCD syndrome. BCOR was originally found as a co-repressor of transcriptional repressor BCL6 (Huynh et al., 2000). BCOR has also been shown to regulate the targets of BCL6 as a component of an 800 kDa complex which contains at least two chromatin modifying factors; the PcG transcriptional repressor RNF2, a histone H2A E3 ubiquitin ligase, and FBXL10/JHDM1B, a Jmjc histone demethylase (Gearhart et al., 2006; Sanchez et al., 2007). Recently, our work using Xenopus embryos revealed that BCOR interacts with BCL6 and blocks the Notch-dependent transcription of the selected target gene, ESR1, to maintain the expression of Pitx2 in the left LPM during the patterning of LR asymmetry (Sakano et al., 2010). The study presented here with these previous data indicates the model mechanism which causes cardiac/laterality defects in OFCD syndrome; dysfunction of the BCL6/BCOR complex by mutations in the BCOR gene induces the expression of ESR1 through abnormal activation of Notch signaling, and binding of ESR1 to the ASE region leads to the suppression of Pitx2 in the left LPM by recruiting HDAC1, resulting in cardiac/laterality defects. It still remains unclear if the suppression of Pitx2 gene induced by dysfunction of BCOR leads to other disorders of OFCD syndrome such as ocular, dental and skeletal anomalies as well as mental retardation. Interestingly, previous studies have shown that knockout of Pitx2 resulted in embryonic lethality as well as ocular, craniofacial, dental, brain, heart, lung, body wall and other systemic defects (Evans and Gage, 2005; Gage et al., 1999; Kitamura et al., 1999; Lin et al., 1999; Liu et al., 2003; Lu et al., 1999; Martin et al., 2004), suggesting that the suppression of Pitx2 gene may cause most defects of OFCD syndrome. However, there may not be only one inhibitory mechanism of Pitx2 gene transcription. Pitx2 gene encodes several different isoforms; these include four, Pitx2A-D, in humans (Cox et al., 2002), three, Pitx2a-c, in mice and frogs (Schweickert et al., 2000) and two, Pitx2a and Pitx2c, in chickens (Yu et al., 2001) and zebrafish (Essner et al., 2000). The isoforms have different N-terminal sequences but share the homeodomain and C-terminal region. While the expression profile of Pitx2 isoforms showed largely overlapping expression patterns, some differences of expression patterns among Pitx2 isoforms were observed (Essner et al., 2000; Liu et al., 2003; Schweickert et al., 2000; Yu et al., 2001). This suggests that some isoforms likely use specific enhancers for transcription initiation. To date, only the Pitx2c expression in the left LPM of Xenopus embryos, which is regulated through the ASE region (Shiratori et al., 2001), has been reported to be regulated by the BCL6/BCOR complex (Hilton et al., 2007; Sakano et al., 2010). Furthermore, BCOR has been shown to bind to some transcriptional factors in addition to BCL6. For instance, the transcriptional regulator AF9/MLLT3 directly interacts with BCOR (Srinivasan et al., 2003). AF9 is a regulator of Hox gene expression and essential for skeletal development (Collins et al., 2002). Therefore, it is possible that unknown inhibitory mechanisms of Pitx2 gene transcription other than the inhibition by uncontrolled Notch activity may be involved in the pathogenesis of OFCD syndrome. Further studies will be necessary to more deeply dissect the molecular pathogenesis of OFCD syndrome.
Xenopus has been a principal animal model system for studies of biomedical science for over 80 years and has contributed to diverse fields including molecular, cell, developmental, stem cell and system biology as well as ion channel physiology and toxicology (Khokha, 2012). Xenopus has many favorable features for in vivo studies such as the production of thousands of embryos with each fecundation (sufficient quantity for biochemical purification and microinjection of nucleic acids), a very rapid developmental time during early development, larger size embryos (the best material for manipulation and microsurgery) and external development. Xenopus also provides abundant totipotent cells that can be transformed into a host of different tissues (useful material to study stem cell biology). Since Xenopus exhibits close homology to human genes with conserved cell signaling and has a similar anatomical structure to humans (both tetrapods), the basic cell, molecular, and developmental mechanisms discovered in Xenopus can generally be applied to studies for human diseases. Furthermore, rapid gain-of-function and loss-of-function assays using microinjection of RNA or antisense morpholino oligonucleotides, one of the favorable features of Xenopus, provide a very useful method to examine the abnormal function of potential responsible genes for human diseases in vivo (Boskovski et al., 2013; Hilton et al., 2007). These gain-of-function and loss-of-function assays have also been used to identify a responsible sequence variant relevant to human diseases among many sequence variants discovered in human patient samples by the benefits of recent sequencing techniques (Fakhro et al., 2011; Pittman et al., 2009). The study presented here using these favorable features of Xenopus described above has shown that Xenopus is very valuable animal model system that can provide insights into the molecular pathogenesis of human diseases. Therefore, the frog Xenopus should be reaffirmed as a suitable animal model system to study human diseases and used more in future studies of human diseases.
Materials and Methods
Embryo Manipulations
Eggs were artificially fertilized by using testis homogenate and cultivated in 0.1× Marc’s Modified Ringer’s solution (MMR) (Peng, 1991). Embryos were staged according to Nieuwkoop and Faber (Nieuwkoop and Faber, 1967).
DNA Constructs
pCS2+ GR-NICD, pCS2+ GR-ESR1, pCS2+ HA-ESR1, and pCS2+ E1A have been described previously (Kato et al., 1999; Sakano et al., 2010). Xenopus FoxH1-VP16, Xenopus FoxH1, or flag-human p300 was sub-cloned into pCS2+ GR or pCS2+ vector, respectively.
Microinjection of Synthetic RNA
Capped synthetic RNAs were generated by in vitro transcription with SP6 polymerase, using the mMessage mMachine kit (Ambion, Inc.). For microinjections, embryos were injected with 5–10 nl of the specified amount of RNA in 3% Ficoll in 0.1 x MMR and cultured in 0.1 x MMR until the desired stage. nucβ-gal RNA for whole mount in situ hybridization was injected as a tracer. For the activation of GR-fused proteins, dexamethasone (DEX: final concentration 200 μM) was added into the culture medium. For inhibition of protein synthesis, cycloheximide (CHX: final concentration 5 μg/ml) was added into the culture medium 30 minutes before DEX was added.
Beta-Galactosidase Staining and Whole Mount In Situ Hybridization
Embryos were fixed with MEMFA (0.1 M MOPS, 2 mM EGTA [pH8.0], 1 mM MgSO4 and 3.7 % formaldehyde) containing 0.02 % Triton-X for 30 minutes at room temperature. Galactosidase activity was visualized with the RedGal substrate (Research Organics) in staining buffer (5 mM K3[Fe(CN)6], 5 mM K4[Fe(CN)6], 2 mM MgCl2 in PBS). After staining, embryos were re-fixed with MEMFA for 30 minutes. Whole mount in situ hybridization was performed as described previously (Harland, 1991; Kiyota et al., 2008; Takada et al., 2005) by using Digoxigenin (Roche Applied Science)-labeled antisense RNA probes and BM purple (Roche Applied Science) for the chromogenic reaction.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed according to Blythe et al. (Blythe et al., 2009). Briefly, stage 10 embryos were fixed with 1% formaldehyde in PBS for 60 minutes at room temperature. After fixation, embryos were homogenized in 600 μl RIPA buffer (50 mM Tris-HCl [pH 7.4], 1 % NP-40, 0.25 % Na-Deoxycholate, 150 mM NaCl, 1 mM EDTA, 0.1 % SDS, 0.5 mM DTT, 5 mM Na-Butyrate, Protease Inhibitor Cocktail, Phosphatase Inhibitor Cocktail), and centrifuged. The pellet was re-homogenized in 650 μl RIPA buffer and crosslinked chromatin is sheared to <1,000-bp fragments by sonication. Sonicated samples were immunoprecipitated by 1 μg α-myc, α-HA, α-HDAC1 (Santa Cruz Biotechnology, Inc) or α-flag (Sigma) antibody with 50 μl Protein G-PLUS agarose beads (Santa Cruz biotechnology, Inc). After immunoprecipitation, eluted DNA was purified by Phenol/Chloroform extraction and analyzed by using PCR with specific primer pair (FoxH1-BS, PCR cycle 40: Forward 5′-AGGATCTAGGGATCTCATTAGCAC-3′, Reverse 5′-AATCTGCTGTGACTGAAAGAAACTT-3′).
Animal Cap Assay and Semi-quantitative RT-PCR Analysis
After synthetic RNAs were injected into animal pole of 2-cell stage embryos, animal caps were dissected from stage 8 embryos. Caps were cultured in 0.5 x MMR until stage 10 and total RNA was isolated from them. For inhibition of HDAC function, Trichostatin A (TSA: final concentration: 50 nM) was added into the culture medium. Total RNA was isolated with TRIzol reagent (Life technologies) according to the manufacturer’s instructions. Semi-quantitative RT-PCR was performed as described previously (Kato et al., 1999). Primer sequences are followed. ODC (PCR cycles 20): Forward 5′-ACATGGCATTCTCCCTGAAG-3′, Reverse 5′-TGGCCCAAGGCTAAAGTTG-3′. Pitx2 (PCR cycles 25): Forward 5′-TGGCTGGGAGTAGAGTTGCT-3′, Reverse 5′-ATCGGTACTGCTGTCCTCGT-3′.
Highlights.
Notch-ESR1 signal directly blocks the left-specific expression of Pitx2.
ESR1 binds to the ASE region of Pitx2 gene and recruits HDAC1 to this region.
ESR1 excludes p300 from the Xnr1-induced transcriptional complex on the ASE region.
Uncontrolled Notch-ESR1 signal may cause some phenotypes of OFCD syndrome.
Acknowledgments
We thank Drs. Whitman and Hamada for plasmids. We also thank S. B. and R. E. for critical suggestions to the manuscript. This work was supported by NIH grant (GM087641).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aalfs CM, Oosterwijk JC, van Schooneveld MJ, Begeman CJ, Wabeke KB, Hennekam RC. Cataracts, radiculomegaly, septal heart defects and hearing loss in two unrelated adult females with normal intelligence and similar facial appearance: confirmation of a syndrome? Clinical dysmorphology. 1996;5:93–103. doi: 10.1097/00019605-199604000-00001. [DOI] [PubMed] [Google Scholar]
- Blum M, Beyer T, Weber T, Vick P, Andre P, Bitzer E, Schweickert A. Xenopus, an ideal model system to study vertebrate left-right asymmetry. Dev Dyn. 2009;238:1215–1225. doi: 10.1002/dvdy.21855. [DOI] [PubMed] [Google Scholar]
- Blythe SA, Reid CD, Kessler DS, Klein PS. Chromatin immunoprecipitation in early Xenopus laevis embryos. Dev Dyn. 2009;238:1422–1432. doi: 10.1002/dvdy.21931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorman CJ, Shimeld SM. The evolution of left-right asymmetry in chordates. Bioessays. 2002;24:1004–1011. doi: 10.1002/bies.10171. [DOI] [PubMed] [Google Scholar]
- Boskovski MT, Yuan S, Pedersen NB, Goth CK, Makova S, Clausen H, Brueckner M, Khokha MK. The heterotaxy gene GALNT11 glycosylates Notch to orchestrate cilia type and laterality. Nature. 2013;504:456–459. doi: 10.1038/nature12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campione M, Steinbeisser H, Schweickert A, Deissler K, van Bebber F, Lowe LA, Nowotschin S, Viebahn C, Haffter P, Kuehn MR, Blum M. The homeobox gene Pitx2: mediator of asymmetric left-right signaling in vertebrate heart and gut looping. Development (Cambridge, England) 1999;126:1225–1234. doi: 10.1242/dev.126.6.1225. [DOI] [PubMed] [Google Scholar]
- Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes & development. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins EC, Appert A, Ariza-McNaughton L, Pannell R, Yamada Y, Rabbitts TH. Mouse Af9 is a controller of embryo patterning, like Mll, whose human homologue fuses with Af9 after chromosomal translocation in leukemia. Molecular and cellular biology. 2002;22:7313–7324. doi: 10.1128/MCB.22.20.7313-7324.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CJ, Espinoza HM, McWilliams B, Chappell K, Morton L, Hjalt TA, Semina EV, Amendt BA. Differential regulation of gene expression by PITX2 isoforms. The Journal of biological chemistry. 2002;277:25001–25010. doi: 10.1074/jbc.M201737200. [DOI] [PubMed] [Google Scholar]
- Davis RL, Turner DL. Vertebrate hairy and Enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene. 2001;20:8342–8357. doi: 10.1038/sj.onc.1205094. [DOI] [PubMed] [Google Scholar]
- Essner JJ, Branford WW, Zhang J, Yost HJ. Mesendoderm and left-right brain, heart and gut development are differentially regulated by pitx2 isoforms. Development (Cambridge, England) 2000;127:1081–1093. doi: 10.1242/dev.127.5.1081. [DOI] [PubMed] [Google Scholar]
- Evans AL, Gage PJ. Expression of the homeobox gene Pitx2 in neural crest is required for optic stalk and ocular anterior segment development. Human molecular genetics. 2005;14:3347–3359. doi: 10.1093/hmg/ddi365. [DOI] [PubMed] [Google Scholar]
- Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AL, Ohsako S, Caudy M. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Molecular and cellular biology. 1996;16:2670–2677. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage PJ, Suh HY, Camper SA. Dosage requirement of Pitx2 for development of multiple organs. Development (Cambridge, England) 1999;126:4643–4651. doi: 10.1242/dev.126.20.4643. [DOI] [PubMed] [Google Scholar]
- Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Molecular and cellular biology. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlin RJ, Marashi AH, Obwegeser HL. Oculo-facio-cardio-dental (OFCD) syndrome. American journal of medical genetics. 1996;63:290–292. doi: 10.1002/(SICI)1096-8628(19960503)63:1<290::AID-AJMG47>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Grbavec D, Lo R, Liu Y, Stifani S. Transducin-like Enhancer of split 2, a mammalian homologue of Drosophila Groucho, acts as a transcriptional repressor, interacts with Hairy/Enhancer of split proteins, and is expressed during neuronal development. European journal of biochemistry / FEBS. 1998;258:339–349. doi: 10.1046/j.1432-1327.1998.2580339.x. [DOI] [PubMed] [Google Scholar]
- Grbavec D, Stifani S. Molecular interaction between TLE1 and the carboxyl-terminal domain of HES-1 containing the WRPW motif. Biochemical and biophysical research communications. 1996;223:701–705. doi: 10.1006/bbrc.1996.0959. [DOI] [PubMed] [Google Scholar]
- Hamada H, Meno C, Watanabe D, Saijoh Y. Establishment of vertebrate left-right asymmetry. Nature reviews. 2002;3:103–113. doi: 10.1038/nrg732. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods in cell biology. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hayward JR. Cuspid gigantism. Oral surgery, oral medicine, and oral pathology. 1980;49:500–501. doi: 10.1016/0030-4220(80)90070-5. [DOI] [PubMed] [Google Scholar]
- Hilton E, Johnston J, Whalen S, Okamoto N, Hatsukawa Y, Nishio J, Kohara H, Hirano Y, Mizuno S, Torii C, Kosaki K, Manouvrier S, Boute O, Perveen R, Law C, Moore A, Fitzpatrick D, Lemke J, Fellmann F, Debray FG, Dastot-Le-Moal F, Gerard M, Martin J, Bitoun P, Goossens M, Verloes A, Schinzel A, Bartholdi D, Bardakjian T, Hay B, Jenny K, Johnston K, Lyons M, Belmont JW, Biesecker LG, Giurgea I, Black G. BCOR analysis in patients with OFCD and Lenz microphthalmia syndromes, mental retardation with ocular anomalies, and cardiac laterality defects. Eur J Hum Genet. 2009;17:1325–1335. doi: 10.1038/ejhg.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton EN, Manson FD, Urquhart JE, Johnston JJ, Slavotinek AM, Hedera P, Stattin EL, Nordgren A, Biesecker LG, Black GC. Left-sided embryonic expression of the BCL-6 corepressor, BCOR, is required for vertebrate laterality determination. Human molecular genetics. 2007;16:1773–1782. doi: 10.1093/hmg/ddm125. [DOI] [PubMed] [Google Scholar]
- Hirata H, Ohtsuka T, Bessho Y, Kageyama R. Generation of structurally and functionally distinct factors from the basic helix-loop-helix gene Hes3 by alternative first exons. The Journal of biological chemistry. 2000;275:19083–19089. doi: 10.1074/jbc.M001075200. [DOI] [PubMed] [Google Scholar]
- Horn D, Chyrek M, Kleier S, Luttgen S, Bolz H, Hinkel GK, Korenke GC, Riess A, Schell-Apacik C, Tinschert S, Wieczorek D, Gillessen-Kaesbach G, Kutsche K. Novel mutations in BCOR in three patients with oculo-facio-cardio-dental syndrome, but none in Lenz microphthalmia syndrome. Eur J Hum Genet. 2005;13:563–569. doi: 10.1038/sj.ejhg.5201391. [DOI] [PubMed] [Google Scholar]
- Howe JA, Mymryk JS, Egan C, Branton PE, Bayley ST. Retinoblastoma Growth Suppressor and a 300-Kda Protein Appear to Regulate Cellular DNA-Synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:5883–5887. doi: 10.1073/pnas.87.15.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh KD, Fischle W, Verdin E, Bardwell VJ. BCoR, a novel corepressor involved in BCL-6 repression. Genes & development. 2000;14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- Janknecht R, Wells NJ, Hunter T. TGF-beta-stimulated cooperation of smad proteins with the coactivators CBP/p300. Genes & development. 1998;12:2114–2119. doi: 10.1101/gad.12.14.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development (Cambridge, England) 2007;134:1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- Kato Y. The multiple roles of Notch signaling during left-right patterning. Cell Mol Life Sci. 2011;68:2555–2567. doi: 10.1007/s00018-011-0695-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Shi Y, He X. Neuralization of the Xenopus embryo by inhibition of p300/ CREB-binding protein function. J Neurosci. 1999;19:9364–9373. doi: 10.1523/JNEUROSCI.19-21-09364.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, Jeffers M, Kumar S, Hackett C, Boldog F, Khramtsov N, Qian X, Mills E, Berghs SC, Carey N, Finn PW, Collins LS, Tumber A, Ritchie JW, Jensen PB, Lichenstein HS, Sehested M. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. The Biochemical journal. 2008;409:581–589. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- Khokha MK. Xenopus white papers and resources: folding functional genomics and genetics into the frog. Genesis. 2012;50:133–142. doi: 10.1002/dvg.22015. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Miura H, Miyagawa-Tomita S, Yanazawa M, Katoh-Fukui Y, Suzuki R, Ohuchi H, Suehiro A, Motegi Y, Nakahara Y, Kondo S, Yokoyama M. Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development (Cambridge, England) 1999;126:5749–5758. doi: 10.1242/dev.126.24.5749. [DOI] [PubMed] [Google Scholar]
- Kiyota T, Kato A, Altmann CR, Kato Y. The POU homeobox protein Oct-1 regulates radial glia formation downstream of Notch signaling. Developmental biology. 2008;315:579–592. doi: 10.1016/j.ydbio.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Lamar E, Kintner C. The Notch targets Esr1 and Esr10 are differentially regulated in Xenopus neural precursors. Development (Cambridge, England) 2005;132:3619–3630. doi: 10.1242/dev.01937. [DOI] [PubMed] [Google Scholar]
- Levin M. Left-right asymmetry in embryonic development: a comprehensive review. Mechanisms of development. 2005;122:3–25. doi: 10.1016/j.mod.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Lin AE, Ticho BS, Houde K, Westgate MN, Holmes LB. Heterotaxy: associated conditions and hospital-based prevalence in newborns. Genetics in medicine : official journal of the American College of Medical Genetics. 2000;2:157–172. doi: 10.1097/00125817-200005000-00002. [DOI] [PubMed] [Google Scholar]
- Lin CR, Kioussi C, O’Connell S, Briata P, Szeto D, Liu F, Izpisua-Belmonte JC, Rosenfeld MG. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- Liu W, Selever J, Lu MF, Martin JF. Genetic dissection of Pitx2 in craniofacial development uncovers new functions in branchial arch morphogenesis, late aspects of tooth morphogenesis and cell migration. Development (Cambridge, England) 2003;130:6375–6385. doi: 10.1242/dev.00849. [DOI] [PubMed] [Google Scholar]
- Logan M, Pagan-Westphal SM, Smith DM, Paganessi L, Tabin CJ. The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell. 1998;94:307–317. doi: 10.1016/s0092-8674(00)81474-9. [DOI] [PubMed] [Google Scholar]
- Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development (Cambridge, England) 2003;130:2303–2316. doi: 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- Lu MF, Pressman C, Dyer R, Johnson RL, Martin JF. Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature. 1999;401:276–278. doi: 10.1038/45797. [DOI] [PubMed] [Google Scholar]
- Marashi AH, Gorlin RJ. Radiculomegaly of canines and congenital cataracts--a syndrome? Oral surgery, oral medicine, and oral pathology. 1990;70:802–803. doi: 10.1016/0030-4220(90)90025-n. [DOI] [PubMed] [Google Scholar]
- Marashi AH, Gorlin RJ. Radiculomegaly of canine teeth and congenital cataracts: confirmation of a syndrome. American journal of medical genetics. 1992;42:143. doi: 10.1002/ajmg.1320420132. [DOI] [PubMed] [Google Scholar]
- Martin DM, Skidmore JM, Philips ST, Vieira C, Gage PJ, Condie BG, Raphael Y, Martinez S, Camper SA. PITX2 is required for normal development of neurons in the mouse subthalamic nucleus and midbrain. Developmental biology. 2004;267:93–108. doi: 10.1016/j.ydbio.2003.10.035. [DOI] [PubMed] [Google Scholar]
- Meno C, Shimono A, Saijoh Y, Yashiro K, Mochida K, Ohishi S, Noji S, Kondoh H, Hamada H. lefty-1 is required for left-right determination as a regulator of lefty-2 and nodal. Cell. 1998;94:287–297. doi: 10.1016/s0092-8674(00)81472-5. [DOI] [PubMed] [Google Scholar]
- Ng D, Thakker N, Corcoran CM, Donnai D, Perveen R, Schneider A, Hadley DW, Tifft C, Zhang L, Wilkie AO, van der Smagt JJ, Gorlin RJ, Burgess SM, Bardwell VJ, Black GC, Biesecker LG. Oculofaciocardiodental and Lenz microphthalmia syndromes result from distinct classes of mutations in BCOR. Nature genetics. 2004;36:411–416. doi: 10.1038/ng1321. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) North-Holland Publishing Co; Amsterdam, The Netherlands: 1967. [Google Scholar]
- Oberoi S, Winder AE, Johnston J, Vargervik K, Slavotinek AM. Case reports of oculofaciocardiodental syndrome with unusual dental findings. American journal of medical genetics Part A. 2005;136:275–277. doi: 10.1002/ajmg.a.30811. [DOI] [PubMed] [Google Scholar]
- Palmer AR. Symmetry breaking and the evolution of development. Science. 2004;306:828–833. doi: 10.1126/science.1103707. [DOI] [PubMed] [Google Scholar]
- Peng HB. Xenopus laevis: Practical uses in cell and molecular biology. Solutions and protocols. Methods in cell biology. 1991;36:657–662. [PubMed] [Google Scholar]
- Piedra ME, Icardo JM, Albajar M, Rodriguez-Rey JC, Ros MA. Pitx2 participates in the late phase of the pathway controlling left-right asymmetry. Cell. 1998;94:319–324. doi: 10.1016/s0092-8674(00)81475-0. [DOI] [PubMed] [Google Scholar]
- Pittman AM, Naranjo S, Webb E, Broderick P, Lips EH, van Wezel T, Morreau H, Sullivan K, Fielding S, Twiss P, Vijayakrishnan J, Casares F, Qureshi M, Gomez-Skarmeta JL, Houlston RS. The colorectal cancer risk at 18q21 is caused by a novel variant altering SMAD7 expression. Genome Res. 2009;19:987–993. doi: 10.1101/gr.092668.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya A, Belmonte JC. Left-right asymmetry in the vertebrate embryo: from early information to higher-level integration. Nature reviews. 2006;7:283–293. doi: 10.1038/nrg1830. [DOI] [PubMed] [Google Scholar]
- Ryan AK, Blumberg B, Rodriguez-Esteban C, Yonei-Tamura S, Tamura K, Tsukui T, de la Pena J, Sabbagh W, Greenwald J, Choe S, Norris DP, Robertson EJ, Evans RM, Rosenfeld MG, Izpisua Belmonte JC. Pitx2 determines left-right asymmetry of internal organs in vertebrates. Nature. 1998;394:545–551. doi: 10.1038/29004. [DOI] [PubMed] [Google Scholar]
- Sakano D, Kato A, Parikh N, McKnight K, Terry D, Stefanovic B, Kato Y. BCL6 canalizes Notch-dependent transcription, excluding Mastermind-like1 from selected target genes during left-right patterning. Developmental cell. 2010;18:450–462. doi: 10.1016/j.devcel.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C, Sanchez I, Demmers JA, Rodriguez P, Strouboulis J, Vidal M. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes & development. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- Schweickert A, Campione M, Steinbeisser H, Blum M. Pitx2 isoforms: involvement of Pitx2c but not Pitx2a or Pitx2b in vertebrate left-right asymmetry. Mechanisms of development. 2000;90:41–51. doi: 10.1016/s0925-4773(99)00227-0. [DOI] [PubMed] [Google Scholar]
- Schweickert A, Vick P, Getwan M, Weber T, Schneider I, Eberhardt M, Beyer T, Pachur A, Blum M. The nodal inhibitor Coco is a critical target of leftward flow in Xenopus. Curr Biol. 2010;20:738–743. doi: 10.1016/j.cub.2010.02.061. [DOI] [PubMed] [Google Scholar]
- Shiratori H, Sakuma R, Watanabe M, Hashiguchi H, Mochida K, Sakai Y, Nishino J, Saijoh Y, Whitman M, Hamada H. Two-step regulation of left-right asymmetric expression of Pitx2: initiation by nodal signaling and maintenance by Nkx2. Molecular cell. 2001;7:137–149. doi: 10.1016/s1097-2765(01)00162-9. [DOI] [PubMed] [Google Scholar]
- Speder P, Petzoldt A, Suzanne M, Noselli S. Strategies to establish left/right asymmetry in vertebrates and invertebrates. Current opinion in genetics & development. 2007;17:351–358. doi: 10.1016/j.gde.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Srinivasan RS, de Erkenez AC, Hemenway CS. The mixed lineage leukemia fusion partner AF9 binds specific isoforms of the BCL-6 corepressor. Oncogene. 2003;22:3395–3406. doi: 10.1038/sj.onc.1206361. [DOI] [PubMed] [Google Scholar]
- Takada H, Hattori D, Kitayama A, Ueno N, Taira M. Identification of target genes for the Xenopus Hes-related protein XHR1, a prepattern factor specifying the midbrain-hindbrain boundary. Developmental biology. 2005;283:253–267. doi: 10.1016/j.ydbio.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Wang L, Grossman SR, Kieff E. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:430–435. doi: 10.1073/pnas.97.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Whitman M. FAST-1 is a key maternal effector of mesoderm inducers in the early Xenopus embryo. Development (Cambridge, England) 1999;126:5621–5634. doi: 10.1242/dev.126.24.5621. [DOI] [PubMed] [Google Scholar]
- Wilkie AO, Taylor D, Scambler PJ, Baraitser M. Congenital cataract, microphthalmia and septal heart defect in two generations: a new syndrome? Clinical dysmorphology. 1993;2:114–119. [PubMed] [Google Scholar]
- Yoshioka H, Meno C, Koshiba K, Sugihara M, Itoh H, Ishimaru Y, Inoue T, Ohuchi H, Semina EV, Murray JC, Hamada H, Noji S. Pitx2, a bicoid-type homeobox gene, is involved in a lefty-signaling pathway in determination of left-right asymmetry. Cell. 1998;94:299–305. doi: 10.1016/s0092-8674(00)81473-7. [DOI] [PubMed] [Google Scholar]
- Yu X, St Amand TR, Wang S, Li G, Zhang Y, Hu YP, Nguyen L, Qiu MS, Chen YP. Differential expression and functional analysis of Pitx2 isoforms in regulation of heart looping in the chick. Development (Cambridge, England) 2001;128:1005–1013. doi: 10.1242/dev.128.6.1005. [DOI] [PubMed] [Google Scholar]