Abstract
Fragile X-associated tremor and ataxia syndrome, Fragile X-associated primary ovarian insufficiency and Fragile X syndrome are Repeat Expansion Diseases caused by expansion of a CGG•CCG-repeat microsatellite in the 5′ UTR of the FMR1 gene. To help understand the expansion mechanism responsible for these disorders we have crossed mice containing ~147 CGG•CCG repeats in the endogenous murine Fmr1 gene with mice containing a null mutation in the gene encoding the mismatch repair protein MSH2. MSH2 mutations are associated with elevated levels of generalized microsatellite instability. However, we show here for the first time that in the FX mouse model all maternally and paternally transmitted expansions require Msh2. Even the loss of one Msh2 allele reduced the intergenerational expansion frequency significantly. Msh2 is also required for all somatic expansions and loss of even one functional Msh2 allele reduced the extent of somatic expansion in some organs. Tissues with lower levels of MSH2 were more sensitive to the loss of a single Msh2 allele. This suggests that MSH2 is rate-limiting for expansion in this mouse model and that MSH2 levels may be a key factor that accounts for tissue-specific differences in expansion risk.
Keywords: Fragile X-related disorders, FXTAS, FXPOI, Triplet repeat expansion, Mismatch repair, MMR, MSH2
Introduction
The Fragile X-related disorders are a group of genetic conditions resulting from expansion of a CGG•CCG-repeat tract in the 5′ UTR of the FMR1 (MIM# 309550) gene. These disorders include a late onset neurodegenerative condition known as Fragile X-associated tremor/ataxia syndrome (FXTAS; MIM# 300623) and Fragile X-associated primary ovarian insufficiency (FXPOI; MIM# 300624), an ovarian disorder that is associated with infertility, menstrual irregularities and menopause before the age of forty. These conditions are seen in carriers of FMR1 Premutation (PM) alleles which have 55–200 repeats. These alleles show a propensity to expand both in the germline and somatic tissue with the expansion risk being related to repeat number. Children who inherit alleles with >200 repeats have Fragile X syndrome (FXS; MIM# 300624), the most common heritable cause of intellectual disability. Repeat expansion is also responsible for a growing number of other human diseases as well, including Myotonic Dystrophy type I (DM1; MIM# 160900), Huntington Disease (HD; MIM# 143100) and Friedreich ataxia (FRDA; MIM# 229300). These disorders are collectively referred to as the Repeat Expansion Diseases (Usdin, 2008; Usdin and Grabczyk, 2000). The expansion mechanism is not known. It is not even known whether different repeats/diseases share a common expansion mechanism or even whether germline and somatic expansions in the same model system do.
Whatever the mechanism or mechanisms involved, expansion is thought to be related to the ability of the disease-associated repeats to form secondary structures which act as substrates for the expansion process. These structures, which include hairpins, tetraplexes and triplexes (Mirkin, 2006; Mitas, et al., 1995a; Mitas, et al., 1995b; Suen, et al., 1999; Yu, et al., 1997; Yu, et al., 1995), could potentially form whenever the DNA is unpaired, for example during transcription or replication.
We have previously developed a mouse model for the Fragile X premutation (FX PM) (Entezam and Usdin, 2008; Entezam and Usdin, 2009). This mouse contains a targeted insertion of CGG•CCG-repeats in the PM range in the endogenous mouse Fmr1 gene. Using this model we have shown that expansion occurs in postmitotic cells such as neurons (Lokanga, et al., 2013) and that oxidative damage exacerbates repeat expansion (Entezam, et al., 2010). This suggests that aberrant DNA damage repair rather than a DNA replication problem may be responsible for expansion in FX.
Experiments with mouse models of DM1, HD and FRDA have shown a variable effect of mutations in other genes involved in DNA replication and repair. Perhaps the most striking of these is the effect of a mutation in the mutS homolog 2 (Msh2; MIM# 609309) gene. The Msh2 gene product is involved in mismatch repair (MMR) as well as a variety of other DNA repair processes including Transcription Coupled Repair (Leadon and Avrutskaya, 1997; Mellon, et al., 1996) and the suppression of homologous recombination (HR), a DNA repair process associated with the repair of double-strand DNA breaks and stalled replication forks (Smith, et al., 2007). Msh2 null mutations abolish 96% of paternally and maternally transmitted expansions in a mouse model of DM1, a disorder that involves CTG/CAG repeats (Foiry, et al., 2006; Savouret, et al., 2003). Msh2 mutations abolished most paternally transmitted expansions in a mouse model of another CTG/CAG-expansion disorder, HD, but did not affect the frequency of the maternally transmitted expansions (Wheeler, et al., 2003). In contrast, in a transgenic mouse model of the GAA/TTC-expansion disorder FRDA, Msh2 mutations reduced somatic (Bourn, et al., 2012) but not germline expansions (Ezzatizadeh, et al., 2012), and protected the parental gamete against repeat contractions. Thus expansion may be generated in a number of different ways, some of which involve Msh2 and some of which do not.
In order to assess the role of MSH2 in CGG/CCG-repeat expansion in FX PM mice we examined the effect of an Msh2 null mutation on expansion in these animals. We show here that the FX PM mouse falls at one end of what could be considered the “MSH2-dependence spectrum” with MSH2 being required for all intergenerational and somatic expansions. Thus our data serves to reinforce the idea of just how variable the consequences of the loss of Msh2 can be for repeat instability in mouse models of different Repeat Expansion Diseases. Furthermore, the effect of the loss of Msh2 in the FX PM mouse is also different in some respects from what is seen even with MSH2-dependent expansions in the other mouse models. Whether this reflects the modifying effects of cis- or transacting factors or a fundamental difference in mechanisms that maintain genomic stability remains to be seen.
Materials and Methods
Mouse maintenance
The generation of the FX PM mice was described previously (Entezam, et al., 2007). Msh2 mutant mice were a kind gift of Tak Mak (University Health Network, Toronto, Canada). Both mice were in a C57BL/6 background. Mice were maintained in accordance with the guidelines of the NIDDK Animal Care and Use Committee and with the Guide for the Care and Use of Laboratory Animals (NIH publication no. 85-23, revised 1996).
Mouse breeding
Crosses were carried out with FX PM mice with ~147 repeats and Msh2 null mice to generate Msh2+/+, Msh2+/−, and Msh2−/− mice carrying a single copy of the PM allele. These mice were then crossed with Msh2+/+, Msh2+/−, and Msh2−/− mice carrying a normal Fmr1 allele. At least 3 different breeding pairs were used for each cross and the data pooled. The relative proportion of expansions and contractions seen in Msh2+/+ mice was consistent with data previously obtained for a large number of breeding pairs (Entezam, et al., 2007; Entezam, et al., 2010; Entezam and Usdin, 2008; Entezam and Usdin, 2009).
Genotyping and analysis of repeat number
Msh2 genotyping was carried out as described previously (Reitmair, et al., 1995). The repeat size was determined using a fluorescent PCR assay and FraxM4 and FraxM5 primer pair as described previously (Lokanga, et al., 2013). The statistical significance of the differences in the number of intergenerational expansions, contractions and unchanged alleles was determined using Fisher’s exact test (QuickCalcs GraphPad Software; www.graphpad.com/quickcalcs/) and pair-wise comparisons of different allele classes. The differences in the distribution of repeats added or lost with intergenerational transmission were evaluated using both the t test (QuickCalcs GraphPad Software) and the Mann-Whitney test (VassarStats; VassarStats.net). To evaluate young animals and to help distinguish what fraction of the PCR products observed represents bona fide expansions and contractions and what are PCR artifacts we compared the PCR profiles generated by GeneMapper for multiple animals of each genotype. The composite PCR profile for each genotype was calculated by comparing the peak height of the PCR products corresponding to −5 to +5 repeats relative to the major allele in the PCR profile of 9–14 animals of each genotype. The threshold for analysis was set at 10% of the height of the highest peak. Peaks higher than this threshold were expressed as a fraction of the major allele. The normalized peak heights for each genotype were averaged and the data plotted. Statistical evaluation was carried out using the t test (QuickCalcs GraphPad Software). The Somatic Instability Index (SII) was calculated as previously described (Lee, et al., 2010) and used to evaluate the extent of somatic expansion in adult mice.
Results
MSH2 is required for both paternally and maternally transmitted expansions in the FX PM mice
In order to assess the role of MSH2 in the expansion of the FX repeat we crossed our FX PM mice, which have ~147 repeats in the endogenous Fmr1 gene, with Msh2−/− mice to generate mice that had one copy of the PM and that were Msh2+/+, Msh2+/− or Msh2−/−. We then crossed these mice with mice that had a normal Fmr1 allele but the same Msh2 genotype. We then examined the repeat number in the progeny of these mice using the DNA from tails taken at 3 weeks of age. We also examined the repeat number in the organs of mice at different ages. The repeat size was measured at single repeat resolution by capillary electrophoresis (CE) of a PCR product generated using a fluorescently labeled primer (24). In 3-week old animals PCR amplification of these repeats produces a symmetrical size distribution centered on the most prominent (major) PCR product. This product is thought to represent the original allele in the offspring since it is also the major PCR product seen in all organs of 2 month old animals (Lokanga, et al., 2013). While in principle, PCR products smaller and larger than this product may reflect contractions and expansions respectively from the original allele, some fraction of these products, particularly those smaller than the major PCR product are thought to be “stutter-products” resulting from strand-slippage during the PCR and the preferential amplification of alleles containing fewer repeats (Hite, et al., 1996; Shinde, et al., 2003). In older animals the PCR profile typically shifts towards larger alleles and broadens as somatic expansions accumulate. For intergenerational transmissions, the major peak in the PCR profile of the offspring in the tail DNA at three weeks of age was compared to the major product seen in the parental PCR profile using DNA taken at the same age. PCR of the offspring and parental DNA was done at the same time and the samples were analyzed in the same CE run. For the assessment of the extent of postnatal somatic expansion, the PCR profile for each organ was compared to the PCR profile in the heart, an organ that we have previously demonstrated to show little or no expansion in adult animals (Lokanga, et al., 2013). The profiles were generated at the same time and analyzed in the same CE run.
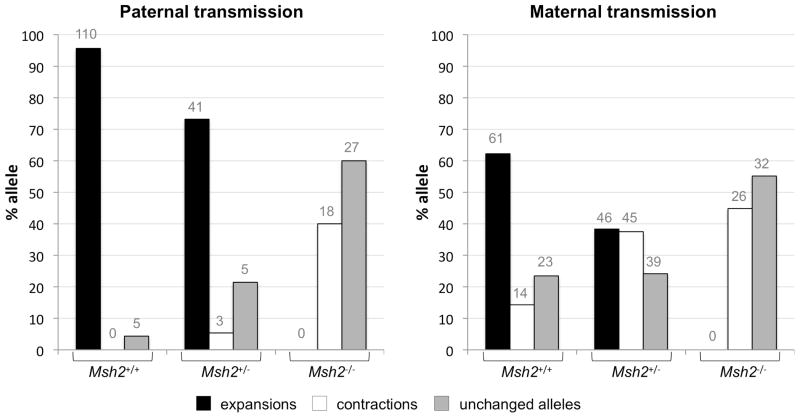
In Msh2+/+ mice expansions predominate when the repeat is transmitted intergenerationally. They are seen in 62% of the offspring of Msh2+/+ females and 96% of the offspring of Msh2+/+ males. In contrast, no expansions were seen in the offspring of Msh2−/− mice irrespective of whether the PM allele was maternally or paternally transmitted (Fig. 1) demonstrating that MSH2 is required for all maternally and paternally transmitted expansions. While the loss of Msh2 eliminated expansions, the frequency of alleles that were the same size as the parental allele or smaller than the parental allele both increased. The increase in contractions seen in Msh2−/− mice suggests that MSH2 is not required for intergenerational repeat contraction, supporting the idea that expansions and contractions of the FX repeat occur via different mechanisms in these animals. However, it remains to be seen whether defective mismatch repair itself contributes to contraction.
Fig. 1. The frequency of expansions, contractions and unchanged alleles in the offspring of Msh2+/+, Msh2+/− and Msh2−/− mice.
The repeat length changes resulting from both maternal and paternal transmissions of the FX PM allele are shown. In all instances the parental repeat number was ~147. All differences between Msh2+/+ and both Msh2+/− and Msh2−/− animals were significant at p<0.05 (Fisher’s exact test), except the difference in the percentage of unchanged alleles in Msh2+/+ and Msh2+/− females and the difference in the percentage of contractions in the offspring of Msh2+/− and Msh2−/− females. The number above each bar corresponds to the number of animals observed in that category.
Even the loss of one Msh2 allele reduces the intergenerational expansion frequency
Female Msh2+/− mice had an intergenerational expansion frequency that was 41% lower than Msh2+/+ animals, while male Msh2+/− mice had an intergenerational expansion frequency that was 25% lower. However, while Msh2+/− parents transmitted expansions less frequently than Msh2+/+ parents, the Msh2+/− and Msh2−/− offspring of Msh2+/− parents had a similar likelihood of having an expansion as their Msh2+/+ littermates (Table 1). This would suggest that intergenerational expansions are sensitive to the parental Msh2 gene dosage but not the offspring’s Msh2 gene dosage.
Table 1.
Expansion frequency in offspring of Msh2+/+, Msh2+/− and Msh2−/− parents.
| Gender of transmitting parent | Parental genotype* | Offspring genotype | % expansions | % deletions | % unchanged |
|---|---|---|---|---|---|
| female | Msh2+/+ | Msh2+/+ | 62 | 14 | 24 |
| Msh2+/− | Msh2+/+ | 43§ | 36 | 21 | |
| Msh2+/− | 36§ | 41 | 23 | ||
| Msh2−/− | 38§ | 31 | 31 | ||
| Msh2−/− | Msh2−/− | 0 | 45 | 55 | |
| male | Msh2+/+ | Msh2+/+ | 96 | 0 | 4 |
| Msh2+/− | Msh2+/+ | 73§ | 7 | 20 | |
| Msh2+/− | 66§ | 7 | 27 | ||
| Msh2−/− | 83§ | 0 | 17 | ||
| Msh2−/− | Msh2−/− | 0 | 40 | 60 |
crosses in each case involved animals with the same Msh2 genotype only one of which contained the PM allele
not statistically significantly different
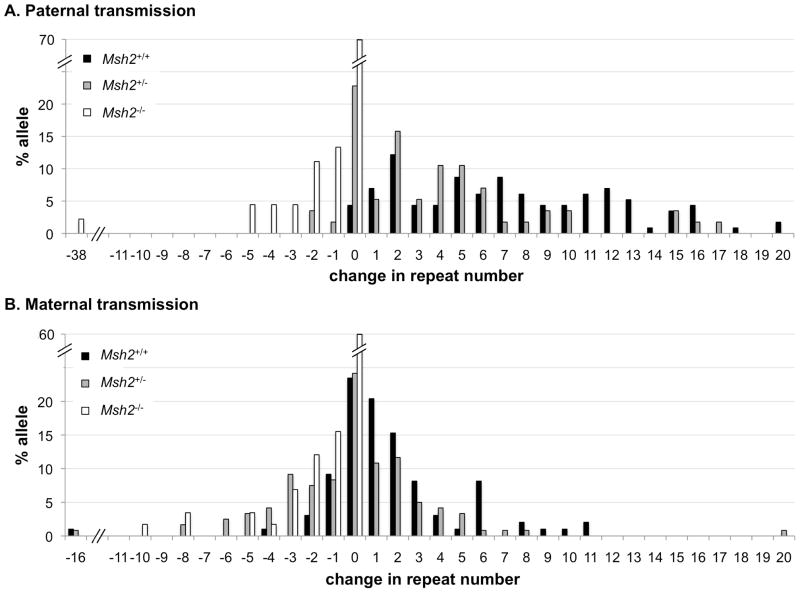
The average size of each expansion was also sensitive to Msh2 gene dosage but only when the PM allele was paternally transmitted (Fig. 2). In Msh2+/+ animals the average number of repeats added was 7.7 when only those transmissions that resulted in expansions were considered. In the case of Msh2+/− parents the average number of repeats added with each paternally transmitted expansion was 5.5 repeats. The difference was significant at the level of p=0.009 (t test) and p=0.007 (Mann-Whitney test). No significant difference in the average expansion size between Msh2+/+ and Msh2+/− animals was seen when the PM allele was maternally transmitted (3.2 vs 3.0 repeats added). No significant difference in the average contraction size was seen in Msh2+/+, Msh2+/− or Msh2−/− animals further supporting the idea that MSH2 is not involved in the contraction process itself in the FX mouse model.
Fig. 2. The effect of Msh2 heterozygosity on the repeat length changes seen on intergenerational transmission of the PM allele.
Graph depicting the percentage of alleles with the indicated change in repeat length for each genotype for paternal (Panel A) and maternal (Panel B) transmissions. Msh2+/+ and Msh2+/− males showed a significant difference in the average number of repeats added when expansions were considered separately from contractions (7.7 vs 5.5 repeats; p=0.009 by t test and p=0.007 by Mann-Whitney). The difference between Msh2+/+ and Msh2+/− females was not significant (3.2 vs 3.0 repeats, p=0.762 by t test and p=0.889 by Mann-Whitney). The number of paternal contractions was too small to evaluate differences between genotypes and no statistically significant difference in the number of repeats lost was seen on maternal transmission from Msh2+/+ and Msh2+/− and Msh2−/− animals (average repeat number lost was 2.5 and 2.9 for Msh2+/+ and Msh2−/− animals respectively, p=0.707 by t test and p=0.105 by Mann-Whitney).
Msh2 is required for early post-natal somatic expansion
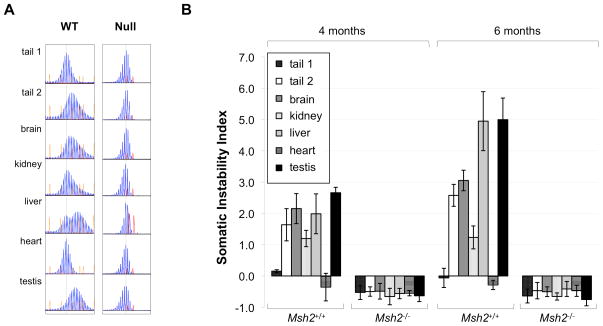
The PCR profile in Msh2+/+ mice changes with age (Figs. 3 and 4), accumulating ever larger alleles. In contrast, the PCR profile of Msh2−/− mice remains constant over the lifetime of the animal. Comparisons of composite profiles of 1 day-old Msh2+/+ mice with either 21 day-old Msh2+/+ mice or 21 day-old Msh2−/− mice showed significant differences in the proportion of PCR products with 2–5 repeats more than the original allele (p=0.004 and p=0.041 respectively; Fig. 3B). The Msh2+/+ and Msh2−/− mice were also significantly different at 21 days of age (p=0.00001). While the phenomenon of PCR “stutter” is known to generate supernumerary peaks from templates containing simple repeats, the fact that statistically significant differences are observed in the composite PCR profiles between genotypes and time points in the fraction of +2 to +5 repeats present, argues strongly that somatic expansions are occurring in Msh2+/+ mice and that early postnatal somatic expansion requiring MSH2 is a feature of the FX PM repeats in this mouse model. Our data suggests that at birth Msh2+/+ animals have already accumulated more expansions than 21 day-old Msh2−/− animals and these expansions continue to accumulate as the Msh2+/+ animals age.
Fig. 3. The effect of the absence of Msh2 on the PCR product profiles of male PM mice.
A) Representative Genemapper profiles from tail DNA of 3 week old Msh2+/+ and Msh2−/− mice and from tail DNA of 1 day old Msh2+/+ animals showing the PCR products corresponding to −5 repeats to +5 repeats from the major allele. Note that the 3 week old Msh2+/+ mouse inherited a parental allele that was one repeat larger than the parental allele inherited by the other two mice. However, to facilitate comparison of the different peak heights the PCR profiles were aligned so that the major peaks coincide. B) The composite PCR profiles for 3 week old Msh2+/+ and Msh2−/− animals and newborn Msh2+/+ pups were determined as described in the Materials and Methods. The error bars indicate the standard deviation. *different from Msh2+/+ at p<0.05; **different from Msh2+/+ at p<0.01.
Fig. 4. The effect of Msh2 gene dosage on somatic expansion in adults.
A) GeneMapper profiles of 6 month old Msh2+/+ and Msh2−/− mice each with a starting allele having ~147 repeats. The dotted line indicates the size of the original allele based on the allele size in heart, an organ that shows little or no somatic instability (Lokanga, et al., 2013). A LIZ 1200 standard and a ROX 500 standard were used for the Msh2+/+ and Msh2−/− mice respectively, but the choice of standard does not affect the profile obtained. Tail 1 refers to the tail DNA sample taken at weaning (3 weeks of age). Tail 2 refers to the tail DNA sample taken at 6 months of age. B) The average somatic instability index (Lee, et al., 2010) was calculated for the indicated organs based on data from 6 Msh2+/+ and 6 Msh2−/− animals at 4 and 6 months of age. The error bars indicate the standard deviation. Tail 1 refers to the tail DNA sample taken at weaning (3 weeks of age). Tail 2 refers to the tail DNA sample taken at 6 months of age.
There were no significant differences between any of the three composite PCR profiles in the region containing 2–5 repeats fewer than the major allele. Whether these smaller PCR products are in vitro artifacts or represent genuine in vivo contraction events or are some combination of both is not known, but since they are same in Msh2+/+ and Msh2−/− animals, whatever their origin, they are MSH2-independent.
Msh2 is also required for adult somatic expansion
We compared the extent of adult somatic expansion by examining the PCR profiles and somatic instability index (SII), a general measure of repeat instability (Lee, et al., 2010), in heart, liver, kidney, testes and brain taken from Msh2+/+ and Msh2−/− mice of different ages. As previously reported, in the initial profiles taken 3 weeks post-partum no significant differences between different organs were observed in Msh2+/+ animals (Lokanga, et al., 2013). However, by 16 weeks of age differences in the profiles could be already be distinguished in many organs including liver, brain, and testis (Fig. 4), with these differences becoming more marked as the animals aged (Lokanga, et al., 2013). In contrast, neither the PCR profile or the SII of the heart changed over time indicating that continued somatic expansion does not occur in the adult heart (Lokanga, et al., 2013).
In Msh2−/− mice the PCR profiles of the organs of older mice were indistinguishable from the PCR profiles of the same organs in young mice. These PCR profiles were also indistinguishable from that of the heart at the same age (Fig. 4). The absence of any effective change in repeat number is reflected in the fact that the SII does not change with age in Msh2−/− mice (note that the instability index in the absence of expansions is negative because the GeneMapper profiles are biased towards smaller products due the presence of PCR artifacts and the potential preferential amplification of alleles with fewer repeats (Lee, et al., 2010)). This suggests that there is no net gain or loss of repeat units in Msh2−/− mice with age.
Msh2 gene dosage also affects the average size of somatic expansions
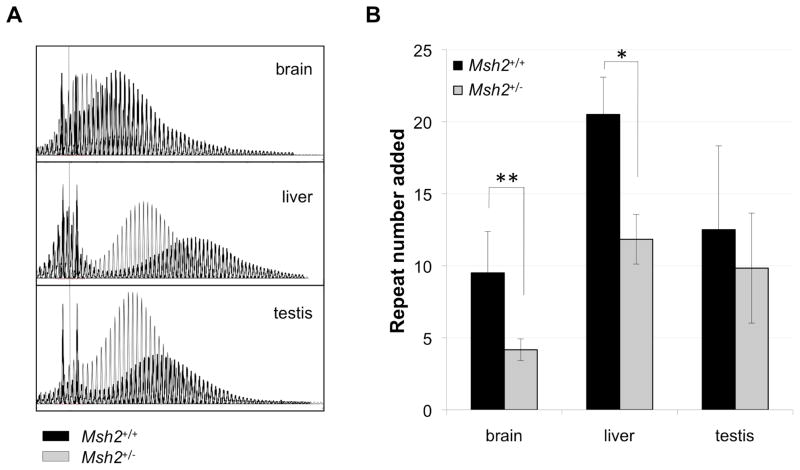
Msh2+/− animals showed less extensive increases in repeat number in adult somatic tissue than Msh2+/+ animals (Fig. 5). In Msh2+/+ animals organs such as brain and liver show saltatory increases in repeat number with age consistent with a series of successive “jumps” in which an ever increasing number of repeats are added to the derivative allele (Lokanga, et al., 2013). At 12 months of age these expanded alleles show a mean of 9.5, 20.5 and 12.5 repeats added in brain, liver and testis respectively. In contrast, in Msh2+/− mice, an average of 4.2, 11.8 and 9.8 repeats were added over the same period of time. The difference in the extent of somatic expansion was significant for brain and liver but not testis (Fig. 5). Thus the loss of even one Msh2 allele has the effect of reducing the number of repeats gained over time in some organs. While it is formally possible that the smaller net gain in repeat number in heterozygous mice reflects some contribution of increased contractions in Msh2+/− animals, the fact that there is no gain in the fraction of PCR products smaller than the major allele in Msh2−/− animals suggests that this may not be the case. The simplest explanation is that the smaller net gain of repeats simply reflects a decrease in the rate at which somatic expansions occur in Msh2-deficient animals.
Fig. 5. Somatic expansion in Msh2+/− mice.
A) Superimposed GeneMapper profiles of the repeats in brain, liver, and testis from a 12-month-old Msh2+/+ and a 12-month-old Msh2+/− mouse, each with a starting allele having ~147 repeats. The dotted line indicates the size of the original allele based on the allele size in heart, an organ that shows little or no somatic instability (Lokanga, et al., 2013). A ROX 500 standard was used for both the Msh2+/+ and Msh2+/− mice. B) The average difference between number of repeats in the original allele (indicated by the dotted line) and the number of repeats in the major derivative allele (repeat number added) for the indicated organs based on data from 6 Msh2+/+ and 6 Msh2+/− animals at 12 months of age. The error bars indicate the standard deviation. Differences in the repeat number added to the original allele in Msh2+/+ and Msh2+/− mice was significant for brain and liver (p=0.0018 and 0.012 respectively). The differences the repeat number added in the testis of Msh2+/+ and Msh2+/− animals was not statistically significant.
Discussion
While MSH2 mutations increase the frequency of generalized microsatellite instability (MSI) characteristic of cancer-predisposition disorders like Lynch Syndrome, we have shown that Msh2 is required for all intergenerational expansions in the FX PM mice (Fig. 1). Thus while MSH2 acts to protect the genome against generalized microsatellite instability, functional MSH2 is required for expansion of the CGG/CCG-repeat tract in the FX PM mouse. The obligatory role of Msh2 is seen irrespective of the gender of the transmitting parent. The loss of even one Msh2 allele caused a significant drop in the expansion frequency
Furthermore, while the Msh2−/− offspring of Msh2−/− mice showed no expansions, i.e., they had an expansion frequency of zero, Msh2−/− offspring of Msh2+/− parents had expansion frequencies of 38 % in the case of maternal transmission and 83% in the case of paternal transmissions i.e., expansion frequencies that were not significantly different from their Msh2+/+ and Msh2+/− littermates (Table 1). Thus expansion frequencies are sensitive to parental gene dosage rather than offspring gene dosage. This suggests that the intergenerational expansion that gives rise to the predominant new allele seen in young FX PM mice occurs prezygotically. This is consistent with data from humans that demonstrates that all organs of young full mutation (FM) fetuses, including testis, have a FM allele (Malter, et al., 1997), indicating that expansion in humans occurs either in the gamete or early in embryogenesis before germ layer differentiation has occurred.
However, while expansion can occur prezygotically, some post-zygotic expansion was also seen (Figs. 3–5). Somatic expansion in juvenile Msh2+/+ animals appears to occur uniformly in all organs. In addition, tissue-specific somatic expansion was also seen in older Msh2+/+ animals. Both juvenile and adult somatic expansions were dependent on Msh2. Heterozygosity for Msh2 reduced the extent of adult expansions in some expansion-prone tissue like brain and liver. However, the loss of one Msh2 allele did not have a significant effect in testis, an organ that is also prone to expansion in Msh2+/+ animals (Fig. 5). The reason for this may lie in the fact that MSH2 levels are much higher in testis than in brain or liver (Lokanga, et al., 2013). It may be that the extent of expansion is dependent in part on the availability of MSH2 to generate expansions and that in adult heterozygous mice, MSH2 is rate-limiting in brain and liver, but not in testis. A similar explanation could account for the fact that in neonatal mice all tissues are equally expansion prone: MSH2 levels are generally higher in neonatal rodents (Marietta, et al., 1998) and may thus not be limiting in any tissue of very young animals. A variety of human MSH2 polymorphisms have been described that affect protein efficiency (Mo, et al., 2012). Furthermore, a variety of factors including estrogen (Miyamoto, et al., 2006), oxidative stress (Mo, et al., 2012) and TGF-β (Yu, et al., 2010) can affect the expression and/or activity of proteins, including MSH2, that are involved in MMR or other MSH2-dependent processes. Thus it is possible that differences in genetic or environment factors such as these that affect the efficiency of MSH2 and its associated proteins could account for the wide variation in somatic instability seen in PM and FM patient cells (Govaerts, et al., 2007; Grasso, et al., 1999; Han, et al., 2006; Mannermaa, et al., 1996; Mila, et al., 1996; Petek, et al., 1999; Taylor, et al., 1999).
While the contraction frequency increased in the offspring of Msh2−/− animals, no change was seen in the somatic instability index or the PCR profiles of Msh2−/− animals with age, suggesting that there is no gain or loss of repeats in these animals and thus that somatic contractions do not accumulate. Thus the loss of Msh2 has different outcomes in gametes and somatic cells. The data we have presented here, along with our previously published data suggest that expansions and contractions involve very different molecular pathways (Entezam, et al., 2010; Entezam and Usdin, 2008; Entezam and Usdin, 2009). It may be that somatic cells have lower levels of the proteins involved in the pathway that generates contractions than gametes or that there is a minimum repeat number required for contractions to occur that is higher in somatic cells.
The percentage of expansions that are MSH2-dependent in mouse models for other REDs ranges from 0–96% depending on the sequence of the repeat, its genomic location and the gender of the transmitting parent (Dragileva, et al., 2009; Ezzatizadeh, et al., 2012; Kovtun and McMurray, 2001; Savouret, et al., 2003; Wheeler, et al., 2003). Thus our data places the FX PM mouse model at one end of the MSH2-dependence spectrum, with MSH2 being required for all maternally and paternally transmitted expansions as well as for all somatic expansions. The loss of even one Msh2 allele has a significant effect on both intergenerational and somatic expansion.
However, even the Msh2-dependent expansions seen in the different mouse models differ from one another in many respects. These differences include differences in the expansion frequency, the cell-type specificity, the sensitivity to Msh2 heterozygosity and the effects of gender (Foiry, et al., 2006; Kovtun, et al., 2004; Manley, et al., 1999; Savouret, et al., 2003; Savouret, et al., 2004; van den Broek, et al., 2002). However, evidence from bacteria, yeast, human cells in tissue culture and work in mice suggests that factors like transcriptional activity, sequence context and strain background can affect repeat instability (Ditch, et al., 2009; Freudenreich, et al., 1997; Mochmann and Wells, 2004; Paiva and Sheardy, 2004; Tome, et al., 2013). More work will be required to understand whether these factors fully explain the differences between the different mouse models or whether different mechanisms are in fact responsible for repeat expansion in different disease models.
Differences also exist between different mouse models in their response to the loss of Msh2. For example, some mouse models of CAG/CTG expansion diseases show an increase in intergenerational contractions in Msh2−/− animals without any significant increase in the frequency of alleles corresponding in size to the original parental allele (Savouret, et al., 2003; Wheeler, et al., 2003). This would be consistent with the idea that all alleles that would have been expanded in the presence of MSH2 are processed to generate contractions in its absence. This is different from the situation in FX PM mice where both unchanged and contracted alleles are seen in Msh2−/− animals. The simplest explanation for this observation is that in the FX PM mice, alleles that would normally be processed by MSH2 to generate expansions in Msh2+/+ mice are, in Msh2−/− mice, processed by other pathways to yield both unchanged alleles and contractions. Why the FX PM mice should differ from the other mouse models in this respect is currently unknown. Perhaps it is related to differences in how different repeats are processed in the absence of Msh2. This in turn could be related to differences in the structures formed by the repeats, the proposed substrates for repeat instability.
Whatever the molecular basis for these differences, it has been suggested that targeting Msh2 or its binding partners may be a valid therapeutic approach to reducing expansion in disorders like HD and DM1 (Dragileva, et al., 2009; Gonitel, et al., 2008; Lopez Castel, et al., 2010). Our data suggests that a similar approach could be useful in the FX-related disorders.
Acknowledgments
Grant Sponsor: Intramural Program of NIH (NIDDK) grant# DK057808-05
The Usdin lab would like to thank everyone involved in the care of the mice used in this study that made this work possible. They would also like to thank Shika Gupta, Wei Yang, Peggy Hsieh and Hui Geng for their valuable insights and Bruce Hayward for his careful reading of our manuscript.
Footnotes
Conflict of Interest
The authors have no conflict of interest to declare.
References
- Bourn RL, De Biase I, Pinto RM, Sandi C, Al-Mahdawi S, Pook MA, Bidichandani SI. Pms2 suppresses large expansions of the (GAA.TTC)n sequence in neuronal tissues. PloS one. 2012;7:e47085. doi: 10.1371/journal.pone.0047085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditch S, Sammarco MC, Banerjee A, Grabczyk E. Progressive GAA.TTC repeat expansion in human cell lines. PLoS Genet. 2009;5:e1000704. doi: 10.1371/journal.pgen.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragileva E, Hendricks A, Teed A, Gillis T, Lopez ET, Friedberg EC, Kucherlapati R, Edelmann W, Lunetta KL, MacDonald ME, et al. Intergenerational and striatal CAG repeat instability in Huntington’s disease knock-in mice involve different DNA repair genes. Neurobiol Dis. 2009;33:37–47. doi: 10.1016/j.nbd.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entezam A, Biacsi R, Orrison B, Saha T, Hoffman GE, Grabczyk E, Nussbaum RL, Usdin K. Regional FMRP deficits and large repeat expansions into the full mutation range in a new Fragile X premutation mouse model. Gene. 2007;395:125–34. doi: 10.1016/j.gene.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entezam A, Lokanga AR, Le W, Hoffman G, Usdin K. Potassium bromate, a potent DNA oxidizing agent, exacerbates germline repeat expansion in a fragile X premutation mouse model. Hum Mutat. 2010;31:611–6. doi: 10.1002/humu.21237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entezam A, Usdin K. ATR protects the genome against CGG.CCG-repeat expansion in Fragile X premutation mice. Nucleic Acids Res. 2008;36:1050–6. doi: 10.1093/nar/gkm1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entezam A, Usdin K. ATM and ATR protect the genome against two different types of tandem repeat instability in Fragile X premutation mice. Nucleic Acids Res. 2009;37:6371–7. doi: 10.1093/nar/gkp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzatizadeh V, Pinto RM, Sandi C, Sandi M, Al-Mahdawi S, Te Riele H, Pook MA. The mismatch repair system protects against intergenerational GAA repeat instability in a Friedreich ataxia mouse model. Neurobiol Dis. 2012;46:165–71. doi: 10.1016/j.nbd.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiry L, Dong L, Savouret C, Hubert L, te Riele H, Junien C, Gourdon G. Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum Genet. 2006;119:520–6. doi: 10.1007/s00439-006-0164-7. [DOI] [PubMed] [Google Scholar]
- Freudenreich CH, Stavenhagen JB, Zakian VA. Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Molecular and cellular biology. 1997;17:2090–8. doi: 10.1128/mcb.17.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonitel R, Moffitt H, Sathasivam K, Woodman B, Detloff PJ, Faull RL, Bates GP. DNA instability in postmitotic neurons. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3467–72. doi: 10.1073/pnas.0800048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govaerts LC, Smit AE, Saris JJ, VanderWerf F, Willemsen R, Bakker CE, De Zeeuw CI, Oostra BA. Exceptional good cognitive and phenotypic profile in a male carrying a mosaic mutation in the FMR1 gene. Clinical genetics. 2007;72:138–44. doi: 10.1111/j.1399-0004.2007.00829.x. [DOI] [PubMed] [Google Scholar]
- Grasso M, Faravelli F, Lo Nigro C, Chiurazzi P, Sperandeo MP, Argusti A, Pomponi MG, Lecora M, Sebastio GF, Perroni L, et al. Mosaicism for the full mutation and a microdeletion involving the CGG repeat and flanking sequences in the FMR1 gene in eight fragile X patients. American journal of medical genetics. 1999;85:311–6. doi: 10.1002/(sici)1096-8628(19990730)85:3<311::aid-ajmg24>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Han XD, Powell BR, Phalin JL, Chehab FF. Mosaicism for a full mutation, premutation, and deletion of the CGG repeats results in 22% FMRP and elevated FMR1 mRNA levels in a high-functioning fragile X male. American journal of medical genetics Part A. 2006;140:1463–71. doi: 10.1002/ajmg.a.31291. [DOI] [PubMed] [Google Scholar]
- Hite JM, Eckert KA, Cheng KC. Factors affecting fidelity of DNA synthesis during PCR amplification of d(C-A)n.d(G-T)n microsatellite repeats. Nucleic acids research. 1996;24:2429–34. doi: 10.1093/nar/24.12.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat Genet. 2001;27:407–11. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- Kovtun IV, Thornhill AR, McMurray CT. Somatic deletion events occur during early embryonic development and modify the extent of CAG expansion in subsequent generations. Hum Mol Genet. 2004;13:3057–68. doi: 10.1093/hmg/ddh325. [DOI] [PubMed] [Google Scholar]
- Leadon SA, Avrutskaya AV. Differential involvement of the human mismatch repair proteins, hMLH1 and hMSH2, in transcription-coupled repair. Cancer research. 1997;57:3784–91. [PubMed] [Google Scholar]
- Lee JM, Zhang J, Su AI, Walker JR, Wiltshire T, Kang K, Dragileva E, Gillis T, Lopez ET, Boily MJ, et al. A novel approach to investigate tissue-specific trinucleotide repeat instability. BMC Syst Biol. 2010;4:29. doi: 10.1186/1752-0509-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokanga RA, Entezam A, Kumari D, Yudkin D, Qin M, Smith CB, Usdin K. Somatic expansion in mouse and human carriers of Fragile X premutation alleles. Hum Mutat. 2013;34:157–166. doi: 10.1002/humu.22177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nature reviews Molecular cell biology. 2010;11:165–70. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- Malter HE, Iber JC, Willemsen R, de Graaff E, Tarleton JC, Leisti J, Warren ST, Oostra BA. Characterization of the full fragile X syndrome mutation in fetal gametes. Nature genetics. 1997;15:165–9. doi: 10.1038/ng0297-165. [DOI] [PubMed] [Google Scholar]
- Manley K, Shirley TL, Flaherty L, Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat Genet. 1999;23:471–3. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- Mannermaa A, Pulkkinen L, Kajanoja E, Ryynanen M, Saarikoski S. Deletion in the FMR1 gene in a fragile-X male. American journal of medical genetics. 1996;64:293–5. doi: 10.1002/(SICI)1096-8628(19960809)64:2<293::AID-AJMG12>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Marietta C, Palombo F, Gallinari P, Jiricny J, Brooks PJ. Expression of long-patch and short-patch DNA mismatch repair proteins in the embryonic and adult mammalian brain. Brain Res Mol Brain Res. 1998;53:317–20. doi: 10.1016/s0169-328x(97)00311-2. [DOI] [PubMed] [Google Scholar]
- Mellon I, Rajpal DK, Koi M, Boland CR, Champe GN. Transcription-coupled repair deficiency and mutations in human mismatch repair genes. Science. 1996;272:557–60. doi: 10.1126/science.272.5261.557. [DOI] [PubMed] [Google Scholar]
- Mila M, Castellvi-Bel S, Sanchez A, Lazaro C, Villa M, Estivill X. Mosaicism for the fragile X syndrome full mutation and deletions within the CGG repeat of the FMR1 gene. Journal of medical genetics. 1996;33:338–40. doi: 10.1136/jmg.33.4.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin SM. DNA structures, repeat expansions and human hereditary disorders. Curr Opin Struct Biol. 2006;16:351–8. doi: 10.1016/j.sbi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Mitas M, Yu A, Dill J, Haworth IS. The trinucleotide repeat sequence d(CGG)15 forms a heat-stable hairpin containing Gsyn. Ganti base pairs. Biochemistry. 1995a;34:12803–11. doi: 10.1021/bi00039a041. [DOI] [PubMed] [Google Scholar]
- Mitas M, Yu A, Dill J, Kamp TJ, Chambers EJ, Haworth IS. Hairpin properties of single-stranded DNA containing a GC-rich triplet repeat: (CTG)15. Nucleic Acids Res. 1995b;23:1050–9. doi: 10.1093/nar/23.6.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T, Shiozawa T, Kashima H, Feng YZ, Suzuki A, Kurai M, Nikaido T, Konishi I. Estrogen up-regulates mismatch repair activity in normal and malignant endometrial glandular cells. Endocrinology. 2006;147:4863–70. doi: 10.1210/en.2006-0632. [DOI] [PubMed] [Google Scholar]
- Mo C, Dai Y, Kang N, Cui L, He W. Ectopic expression of human MutS homologue 2 on renal carcinoma cells is induced by oxidative stress with interleukin-18 promotion via p38 mitogen-activated protein kinase (MAPK) and c-Jun N-terminal kinase (JNK) signaling pathways. The Journal of biological chemistry. 2012;287:19242–54. doi: 10.1074/jbc.M112.349936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochmann LH, Wells RD. Transcription influences the types of deletion and expansion products in an orientation-dependent manner from GAC*GTC repeats. Nucleic Acids Res. 2004;32:4469–79. doi: 10.1093/nar/gkh787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva AM, Sheardy RD. Influence of sequence context and length on the structure and stability of triplet repeat DNA oligomers. Biochemistry. 2004;43:14218–27. doi: 10.1021/bi0494368. [DOI] [PubMed] [Google Scholar]
- Petek E, Kroisel PM, Schuster M, Zierler H, Wagner K. Mosaicism in a fragile X male including a de novo deletion in the FMR1 gene. American journal of medical genetics. 1999;84:229–32. [PubMed] [Google Scholar]
- Reitmair AH, Schmits R, Ewel A, Bapat B, Redston M, Mitri A, Waterhouse P, Mittrucker HW, Wakeham A, Liu B, et al. MSH2 deficient mice are viable and susceptible to lymphoid tumours. Nat Genet. 1995;11:64–70. doi: 10.1038/ng0995-64. [DOI] [PubMed] [Google Scholar]
- Savouret C, Brisson E, Essers J, Kanaar R, Pastink A, te Riele H, Junien C, Gourdon G. CTG repeat instability and size variation timing in DNA repair-deficient mice. Embo J. 2003;22:2264–73. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savouret C, Garcia-Cordier C, Megret J, te Riele H, Junien C, Gourdon G. MSH2-dependent germinal CTG repeat expansions are produced continuously in spermatogonia from DM1 transgenic mice. Mol Cell Biol. 2004;24:629–37. doi: 10.1128/MCB.24.2.629-637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde D, Lai Y, Sun F, Arnheim N. Taq DNA polymerase slippage mutation rates measured by PCR and quasi-likelihood analysis: (CA/GT)n and (A/T)n microsatellites. Nucleic acids research. 2003;31:974–80. doi: 10.1093/nar/gkg178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, Bannister LA, Bhattacharjee V, Wang Y, Waldman BC, Waldman AS. Accurate homologous recombination is a prominent double-strand break repair pathway in mammalian chromosomes and is modulated by mismatch repair protein Msh2. Molecular and cellular biology. 2007;27:7816–27. doi: 10.1128/MCB.00455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen IS, Rhodes JN, Christy M, McEwen B, Gray DM, Mitas M. Structural properties of Friedreich’s ataxia d(GAA) repeats. Biochim Biophys Acta. 1999;1444:14–24. doi: 10.1016/s0167-4781(98)00267-x. [DOI] [PubMed] [Google Scholar]
- Taylor AK, Tassone F, Dyer PN, Hersch SM, Harris JB, Greenough WT, Hagerman RJ. Tissue heterogeneity of the FMR1 mutation in a high-functioning male with fragile X syndrome. Am J Med Genet. 1999;84:233–9. [PubMed] [Google Scholar]
- Tome S, Manley K, Simard JP, Clark GW, Slean MM, Swami M, Shelbourne PF, Tillier ER, Monckton DG, Messer A, et al. MSH3 polymorphisms and protein levels affect CAG repeat instability in Huntington’s disease mice. PLoS genetics. 2013;9:e1003280. doi: 10.1371/journal.pgen.1003280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usdin K. The biological effects of simple tandem repeats: lessons from the repeat expansion diseases. Genome Res. 2008;18:1011–9. doi: 10.1101/gr.070409.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usdin K, Grabczyk E. DNA repeat expansions and human disease. Cell Mol Life Sci. 2000;57:914–31. doi: 10.1007/PL00000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Broek WJ, Nelen MR, Wansink DG, Coerwinkel MM, te Riele H, Groenen PJ, Wieringa B. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum Mol Genet. 2002;11:191–8. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- Wheeler VC, Lebel LA, Vrbanac V, Teed A, te Riele H, MacDonald ME. Mismatch repair gene Msh2 modifies the timing of early disease in Hdh(Q111) striatum. Hum Mol Genet. 2003;12:273–81. doi: 10.1093/hmg/ddg056. [DOI] [PubMed] [Google Scholar]
- Yu A, Barron MD, Romero RM, Christy M, Gold B, Dai J, Gray DM, Haworth IS, Mitas M. At physiological pH, d(CCG)15 forms a hairpin containing protonated cytosines and a distorted helix. Biochemistry. 1997;36:3687–99. doi: 10.1021/bi9625410. [DOI] [PubMed] [Google Scholar]
- Yu A, Dill J, Mitas M. The purine-rich trinucleotide repeat sequences d(CAG)15 and d(GAC)15 form hairpins. Nucleic Acids Res. 1995;23:4055–7. doi: 10.1093/nar/23.20.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Wang Y, Ren X, Tsuyada A, Li A, Liu LJ, Wang SE. Context-dependent bidirectional regulation of the MutS homolog 2 by transforming growth factor beta contributes to chemoresistance in breast cancer cells. Molecular cancer research : MCR. 2010;8:1633–42. doi: 10.1158/1541-7786.MCR-10-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]