Abstract
The Hippo pathway controls metazoan organ growth by regulating cell proliferation and apoptosis. Many components have been identified, but our knowledge of the composition and structure of this pathway is still incomplete. Using existing pathway components as baits, we generated by mass spectrometry a high-confidence Drosophila Hippo protein-protein interaction network (Hippo-PPIN) consisting of 153 proteins and 204 interactions. Depletion of 67% of the proteins by RNA interference regulated the transcriptional coactivator Yorkie (Yki) either positively or negatively. We selected for further characterization a new member of the alpha-arrestin family, Leash, and show that it promotes degradation of Yki through the lysosomal pathway. Given the importance of the Hippo pathway in tumor development, the Hippo-PPIN will contribute to our understanding of this network in both normal growth and cancer.
Central to the Hippo pathway are the two protein kinases Hippo and Warts (Wts) kinases that regulate the Yki transcriptional coactivator (1–3). Yki together with the TEA/AATS domain (TEAD) transcription factor Scalloped induces the transcription of a number of targets that promote cell proliferation and survival. Hippo activates Wts by phosphorylation, which in turn inhibits Yki by phosphorylation, causing its cytoplasmic retention by association with 14-3-3. Hippo signaling is regulated by the protocadherins Fat (Ft) and Daschous (Ds) (1–3). Ft regulates Wts independently of Hpo by affecting the stability of Wts through the unconventional myosin Dachs (D) (4). A number of other proteins, including Merlin (Mer), Kibra, Tao, Salt-inducible kinases, and cell polarity proteins, also regulate the core kinases (5, 6).
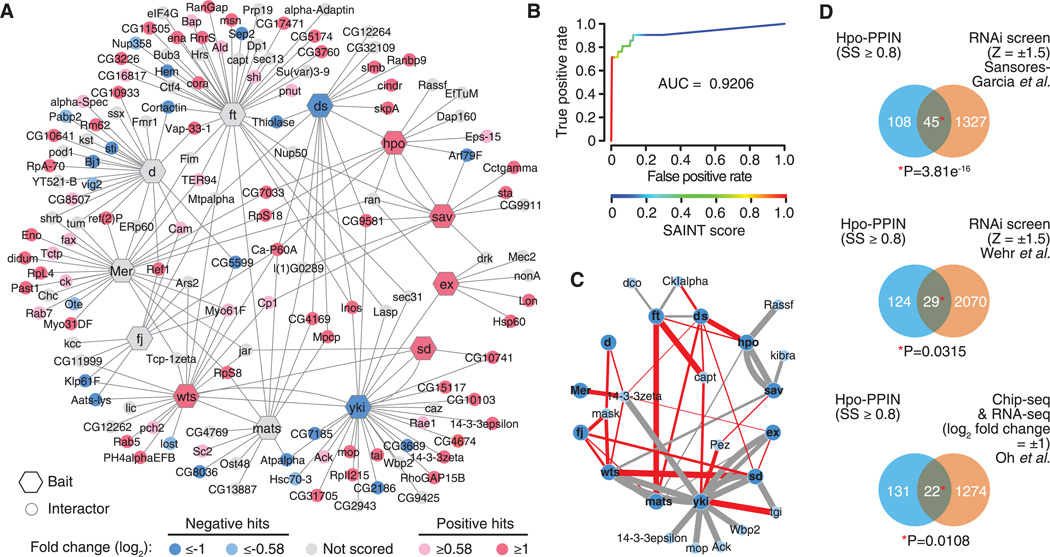
Most components of the Hippo pathway have been identified from genetic screens in Drosophila. To complement these approaches and gain information on the organization of the signaling network, we constructed a PPIN centered on the Drosophila Hippo canonical pathway components by affinity purification and mass spectrometry (AP/MS) from Drosophila S2R+ cells (7, 8). In total, we identified an unfiltered network of 4560 interactions (edges) among 1518 proteins (nodes). The total spectral counts for each identified protein were processed using the Significance Analysis of Interactome (SAINT) algorithm that confers a confidence score for PPI data obtained with AP/MS (9). Evaluation of the network by SAINT score (SS) performance test reveals that it is of high quality (Fig. 1B). PPIs with SS ≥ 0.8 (false positive rate < 3%; 204 interactions and 153 proteins) are shown in Fig. 1A (see table S1 for the full list of interactions and table S2 for the list of human orthologs of proteins with SS ≥ 0.8). We also compared the Hippo-PPIN with literature-based physical and genetic interaction networks (7). The overlap between Hippo-PPIN and the literature-based networks increased along with the increase of SS (fig. S1 and table S3), indicating that SS reliably represents the confidence of PPIs. Furthermore, we experimentally validated 23 out of 26 PPIs (~13% of the high-confidence PPIs; SS ≥ 0.8) with coimmunoprecipitation (co-IP) assay (fig. S2), leading to an estimate of ~11.5% experimental false positive rate with SS ≥ 0.8.
Fig. 1. Proteomic identification of PPIs surrounding the Hippo pathway.
(A) Network representation of the functional Hippo-PPIN. PPIs with SS ≥ 0.8 are shown. Node color indicates the RNAi screen results. (B) Receiver operating characteristic (ROC) curve showing the performance of SAINT score. The true-positive rate and false-positive rate are computed at various SAINT score cutoffs. The area under curve (AUC) is 0.9206. (C) Recovered PPIs among known components from the PPIN. Gray edges indicate the known interactions recapitulated from the PPIN. Red edges indicate novel interactions recovered from the PPIN. Edge thickness corresponds to SS. (D) Comparison of Hippo-PPIN with the published RNAi screen, Chip-sequencing, and RNA-sequencing data sets. Yki targets correspond to the genes identified by both Yki-Chip and wtsP2-RNA sequencing data.
Of known biochemical interactions between Hippo pathway components, 16 out of 31 expected interactions (table S4) with the baits (~52% coverage) were represented in the Hippo-PPIN (Fig. 1C, gray edges). Importantly, our Hippo-PPIN uncovered a number of new interactions between known components, including Ft-Capulet (Capt), Ft-Mob as tumor suppressor (Mats), Ds-Hippo, and Yki-Tondu-domain–containing growth inhibitor (Tgi) (Fig. 1C, red edges). Moreover, the Hippo-PPIN was significantly enriched for the hits from the published RNA interference (RNAi) screens (5, 10) and the potential transcriptional targets of Yki (11) (Fig. 1D and fig. S3).
To address the contribution of each node to the regulation of the downstream transcription factor Yki, we performed a focused-RNAi screen for 98% of the high-confidence nodes (SS ≥ 0.8) (see supplementary materials) (Fig. 2A). This screen efficiently recovered multiple known pathway components (Fig. 2B) and provided a functional validation for 67% of the high-confidence nodes, comprising 77 negative and 25 positive regulators with fold-change (log2) cutoff ± 0.58 (see supplementary materials) (Fig. 1A and table S5).
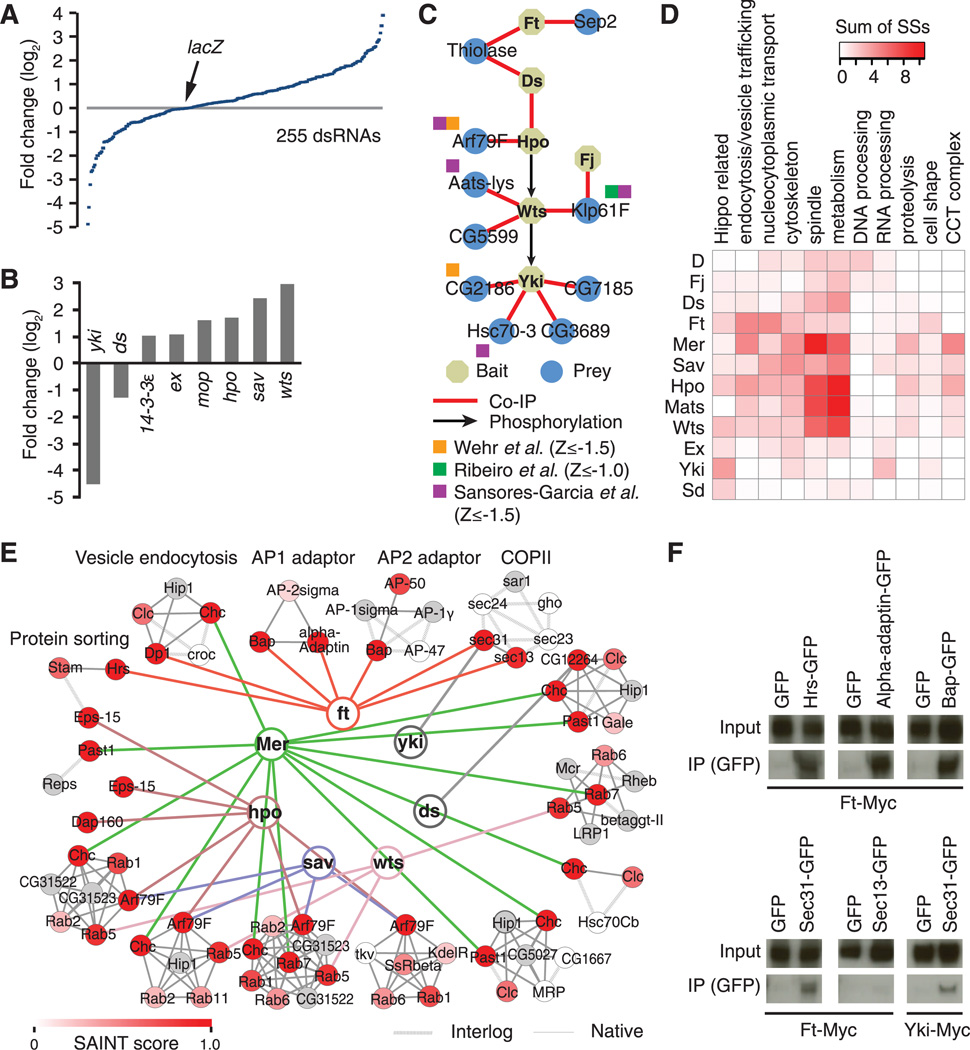
Fig. 2. Validation of Hippo-PPIN with functional RNAi screen and co-IP.
(A) Distribution of Yki-reporter values for individual double-stranded RNAs (dsRNAs) in our focused RNAi screen. About 70% of genes are covered by two dsRNAs. (B) Recovery of Hippo pathway components from RNAi screen [fold-change (log2) cutoff ± 1]. (C) The positive regulatory network validated with co-IP. (D) Heat map, based on COMPLEAT analysis (fig. S5 and table S7), showing interaction between baits (left side) and selected cellular processes (top). The sum of SSs for the PPIs between bait and subcomplexes in a biological process is used to show the interaction strength. (E) Interaction network with subcomplexes involved in endocytosis and vesicle trafficking. All edges with baits have SS ≥ 0.8. Node color represents the highest SS. (F) Co-IP validation of novel PPIs (SS ≥ 0.8) with subcomplexes related to endocytosis and vesicle trafficking.
Existing Hippo pathway components are enriched for negative effectors of Yki because their overgrowth phenotypes and increase of yki-reporter activity in vivo are readily detectable. By combining proteomics and functional genomics data, we identified 19 positive effectors of yki with fold-change (log2) ≤ −1. We tested 15 PPIs with the positive effectors based on the availability of cDNA clones and validated 13 PPIs with co-IP (Fig. 2C and fig. S4). Additionally, positive effectors such as Arf79F, CG2186, Aats-lys, Hsc70-3, and Klp61F were independently identified in (5, 10, 12) (Fig. 2C).
To gain further insights on the organization of Hippo-PPIN, we used the Protein Complex Enrichment Analysis Tool (COMPLEAT) (13), which identifies enriched known protein complex in a data set based on a comprehensive protein complex resource. The confidence of the interaction between a complex and Hippo pathway components over the entire unfiltered PPIN was represented with interquartile means of SSs for nodes in a complex (Complex Score). From this analysis, we identified 299 subcomplexes from the entire network (table S6) that revealed potential links with multiple biological processes impinging on Hippo signaling (Fig. 2D, fig. S5, and table S7). Notably, consistent with the observation that multiple actin regulators affect Hippo signaling (10, 14), interactions with cytoskeletal complexes were prominent (fig. S6). Further, reminiscent of the function of a number of Hippo pathway components in the regulation of cell division orientation (15) and positioning anaphase spindle and astral microtubules (16), many complexes related to spindle organization were identified (fig. S6). Strong association with endocytosis and vesicle trafficking complexes was also uncovered (Fig. 2E). Among the high-confidence PPIs, we validated 5 PPIs between Ft and endocytic components, and PPIs of Yki-Sec31 and Hpo-Arf79F (Fig. 2F and fig. S4). Furthermore, our RNAi screen validated a few high-confidence nodes as positive and negative effectors of Yki (fig. S6E). Given the active involvement of intracellular vesicles in multiple signaling events, our results implicate potential roles for vesicle trafficking in various aspects of Ft and Hippo signaling.
The Hippo-PPIN contains many potential new components of Hippo signaling. To initiate their characterization, we focused on an interactor of Yki, CG4674, which belongs to the arrestin domain containing (Arrdc) protein family. CG4674 is of particular interest because its mammalian orthologs, including Arrdc3, are implicated in tumor suppression by regulating cell proliferation and survival (17).
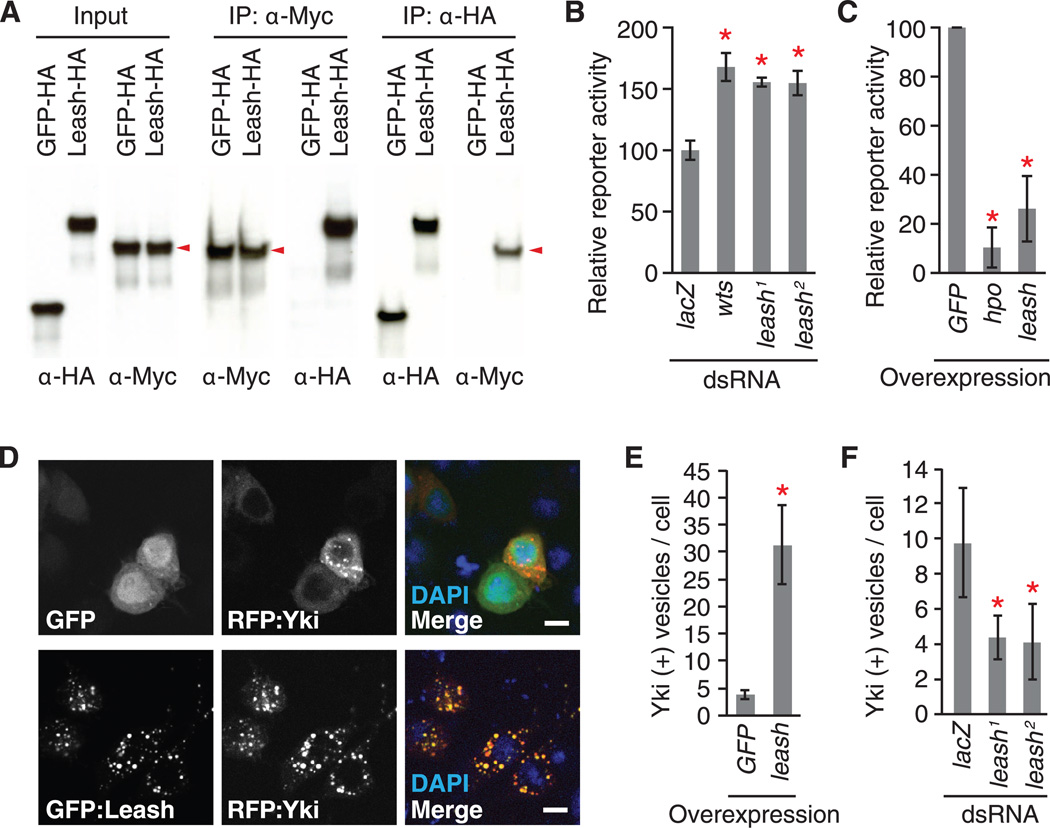
Co-IP (Fig. 3A) and reciprocal AP/MS (table S8) further established the association between CG4674 and Yki. Depletion of CG4674 by RNAi increased Yki-reporter activity (Fig. 3B), and overexpression had the opposite effect (Fig. 3C), suggesting that CG4674 restrains Yki activity (thus leading us to refer to CG4674 as leash). Although Yki was found in the cytoplasm, nucleus, and cytoplasmic vesicles, Leash predominantly localized to cytoplasmic vesicles. However, when Yki and Leash were expressed together, strong co-localization was observed (Fig. 3D). Importantly, overexpression and RNAi of leash increased and decreased vesicular localization of Yki, respectively (Fig. 3, E and F). Further, the late endosome marker, Rab9, colocalized with a subset of Yki-positive vesicles, and the lysosomal marker, Lamp1, encircled some Yki-positive punctae (fig. S7). Overexpression of wts significantly increased the abundance of Yki-containing vesicles (fig. S8), suggesting that Hippo signaling regulates the vesicular localization of Yki. Arrdc proteins have conserved arrestin domains at their N-termini that interact with their cargos. Expression of Leash N-terminal arrestin domains alone was sufficient to regulate Yki activity by forming a complex (fig. S9), indicating that Yki is a Leash cargo.
Fig. 3. Identification of Leash as a regulator of Yki.
(A) co-IP. Arrowheads indicate Yki-Myc. (B and C) Yki-reporter assay with knockdown (B) or overexpression (C) of leash. Nonoverlapping dsRNAs against leash (1, DRSC15617; 2, DRSC28712) were used. (D) Induction of vesicular localization of Yki by Leash. (E) Quantification of Yki-containing vesicles following leash expression. (F) Quantification of Yki-containing vesicles after leash knockdown. Mean ± SDs are shown. *P ≤ 0.05, Student’s t test.
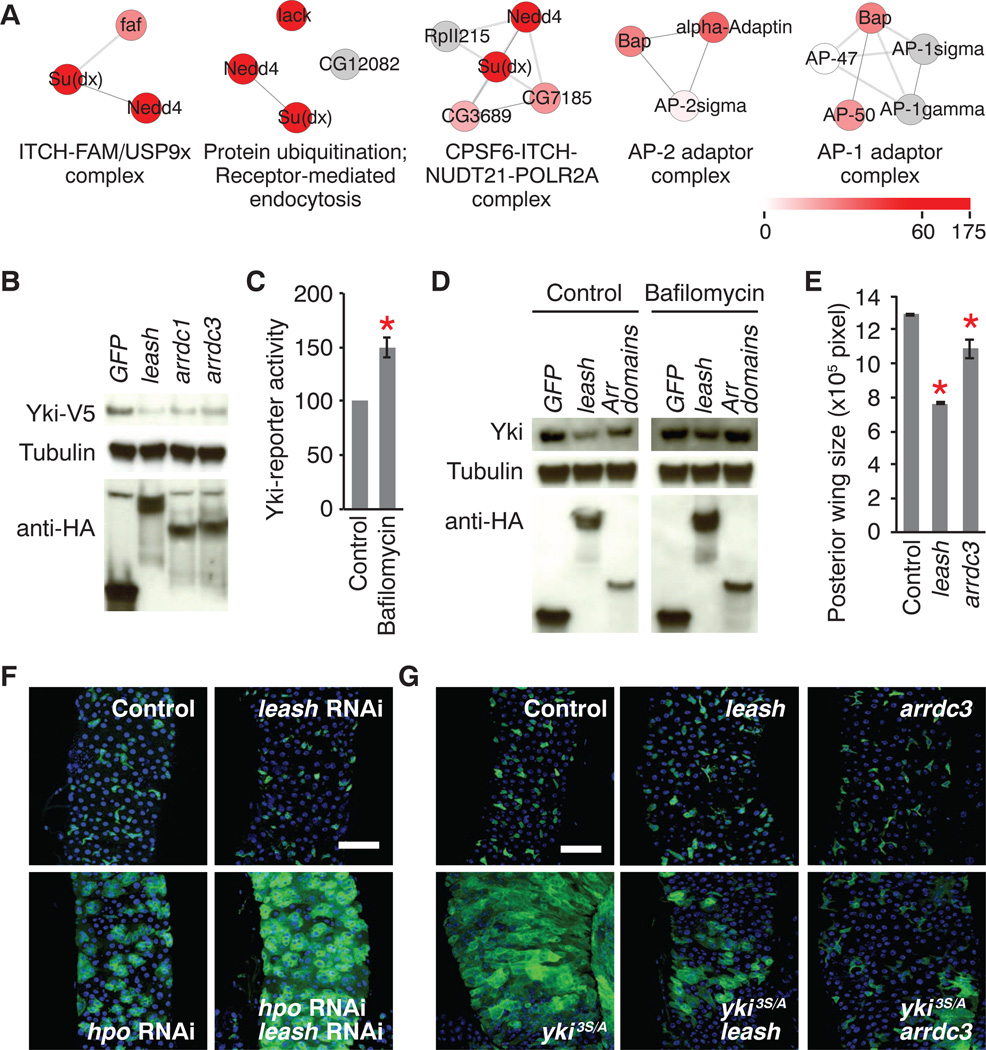
We also identified additional Leash interactors by AP/MS (tables S8 and S9) and found that Leash strongly binds to Nedd4 and a few other HECT (homologous to E6-AP carboxyl terminus) ubiquitin ligases (Fig. 4A). Moreover, we identified three ubiquitinated lysines on Leash that were important for complex formation with Yki (fig. S10). To address whether human Arrdcs could down-regulate Yki, we tested five human Leash orthologs (fig. S11). Expression of Arrdc1 or Arrdc3 reduced Yki-reporter activity (fig. S11B) and Yki protein abundance (Fig. 4B). Because Nedd4 induces degradation of its substrate through the endosomallysosomal pathway (18), we tested the effect of Bafilomycin, an inhibitor of lysosome acidification. Bafilomycin treatment increased Yki activity (Fig. 4C) and reversed the effect of overexpression of full-length Leash or Arrestin domains (Fig. 4D), indicating that Leash decreases Yki abundance through the lysosomal degradation pathway.
Fig. 4. Mechanism of Yki inhibition by Leash.
(A) Subcomplexes identified from the Leash-interactome. Legend shows the sum of spectral counts in triplicates. (B) Yki protein level. (C) Yki-reporter activity. Bafilomycin (0.01 µM) was treated for 2 days. (D) Suppression of Leash-induced Yki degradation after Bafilomycin treatment. “Arr domains” corresponds to amino acids 1 to 305 of Leash (fig. S9A). (E) Quantification of wing size. (F) Enhancement of Hippo knockdown phenotype by leash knockdown in gut stem cells. Stem cells and enteroblasts are marked with green fluorescent protein (GFP). Transgenes were induced for 8 days. (G) Suppression of yki3S/A-induced proliferation by overexpression of leash or arrdc3. Transgenes were induced for 6 days. Mean ± SDs are shown. *P ≤ 0.05, Student’s t test.
Yki activity is a critical determinant of growth, and Hippo signaling restricts growth by suppressing Yki. Consistent with the role of Leash in inhibiting Yki, overexpression of leash or arrdc3 reduced wing size significantly (Fig. 4E and fig. S12). Further, depletion of leash in wing discs affected neither wing size nor Yki protein abundance significantly, suggesting functional redundancy between family members or additional regulatory mechanisms. In the midgut, under normal homeostasis, Yki is inhibited. However, when an active form of yki (yki3S/A) is expressed or an upstream regulator is inactivated, midgut stem cells overproliferate (19, 20). Depletion of leash enhanced proliferation induced by hippo knock-down without affecting proliferation under normal homeostasis (Fig. 4F). Furthermore, overexpression of leash or arrdc3 suppressed yki3S/A-induced proliferation (Fig. 4G). Altogether, these results indicate that leash is a negative regulator of Yki.
In summary, we generated a PPIN for the Hippo pathway. Analysis of the interactions using functional RNAi screen and COMPLEAT revealed a snapshot of the overall organization of the Hippo signaling network. Further, we characterized Leash, a novel component of the Hippo pathway, and showed that it down-regulates Yki through lysosomal degradation. Altogether, the Hippo-PPIN provides a resource for further in-depth characterization of new and existing components of the Hippo pathway.
Supplementary Material
Acknowledgments
We thank the Transgenic RNAi Project, Drosophila RNAi Screening Center, and Bloomington Stock Center for fly stocks and reagents; C. Villata, E. Heffern, and R. Binari for technical support; L. Sansores-Garcia and G. Halder for help with the Yki-reporter assay; S. Blair and K. Irvine for reagents; and P. Karpowicz and R. Sopko for comments. Y.K. was supported by the Damon Runyon Cancer Research Foundation. This work was supported by R01-DK088718 and P01-CA120964 to N.P. N.P. is an Investigator of the Howard Hughes Medical Institute. AP/MS data are presented in the supplementary materials. RNAi screen data are available at Drosophila RNAi Screening Center.
Footnotes
Supplementary Materials
www.sciencemag.org/content/342/6159/737/suppl/DC1
Materials and Methods
Figs. S1 to S12
Tables S1 to S9
References (21–25)
References and Notes
- 1.Pan D. Dev. Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Staley BK, Irvine KD. Dev. Dyn. 2012;241:3–15. doi: 10.1002/dvdy.22723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halder G, Johnson RL. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho E, et al. Nat. Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- 5.Wehr MC, et al. Nat. Cell Biol. 2013;15:61–71. doi: 10.1038/ncb2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu FX, Guan KL. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman AA, et al. Sci. Signal. 2011;4:rs10. doi: 10.1126/scisignal.2002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kyriakakis P, Tipping M, Abed L, Veraksa A. Fly (Austin) 2008;2:229–235. doi: 10.4161/fly.6669. [DOI] [PubMed] [Google Scholar]
- 9.Choi H, et al. Nat. Methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sansores-Garcia L, et al. EMBO J. 2011;30:2325–2335. doi: 10.1038/emboj.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh H, et al. Cell Rep. 2013;3:309–318. doi: 10.1016/j.celrep.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ribeiro PS, et al. Mol. Cell. 2010;39:521–534. doi: 10.1016/j.molcel.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Vinayagam A, et al. Sci. Signal. 2013;6:rs5. doi: 10.1126/scisignal.2003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halder G, Dupont S, Piccolo S. Nat. Rev. Mol. Cell Biol. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- 15.Mao Y, et al. Genes Dev. 2011;25:131–136. doi: 10.1101/gad.610511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rock JM, et al. Science. 2013;340:871–875. doi: 10.1126/science.1235822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Draheim KM, et al. Oncogene. 2010;29:5032–5047. doi: 10.1038/onc.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tofaris GK, et al. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17004–17009. doi: 10.1073/pnas.1109356108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karpowicz P, Perez J, Perrimon N. Development. 2010;137:4135–4145. doi: 10.1242/dev.060483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren F, et al. Proc. Natl. Acad. Sci. U.S.A. 2010;107:21064–21069. doi: 10.1073/pnas.1012759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.