Abstract
Autophagy is an evolutionarily conserved catabolic process that directs cytoplasmic proteins, organelles and microbes to lysosomes for degradation. Autophagy acts at the intersection of pathways involved in cellular stress, host defense, and modulation of inflammatory and immune responses; however, the details of how the autophagy network intersects with these processes remain largely undefined. Given the role of autophagy in several human diseases, it is important to determine the extent to which modulators of autophagy also modify inflammatory or immune pathways, and whether it is possible to modulate a subset of these pathways selectively. Here, we identify small-molecule inducers of basal autophagy (including several FDA-approved drugs) and characterize their effects on IL-1β production, autophagic engulfment and killing of intracellular bacteria, and development of Treg, TH17, and TH1 subsets from naïve T cells. Autophagy inducers with distinct, selective activity profiles were identified that reveal the functional architecture of connections between autophagy, and innate and adaptive immunity. In macrophages from mice bearing a conditional deletion of the essential autophagy gene Atg16L1, the small molecules inhibit IL-1β production to varying degrees suggesting that individual compounds may possess both autophagy-dependent and autophagy-independent activity on immune pathways. The small molecule autophagy inducers constitute useful probes to test the contributions of autophagy-related pathways in diseases marked by impaired autophagy or elevated IL-1β, and to test novel therapeutic hypotheses.
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved catabolic process that directs cytoplasmic proteins and organelles to the lysosome, where they are degraded for subsequent recycling. Autophagy plays important roles in normal development and differentiation, and helps maintain homeostasis in the face of nutrient and other cellular stresses. Autophagy has also been implicated in a wide variety of diseases, including cancer, neurodegenerative disease, susceptibility to pathogens, and inflammatory bowel disease.(1) Consistent with this broad disease involvement, autophagy (or individual autophagy proteins) exerts functional connections to many other cell processes, including host responses to intracellular bacteria, inflammatory signaling, and innate and adaptive immunity.(2, 3) For instance, autophagy leads to the killing of intracytoplasmic group A Streptococcus in an Atg5-dependent fashion,(4) and induction of autophagy decreases intracellular survival of M. tuberculosis. (5) Individual autophagy proteins (such as Atg5) can also mediate host defense against intracellular pathogens (such as L. Monocytogenes and the protozoan T. Gondii) in an autophagy-independent manner, highlighting that autophagy proteins may contribute to host defense through multiple mechanisms.(6)
Autophagy also modulates inflammation and adaptive immunity. In myeloid cells, activation of the NLRP3 (NOD-like receptor family pyrin containing 3) inflammasome complex leads to caspase-1-mediated release of cytokines such as IL-1β, IL-18, and IL-33. Murine macrophages lacking the essential autophagy gene Atg16L1 produce increased amounts of IL-1β upon stimulation with the TLR4 ligand lipopolysaccharide (LPS).(7) Autophagy proteins also contribute to antigen presentation to CD4 and CD8 T cells. For instance, in dendritic cells, basal autophagy, as well as autophagy induced by the intracellular pattern recognition receptor NOD2, stimulates MHC class II presentation to CD4+ T cells, and antigen-specific CD4+ T cell proliferation.(8, 9)
The pathways that mediate autophagy (and connect autophagy with inflammation and innate and adaptive immunity) likely involve a network of hundreds of proteins. Multiple steps of autophagy (including nucleation and elongation of an isolation membrane, fusion of the edges of the isolation membrane to form an autophagosome containing the cytoplasmic cargo, and fusion of the autophagosome with a lysosome to form an autolysosome) are mediated by multi-protein complexes and subject to regulatory inputs. A proteomic study of basal autophagy identified a network of 751 protein/protein interactions among over 400 proteins.(10) Genome-wide siRNA screens for modulators of basal or viral autophagy also support the notion of an autophagy network involving hundreds of proteins.(11, 12)
To complement these genetic screens, and to better define the functional connections associated with autophagy, host response to pathogens, inflammation, and immunity, we undertook a chemical biology approach using FDA-approved drugs and bioactive compounds with predominantly known targets and mechanisms. We identified small molecules that induce basal autophagic flux, using image-based quantitation of autophagosomes and autolysosomes. Autophagic inducers were further characterized for their ability to reduce production of IL-1β by bone marrow-derived macrophages; enhance localization of the intracellular bacterium S. Typhimurium to autophagosomes in a manner that correlates with subsequent killing; and modulate differentiation of naive T-cells into Treg, TH17 and TH1 subsets. We find that compounds partition themselves into a variety of classes with distinct, selective activity profiles across these assays. Small molecules were further examined in macrophages from mice bearing a conditional knockout of the essential autophagy gene Atg16L1, where their ability to inhibit IL-1β suggests their potential utility in a variety of disease models.
RESULTS AND DISCUSSION
Small-molecule screen to identify inducers of basal autophagy
Our goal was to identify small molecules that induced basal autophagy, and to partition them into different subcategories of activity with respect to inflammation, and innate and adaptive immunity (Figure 1a). We undertook an initial screen of ~3,700 known bioactive compounds, including FDA-approved drugs and tool compounds with known mechanisms.
Figure 1.
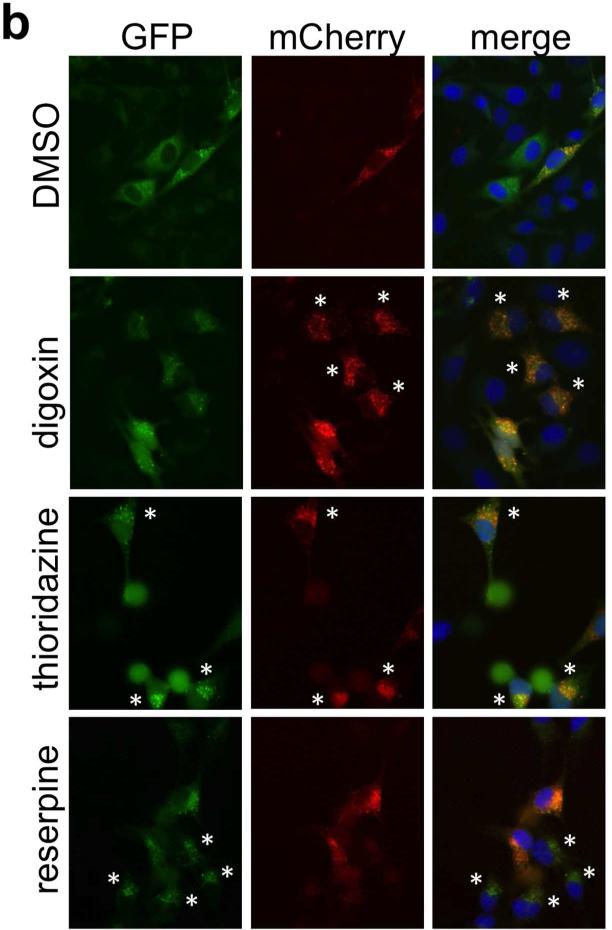
Overview of autophagy screen and inflammatory and immune assays. a. Summary of initial small molecule screen, Compound Set Enrichment Analysis, and associated inflammatory and immune assays. b. Representative images from small molecule screen. DMSO wells show occasional cells with punctae but also homogeneous cytoplasmic LC3 signal, whereas other compounds show increases in GFP+ punctae, mCherry+ punctae, or both. Cells that exemplify autophagosome or autolysosome enrichment patterns typical of the small molecule class (see Figure 2) are marked by asterisks. (Note images do not correspond directly to CSEA analysis, which analyzed GFP+ and mCherry+ GFP- punctae.)
We used a dual fluorescent reporter for the essential autophagosome membrane protein LC3 (mCherry-GFP-LC3) in our screen.(13, 14) This assay can better distinguish between small molecules that increase GFP+ punctae via increased autophagic flux (our desired hits) from those that impair the ability of autophagy to proceed to completion (for example, by impairing autophagosome maturation into autolysosomes); because the acidic lysosomal pH quenches the GFP signal, autolysosomes may appear as mCherry+ GFP- punctae.
A HeLa cell line stably expressing mCherry-GFP-LC3 was treated with 3,713 small molecules in duplicate for eight hours in the presence of nutrient-rich media, and then analyzed by fluorescence microscopy. We performed automated image analysis using the open source software CellProfiler,(15) based on four 20× fields imaged per well; for each image, we captured three fluorescence channels corresponding to Hoechst nuclear signal, mCherry, and GFP (Figure 1b). For the mCherry and GFP channels, we quantitated punctae-per-cell as a Z-score, relative to the distribution of punctae-per-cell observed in the presence of DMSO alone. Specifically, we scored each well based on the fraction of cells containing punctae in numbers that were at least three standard deviations greater than the mean number of punctae-per-cell observed in DMSO-treated wells.
Because we assessed LC3-associated punctae at a single time point, compounds that induce autophagy could result in increased numbers of autophagosomes, but also potentially decreased autophagosomes (e.g., if autophagosome maturation to autophagolysosomes is rapid). An increase in autolysosomes (mCherry+ GFP-) is suggestive of bona fide activation of autophagy. However, electron microscopy has confirmed that GFP+ punctae can in fact be autolysosomes, imaged presumably before the GFP is quenched.(12) Because of these subtleties in interpretation of the mCherry-GFP-reporter, we analyzed our screen for different image-based endpoints that could indicate enhanced basal autophagy: compounds that increased or decreased the number of GFP+ punctae per cell (autophagosomes or autolysosomes), or that increased numbers of mCherry+ GFP- autolysosomes per cell (autolysosomes).
Compound set enrichment analysis (CSEA) to identify compound classes that induce basal autophagy
We analyzed our small-molecule screening data using compound set enrichment analysis (CSEA), which evaluates whether a set of functionally or structurally related compounds is statistically enriched among top-scoring compounds in an assay.(16) The output of CSEA is a Normalized Enrichment Score (NES) for each compound set; statistical significance of the enrichment can be evaluated by 1000 random permutations of compound set membership. CSEA is statistically analogous to gene set enrichment analysis (GSEA), which is widely used to detect concordant patterns of increased or decreased gene expression among related genes. By analyzing the performance of an entire set of related compounds in the assay, CSEA can improve the robustness of choosing compounds for follow up studies, compared to choosing “hits” in isolation.
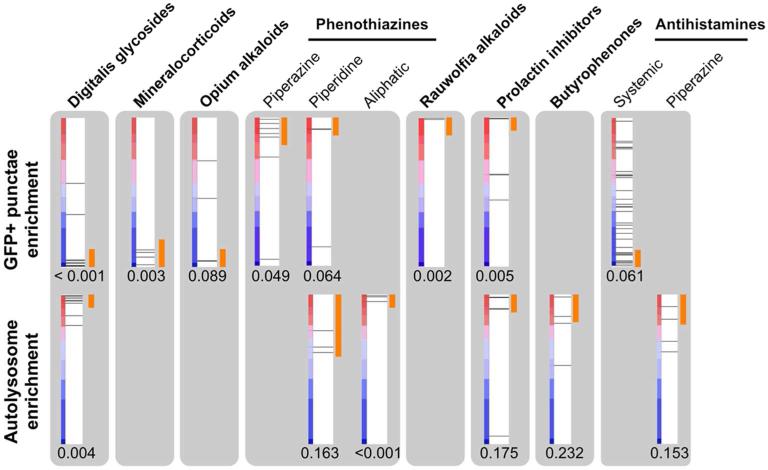
Based on CSEA, we selected 16 compound sets that were top-scoring among the following classes: increased GFP+ punctae (positive NES for GFP+ punctae), decreased GFP+ punctae (negative NES for GFP+ punctae), or increased mCherry+ GFP- punctae (canonical autolysosomes; positive NES for mCherry+ GFP- punctae). Sets were chosen for follow-up based on one or more CSEA criteria: permutation p-value < 0.05; the same set scored well for both GFP+ punctae and canonical autolysosomes; multiple sets that were structurally or functionally related scored well; or all of the set's compounds scored in the same half (upper or lower) of the ranked list of all compounds (Supplementary Figure S1; Supplementary Table S1, Data File S1).
Representative compounds from these sets were further evaluated in autophagic flux assays, consisting of Western blot analysis to detect the phosphatidylethanolamine-conjugated form of LC3 (LC3-II).(14) Each compound was evaluated in the presence and absence of E64D/pepstatin A (which inhibit lysosomal proteases); compounds that induce autophagic flux would be expected to show a further increase in LC3-II by Western blotting in the presence of E64D/pepstatin A, whereas compounds that inhibit autophagosome maturation (e.g., by inhibiting lysosomes) would not. Twelve out of 16 representative compounds evaluated by Western blot were thus confirmed to induce autophagic flux (Supplementary Figure S2; Supplementary Table S1); their CSEA profiles are shown in Figure 2. Autophagy inducers that increase GFP+ punctae include reserpine (from the set Rauwolfia alkaloids, NES 1.46, p = 0.002), metergoline (prolactin inhibitors, NES = 1.69, p = 0.005), and the phenothiazine thioridazine (phenothiazines with piperidine structure, NES 1.47, p = 0.064). Of the compound sets that decrease GFP+ punctae, increased autophagic flux was confirmed for digoxin (digitalis glycosides, NES −2.46, p < 0.001), fludrocortisone (mineralocorticoids, NES −1.68, p = 0.003), noscapine (opium alkaloids, NES = −1.39, p = 0.089), and clemastine (antihistamines for systemic use, NES −1.29, p = 0.061). (Figure 2; Supplementary Table S1)
Figure 2.
CSEA-identified compound sets enriched for enhancers of basal autophagy. Each red-blue bar depicts the list of screened compounds, ranked according to their score for GFP+ punctae (top) or mCherry+ GFP- autolysosomes (bottom). Horizontal lines show the position within the ranked list of a member of a compound set. Orange segments denote the set members that contributed to enrichment by the CSEA algorithm. The permutation p-value for enrichment is shown below each bar.
Compounds were also associated with increased mCherry+ GFP- punctae and confirmed to increase autophagic flux, especially digoxin (NES 2.26, p = 0.004), and two related phenothiazines, chlorpromazine and thioridazine (phenothiazines with aliphatic or piperidine substituents, respectively; NES 1.88, p < 0.001 and NES = 1.29, p = 0.163). Compound sets including the digitalis glycosides, phenothiazines, and selected subsets of antihistamines scored well by CSEA according to both GFP+ and autolysosome criteria (Figure 2).
Inhibition of IL-1β production
We next evaluated compounds confirmed to increase autophagic flux for their ability to inhibit IL-1β production, a phenotype related to innate immunity and inflammation. Bone marrow-derived macrophages (BMDMs) were primed with IFN-γ, and stimulated with LPS (a TLR4 agonist) and muramyl dipeptide (MDP, a peptidoglycan component of bacterial cell walls that activates the NLRP3 inflammasome).(17) The majority of compounds significantly decreased IL-1β production; the exceptions were some but not all of the digitalis glycosides (Figure 3). At the Bonferroni threshold for significance of 0.0033 (which is overly stringent because the five digitalis glycosides are not independent), all of the digitalis glycosides, cetirizine, and reserpine did not meet statistical significance. Testing at multiple doses confirmed that reductions in IL-1β production were observed at a dose (5 μM) that did not decrease cell viability (Supplementary Figure S3). In general, we observed minimal correlation between small molecules’ effects on inhibition of IL-1β release and reactive oxygen species (ROS) production (Supplementary Figure S4).
Figure 3.
Effects of small-molecule enhancers of autophagy on IL-1β production from BMDMs. Compounds were all tested at 5 μM, and IL-1β production was normalized to stimulated DMSO control (treated with IFNγ, LPS and MDP in the presence of DMSO). P-values are shown for the comparison of each compound with stimulated DMSO control.
When BMDMs are stimulated with ATP (which does not act through Nod2), we observe that several compounds significantly inhibit IL-1β production in response to both ATP or MDP, whereas others inhibit IL-1β production only in response to MDP (Supplementary Figure S5). Thus, our autophagy inducers may be partitioned into those that inhibit both Nod2-dependent and Nod2-independent inflammasome-mediated IL-1β production, and those that inhibit only Nod2-dependent IL-1β production.
Bacteria-induced autophagy and killing
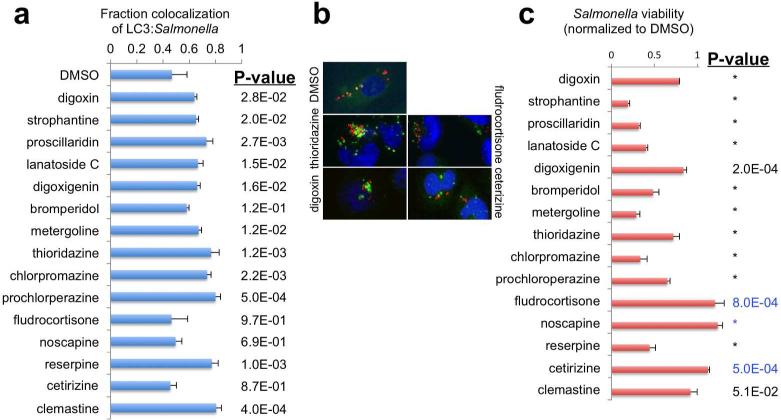
To evaluate the impact of our autophagy inducers on host defense mechanisms against intracellular bacteria, we quantitated the effect of compounds on co-localization of GFP-LC3-labelled autophagosomes and DsRed-expressing S. Typhimurium. The majority of the compounds showed significant, dose-dependent increases in LC3:Salmonella colocalization; in contrast, bromperidol, fludrocortisone, noscapine and cetirizine did not reach statistical significance (Figure 4a, 4b; Supplementary Figure S6). At a Bonferroni-corrected threshold (0.0033), proscillaridin (alone among the digitalis glycosides), thioridazine, chlorpromazine, prochloperazine, reserpine and clemastine caused statistically significant increases in bacterial autophagy without significantly affecting cell viability (Supplementary Figure S6).
Figure 4.
Effects of small-molecule enhancers of autophagy on S. typhimurium autophagy and killing. a. Fractional co-localization of LC3 punctae with DsRed S. Typhimurium. All compounds were evaluated at 5 μM. b. Representative images showing increased (thioridazine, digoxin) or unchanged (fludrocortisone, cetirizine) co-localization of LC3 punctae with DsRed S. Typhimurium. c. S. Typhimurium survival, as assessed by a bacterial bioluminescence assay. P-values in panels a and c are for the comparison with DMSO control. * denotes p < 1.0 ×10−4. (Blue p-values represent comparisons where compound wells showed a modest increase in Salmonella compared to DMSO.)
We further tested whether co-localization led to increased Salmonella killing, by quantitating surviving bacteria using a bacterial bioluminescence assay. In general, bacterial killing showed an excellent correlation with increased LC3-colocalization. All of the compounds that significantly increased LC3:bacterial colocalization also enhanced bacterial killing (Figure 4c). Of the compounds that failed to significantly increase LC3: bacterial colocalization, bromperidol is the only one that increased bacterial killing (this discrepancy may be due simply to the 5 μM dose selected for testing, as bromperidol caused a significant increase in LC3:Salmonella colocalization at 10 μM; Supplementary Figure S6). This suggests that the autophagosomes induced by our small molecules have a high probability of maturing into autolysosomes, and killing their bacterial cargo. We confirmed that enhanced bacterial killing was not due to direct bactericidal activity of the small molecules in the absence of host cells (Supplementary Figure S7).
We also noted within-class heterogeneity for the digitalis glycosides (Figure 4C); detailed dose titration shows that digoxin and digoxigenin show more modest effects on bacterial killing, while other glycosides (including lanatoside C, strophantine, and proscillaridin) show greater effects (Supplementary Figure S8).
T cell differentiation
We next sought to define how the autophagy inducers modulated development of T cell subsets from naïve T cells. Compounds were incubated with naïve wild-type murine T cells in the presence of suboptimal concentrations of cytokines that bias differentiation towards Treg, TH17 or TH1 subsets; compound effects on each T cell subset were scored on a normalized effect scale ranging from 0 (percentage observed under control conditions) to 1 (percentage observed under maximal stimulation, or positive control, conditions).
Compounds from multiple functional classes caused dose-dependent changes in different T cell subsets (Figure 5; Supplementary Figure S9a-c). For instance, thioridazine, prochlorperazine and proscillaridin selectively increased Treg development. Digoxin, chlorpromazine and bromperidol caused significant dose-dependent decreases in TH17 development, with minimal effect on other lineages. Fludrocortisone markedly decreased TH17 and TH1 lineages concordantly. Noscapine caused an increase in Treg cells as well as a modest increase in TH17 cells. We confirmed that these effects all occurred at doses without significant cell toxicity or death. By comparison, none of the compounds tested affected development of the TR1 anti-inflammatory cell population (expressing both IFN-γ and IL-10), providing additional evidence that the observed effects on T cell subsets are not non-specific compound effects (Figure 5).
Figure 5.
Effects of small-molecule enhancers of autophagy on T cell subset differentiation. Data are presented as normalized fraction of T cell subset prevalence under maximal stimulation. Blue, orange and black outlined panels indicate predominantly Treg , TH17, or mixed effects, respectively. G+ 10+ data points (green) refer to the TR1 anti-inflammatory cell population (expressing both IFN-γ and IL-10).
Small molecule effects on BMDMs from Atg16L1−/− mice: IL-1β production
Our findings that many autophagy inducers can also modulate processes associated with innate and adaptive immunity raise the hypothesis that these small molecules may suppress adverse effects in genetically defined models of autophagy and disease. To address this, we examined IL-1β production in murine BMDMs bearing a floxed allele of the essential autophagy gene Atg16L1 (i.e., containing one deleted allele and one floxed allele); in the absence of Cre expression, the BMDMs are heterozygous for Atg16L1 (analogous to hypomorphous Atg16L1 function), whereas expression of Cre recombinase results in Atg16L1-null BMDMs.
In BMDMs heterozygous for Atg16L1 deletion, the small-molecule effects on IL-1β production corresponded closely to those in cells wild type for Atg16L1; compounds that inhibited IL-1β production in wild-type cells (Figure 3) also inhibited in Atg16L1 heterozygous cells, with a high degree of statistical significance (Figure 6a). Thus, our identified autophagy inducers can still significantly inhibit IL-1β production in cells haploinsufficient for an essential autophagy gene, Atg16L1.
Figure 6.
Inhibition of IL-1β production in BMDMs deficient in Atg16L1. a. IL-1β production in BMDMs bearing one functional Atg16L1 allele (KO/wt). P-values are for the comparison between compound treatment and stimulated DMSO control (“stim” = stimulated control cells treated with IFNγ, LPS and MDP in the presence of DMSO; “no stim” = treated with IFNγ but neither LPS nor MDP; see Methods). b. IL-1β production in BMDMs either heterozygous (KO/wt) or null (KO/flox) for Atg16L1, expressed as a normalized ratio of the maximal stimulated value. P-values are for the comparison between the compound's normalized effect in Atg16L1-heterozygous vs.-null cells. c. Compound effects in Atg16L1 null cells (KO/flox). P-values are for the comparison between compound and stimulated DMSO control (“stim”). P-values in blue denote IL-1β values that are mildly increased vs. control.* denotes p < 1.0 ×10−4. Absolute levels of IL-1β production for KO/wt or KO/flox cells are shown in panels a and c, respectively, whereas normalized IL-1β levels are shown in panel b to facilitate comparison between KO/wt and KO/flox cells.
For the compounds that inhibited IL-1β production in wild-type or Atg16L1 heterozygous cells, complete deletion of Atg16L1 causes a marked, statistically significant increase in IL-1β production in the presence of compound, compared to Atg16L1 heterozygous cells (Figure 6b). This suggests that the compounds’ inhibition of IL-1β production has a significant component that is dependent either upon the process of autophagy, or on Atg16L1 itself.
Absolute levels of IL-1β production in Atg16L1-null BMDMs are dramatically increased, both at baseline, and following LPS/MDP stimulation (compared to Cre+ control cells bearing a functioning Atg16L1 allele) (Fig. 6b, c). Upon LPS/MDP stimulation, all of the small molecules that previously inhibited IL-1β production (with the exception of noscapine) also significantly decreased IL-1β production in Atg16L1−/− cells (Fig. 6c), although to a more modest extent compared to in Atg16L1 heterozygous cells. To the extent that the compounds show modest inhibition of IL-1β production in the absence of Atg16L1, this suggests that certain compounds possess varying degrees of autophagy-independent effects on IL-1β production as well. A two-way ANOVA for IL-1β production shows a significant effect of Atg16L1 genotype (p < 1 × 10−04), a significant effect of the small molecules (p < 1 × 10−04), and a significant interaction between Atg16L1 genotype and the small molecules (p < 1 × 10−04).
Because the conditional Cre expression does not delete Atg16L1 in epithelial cells, we used siRNA knockdown to decrease Atg16L1 expression in HeLa cells by approximately 80% (Supp. Fig. S10a). When anti-Salmonella autophagy was examined in these Atg16L1 knockdown cells, only digoxin and strophantine showed significant decreases in Salmonella viability (Supp. Fig. S10b). This implies that the majority of our compounds enhance anti-bacterial autophagy in an autophagy-dependent manner. However, because the cells retain significant Atg16L1 expression, we cannot exclude the possibility that even digoxin and strophantine also require autophagy for their effects; this question can be further examined in Atg16L1-null epithelial cells.
Selective small molecule activity profiles reveal architecture of autophagy and inflammatory and immune pathways
The data presented here improve our understanding of the architecture underlying connections between autophagy and inflammatory and immune-related pathways. We have partitioned small molecules that induce basal autophagy into several distinct functional classes based on their effects on IL-1β production, intracellular bacterial autophagy and killing, and T cell development. (Figure 7a) These patterns of small molecule activity profiles define potential genetic entry points into autophagy and related pathways, corresponding to nodes of intersection between autophagy, inflammation and innate and adaptive immunity (Figure 7b). By using Compound Set Enrichment Analysis (CSEA) to prioritize compounds that induce basal autophagy, we identified compounds not previously known to induce autophagy, compounds previously annotated as autophagy inhibitors rather than inducers, as well as previously identified inducers. For instance, chlorpromazine and fludrocortisone, both confirmed to increase autophagic flux here, were previously characterized as autophagy inhibitors based on GFP+ punctae.(18) To our knowledge, bromperidol has not been previously associated with autophagy.
Figure 7.
Activity profiles of small molecule autophagy enhancers. a. Heatmap for IL-1β inhibition, enhanced Salmonella:LC3 co-localization, and increased Salmonella killing, depicting the negative log of the P-value for the comparison of the compound with DMSO control. Because small molecule effects could result in either direction of effect for T cell subsets, effects were noted as +1 (increased), 0 (unchanged), or −1 (decreased) if a compound altered T cell subset development at any concentration. b. Diagram of activity profiles of small molecules (colored circles) across autophagy and associated assays (grey circles). The thickness of the edges reflects the number of compounds showing these relationships; note digoxin is the only cardiac glycoside depicted for clarity.
Small-molecule modulators of autophagy (18-24) have largely been studied in individual biological contexts (e.g., cytokine production, pathogen killing, or degradation of intracellular protein aggregates). The extent to which genetic or small molecule perturbation could coordinately or selectively modulate multiple processes was not obvious a priori. Our data show convincingly that many of these functional connections appear to be amenable to specific, selective modulation by small molecules. Further studies will help clarify the mechanisms by which our small molecules modulate basal autophagy, IL-1β production, bacterial autophagy or T cell differentiation, all of which are subject to multiple regulatory inputs.
For instance, one cluster of activities comprises the cardiac glycosides, exemplified by the widely used drug digoxin, and reserpine (an antipsychotic and antihypertensive drug) (Compound structures are shown in Supplementary Figure S11). Despite acting primarily through distinct protein targets (the Na+-K+ ATPase and vesicular monoamine transporter, respectively) these compounds do not inhibit IL-1β production, yet significantly enhance autophagy and killing of Salmonella. Another cluster includes drugs from different structural groups that share activity against dopamine and serotonin receptors (25): thioridazine, chlorpromazine, bromperidol and metergoline (an ergoline that is also used to treat hyperprolactinemia). (Figure 7a) These compounds show autophagy-dependent inhibition of IL-1β production, and enhanced Salmonella-induced autophagy and killing. Other compounds inhibited IL-1β production with no effect on Salmonella autophagy or killing (noscapine, fludrocortisone). Overall, we note several examples of small molecules that share autophagy-related activity profiles despite acting through distinct targets, suggesting either a shared secondary target, or distinct targets that occupy analogous functional roles (or network connections) in autophagy regulation.
Deletion of the essential autophagy gene Atg16L1 causes a significant diminution in the ability of most of these compounds to inhibit IL-1β production, but still reveals modest inhibitory effects of the compounds. This suggests that, while many of our compounds act through mechanisms dependent on the process of autophagy (or the action of Atg16L1), we cannot exclude the possibility that some of these phenotypes are mediated by small molecules acting on one or more protein targets independent of autophagy. For instance, digoxin has been reported to inhibit TH17 differentiation by binding to and inhibiting the transcriptional activity of the retinoic acid-related orphan nuclear receptor γt (RORγt).(26). Furthermore, phenothiazines inhibit growth of M. tuberculosis, attributed potentially to inhibition of calmodulin (27) or PI3K/Akt/mTor (28) signaling. While fludrocortisone possesses only minimal glucocorticoid activity, it is possible that the ability of glucocorticoids such as dexamethasone to inhibit IL-1β expression through post-transcriptional mechanisms (29) could contribute to autophagy-independent effects. Future studies will be necessary to systematically determine the autophagy-independent effects of our compounds on the T cell differentiation pathway as well as on intracellular Salmonella killing.
Small molecule activity profiles suggest therapeutic hypotheses
Identifying compounds that selectively modulate basal autophagy, IL-1β production, as well as additional autophagy-related inflammatory or immune processes constitutes a proof-of-concept for the future development of therapeutics that exploit these pathways. For instance, Crohn's disease (a form of inflammatory bowel disease) susceptibility alleles have been found in essential autophagy genes such as ATG16L1 and IRGM, the bacterial intracellular pattern recognition receptor NOD2 (which can trigger autophagy of intracellular bacteria), and the inflammasome component NLRP3.(30, 31) RNAi knockdown of ATG16L1 impairs anti-bacterial autophagy.(30, 32) Elevated levels of IL-1β have been reported in mucosal biopsy specimens from IBD patients,(33-35) and from macrophages from chimeric Atg16L1 mutant mice that also develop a severe experimental colitis.(7) Crohn's disease is also associated with increased TH17 and decreased Treg activity.(36) Increased IL-1β signaling also underlies a group of autoinflammatory syndromes, many of which respond clinically to IL-1β blockade.(37, 38) These include monogenic conditions such as cryopyrin associated periodic syndrome, as well as genetically complex diseases such as systemic juvenile idiopathic rheumatoid arthritis and Still's disease.
These reports, combined with our data, provide a strong rationale for further evaluation of our FDA-approved autophagy inducers in disease models of autophagy deficiency or IL-1β excess. Several of the FDA-approved drugs identified here demonstrate a constellation of potentially beneficial effects for controlling pathogen infection, or auto-immune or auto-inflammatory diseases: inducing autophagy, especially of intracellular bacteria; inhibiting IL-1β production; decreasing TH17 differentiation; and increasing Treg differentiation. For instance, several small molecules all inhibit IL-1β production and enhance killing of intracellular bacteria; of these, bromperidol also decreases TH17 differentiation, thioridazine and prochlorperazine enhance Treg differentiation, and metergoline exerts minimal effects on T cell development. The cardiac glycoside digoxin has favorable effects on bacterial autophagy and killing, and downregulates TH17 differentiation. Many of our autophagy inducers can inhibit IL-1β production in cells haploinsufficient or even null for Atg16L1. Furthermore, there is an unmet need for therapies for auto-inflammatory diseases such as Crohn's disease that are not associated with opportunistic infections, and broad-based suppression of immune function. The autophagy inducers identified here are approved for a variety of psychiatric, cardiac, endocrine, or allergic indications, and none are associated with generalized immunodeficiency. More generally, these compounds may inform therapeutic development of antibacterial agents, as well as drugs for auto-immune or auto-inflammatory diseases.
In addition to therapeutic trials, our compound classes with distinct activities can clarify the mechanistic contributions of different autophagy-related pathways on cellular phenotypes and mouse disease models. It will be informative to integrate data from chemical biology studies (including this study) with systematic RNAi data for the same inflammatory and immune assays described here to address these questions.(24, 39)
METHODS
Screen for inducers of basal autophagy
HeLa Cells stably expressing mCherry-GFP-LC3 (8000 per well) were plated in Matrical 96-well glass bottom plates overnight in Iscove's Modified Dulbecco's Medium (IMDM) media. Compounds were pinned the following morning (in duplicate) using the CyBIO CyBi Well Vario (96-well pintool). Cells were treated for 8 hours, washed 3X in PBS, fixed in 4% paraformaldehyde (PFA) for 15 minutes, washed 3X in PBS, and stained with Hoechst dye for 15 minutes. Imaging (Molecular Devices ImageXpress Micro) was performed the following day, with four 20× images taken with DAPI/FITC/TRITC filters (at fixed exposures and brightness throughout).
Data analysis used a CellProfiler module that calculates number of punctae per individual cell across an image. We calculated GFP+ punctae per cell, and mCherry+ GFP- punctae per cell (canonical autolysosomes). For each of these punctae species, we first calculated the average and standard deviation (SD) of punctae per cell for ~160 wells treated only with DMSO. We defined a cutoff number of punctae per cell as the mean + 3 SD punctae for DMSO wells. For each compound (and each of the GFP+ and mCherry+ GFP- punctae species), we calculated the percent of cells with punctae exceeding the cutoff value. Z-Scores were then calculated for each compound and for each punctae species (Z = (compound mean - DMSO mean)/DMSO SD).
Compound Set Enrichment Analysis (CSEA) of basal autophagy screen
CSEA was performed as described previously (16) to detect three types of enrichment: (i) increased GFP+ punctae per cell (i.e., positive enrichment within the GFP+ punctae distribution) ; (ii) decreased GFP+ punctae per cell (i.e., negative enrichment within the GFP+ punctae distribution); (iii) increased mCherry+ GFP- punctae per cell.
IL-1β assay in bone marrow-derived macrophages (BMDMs)
Bone marrow cells from femurs and tibia of C57 BL/6, Atg16L1KO/wt × Tie2-cre, and Atg16L1KO/f × Tie2-cre mice were isolated and cultured at 37°C in RPMI 1640 containing 10% FBS, 100U/ml penicillin/streptomycin, and 25ng/ml M-CSF (Peprotech). On Day 5, 1.5×105 BMDMs/well were plated in 96-well plates (Corning, 3904) and all conditions (including non-stimulated cells) were treated with 100ng/ml IFNγ for 16h. Stimulated BMDMs were then treated with 2ng/ml LPS and 10μg/ml MDP for 24h. IFNγ and LPS/MDP treatments were performed in the presence of compound. IL1-β release was measured from cell supernatants by ELISA (BD Biosciences, 559603). Mouse maintenance and cell isolation was performed under protocols approved by the Massachusetts General Hospital Subcommittee on Research Animal Care (SRAC), in conformance with the NIH Guide for the Care and Use of Laboratory Animals.
LC3: Salmonella co-localization assay
Bacterial autophagy assays were performed as described, with minor modifications.(30, 32) Details are in Supporting Information.
Bioluminescent Salmonella replication assay
1×104 HeLa cells/well were plated in 50μl antibiotic-free IMDM in 96-well plates (Corning, 3904) and incubated at 37°C. S. typhimurium expressing the Photorhabdus luminescens lux operon (Xen26, PerkinElmer, 119230) were grown in Luria Broth (LB) with 30 μg/mL kanamycin for 12-16 hours then diluted (1:30) into fresh LB + kanamycin and incubated for an additional 4 h. The bacterial culture was subsequently diluted (1:40) in antibiotic-free IMDM, and 100μl/well was added to HeLa cells. Following infection (37°C for 30 minutes), cells were washed with IMDM containing 20 μg/ mL gentamicin and incubated with IMDM containing compound and gentamicin. Luminescence was measured with a BioTek Synergy H4 plate reader.
T cell differentiation
Balb/c mice (Jackson Laboratories) were euthanized and splenic CD4+CD62L+ T cells were magnetically sorted (Miltenyi Biotec, USA) to >95% purity and cultured in DMEM media on plates pre-coated with anti-CD3 and anti-CD28 antibodies (clones 145-2C11 and 37.51, BioXCell, USA). Media was supplemented with neutralizing antibodies (BioXCell, USA) at 2μg/ml as follows: all Treg and TH17 conditions, anti-IL-4 (clone 11B11), anti-IL-12 (clone C17.8) and anti-IFNγ (clone XMG1.2); all TH1 conditions, anti-IL-4. Media was also supplemented with cytokines (Peprotech, USA) as follows: Treglow and Treghigh, TGFβ (2ng/ml and 10ng/ml, respectively); TH1low and TH1high, IL-12 (0.05ng/ml and 10ng/ml, respectively); TH17low, TGFβ (0.5ng/ml) and IL-6 (5ng/ml); TH17high, TGFβ (0.5ng/ml), IL-6 (20ng/ml) and IL-1β (20ng/ml). Small molecules were added at day 0. Cultures were fed with DMEM + IL-2 (10ng/ml, Peprotech) containing small molecules at day 2, split 1:2 into DMEM + IL-2 at day 3 and analyzed at day 4. 5 hours prior to analysis, TH1 and TH17 cultures were restimulated using Golgistop (BD Biosciences, USA), PMA and ionomycin (50 and 500 ng/ml respectively, Sigma Aldrich, USA). Flow cytometry quantitation of T cell subsets is described in Supporting Information.
Sytox green assay
(Invitrogen) was performed according to manufacturer instructions.
Statistical analysis
Compound effects were compared with DMSO using two-way student's t-test; Bonferroni corrections for multiple hypothesis testing were also applied. Two-way ANOVA (GraphPad Prism 5) was used to evaluate the combined effects of Atg16L1 genotype and compounds.
Supplementary Material
Acknowledgements
This work was supported with funds from NIH grants DK D43351, DK 097485 and DK092405 (RJX), HHSN268201000044C (SYS), and the Helmsley Trust (RJX and SLS).
Footnotes
Supporting Information Available
Additional methods, tables and figures may be found in Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare that they have no competing interests.
REFERENCES
- 1.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuballa P, Nolte WM, Castoreno AB, Xavier RJ. Autophagy and the immune system. Annu Rev Immunol. 2012;30:611–646. doi: 10.1146/annurev-immunol-020711-074948. [DOI] [PubMed] [Google Scholar]
- 4.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, Hamada S, Yoshimori T. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 5.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, Schmidt RE, Mizushima N, Deretic V, Sibley LD, Virgin HW. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, Tanaka K, Kawai T, Tsujimura T, Takeuchi O, Yoshimori T, Akira S. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 8.Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 10.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipinski MM, Hoffman G, Ng A, Zhou W, Py BF, Hsu E, Liu X, Eisenberg J, Liu J, Blenis J, Xavier RJ, Yuan J. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev Cell. 2010;18:1041–1052. doi: 10.1016/j.devcel.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orvedahl A, Sumpter R, Jr., Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, Forst CV, Wrana JL, Zhang YE, Luby-Phelps K, Xavier RJ, Xie Y, Levine B. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaw SY, Blodgett DM, Ma MS, Westly EC, Clemons PA, Subramanian A, Schreiber SL. Disease allele-dependent small-molecule sensitivities in blood cells from monogenic diabetes. Proc Natl Acad Sci U S A. 2011;108:492–497. doi: 10.1073/pnas.1016789108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plantinga TS, Crisan TO, Oosting M, van de Veerdonk FL, de Jong DJ, Philpott DJ, van der Meer JW, Girardin SE, Joosten LA, Netea MG. Crohn's disease-associated ATG16L1 polymorphism modulates pro-inflammatory cytokine responses selectively upon activation of NOD2. Gut. 2011;60:1229–1235. doi: 10.1136/gut.2010.228908. [DOI] [PubMed] [Google Scholar]
- 18.Hundeshagen P, Hamacher-Brady A, Eils R, Brady NR. Concurrent detection of autolysosome formation and lysosomal degradation by flow cytometry in a high-content screen for inducers of autophagy. BMC Biol. 2011;9:38. doi: 10.1186/1741-7007-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, Jahreiss L, Fleming A, Pask D, Goldsmith P, O'Kane CJ, Floto RA, Rubinsztein DC. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renna M, Jimenez-Sanchez M, Sarkar S, Rubinsztein DC. Chemical inducers of autophagy that enhance the clearance of mutant proteins in neurodegenerative diseases. J Biol Chem. 2010;285:11061–11067. doi: 10.1074/jbc.R109.072181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balgi AD, Fonseca BD, Donohue E, Tsang TC, Lajoie P, Proud CG, Nabi IR, Roberge M. Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS One. 2009;4:e7124. doi: 10.1371/journal.pone.0007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, Yu J, Pan H, Hu P, Hao Y, Cai W, Zhu H, Yu AD, Xie X, Ma D, Yuan J. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci U S A. 2007;104:19023–19028. doi: 10.1073/pnas.0709695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fleming A, Noda T, Yoshimori T, Rubinsztein DC. Chemical modulators of autophagy as biological probes and potential therapeutics. Nat Chem Biol. 2011;7:9–17. doi: 10.1038/nchembio.500. [DOI] [PubMed] [Google Scholar]
- 24.Sundaramurthy V, Barsacchi R, Samusik N, Marsico G, Gilleron J, Kalaidzidis I, Meyenhofer F, Bickle M, Kalaidzidis Y, Zerial M. Integration of Chemical and RNAi Multiparametric Profiles Identifies Triggers of Intracellular Mycobacterial Killing. Cell Host Microbe. 2013;13:129–142. doi: 10.1016/j.chom.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 25.Goodman LS, Hardman JG, Limbird LE, Gilman AG. Goodman & Gilman's the pharmacological basis of therapeutics. 2001;xxvii:2148. [Google Scholar]
- 26.Huh JR, Leung MW, Huang P, Ryan DA, Krout MR, Malapaka RR, Chow J, Manel N, Ciofani M, Kim SV, Cuesta A, Santori FR, Lafaille JJ, Xu HE, Gin DY, Rastinejad F, Littman DR. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORgammat activity. Nature. 2011;472:486–490. doi: 10.1038/nature09978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roufogalis BD, Minocherhomjee AM, Al-Jobore A. Pharmacological antagonism of calmodulin. Can J Biochem Cell Biol. 1983;61:927–933. doi: 10.1139/o83-118. [DOI] [PubMed] [Google Scholar]
- 28.Kang S, Dong SM, Kim BR, Park MS, Trink B, Byun HJ, Rho SB. Thioridazine induces apoptosis by targeting the PI3K/Akt/mTOR pathway in cervical and endometrial cancer cells. Apoptosis. 2012;17:989–997. doi: 10.1007/s10495-012-0717-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kern JA, Lamb RJ, Reed JC, Daniele RP, Nowell PC. Dexamethasone inhibition of interleukin 1 beta production by human monocytes. Posttranscriptional mechanisms. J Clin Invest. 1988;81:237–244. doi: 10.1172/JCI113301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, Shugart YY, Griffiths AM, Targan SR, Ippoliti AF, Bernard EJ, Mei L, Nicolae DL, Regueiro M, Schumm LP, Steinhart AH, Rotter JI, Duerr RH, Cho JH, Daly MJ, Brant SR. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villani AC, Lemire M, Fortin G, Louis E, Silverberg MS, Collette C, Baba N, Libioulle C, Belaiche J, Bitton A, Gaudet D, Cohen A, Langelier D, Fortin PR, Wither JE, Sarfati M, Rutgeerts P, Rioux JD, Vermeire S, Hudson TJ, Franchimont D. Common variants in the NLRP3 region contribute to Crohn's disease susceptibility. Nat Genet. 2009;41:71–76. doi: 10.1038/ng285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn's disease associated ATG16L1 variant. PLoS One. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dionne S, Hiscott J, D'Agata I, Duhaime A, Seidman EG. Quantitative PCR analysis of TNF-alpha and IL-1 beta mRNA levels in pediatric IBD mucosal biopsies. Dig Dis Sci. 1997;42:1557–1566. doi: 10.1023/a:1018895500721. [DOI] [PubMed] [Google Scholar]
- 34.Casini-Raggi V, Kam L, Chong YJ, Fiocchi C, Pizarro TT, Cominelli F. Mucosal imbalance of IL-1 and IL-1 receptor antagonist in inflammatory bowel disease. A novel mechanism of chronic intestinal inflammation. J Immunol. 1995;154:2434–2440. [PubMed] [Google Scholar]
- 35.Brynskov J, Tvede N, Andersen CB, Vilien M. Increased concentrations of interleukin 1 beta, interleukin-2, and soluble interleukin-2 receptors in endoscopical mucosal biopsy specimens with active inflammatory bowel disease. Gut. 1992;33:55–58. doi: 10.1136/gut.33.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dinarello CA. Interleukin-1beta and the autoinflammatory diseases. N Engl J Med. 2009;360:2467–2470. doi: 10.1056/NEJMe0811014. [DOI] [PubMed] [Google Scholar]
- 38.Dinarello CA, Simon A, van der Meer JW. Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nat Rev Drug Discov. 2012;11:633–652. doi: 10.1038/nrd3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar D, Nath L, Kamal MA, Varshney A, Jain A, Singh S, Rao KV. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell. 2010;140:731–743. doi: 10.1016/j.cell.2010.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.