SUMMARY
Staphylococcus aureus encodes the specialized ESAT-6 Secretion System (ESS). EsxA and EsxB are secreted by the ESS pathway, and share sequence features of ESAT-6 and CFP-10 of the Type VII Secretion System (T7SS) of Mycobacterium tuberculosis. Unlike ESAT-6 and CFP-10, EsxA and EsxB do not interact. Instead, EsxB associates with a novel substrate, EsxD, and EsxA dimerizes with itself or EsxC (EsaC). Unlike EsxA and EsxB, EsxC and EsxD do not share obvious sequence features of WXG100 proteins nor PE/PPE and Esp families of proteins, all of which belong to the pfam EsxAB clan of mycobacterial T7SS. EsxD carries the C terminal motif YxxxD/E that has been proposed to target T7 substrates for secretion in mycobacteria. Here, we find that deletion, but not amino acid substitutions, in this motif prevent secretion of EsxA and EsxC but not EsxB or EsxD. This is unlike the genetic inactivation of esxA, esxB, esxC or esxD that leads to loss of secretion of all four substrates. Thus, substrate secretion can be uncoupled by deleting the last six amino acids of EsxD. The physical association of EsxC and EsxD with canonical WXG100 proteins suggests that these proteins belong to the EsxAB clan.
INTRODUCTION
Staphylococcal EsxA and EsxB are small, secreted proteins, that lack a canonical topogenic sequence. Typically, secreted proteins are synthesized as precursor proteins with an N-terminal type I or II leader sequence and are directed to the general Sec machinery. Leader sequences are cleaved following translocation of precursor proteins across the plasma membrane (Schneewind & Missiakas, 2012, Driessen & Nouwen, 2008). EsxA and EsxB belong to the WXG100 family of proteins named after the amino acid sequence motif WXG lying roughly in the center of a 100-amino acid long domain (Pallen, 2002). Genetic disruption of several genes has been shown to affect the synthesis and secretion of EsxA and EsxB. These genes are found in the so-called ESS cluster along with esxA and esxB and define the ESAT-6 Secretion System (ESS) of S. aureus (Burts et al., 2005, Pallen, 2002, Burts et al., 2008, Anderson et al., 2011). This nomenclature is based on sequence features shared with Mycobacterium tuberculosis Mt-EsxA also known as Early Secreted Antigen 6 kDa, ESAT-6 and secreted by the ESX-1 pathway (Abdallah et al., 2007). M. tuberculosis encodes five clusters, ESX-1 through ESX-5 specifying WXG100 proteins and their cognate secretion apparatus (Stoop et al., 2012). ESAT-6 and CFP-10 form a tight dimer (ESAT-6•CFP-10) and CFP-10 has been shown to carry a C-terminal topogenic sequence that marks ESAT-6•CFP-10 for secretion (Champion et al., 2006). The secretion signal of staphylococcal EsxA and EsxB is unknown but genetic deletion of one impairs the secretion of both gene products, suggesting that perhaps the proteins assemble into a complex (Burts et al., 2005). However, biochemical reconstitution of such a complex could not be achieved. Instead, an EsxA•EsxA homodimer was crystallized and the structure solved by X-ray (Sundaramoorthy et al., 2008). Each subunit folds into an elongated cylinder of two helices bent by a hairpin carrying the WXG motif. This structure is reminiscent of other WXG100 protein pairs including ESAT-6•CFP-10 and EsxG•EsxH of M. tuberculosis ESX-1 and ESX-3 secretion systems, respectively (Renshaw et al., 2005, Ilghari et al., 2011) and the more unusual EsxR•EsxS pair composed of two four-helix bundles that assemble like two linked ESAT-6•CFP-10 structures (Arbing et al., 2010). The notion that recombinant EsxA from S. aureus forms a homodimer leaves unresolved several genetic observations, including its requirement for secretion of EsxB. To address this problem, we sought to examine interacting partners of EsxA and EsxB in S. aureus. To this end, ESS-dependent secretion was examined in the clinical strain USA300, the current community acquired Methicillin Resistant Staphylococcus aureus isolate (MRSA) for which we previously reported increased ESS activity as compared to strain Newman. In this study, we resolve a previous conundrum by showing that the low ESS activity in strain Newman can be explained by the constitutive allele of the SaeR signaling kinase. Using strain USA300, we identify a new factor that interacts with EsxB, designated herein as EsxD. We find that EsxD is encoded within the ESS cluster and is secreted in an ESS-dependent manner although it lacks the WXG100 signature motif. Deletion of esxD abrogated the production of EsxB altogether and affected the secretion, but not the production, of EsxA and EsxC (formerly EsaC (Burts et al., 2008)). Our data suggest that EsxA and EsxC form both homo- and heterodimers whereas EsxB and EsxD appear to function as a heterodimer exclusively. Interestingly, EsxD carries the C-terminal sequence motif previously proposed to represent the universal secretion signal for mycobacterial T7 substrates (Daleke et al., 2012). Alteration of this motif however did not prevent substrate secretion whereas deletion of the last six amino acids blocked secretion of EsxA and EsxC but not EsxB and EsxD. Together, the data suggest that staphylococci encode at least four secreted substrates that can assemble in four pairs containing canonical and non-canonical WXG100 partners and each pair plays an intricate contribution to the integrity of the ESS pathway.
RESULTS
The two-component system SaeRS represses expression to the ess gene cluster
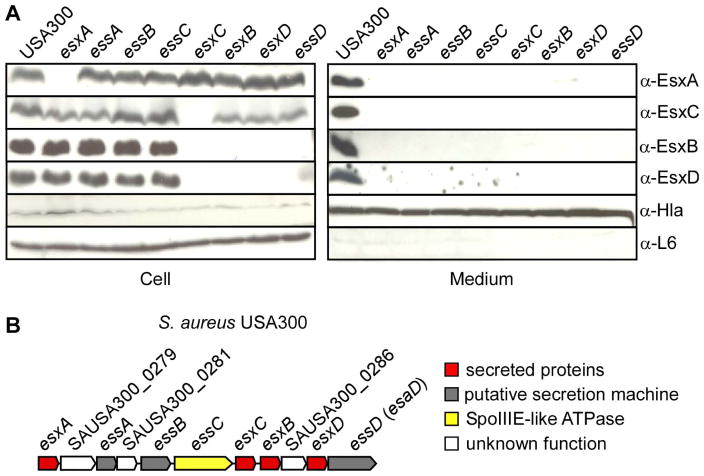
The S. aureus ESS secretion cluster has been described previously using strain Newman (Burts et al., 2005). However, EsxA and EsxB are produced and secreted at a low level in this strain, as compared to strains USA300 and USA400 (Burts et al., 2008). In strain Newman but not USA300, EsxC (EsaC) is produced only when esaB is deleted (Burts et al., 2008). Further, EsxA and EsxB proteins are unstable making it difficult to assign a regulatory or translocation function to specific genes in the ESS cluster (Burts et al., 2008). The secretome of S. aureus is controlled at the transcriptional level by several regulators to coordinate environmental cues and virulence (Novick & Geisinger, 2008, Cheung et al., 2004). In particular, the two-component system SaeRS of which SaeS is the signaling kinase and SaeR the response regulator (Giraudo et al., 1994, Giraudo et al., 1997) controls a large set of secreted virulence factors. Strain Newman carries an SaeS allele (L18P) with altered signaling property that renders SaeS constitutively active (Adhikari & Novick, 2008). We hypothesized that perhaps SaeRS is a negative regulator of the Ess cluster, and thus, the cluster would be largely repressed in strain Newman carrying the constitutive SaeS L18P allele. To test this hypothesis, levels of EsxA protein were compared in S. aureus strain Newman either wild-type or with bursa aurealis transposon insertions in the saeR or saeS genes (Fig 1A). EsxA production was increased in these mutants as compared to Newman. Increased EsxA as well as EssB and EssD production was also observed in strain USA300 carrying saeRS bursa aurealis alleles (Figure 1B). Thus, this finding indicates that the SaeRS locus negatively regulates the expression of the ess cluster. For this reason, we selected strain USA300 for subsequent studies on the Ess pathway.
Figure 1. Mutations in the SaeRS two component system alter production of EsxA, EssB and EssD.
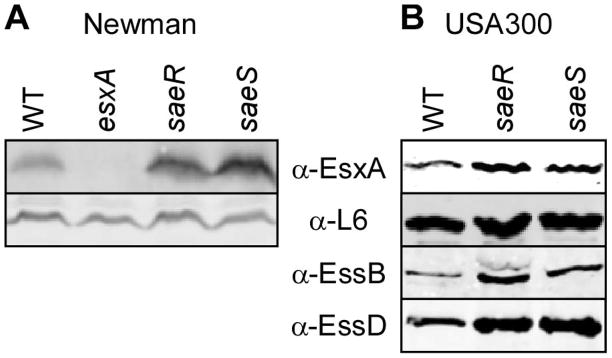
Bacterial cultures of wild type and isogenic variants of strain Newman (A) or USA300 (B) grown overnight were diluted 1:100 in fresh medium supplemented with naïve horse serum to a final concentration of 0.2% and cultures were incubated at 37°C for 2.5 h. Proteins in lyzed culture extracts were precipitated with trichloroacetic acid, separated by SDS-PAGE and detected by immunoblotting with specific antibodies (α-EsxA, α-EssB, α-EssD or α-L6 as a loading control).
EsxB interacts with EsxD (SAUSA300_0287)
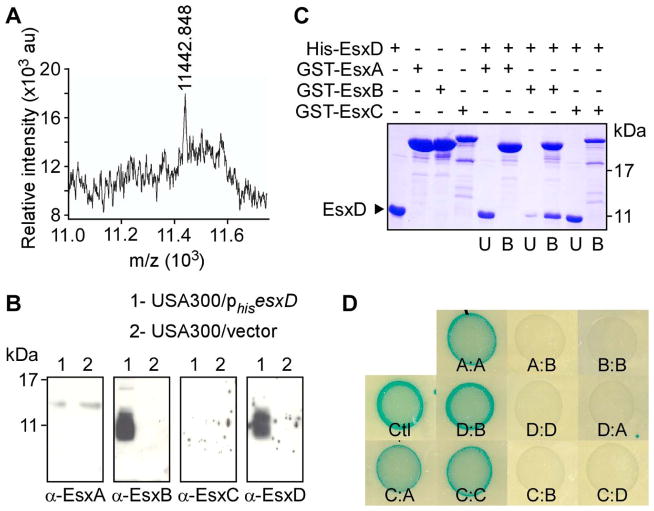
In S. aureus, EsxA and EsxB are the only two proteins with an obvious WXG100 domain. Genetic disruption of either gene disrupted the production of both gene products in strain Newman leading to the hypothesis that perhaps the proteins interact physically (Burts et al., 2005). Sundaramoorthy et al. (Sundaramoorthy et al., 2008) reported that EsxA and EsxB purified from E. coli do not form a complex, an observation that we were able to confirm independently (Suppl. Figure 1S). These authors obtained crystals of EsxA (but not EsxB) and solved the structure of this recombinant EsxA homodimer (Sundaramoorthy et al., 2008). We wondered whether EsxA and EsxB interact in their natural environment. To this effect, we developed a strain of USA300 that produces histidine-tagged EsxB from a plasmid and purified the tagged protein from culture supernatants using Ni-NTA affinity chromatography. Candidate interacting proteins were eluted in buffer containing 8 M urea. The samples were desalted and subjected to mass spectrometry analysis. This approach consistently yielded a peak with mass 11,442.848, however tryptic digestion did not provide additional information toward the identification of this polypeptide (Figure 2A). A close look at the ESS cluster suggested a possible candidate, SAUSA300_0287, located two genes downstream of esxB. SAUSA300_0287 is predicted to encode a polypeptide of 105 amino acids with a calculated mass of 11,414.79, and a high α-helical content. The SAUSA300_0287 protein contains nine lysine residues, thus it would be poorly amenable to mass spectrometry analyses of tryptic digests. The difference in measured and predicted mass could be explained by formylation of the initiator methionine. To test our hypothesis, SAUSA300_0287 was cloned and expressed in strain USA300 with a histidine tag. The corresponding polypeptide was purified over Ni-NTA from the spent medium of bacterial cultures and bound proteins were eluted with imidazole, separated by SDS-PAGE and transferred to PVDF membrane for western blot analyses (Figure 2B). A strain, carrying vector alone, was used for control experiments. Immunoblot analyses revealed the presence of EsxB but not EsxA or EsxC in eluted fractions [Note that EsxC is a new name to designate EsaC, a protein that we described earlier as a non canonical ESS-dependent secreted substrate (Burts et al., 2008)]. Further, histidine-tagged SAUSA300_0287 was identified with a polyclonal antibody raised against the recombinant protein and tentatively designated as EsxD (Figure 2B). To confirm this interaction, recombinant histidine-tagged EsxD (His-EsxD) purified from E. coli was applied over GST-glutathione beads preloaded with GST-EsxA, GST-EsxB or GST-EsxC. Aliquots of unbound (U) and bound (B) proteins were separated by SDS-PAGE for Coomassie staining (Figure 2C). Most of His-EsxD eluted in the flow through (U) of the GST-EsxA and GST-EsxC columns, whereas most of it remained bound (B) to GST-EsxB. Interactions between the four proteins were also examined by using a bacterial two-hybrid system that monitored reconstitution of adenylate cyclase activity as increased expression of LacZ β-galactosidase. Candidate interacting pairs yielding active adenylate cyclase were monitored on agar plates containing the chromogenic substrate X-gal (Karimova et al., 1998). As expected, when both fragments of adenylate cyclase were fused to EsxA, bacteria colonies (A:A) turned dark blue however candidate pairs EsxA/EsxB (A:B) or EsxB/EsxB (B:B) did not yield a significant change in color, suggesting preferred self-association for EsxA (Figure 2D; top row). Adenylate cyclase activity was also achieved upon expression of both esxB and esxD (D:B) but not when esxD was expressed in combination with esxA (D:A) or with itself (D:D) (Figure 2D; middle row). EsxC was also included in this analysis and the results indicate that like EsxA, EsxC is capable of self-association leading to active adenylate cyclase. Functional reconstitution also occurred for the EsxC and EsxA pair but not when EsxC was paired with EsxB or EsxD (Figure 2D; lower row). Together, the data suggest that EsxB associates preferentially with EsxD while EsxA and EsxC interact as homo- or hetero-oligomers.
Figure 2. Identification of EsxD as an interaction partner of EsxB.
(A) Mass spectrometry analysis of sample enriched for a protein of 11442.848 mass that co-purifies with EsxB in S. aureus USA300. (B) EsxB co-purifies with EsxD in S. aureus USA300. Wild type USA300 cultures carrying a plasmid expressing histidine tagged esxD (phisesxD) or control vector were spun and secreted proteins purified over Ni-NTA agarose. Bound proteins were eluted with imidazole, separated on SDS-PAGE and detected by immunoblotting with specific antibodies (α-EsxA, α-EsxB, α-EsxC, α-EsxD). (C) EsxD interacts with EsxB in vitro but not with EsxA or EsxC. Purified GST-EsxA, GST-EsxB and GST-EsxC were loaded on glutathione sepharose and incubated with purified His-EsxD. U and B denote unbound proteins in the flow through and bound proteins interacting with GST hybrids, respectively. Samples were separated by SDS-PAGE and stained with coomassie. (D) Protein interactions examined with the bacterial two-hybrid system. Bacterial cultures were spotted on solid medium containing Xgal, and the blue color indicates a positive interaction as seen for the control (Ctl, middle row left). Interactions were tested for the following pairs: (1) top row, EsxA:EsxA (A:A), EsxA:EsxB (A:B) and EsxB:EsxB (B:B); (2) middle row: positive control (Ctl), EsxD:EsxB (D:B), EsxD:EsxD (D:D), EsxD:EsxA (D:A); (3) lower row: EsxC:EsxA (C:A), EsxC:EsxC (C:C), EsxC:EsxB (C:B), EsxC:EsxD (C:D).
EsxD is secreted by the ESS pathway
An immunoblotting approach was used to identify EsxD in bacterial extracts. A culture of S. aureus USA300 was grown to mid-logarithmic phase and fractionated by sedimentation to separate intact cells from medium. Cells were lysed with lysostaphin. Trichloracetic acid was added to both fractions (cell and medium) to precipitate proteins for subsequent separation and visualization of sample aliquots by SDS-PAGE and immunoblotting. EsxD was found in both cell and medium fractions of wild type bacteria (Figure 3A). Next, we examined whether the presence of EsxD in these fractions required an intact ESS gene cluster (Figure 3B). Previous studies indicated that at least four proteins EssA, EssB, EssC and EssD (formerly EsaD) are required for secretion of EsxA and EsxB. EssB and EssD have been shown to localize to the membrane (Burts et al., 2005, Anderson et al., 2011, Chen et al., 2012). However, most analyses of the ESS gene cluster were performed in strain Newman using transposon disruption of individual genes, and we deemed it necessary to reassess the function of these genes in strain USA300. Thus, in-frame deletions were generated for the following genes, essA, essC, esxA, esxB and esxC as well as the newly identified esxD gene. Isogenic USA300 mutants in essB and essD (esaD) have been described (Chen et al., 2012, Anderson et al., 2011). The fractionation pattern of EsxD was examined systematically and compared to that of the other known secreted substrates, EsxA, EsxB and EsxC (Figure 3A). Cytosolic ribosomal protein L6 and secreted α-hemolysin (Hla) served as loading and fractionation controls, respectively (Figure 3A). Interestingly, EsxB and EsxD seemed to be unstable in mutants lacking esxC, esxB, esxD or essD and secretion was abolished in all the mutants (Figure 3A). Loss of stability is in agreement with the notion that EsxB and EsxD form a tight complex. Similarly, secretion of EsxA and EsxC was also abolished in all the mutants (essABCD, esxABCD). But unlike EsxB and EsxD, EsxA and EsxC accumulated in the cell pellets when not secreted and were stable in the absence of other substrates. This observation is in good agreement with the notion that EsxA and EsxC may engage in both homo- and hetero-complex formation unlike EsxB and EsxD. In light of these new data, we think it logical to rename EsaC and EsaD, to reflect their function as secreted substrate, EsxC (EsaC), and membrane-embedded component of the secretion machine, EssD (EsaD).
Figure 3. Genetic requirements for secretion of Esx proteins.
(A) Schematic representation of the ESS gene cluster. Proteins with defined functions are indicated as follows: secreted proteins (red), FtsK SpoIIIE-like ATPase (yellow), proposed secretion machinery (grey), not known (white). The locus is annotated for strain USA300. Acronyms Esx and Ess refer to ESAT-6 like secretion extracellular and ESAT-6 like secretion system. Genes previously referred as ESAT-6 secretion accessory (EsaC, EsaD) have been renamed to better reflect their contribution to the secretion system. (B) Bacterial cultures of S. aureus USA300 (WT) or isogenic mutants, esxA, esxB, esxC, esxD and essA, essB, essB, essC were fractionated into cells and medium. Proteins in all fractions were precipitated with trichloroacetic acid, separated by SDS-PAGE and detected by immunoblotting with antibodies specific for secreted substrates (α-EsxA, α-EsxB, α-EsxC, α-EsxD) and cytosolic and secreted protein controls (α-L6, α-Hla).
EssD is unstable in a strain lacking esxD
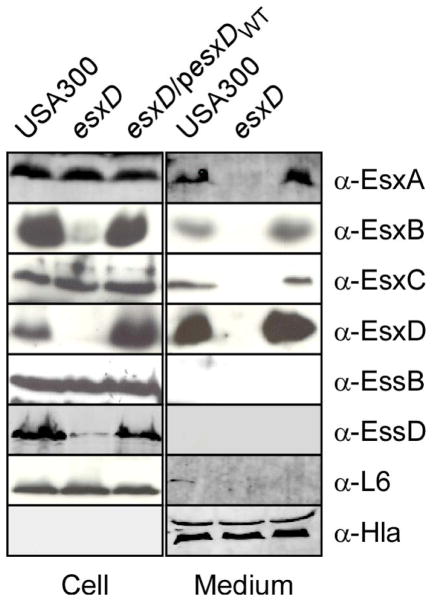
We wondered how loss of EsxD could affect the secretion of EsxA and EsxC. First, we asked if secretion of substrates could be restored, by expressing esxD on a plasmid (esxD/pesxDWT). Fractionation of cell cultures (wild type, esxD and esxD/pesxDWT) followed by immunoblotting showed that production and secretion of all four substrates was restored, suggesting that deletion of esxD did not have a polar effect on the downstream gene essD (Figure 4). We wondered whether loss of esxD might affect components of the secretion machine. We surmise that EssA, EssB, EssC and EssD are part of this machine and the fates of EssB and EssD for which we have polyclonal antibodies could be easily addressed (Chen et al., 2012, Anderson et al., 2011). Immunoblotting of cellular extracts suggested that EssD, but not EssB, was clearly affected by the deletion of esxD. This phenotype could be complemented by plasmid-encoded esxD (Figure 4). It is interesting that the three proteins EsxB, EsxD and EssD, all encoded at the end of the ESS cluster have a triangular relationship in that deleting any one of the cognate genes affects the production or stability of all three proteins as judged by immunoblot (Figs. 3A; 4). Thus, the data suggest that the secretion defect observed in the esxD mutant could be attributed to a reduction in the levels of EssD.
Figure 4. Expression of esxD in trans is sufficient to restore the secretion defect of the esxD mutant strain.
Bacterial cultures of S. aureus USA300 or esxD mutants harboring or not a complementing plasmid (pesxD) were grown to OD600nm of 1.0 and separated into cell and medium fractions. Proteins were precipitated with trichloroacetic acid, separated by SDS-PAGE and detected by immunoblotting with specific antibodies (α-EsxA, α-EsxB, α-EsxC, α-EsxD, α-EssB, α-EssD, and α-Hla or α-L6 for fractionation and loading controls).
Contribution of the YxxxE motif at the C-terminus of EsxD
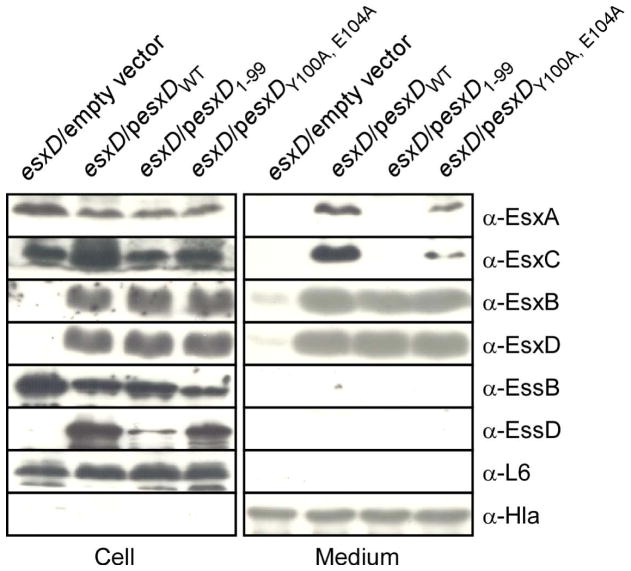
Previously, Champion and colleagues showed that the last 7 amino acids of CFP-10 (Mt-EsxB) are necessary and sufficient for secretion of ESAT-6•CFP-10 (Champion et al., 2006). However, this study did not define a consensus sequence. Recently, Daleke and colleagues showed that C-terminal truncations of PE25 encoded within the ESX-5 cluster of Mycobacterium marinum abolished secretion of PE25 and its interacting partner PPE41 (Daleke et al., 2012). In this study, alanine mutagenesis identified tyrosine and glutamic acid at positions 87 and 91 of the PE25 polypeptide, as key residues for secretion. A more extensive analysis identified the YxxxD/E motif in several ESX-1 and ESX-5 substrates of M. tuberculosis and M. marinum, including the WXG100 CFP-10 protein and EspB and in some instances an experimental validation was also provided (Daleke et al., 2012). Interestingly, the last 6 amino acids of EsxD, Y100yniE104G, also encompass this motif. This prompted us to evaluate the universality of the sequence feature for secretion in Firmicutes. Briefly, the complementing plasmid (pesxDWT) was modified to encode truncated EsxD lacking the last 6 amino acids (pesxD1-99), or a variant with two alanine substitutions at tyrosine 100 and glutamic acid 104 (pesxDY100A, E104A). The plasmids were transferred in the mutant strain lacking chromosomal esxD and cultures were fractionated to evaluate the synthesis and localization of secreted substrates by immunoblot. In this assay, EsxDY100A, E104A behaved similarly to full length EsxDWT. However, deletion of the last 6 amino acids, EsxD1-99, failed to restore secretion of EsxA and EsxC but surprisingly was dispensable for secretion of EsxB and EsxD (Figure 5). Examination of the machine components EssB and EssD by immunoblot revealed that plasmid encoded EsxD1-99, could not restore wild type levels of EssD. In this strain (esxD/pesxD1-99), EssD accumulated at about 20% of the level obtained with the complementing plasmid pesxDWT. For comparison, expression of EsxDY100A, E104A restored levels of EssD to about 75% (pesxDY100A, E104A as compared to pesxDWT) (Figure 5). As before, levels of EssB were not altered (Figure 5). The data suggest that EssD is a limiting factor for secretion of EsxA and EsxC. Thus, the last 6 amino acids of EsxD contribute to the stability of EssD and hence influence secretion of other substrates, suggesting perhaps an ordered sequence of secretion events or the existence of discrete sub-translocons for the secretion of substrates.
Figure 5. The last six amino acids of EsxD are important for secretion of EsxA and EsxC.
Bacterial cultures of the S. aureus esxD mutant harboring either the empty vector or vector with wild type esxD (pesxDWT), truncated esxD (pesxD1-99) or the esxD variant with double alanine substitutions (pesxDY100A, E104A) were grown to OD600nm of 1.0 and separated into cell and medium fractions. Proteins were precipitated with trichloroacetic acid, separated by SDS-PAGE and detected by immunoblotting with specific antibodies (α-EsxA, α-EsxC, α-EsxB, α-EsxD, α-EssB, α-EssD, and α-Hla or α-L6 for fractionation and loading controls).
DISCUSSION
Sec-dependent secretion is a pathway that accounts for the translocation or insertion of proteins across the plasma membrane of bacteria. The genetic requirements for this pathway and the molecular intricacies leading to substrate recognition have been examined extensively (Schneewind & Missiakas, 2012, Chatzi et al., 2013). The Sec pathway is thought to be absolutely essential for all bacterial growth. Many bacterial pathogens have evolved specialized secretion systems for the regulated or coordinated secretion of subsets of proteins that are generally dispensable for their growth in the laboratory, but critical for virulence and replication in infected hosts (Lee & Schneewind, 2001, Thanassi et al., 2012). Gram-positive bacteria in the phylum Firmicutes encode a genetic cluster that bears some similarities with the well-characterized T7SS of acid-fast bacteria M. tuberculosis, M. smegmatis and M. marinum (Abdallah et al., 2007, Bitter et al., 2009, Stoop et al., 2012). The salient feature of T7-like secretion systems includes the presence of small proteins of the WXG100 and SpoIIIE-FtsK-like ATPase family and was first noticed using a bioinformatic approach (Pallen, 2002). Shortly following this observation, experimental validations provided confirmation that proteins with a WXG100 domain are secreted with the help of several gene products that represent novel secretion systems (Pym et al., 2003, Stanley et al., 2003, Burts et al., 2005). S. aureus encodes the ESS cluster that includes two WXG100 proteins, EsxA and EsxB, and one protein with two FtsK-SpoIIIE domains, EssC (Burts et al., 2005). Several proteins in the ESS cluster, including EssC, are required for secretion of EsxA and EsxB but none, other than these three, shares homology with proteins of T7SS in mycobacteria. Further, the protein EsaC, whose sequence features are only conserved in staphylococci, is also secreted in an ESS-dependent manner (Burts et al., 2008). Here, we show that a fourth protein encoded by SAUSA300_0287 immediately downstream of esxB is secreted in the culture medium. Importantly, this protein was identified through its association with EsxB. We hypothesize that such an association occurs in a structural manner that is reminiscent of well-characterized WXG100 homo- and hetero-dimers and thus, we have named this protein EsxD for ESAT-6 secreted extracellular protein D. Further experimentations prompted us to rename EsaC as EsxC and EsaD as EssD (Figure 3B). This rationale is based on the following observations. First, the genetic analysis of the ess gene cluster in strain USA300 clearly demonstrates that the capacity to produce EsxB and EsxD, but not EsxA and EsxC, is tightly linked to production of all four proteins. Second, deletion of any one of the four esx genes prevents the secretion of all four substrates. Third, ESS-dependent secretion is not observed in mutants carrying in-frame deletion in the essA, essB, essC or essD genes. Fourth, biochemical and bacterial two hybrid studies demonstrate the existence of the following pairs EsxB•EsxD, EsxA•EsxC, EsxA•EsxA and EsxC•EsxC. Finally, unlike EsxA and EsxC, the EsxB and EsxD proteins appear to be unstable as reflected by the loss of all immune reactive signals when bacteria lack esxC, esxB, esxC or essD. Thus, the data suggest that in S. aureus, the WXG100 protein EsxB is unstable and cannot be secreted unless it is interacting with the non-WXG100 protein EsxD.
Mycobacteria secrete several substrates that have been classified into the EsxAB clan CL0352 (pfam.janelia.org/clan/EsxAB) and encompass pfam06013 (WXG100), pfam00934 (PE), pfam00823 (PPE) and pfam10824 (Esp). While not found in firmicutes, PE and PPE proteins are named after the Pro-Glu (PE) and Pro-Pro-Glu (PPE) motifs located in conserved domains of approximately 110 (PE) and 180 (PPE) amino acids and their function is unknown (Cole et al., 1998). A variable domain is sometimes fused at the C-terminus of PE or PPE proteins (Cole et al., 1998). Experimental evidence suggests that pairs of PE and PPE proteins interact as heterodimers and individual proteins may not be stable when expressed in isolation (Strong et al., 2006). The X-ray structure of one such pair, M. tuberculosis Rv2431c (PE) and Rv2430c (PPE) shows that Rv2431c is made of two α-helices that run antiparallel to each other and cradle against two antiparallel α-helices of the PPE protein Rv2430c. This structural organization is reminiscent of WXG100 dimers and may represent a structure uniquely compatible for the T7 translocon. The pfam10824 group includes the ESX secretion-associated proteins, EspA, EspC, EspD encoded in an operon. Any genetic disruption in this operon affects the secretion of ESAT-6 and CFP-10 (Fortune et al., 2005, MacGurn et al., 2005, DiGiuseppe Champion et al., 2009). It has been shown that EspC carries a C-terminal secretion signal that is interchangeable with that of CFP-10 and that it pairs with EspA (DiGiuseppe Champion et al., 2009). A structure for EspA, EspC or EspD is not yet available and homologues are not present in the genomes of Firmicutes. When the primary sequence of EsxD was queried using the Protein Model Portal (www.proteinmodelportal.org), a structural match with 3gwkC was retrieved from the Protein Data Bank (PDB). Although the two proteins, query and structural model, are only 14% identical at the amino acid level, 3gwkC is the crystal structure of the uncharacterized protein SAG1039 of Streptococcus agalactiae 2603 V/R that forms a homodimer and bears a canonical WXG100 domain signature. A similar structure has been deduced for the same protein from another strain, GBS1074 from Streptococcus agalactiae NEM316 (Shukla et al., 2010).
Like EsxC, EsxD appears to be species specific and homologues of these proteins are not encountered outside of the genus Staphylococcus. Nonetheless, the genetic organization shown in Figure 3B is conserved in Firmicutes and members of the genera Bacillus, Streptococcus and Listeria, which carry genes of similar size at the locations of esxC and esxD. Recently, it has been proposed that substrates of T7SS share the C-terminal motif YxxxE/D for secretion in mycobacteria (Daleke et al., 2012). This motif may only be carried by one of the partners in heterodimeric pairs. It was also speculated that the sequence motif could be FxxxD/E to include the CFP-10 protein of other high GC bacteria such as Nocardia farcinica, Corynebacterium diphtheriae and Streptomyces coelicolor (Daleke et al., 2012). An examination of staphylococcal substrates revealed the presence of two such motifs in the sequence of EsxD, F85teaD89 and Y100yniE104. Y100yniE104 represented the experimentally validated motif of Mycobacteriaceae and was further investigated. While, alanine substitutions in this motif did not alter substrate secretion, deletion of the last six amino acids (YnieEg) affected the stability of EssD, a candidate component of the ESS translocon and impaired the secretion of EsxA and EsxC. It is tempting to speculate that perhaps EsxD is a morphogenic substrate of the ESS translocon and may govern selective or sequential secretion of substrates.
In this work, we also provide some preliminary evidence for a contribution of the SaeRS two-component regulatory system in controlling expression of the ESS pathway. While, we have not performed an in-depth analysis of this regulation, this information has been useful to rule out the use of strain Newman for the study of ESS secretion. The regulation of the ess cluster is likely to be complex as is the case for many virulence factors in S. aureus (Cheung et al., 2004, Felden et al., 2011, Novick, 2003). A recent analysis suggested that the monocistronic esxA transcript is driven by a sigma A (σA) promoter, and a putative and inactive σB promoter in strain Newman (Schulthess et al., 2012). This analysis also revealed that esxA transcripts accumulate in sarA mutants, but are decreased in agr, arlR and spoVG mutants. Thus, esxA expression is regulated by several transcription factors known to fine-tune the secretome, in order to coordinate pathogenic strategies with bacterial cell density and environmental cues.
It is unclear why some Esx proteins self associate and others interact as heterodimers. Obviously, association may be a feature of secretion and subunits may not remain associated in the host. For example, lowering the pH results in dissociation of the ESAT-6•CFP-10 complex (de Jonge et al., 2007). In M. tuberculosis and M. marium ESX-1-secreted ESAT-6 may insert into host membranes and form pores to escape intracellular compartments (Smith et al., 2008, De Leon et al., 2012, Hsu et al., 2003). Gram-positive bacteria and acid fast staining bacteria appear to have championed Sec-independent secretion through the evolution of several modules. The fact that these proteins share very little sequence identity, both between and within species, but seem to share the same fold, is interesting. It seems reasonable to infer that both the WXG100 fold and dimerization may be pre-requisites for secretion. Sequence diversity may represent an immune escape mechanism reminiscent of antigenic variation. Further, the structural fold may be uniquely refractory to elicitation of broadly neutralizing antibodies. In S. aureus, the ESS pathway is required for persistence of deep-seated abscesses in infected animals (Anderson et al., 2011, Burts et al., 2008). Four secreted proteins must contribute to this process and their exact function and host targets remain to be identified.
EXPERIMENTAL PROCEDURES
Growth conditions
Staphylococci were grown in Tryptic Soy Broth (TSB). Chloramphenicol (Cm) was added to a final concentration of 20 μg/ml for plasmid selection and anhydrotetracycline was used at 50 ng/ml for allelic replacements of target genes (Bae & Schneewind, 2006). To assay for protein production and secretion, cultures were grown with vigorous shaking at 37°C for 2.5 h and the medium was supplemented with naïve horse serum to a final concentration of 0.2%. E. coli was grown in Luria Bertani medium at 37° C. When necessary, ampicillin (Amp) and kanamycin (Kan) were used at 100 μg/ml and 50 μg/ml, respectively. Isopropyl β-D-1-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-indolyl-β-D- galactopyranoside (X-gal) were used at 0.5 mM and 40 μg/ml, respectively.
Cloning and strains
In frame deletion mutants were generated in essA, essB, essC, esxC, esxB, essE and esxD using the pKOR1 recombination plasmid as described (Bae & Schneewind, 2006). Briefly, 1kbp-flanking regions of DNA upstream and downstream of the target gene were ligated together using a BglII restriction enzyme site and the resulting 2kbp fragment of DNA was cloned into the pKOR1 plasmid vector using the Gateway BP Clonase II enzyme from Invitrogen (Cat #11789-020). For each construct, the upstream and downstream pieces of DNA contain the first and last 15 codons, respectively, of the gene targeted for deletion. Thus, the two DNA fragments are fused together to create a pseudogene of 30 codons. All cloning steps were performed in E. coli and plasmid DNA was first electroporated into strain S. aureus RN4220 prior to its introduction into strain USA300 at 30°C in the presence of chloramphenicol. pKOR1 does not replicate at temperatures above 30°C and integrates into the chromosome when cells are grown at 42°C. Selection for homologous recombination, and thus allelic replacement, was performed upon induction of the counter-selection system with anhydrotetracyline as described (Bae & Schneewind, 2006). Strains used in this study are listed in Table S1. For complementation studies, the minimal coding sequence of the esxD gene was cloned in plasmid pWWW412 derived from plasmid pOS1 (Bubeck Wardenburg et al., 2006). In this construct, transcription of esxD is driven by the constitutive promoter of hprK (Bubeck Wardenburg et al., 2006). This plasmid was also used to clone esxB and esxD with a six-histidine repeat at the N-terminal for purification from S. aureus extracts. For protein production and purification in E. coli, the minimal coding sequence for EsxA and EsxB were cloned into plasmid pGEX-2TK (GE Healthcare) with N-terminal fusions to Glutathione S-transferase (GST) and esxD was cloned into plasmid pET15b (Novagen) resulting in a protein with an N-terminal six-histidine. For bacterial two-hybrid analyses, the minimal coding sequences of esxA, esxB, esxC, and esxD were cloned into the pCR®II-Blunt-TOPO® vector (Invitrogen). DNA fragments were excised using XbaI and KpnI followed by subsequent ligation into pT18 or pT25 vectors (Euromedex, France) pre-cut with XbaI and KpnI, resulting in plasmids pT18esxA, pT25esxA, pT18esxB, pT25esxB, pT18esxC, pT25esxC, pT18esxD and pT25esxD, respectively. All primers required for cloning are listed in Table S2.
Protein purification and mass measurement
Recombinant proteins were produced in E. coli BL21 (DE3). Overnight E. coli cultures were refreshed 1:100 in LB and incubated at 30°C until they reached an optical density at 600 nm (OD600nm) of 0.5 at which time IPTG was added to the culture to a final concentration of 1 mM and incubated for 4 h longer. Cells were sedimented by centrifugation, suspended in 50 mM Tris-HCl pH7 buffer A containing 150 mM NaCl, 5% glycerol and frozen at −20°C overnight. Cell pellets were then thawed and lysed in a French pressure cell (Thermo). Crude lysates were centrifuged at 100,000 ×g and the filtered supernatant of extracts producing GST-EsxA or GST-EsxB was subjected to affinity purification using glutathione S-transferase affinity chromatography as described by the manufacturer (GE Healthcare). Proteins were either eluted with 10 mM glutatione in buffer A. When necessary, the N-terminal GST tag was cleaved with thrombin and thrombin removed by incubation with benzamadine sepharose beads per manufacturer’s recommendations (GE Healthcare). His-tagged EsxD was found in the pellet following sedimentation of crude E. coli lysates at 100,000 ×g suggesting that recombinant EsxD is prone to aggregation. The pellets were suspended in buffer A containing 8 M urea and the samples were centrifuged once more at 100,000 ×g for 40 min. Solubilized His-EsxD was applied to a gravity flow column packed with Ni-NTA agarose beads. The column was washed with 2 column volumes of buffer A containing 8 M urea followed by elution with an imidazole gradient. All recombinant proteins were dialyzed in buffer A and stored at −80°C with the exception of denatured His-EsxD that was desalted in buffer A containing 8 M urea and used immediately to assess its ability to interact with GST-EsxA or GST-EsxB. For this assay, His-EsxD was diluted 10-fold in buffer A to bring the concentration of urea to 0.8M, a non-chaotropic concentration that favored refolding of His-EsxD. The renatured protein was subjected to centrifugation at 100,000 × g to remove any insoluble protein and a 1-ml volume was applied to gravity flow columns packed with glutatione beads preincubated with GST-EsxA or GST-EsxB. The 1-ml flow through samples were collected for analysis of unbound proteins whereas the beads were washed with 20 column volumes of buffer A and bound proteins eluted in 1-ml buffer A containing 10 mM glutathione. Proteins in the bound and unbound fractions were subjected to SDS-PAGE and western blot analysis as described below. For purification of His-EsxB and His-EsxD from S. aureus, exponentially growing cultures (twice 5 L and 100 ml, for His-EsxB and His-EsxD respectively) were spun and the filtered supernatant was applied to gravity flow columns packed with Ni-NTA agarose beads. The columns were washed with 10-column volumes of buffer A and interacting proteins eluted in buffer A containing 8 M urea. The material was desalted using Sephadex G-25 (GE Healthcare) in buffer A prior to analyses by mass spectrometry and SDS-PAGE followed by immunoblotting.
Animals for the generation of antisera
Experiments involving the generation of polyclonal antibodies using New Zealand white rabbits purchased from Harlan Sprague Dawley, followed protocols that were reviewed, approved and performed under the regulatory supervision of The University of Chicago’s Institutional Animal Care and Use Committee (IACUC). Animals were managed by the University of Chicago Animal Resource Center, which is accredited by the American Association for Accreditation of Laboratory Animal Care and the Department of Health and Human Services (DHHS number A3523-01). Animals were maintained in accordance with the applicable portions of the Animal Welfare Act and the DHHS “Guide for the Care and Use of Laboratory Animals”. Veterinary Care was under the direction of full-time resident veterinarians boarded by the American College of Laboratory Animal Medicine. Specific antisera were generated by emulsifying 50 μg of purified, renatured His-EsxD and dissolved in 500 μL of PBS with 500 μL of complete Freund’s adjuvant (Difco) by sonication and subcutaneous inoculation of female New Zealand white rabbits. Two subsequent boosts were performed by emulsifying protein with incomplete Freund’s adjuvant. Serum was harvested by cardiac puncture and stored at −80°C with 0.02% sodium azide.
Fractionation of bacterial cultures and immunoblotting
To assess production of proteins in S. aureus, aliquots of cultures were lyzed directly with lysostaphin. To assess secretion, culture aliquots were spun to separate proteins in the medium and the cells. Cells from bacterial pellets were washed and lyzed using lysostaphin. Proteins in these suspensions (lysed cultures or cells and medium) were precipitated by addition of 10 % final concentration trichloroacetic acid, washed in cold acetone, solubilized in 100 μl of buffer B (0.5 M Tris-HCl (pH 8.0), 4% SDS) and heated at 90 °C for 10 min. To assess proteins following purification over Ni-NTA or glutathione agarose beads, 100 μl sample aliquots were mixed with 100 μl of buffer B twice concentrated (2X). Proteins were separated on SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membrane for immunoblot analysis. Proteins were detected with specific rabbit polyclonal antibodies raised against purified protein at a dilution of 1:5,000 (α-EsxA, α-EsxB, α-EsxC, α-EsxD, α-EssB, α-EssD,) or 1:20,000 (α-L6, α-Hla). Immunoreactive products were revealed by chemiluminescent detection after incubation with an anti-rabbit HRP-conjugate secondary antibody (1:20,000, Cell Signaling Technology).
Bacterial two-hybrid analyses
Genes esxA, esxB, esxC or esxD cloned in pT18 or pT25 were transformed into E. coli strain BTH101 (Euromedex, France) on agar containing kanamycin and ampicillin. Isolated colonies were used to inoculate LB medium supplemented with kanamycin and ampicillin and grown for 16 h at 37°C. Cultures were diluted to a final OD600 0.5 and 10 μl plated on solid medium supplemented with kanamycin, ampicillin, IPTG and X-Gal. The plates were incubated at 37°C for 24 h to visualize any color change from white to blue as a reflection of positive interaction.
Supplementary Material
Acknowledgments
The authors thank Olaf Schneewind for careful reading of the manuscript, and members of the Schneewind and Missiakas laboratory for suggestions and discussions. We thank the referees of our manuscript for their input and suggestions. Mark Anderson acknowledges support by the Biodefense Training Grant in Host-Pathogen Interactions T32 AI065382 at the University of Chicago and American Heart Association award 11PRE7600117. This work was supported by the National Institute of Allergy and Infectious Diseases (AI 75258).
References
- Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, Vandenbroucke-Grauls CM, Appelmelk BJ, Bitter W. Type VII secretion--mycobacteria show the way. Nat Rev Microbiol. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- Adhikari RP, Novick RP. Regulatory organization of the staphylococcal sae locus. Microbiology. 2008;154:949–959. doi: 10.1099/mic.0.2007/012245-0. [DOI] [PubMed] [Google Scholar]
- Anderson M, Chen YH, Butler EK, Missiakas DM. EsaD, a secretion factor for the Ess pathway in Staphylococcus aureus. J Bacteriol. 2011;193:1583–1589. doi: 10.1128/JB.01096-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbing MA, Kaufmann M, Phan T, Chan S, Cascio D, Eisenberg D. The crystal structure of the Mycobacterium tuberculosis Rv3019c-Rv3020c ESX complex reveals a domain-swapped heterotetramer. Protein Sci. 2010;19:1692–1703. doi: 10.1002/pro.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Bitter W, Houben EN, Bottai D, Brodin P, Brown EJ, Cox JS, Derbyshire K, Fortune SM, Gao LY, Liu J, Gey van Pittius NC, Pym AS, Rubin EJ, Sherman DR, Cole ST, Brosch R. Systematic genetic nomenclature for type VII secretion systems. PLoS pathogens. 2009;5:e1000507. doi: 10.1371/journal.ppat.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubeck Wardenburg J, Williams WA, Missiakas D. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc Natl Acad Sci U S A. 2006;103:13831–13836. doi: 10.1073/pnas.0603072103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burts ML, DeDent AC, Missiakas DM. EsaC substrate for the ESAT-6 secretion pathway and its role in persistent infections of Staphylococcus aureus. Molecular microbiology. 2008;69:736–746. doi: 10.1111/j.1365-2958.2008.06324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burts ML, Williams WA, DeBord K, Missiakas DM. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc Natl Acad Sci U S A. 2005;102:1169–1174. doi: 10.1073/pnas.0405620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion PA, Stanley SA, Champion MM, Brown EJ, Cox JS. C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science. 2006;313:1632–1636. doi: 10.1126/science.1131167. [DOI] [PubMed] [Google Scholar]
- Chatzi KE, Sardis MF, Karamanou S, Economou A. Breaking on through to the other side: protein export through the bacterial Sec system. Biochem J. 2013;449:25–37. doi: 10.1042/BJ20121227. [DOI] [PubMed] [Google Scholar]
- Chen YH, Anderson M, Hendrickx AP, Missiakas D. Characterization of EssB, a protein required for secretion of ESAT-6 like proteins in Staphylococcus aureus. BMC Microbiol. 2012;12:219. doi: 10.1186/1471-2180-12-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AL, Bayer AS, Zhang G, Gresham H, Xiong YQ. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol Med Microbiol. 2004;40:1–9. doi: 10.1016/S0928-8244(03)00309-2. [DOI] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Daleke MH, Ummels R, Bawono P, Heringa J, Vandenbroucke-Grauls CM, Luirink J, Bitter W. General secretion signal for the mycobacterial type VII secretion pathway. Proc Natl Acad Sci U S A. 2012;109:11342–11347. doi: 10.1073/pnas.1119453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge MI, Pehau-Arnaudet G, Fretz MM, Romain F, Bottai D, Brodin P, Honore N, Marchal G, Jiskoot W, England P, Cole ST, Brosch R. ESAT-6 from Mycobacterium tuberculosis dissociates from its putative chaperone CFP-10 under acidic conditions and exhibits membrane-lysing activity. J Bacteriol. 2007;189:6028–6034. doi: 10.1128/JB.00469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leon J, Jiang G, Ma Y, Rubin E, Fortune S, Sun J. Mycobacterium tuberculosis ESAT-6 exhibits a unique membrane-interacting activity that is not found in its ortholog from non-pathogenic Mycobacterium smegmatis. J Biol Chem. 2012 doi: 10.1074/jbc.M112.420869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiuseppe Champion PA, Champion MM, Manzanillo P, Cox JS. ESX-1 secreted virulence factors are recognized by multiple cytosolic AAA ATPases in pathogenic mycobacteria. Molecular microbiology. 2009;73:950–962. doi: 10.1111/j.1365-2958.2009.06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- Felden B, Vandenesch F, Bouloc P, Romby P. The Staphylococcus aureus RNome and its commitment to virulence. PLoS pathogens. 2011;7:e1002006. doi: 10.1371/journal.ppat.1002006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune SM, Jaeger A, Sarracino DA, Chase MR, Sassetti CM, Sherman DR, Bloom BR, Rubin EJ. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc Natl Acad Sci U S A. 2005;102:10676–10681. doi: 10.1073/pnas.0504922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudo AT, Cheung AL, Nagel R. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch Microbiol. 1997;168:53–58. doi: 10.1007/s002030050469. [DOI] [PubMed] [Google Scholar]
- Giraudo AT, Raspanti CG, Calzolari A, Nagel R. Characterization of a Tn551-mutant of Staphylococcus aureus defective in the production of several exoproteins. Can J Microbiol. 1994;40:677–681. doi: 10.1139/m94-107. [DOI] [PubMed] [Google Scholar]
- Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, Morin PM, Marks CB, Padiyar J, Goulding C, Gingery M, Eisenberg D, Russell RG, Derrick SC, Collins FM, Morris SL, King CH, Jacobs WR., Jr The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci USA. 2003;100:12420–12425. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilghari D, Lightbody KL, Veverka V, Waters LC, Muskett FW, Renshaw PS, Carr MD. Solution structure of the Mycobacterium tuberculosis EsxG.EsxH complex: functional implications and comparisons with other M. tuberculosis Esx family complexes. J Biol Chem. 2011;286:29993–30002. doi: 10.1074/jbc.M111.248732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee VT, Schneewind O. Protein secretion and the pathogenesis of bacterial infections. Genes Dev. 2001;15:1725–1752. doi: 10.1101/gad.896801. [DOI] [PubMed] [Google Scholar]
- MacGurn JA, Raghavan S, Stanley SA, Cox JS. A non-RD1 gene cluster is required for Snm secretion in Mycobacterium tuberculosis. Molecular microbiology. 2005;57:1653–1663. doi: 10.1111/j.1365-2958.2005.04800.x. [DOI] [PubMed] [Google Scholar]
- Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Molecular microbiology. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- Pallen MJ. The ESAT-6/WXG100 superfamily -- and a new Gram-positive secretion system? Trends Microbiol. 2002;10:209–212. doi: 10.1016/s0966-842x(02)02345-4. [DOI] [PubMed] [Google Scholar]
- Pym AS, Brodin P, Majlessi L, Brosch R, Demangel C, Williams A, Griffiths KE, Marchal G, Leclerc C, Cole ST. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat Med. 2003;9:533–539. doi: 10.1038/nm859. [DOI] [PubMed] [Google Scholar]
- Renshaw PS, Lightbody KL, Veverka V, Muskett FW, Kelly G, Frenkiel TA, Gordon SV, Hewinson RG, Burke B, Norman J, Williamson RA, Carr MD. Structure and function of the complex formed by the tuberculosis virulence factors CFP-10 and ESAT-6. Embo J. 2005;24:2491–2498. doi: 10.1038/sj.emboj.7600732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneewind O, Missiakas DM. Protein secretion and surface display in Gram-positive bacteria. Philos Trans R Soc Lond B Biol Sci. 2012;367:1123–1139. doi: 10.1098/rstb.2011.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulthess B, Bloes DA, Berger-Bachi B. Opposing roles of sigmaB and sigmaB-controlled SpoVG in the global regulation of esxA in Staphylococcus aureus. BMC Microbiol. 2012;12:17. doi: 10.1186/1471-2180-12-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Pallen M, Anthony M, White SA. The homodimeric GBS1074 from Streptococcus agalactiae. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:1421–1425. doi: 10.1107/S1744309110036286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Manoranjan J, Pan M, Bohsali A, Xu J, Liu J, McDonald KL, Szyk A, LaRonde-LeBlanc N, Gao LY. Evidence for pore formation in host cell membranes by ESX-1-secreted ESAT-6 and its role in Mycobacterium marinum escape from the vacuole. Infect Immun. 2008;76:5478–5487. doi: 10.1128/IAI.00614-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Raghavan S, Hwang WW, Cox JS. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc Natl Acad Sci USA. 2003;100:13001–13006. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoop EJ, Bitter W, van der Sar AM. Tubercle bacilli rely on a type VII army for pathogenicity. Trends Microbiol. 2012 doi: 10.1016/j.tim.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy R, Fyfe PK, Hunter WN. Structure of Staphylococcus aureus EsxA suggests a contribution to virulence by action as a transport chaperone and/or adaptor protein. J Mol Biol. 2008;383:603–614. doi: 10.1016/j.jmb.2008.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanassi DG, Bliska JB, Christie PJ. Surface organelles assembled by secretion systems of Gram-negative bacteria: diversity in structure and function. FEMS Microbiol Rev. 2012;36:1046–1082. doi: 10.1111/j.1574-6976.2012.00342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.