Abstract
Autophagy is important in the heart for maintaining homeostasis when changes in nutrient levels occur. Autophagy is involved in the turnover of cellular components, and is rapidly upregulated during stress. Studies have found that autophagy is reduced in metabolic disorders including obesity and diabetes. This leads to accumulation of protein aggregates and dysfunctional organelles, which contributes to the pathogenesis of cardiovascular disease. Autophagy is primarily regulated by two components: the mammalian target of rapamycin (mTOR) and AMP-activated protein kinase (AMPK). While mTOR integrates information about growth factors and nutrients and is a negative regulator of autophagy, AMPK is an energy sensor and activates autophagy when energy levels are low. These pathways therefore present targets for the development of autophagy-modulating therapies.
Alterations in Metabolism Impact Autophagy in the Heart
Autophagy (see Glossary) is an important mechanism that tissues utilize to maintain cellular homeostasis, and defective autophagy has been implicated in a wide range of pathologies including heart disease. The heart utilizes autophagy to maintain cellular homeostasis under both baseline conditions and in response to stress [1-4]. While many tissues of the body are susceptible to changes in nutrient supply, it is particularly important for cardiomyocytes to adapt to changes in metabolite supply to sustain contraction. Changes to nutrients, energy status, oxygen levels or external stresses all have the potential to disrupt heart function, and yet it is critical that the heart continues to function despite these major metabolic changes. Changes in metabolism that can directly affect autophagy in the heart include both chronic and acute conditions. Chronic conditions include obesity and metabolic syndrome, resulting in elevated circulating lipid and insulin levels [5]. Acute events such as a myocardial infarction (MI) result in insufficiency of oxygen and glucose supply to a region of the heart and many studies suggest that upregulation of autophagy in response to acute cardiac stress is cardioprotective and important for minimizing myocardial damage [6,7]. In contrast, the reduced autophagic flux that is observed in mouse models of obesity [8], diabetes [9], and metabolic syndrome [10] is thought to contribute to disease pathology, including development of heart failure. Long-term dyslipidemia, defective insulin signaling, and other chronic metabolic changes therefore impact the heart’s cellular stress response in ways distinct from acute damage.
In this review, we discuss how changes in energy levels, nutrients and growth factor availability regulate autophagy in the heart, as well as how repression of autophagy in disease states contributes to cardiovascular diseases. We also address the feasibility of specifically targeting autophagy, and how this represents a new avenue in the treatment or prevention of heart disease.
Molecular Pathways Involved in Autophagy
Autophagy begins with phagophore nucleation that is promoted by a complex comprised of three proteins: Beclin 1, vacuolar protein sorting (VPS)34, and VPS15. The phagophore then elongates via a mechanism that depends on autophagy (Atg) proteins and microtubule-associated protein 1 light chain 3 (LC3) [5,11]. The mature autophagosome engulfs its target material before fusing with a lysosome and degrading the cargo. The amino acids and other components of the degraded material are then transported to the cytosol and reused. Autophagy occurs continuously under baseline conditions in the heart, and impairment of this process results in rapid accumulation of protein aggregates and dysfunctional organelles, leading to heart failure [1,2,7]. Autophagy is rapidly upregulated in the myocardium in response to stress or changes to nutrient supply, in an effort to maintain homeostasis [3,4,6,7]. In addition, mitochondria are responsible for the production of ATP for cellular energy through oxidative phosphorylation, but dysfunctional mitochondria generate excessive reactive oxygen species (ROS) and can promote cell death by releasing death-promoting factors. Degradation of dysfunctional mitochondria by autophagy not only prevents cell damage [7,8,12], but also streamlines energy production and utilization [13,14], which is particularly important in cardiomyocytes that are densely packed with mitochondria.
The Mammalian Target of Rapamycin is a Central Regulator of Autophagy and Metabolism
The mammalian target of rapamycin (mTOR) is a conserved serine/threonine kinase that regulates cell growth and autophagy by integrating growth factors and nutrient signals [15]. mTOR is activated under nutrient-rich conditions and promotes cell growth in part by suppressing autophagy. Insufficiency of nutrients or growth factors results in mTOR inactivation and induction of autophagy. mTOR is part of the mTORC1 signaling complex which includes the scaffolding protein regulatory-associated protein of mTOR (RAPTOR), as well as proline-rich Akt substrate 40 kDa (PRAS40), DEP domain-containing mTOR-interacting protein (DEPTOR), and mTOR associated protein, LST8 homolog (MLST8).
mTOR regulates metabolism by activating specific transcription factors that regulate expression of enzymes involved in metabolic pathways. For instance, mTORC1 activates peroxisome proliferator-activated receptor γ (PPARγ), a nuclear receptor that regulates expression of genes that are involved in fatty acid synthesis and uptake [16]. mTORC1 also activates hypoxia-inducible factor 1α (HIF1α), which promotes the expression of genes that regulate glucose transport and glycolysis [17]. In addition, mTORC1 activates PPARγ coactivator-1 (PGC1α), a nuclear cofactor that regulates mitochondrial biogenesis and the expression of proteins involved in mitochondrial metabolism [18].
Growth Factor-Dependent mTOR Activation
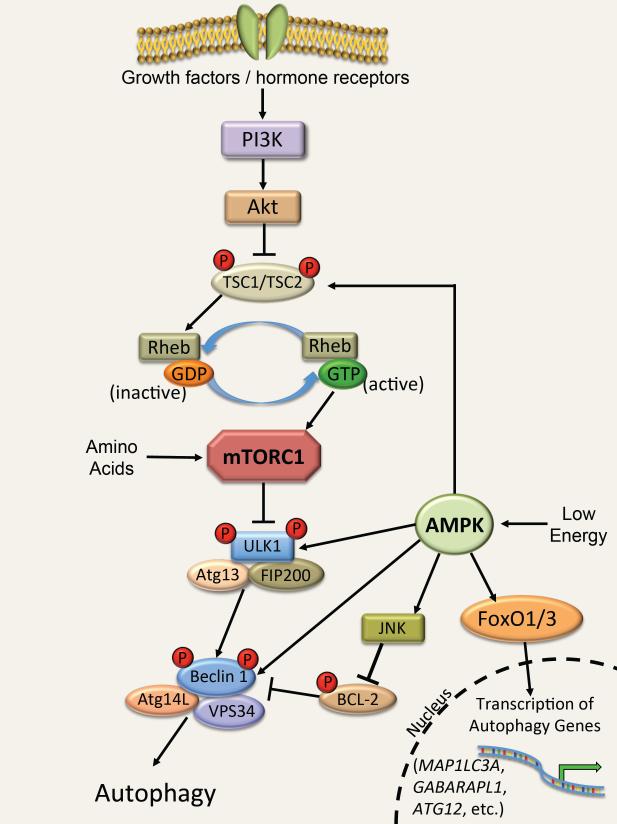
The presence and binding of growth factors and/or insulin to their membrane receptors activates signaling pathways that lead to mTORC1 activation and subsequent inactivation of autophagy. The activation of mTORC1 can occur through an AKTTSC1/2 or an AKT-PRAS40 dependent manner. In the former, Akt phosphorylates and inactivates the tuberous sclerosis (TSC) 1/2 complex [19]. The TSC1/2 complex is a GTPase-activating protein that promotes the conversion of active, GTP-bound Ras homolog enriched in brain (RHEB) to the inactive GDP-bound RHEB. GTP-bound RHEB is a critical activator of mTORC1, and inhibition of TSC1/2 by Akt results in increased GTP-bound RHEB, activation of mTORC1 and suppression of autophagy (Figure 1). Akt can also promote activation of mTORC1 in a TSC1/2-independent manner. PRAS40 is a RAPTOR-interacting protein and a constitutive inhibitor of mTORC1. Phosphorylation of PRAS40 by Akt leads to dissociation of PRAS40 from RAPTOR, and subsequent activation of mTORC1 [20].
Figure 1.
Regulation of autophagy by mTOR and AMPK
Insulin and growth factors signal through phosphoinositide 3-kinase (PI3K) and Akt to inhibit the tuberous sclerosis (TSC)1/2 complex. Inhibition of this complex allows active Ras homolog enriched in brain (RHEB)-GTP to activate mTOR in the mTORC1 complex, leading to phosphorylation of the ULK1 complex and inhibition of autophagy. Amino acids can also activate the mTORC1 complex by a separate mechanism. AMPK responds to low cellular energy levels by activating ULK1 via phosphorylation, resulting in activation of autophagy. Additionally, AMPK can initiate autophagy by disrupting Bcl-2 inhibition of Beclin 1, by activating forkhead box protein O (FoxO) transcription factors, and by directly activating TSC1/2 and Beclin 1.
Amino Acid-Dependent mTOR Activation
The activity of mTORC1 is also independently regulated by intracellular levels of amino acids [21]. When the levels of amino acids present in the cell are sufficient, mTORC1 receives signals that promote its activity and suppress autophagy [22]. In contrast to growth factors, amino acids do not rely on inhibition of TSC1/2 to activate mTORC1 [23]. Instead, the Ras-related GTP-binding (RAG) guanosine triphosphatases (GTPases) play a key role in transducing amino acid signaling to mTORC1 [21]. In the presence of amino acids, active RAG GTPases recruit mTORC1 to the exterior of the lysosome, where it is activated by RHEB [24]. RHEB is responsible for coupling of both growth factor signaling and amino acid levels to mTORC1 [25]. However, growth factors in the absence of amino acids do not induce translocation of mTORC1 to lysosomes, and RNAi-mediated knockdown of RHEB abrogates mTORC1 activation by amino acids but does not interfere with the amino acid-induced movement of mTOR to lysosomes [24]. This suggests that translocation of mTORC1 to the lysosomal surface occurs independently of mTORC1 activity and does not require RHEB or growth factors.
mTORC1 Activity is Critical for Preservation of Heart Function
The importance of amino acid-dependent activation of mTORC1 was recently demonstrated by Efeyan et al. [26]. Upon disruption of the maternal nutrient supply at birth, neonates activate autophagy to adapt to starvation until suckling is established [27]. Mice expressing a constitutively active RAG GTPase have reduced early postnatal survival due to a failure to induce autophagy in tissues including the heart [26]. This suggests that constitutive RAG GTPase activity prevents mTORC1 inhibition, leading to defective autophagy and insufficient amino-acid production. Riehle et al. recently linked the insulin/IGF-1/mTOR pathway to the suppression of autophagy in the neonatal heart once feeding is established after birth [28]. This study found that cardiac specific deletion of the insulin receptor genes Isr1 and Irs2 in myocytes led to reduced mTOR signaling, persistent activation of autophagy and subsequent development of heart failure. Thus, although inhibition of mTORC1 and activation of autophagy in the heart upon birth is important for early postnatal survival [26], subsequent survival depends on activation of mTORC1 to inhibit autophagy and promote cardiac growth [28].
Many other studies have investigated the importance of mTORC1 in the heart. Zhu et al. found that mTOR is essential for cardiac development and function, where cardiomyocyte-specific deletion of mTOR leads to >90% embryonic lethality [29]. Surviving mice have severely impaired cardiac function and 100% mortality by 10 weeks of age [29]. This study reported that mTOR-deficiency results in reduced proliferation and loss of myocytes. However, they did not investigate whether this was due to excessive or inappropriate induction of autophagy in the myocytes during embryonic development. In addition, cardiac specific deletion in adult mice of the gene encoding RAPTOR causes severe heart failure with high mortality within 6 weeks after gene ablation [30]. In RAPTOR-deficient mice, cardiac pressure overload by aortic banding results in rapid development of heart failure without first undergoing adaptive hypertrophy [30]. This study found that lack of adaptive myocyte growth is due to reduced protein synthesis, alterations in metabolism and increased autophagy. mTORC1 thereby also plays a critical role in regulating cardiac hypertrophy. These studies suggest that proper regulation of autophagy and protein synthesis via mTORC1 activation plays an important role in maintaining homeostasis in the heart under both physiological and pathological conditions. Clearly, mTORC1 has a broad range of functions in the heart that are critical to maintaining myocardial health.
Alterations in mTOR and Autophagy Signaling in Cardiovascular Diseases
Autophagy in the Diabetic Heart
Diabetic patients are at increased risk of developing cardiomyopathy [31,32]. These patients also have increased injury and poor prognosis after myocardial infarction [33,34]. The exact mechanisms for the development of cardiomyopathy are unclear, but reduced cardiac autophagy is thought to be one of the factors since restoration of autophagy rescues cardiac function in diabetic mice [35]. Hyperglycemia plays a key role in the development of diabetic cardiomyopathy [36] and has been reported to inhibit autophagy in myocytes [37]. This reduction in autophagy leads to accumulation of cytotoxic protein aggregates and dysfunctional organelles, which in turn lead to activation of apoptosis and loss of myocytes. Interestingly, some studies have found that the impaired cardiac autophagy in hearts of mouse models of Type 1 diabetes (T1D) are compensatory rather than detrimental [9,38]. For instance, restoration of autophagy levels by overexpression of the autophagy protein Beclin 1 in mice decreased cardiac function in diabetic mice, compared to diabetic mice with impaired autophagy or Beclin 1+/− mice [9]. This study found that attenuated autophagy in diabetic mouse hearts was associated with elevated markers of mitochondrial autophagy [9]. Therefore, it may be more beneficial to modulate specific forms of autophagy in the diabetic heart, such as mitochondrial autophagy, rather than enhance general, untargeted autophagy. However, the diminished autophagy levels found in most other tissues in diabetic animals is thought to contribute to pathology, rather than help prevent it [38]. Therefore, maintaining adequate levels of autophagy is important for clearing dysfunctional cell components in the diabetic heart, but overactivation of autophagy may negatively affect heart function in models of diabetes.
The interplay between diabetes and autophagy signaling therefore appears particularly complex. Overactivation of autophagy in diabetes is clearly disadvantageous and possibly causes excessive cell stress that contributes to the development of cardiomyopathy [39]. However, restoration of proper levels of autophagy may be beneficial in preventing development of cardiomyopathy or other complications of diabetes. Clearly, additional studies are needed before autophagy-modulating therapies are clinically applied to diabetic patients. It is necessary to more clearly define when autophagy is beneficial versus deleterious in diabetic cardiomyopathy.
Autophagy and Obesity
Obesity also has suppressive effects on autophagy and is a known risk factor for developing diabetes and heart failure [40,41]. In obesity and metabolic disorder, elevated circulating lipid, amino acid, and insulin levels all contribute to decreased autophagy [8,10,42]. In addition, Yang et al. found that impaired autophagy might contribute to the insulin resistance observed in obesity. Obesity is characterized by the downregulation of the autophagy protein Atg7 in the liver, and restoration of Atg7 improves glucose tolerance in mice [43]. Moreover, mice fed a high fat diet (HFD) increase mTORC1 activity and suppress autophagy in their cardiomyocytes through RHEB activity [44]. Interestingly, mice fed HFD fail to deactivate mTORC1 and initiate autophagy following myocardial ischemia, confirming that a complex network of signaling pathways is in place to sense a wide array of nutrient and cell statuses [44]. In this case, it appears that overactivation of one negatively regulating pathway preempts activation of autophagy by sensors of acute stress such as ischemia [44].
Autophagy in the Aging Heart
Autophagy is also reduced with age and is thought to contribute to the aging process [45-47]. Knockouts of essential autophagy genes result in the manifestation of multiple age-associated problems, such as accumulation of ubiquitinated proteins, lipids, and lysosomes containing lipofuscin, as well as disorganized mitochondria and increased oxidative stress [1,48-50]. In the heart, autophagic activity is reduced with age, and the autophagy deficiency seen in cardiac-specific Atg5 knockout mice resembles an accelerated cardiac aging phenotype [47]. In contrast, increased autophagy delays aging and extends longevity. For instance, reduction in mTORC1 activity has been shown to prolong life in mice [51]. Caloric restriction (CR) is a potent physiological inducer of autophagy in the hearts of rats [45,52], and is well known to extend life span in animals and potentially also humans [53,54]. CR reduces the incidence of diabetes, cardiovascular disease, cancer, and brain atrophy [53,55]. Interestingly, inhibition of autophagy abrogates the anti-aging effects of CR [52]. Thus, these studies suggest that CR may contribute to maintaining sufficient levels of autophagy and helps to preserve heart function in aging individuals by promoting the clearance of protein aggregates and dysfunctional organelles.
Activation of Autophagy by the Energy Sensor AMPK
AMPK is a highly conserved serine/threonine kinase that functions as a key metabolic sensor in cells [56]. AMPK is rapidly activated by conditions that increase the AMP/ATP ratio such as exercise, ischemia, or lack of glucose [57]. To maintain energy homeostasis, AMPK inhibits metabolic pathways that consume energy and increases pathways such as autophagy that produce energy.
Regulation of Autophagy by AMPK
AMPK can activate autophagy via at least five different mechanisms (Figure 1). First, activation of AMPK stimulates JNK1, which mediates BCL-2 phosphorylation and subsequent Beclin 1–BCL-2 dissociation [58]. The dissociation of Beclin 1 from BCL-2 allows Beclin 1 to form a complex with VPS34, an essential component of the autophagy machinery, to initiate phagophore formation. Mice with a conditional cardiac VPS34 knockout develop cardiac hypertrophy and have impaired contractile function [59]. Histological examination of the hearts of VPS34 knockout mice showed an accumulation of abnormal mitochondria, indicating a failure to clear these dysfunctional organelles [59]. Second, AMPK controls the expression of the FoxO transcription factors that induce expression of autophagy-related genes; AMPK activates FoxO1 and FoxO3 in skeletal muscle and cardiomyocytes which in turn increase the expression of microtubule-associated proteins 1A/1B light chain 3A (MAPLC3A), GABA(A) receptor-associated protein-Like 1 (GABARAPL1), and Atg12 [60,61]. AMPK can also indirectly modulate FoxO transcriptional activity by activating the histone deacetylase SIRT1 under metabolic stress [62,63]. SIRT1 deacetylates a number of targets in response to various intracellular stresses. AMPK activation results in increased NAD+ levels that lead to SIRT1 activation and deacetylation-induced activation of its downstream targets, including FoxO1 and FoxO3 [62,64]. Interestingly, FoxO transcription factors are aberrantly active in hearts of diabetic mice and mice fed HFD [65]. Moreover, the FoxOs appear to actually contribute to the progression of diabetic cardiomyopathy, as FoxO1 single knockout and FoxO1/FoxO3 double-knockout mice are protected against HFD-induced cardiac hypertrophy and diabetes-induced loss of contractility [65]. A third way by which AMPK induces autophagy is via phosphorylation and activation of ULK1, a serine/threonine-protein kinase that is involved in autophagy in response to starvation [66,67]. Finally, AMPK can also activate autophagy through TSC2 phosphorylation [68] and by directly phosphorylating Beclin 1 [69]. AMPK is therefore a critical junction in the integration of upstream energy sensing with downstream autophagy activation.
AMPK Action in Cardiac Autophagy
AMPK is activated in the heart during stress to induce autophagy. For instance, during myocardial ischemia, defective mitochondrial oxidative phosphorylation results in a drop in intracellular ATP, AMPK activation and increased autophagy [3]. Matsui et al. reported that transgenic mice overexpressing a dominant negative AMPK mutant have reduced autophagy and increased injury after ischemia, suggesting that AMPK-mediated activation of autophagy is a protective response [3]. Consistent with this finding, recent studies have found that activation of autophagy after a myocardial infarction protects against ischemic injury by removing dysfunctional organelles [7] and preserving energy levels [6]. In contrast, the diabetic cardiomyopathy seen in a mouse model of severe early-onset T1D, is associated with suppression of autophagy and increased apoptosis in myocytes [35]. AMPK activity is decreased in the hearts of these mice, which may account for impaired autophagy, and restoration of AMPK activity with metformin prevents the development of cardiomyopathy [35]. The activation of AMPK and subsequent increase in cardiac autophagy results in reduced myocyte apoptosis in diabetic mouse hearts [58]. Thus, enhancing cardiac autophagy in patients with diabetes could protect against development of diabetic cardiomyopathy.
Upstream Sensors mTORC1 and AMPK Transduce Signals to ULK1
Both mTORC1 and AMPK regulate autophagy via the ULK1 protein kinase in cells including cardiomyocytes (Figure 1). ULK1 forms a complex with ATG13 and FIP200, both essential autophagic proteins, and responds to changes in nutrients and energy signals via a change of its serine phosphorylation status [70]. mTOR has been reported to directly phosphorylate ULK1 and ATG13, which leads to inhibition of the ULK1 complex [70,71]. In contrast, AMPK-mediated phosphorylation leads to activation of the ULK1 complex [66,67]. Kim et al. demonstrated that glucose starvation in cells results in AMPK-dependent phosphorylation of ULK1 on Ser317 and Ser777 sites and activation of autophagy [67]. They also showed that mTORC1 phosphorylates ULK1 on Ser757 (Ser758 in human), which precludes its activation by AMPK [67]. Thus, ULK1 activity appears to depend on which residues are phosphorylated; Ser757 phosphorylation inhibits autophagy, while Ser317 and Ser777 phosphorylation enhances autophagy. mTORC1 also phosphorylates ATG13, thereby blocking formation of the ATG13/ULK1 complex and initiation of autophagy [71]. Inhibition of mTOR by rapamycin, nutrient deprivation, or insufficient insulin signaling therefore leads to upregulation of autophagy via activation of ULK1.
One mechanism by which the ULK1 complex transduces upstream signals to the autophagy pathway was recently elucidated. Russell et al. discovered that ULK1 directly phosphorylates Beclin 1 on Ser14, thereby activating the VPS34-Beclin 1 complex to promote autophagy induction and maturation [72]. However, because ULK1 contains at least 30 phosphorylation sites and many of the responsible kinases and the functions of these phosphorylation events remain to be identified [73], the regulation of ULK1 activity still requires substantial investigation before considering ULK1 as a target for therapeutic intervention.
Therapies and Treatments: Their Effects on Autophagy
Obesity and sedentary lifestyle are two major health risks for cardiovascular disease, diabetes, and metabolic disorder [74,75]. Current treatments options work by targeting a wide range of pathways, and many are very effective at controlling health risks. Diet changes and exercise are always the first line of recommended therapy. Many reports have linked CR and exercise to elevated autophagic flux, and studies suggest that upregulating autophagy can increase lifespan, stabilize atherosclerotic plaques, and reduce the risk of stroke and cardiovascular disease [45,76]. However, lifestyle changes are not always effective in treating obesity or diabetes.
The current standard therapy in reducing cholesterol levels and decreasing the risk of an adverse cardiac event includes treatments with statins [77]. Statins target and inhibit HMG-CoA reductase, the rate-limiting enzyme in the production of cholesterol by the liver. Simvastatin was recently shown to increase autophagy and the specific degradation of mitochondria by autophagy in mouse hearts by decreasing mTOR activity [78]. This effect is directly related to both HMG-CoA reductase inhibition and inactivation of Akt. Moreover, simvastatin is effective in reducing infarct size following myocardial infarction in mice via a mechanism involving autophagy of mitochondria [78]. This emphasizes the cardioprotective effects of autophagy following acute damage.
Metformin is an anti-diabetic drug commonly administered to patients with T2D. It reduces hepatic glucose output, thereby lowering circulating blood glucose. In addition, metformin increases insulin sensitivity and binding of insulin to its receptor. Metformin-mediated activation of AMPK is important for its effects on both hepatic glucose production and insulin sensitization [79]. Treatment of diabetic mice with metformin restores autophagy levels in cardiac tissue and significantly improves cardiac function [35]. Metformin treatment of diabetic mice also reduces cardiomyocyte apoptosis and protects against development of diabetic cardiomyopathy [58]. Thus, activation of autophagy is cardioprotective across a range of disease states (Figure 2). However, it is important to note that chronic or overactive autophagy can have severely detrimental effects in the heart and contributes to pathogenesis in diabetic cardiomyopathy and in response to cardiac loading [9,80]. It is therefore critical to strike a balance between sufficient autophagy to adapt to changing metabolic factors and excessive autophagy that can cause excessive cell stress.
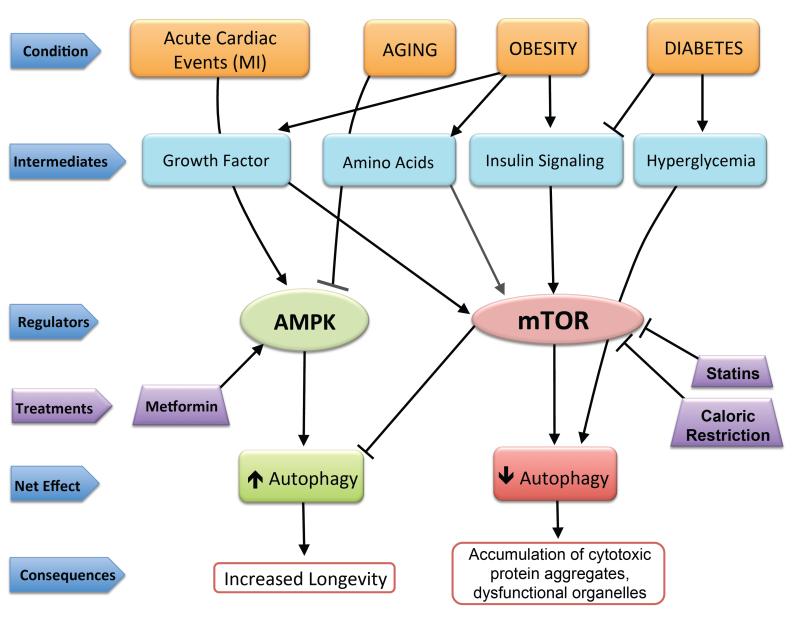
Figure 2.
Effects of various disease conditions and treatments on autophagy regulation. Acute events, such as a myocardial infarction (MI), increase AMPK activity to upregulate autophagy. In contrast, aging suppresses autophagy via AMPK inhibition. Autophagy is affected by conditions such as obesity and diabetes through activation or deactivation of mTOR. Obesity results in impaired autophagy through increased signaling of insulin, amino acid, and growth factor sensing pathways and subsequent mTOR activation. Diabetes results in impaired insulin signaling, but hyperglycemia can directly negatively regulate autophagy. Treatment options such as statins or caloric restriction increase autophagy by inhibiting mTOR, whereas metformin enhances AMPK activity to elevate autophagy levels and promote longevity by enhancing clearance of protein aggregates and dysfunctional organelles.
Concluding Remarks and Future Perspectives
Autophagy helps to adjust to acute metabolic alterations both in the heart and across many tissues of the body. Decreased autophagy is observed in diabetes, obesity, and metabolic disorder [43,81,82]. In nearly all cases, there is evidence that restoring autophagic flux improves outcomes, longevity and quality of life. However, often it is unclear whether impaired autophagy is the cause or consequence of pathology. For instance, reduced levels of autophagy have been implicated as a cause of accumulating dysfunctional mitochondria in nerve, muscle, cardiac, and other tissues, in diabetes [35,38,83], resulting in elevated production of ROS and oxidative damage. Yet, reduced autophagy has also been shown to contribute to development of diabetes [8,81,84]. It is unclear whether impaired autophagy precedes onset of diabetes, or whether it is a consequence of metabolic changes (See Outstanding Questions Box).
Box 1. Outstanding Questions.
Exactly how do chronic metabolic alterations impair autophagy in myocytes?
Why is autophagy cardioprotective in some instances, and detrimental in other cases?
What is the mechanism for amino acid level sensing by RAG GTPases?
How does hyperglycemia overcome impaired insulin signaling to decrease autophagy in the diabetic heart?
Which of the >30 phosphorylation sites of ULK1 are important for its autophagy-regulating function? Which kinases are responsible for phosphorylating these sites, and what is their significance?
Are other statins more effective than simvastatin at activating autophagy and protecting the myocardium during ischemia?
How can mitochondrial autophagy be specifically upregulated by pharmacotherapy to treat or prevent diabetic cardiomyopathy?
As interest in autophagy as a therapy grows, the specificity of drugs that target autophagy will increase, and we may find that these modern drugs are not as effective as interventions that affect multiple pathways such as exercise. The contributions of other systems and pathways that are currently not being considered may become more apparent as drugs become more specific. The complexity of autophagy regulating pathways, with overlapping and distinct signaling pathways, suggests a careful balance in the status of autophagy activation. The sum of inputs from growth factors, insulin, amino acids, and energy status can tip the balance towards either autophagy induction or inhibition. Often, multiple pathways converge to convey the same message of nutrient sufficiency or insufficiency. However, sometimes a glaring lack of a metabolite, for instance amino acid deficiency, can overcome other signals to indicate a need for autophagy-mediated recycling. In this manner, the heart can adapt to ever-changing supply levels and steadfastly continue contracting. Future studies of the pathways involved in the regulation of autophagy are warranted if autophagy is to be explored as a new therapy for the treatment of heart disease.
Highlights.
Autophagy is critical for cardiac homeostasis.
Cardiac autophagy is decreased in obesity, metabolic syndrome and diabetes.
Reduced autophagy leads to myocyte damage, cell death, and contractile dysfunction.
Treatments enhancing cardiac autophagy may prevent diabetic cardiomyopathy.
Table 1.
Autophagy mouse models and phenotype.
| Gene Name |
Function | Mouse model | Defect | Phenotype | Ref |
|---|---|---|---|---|---|
| Atg5 | Important in autophagosome elongation and maturation |
Conditional cardiac- specific Atg5-deficient mice |
Impaired autophagy |
Deletion in adult heart causes cardiomyopathy |
[1] |
| Raga | A GTPase responsible for recruiting mTORC1 to lysosomes |
Knock-in mice expressing a constitutively active form of RagA |
Prevents inhibition of mTORC1 during nutrient deprivation |
Neonates are unable to induce autophagy in tissues at birth; results in neonatal death |
[26] |
| Rheb | Activates mTORC1 |
Cardiac-specific Rheb-deficient mice |
Failure to activate mTORC1 and induce autophagy once feeding is established. |
Defective cardiac hypertrophic growth; mice die 8-10 days after birth. |
[85] |
|
Irs1
and Irs2 |
Activate the PI3K/Akt pathway |
Cardiac-specific deletion of Irs1 and Irs2 |
Persistent activation of cardiac autophagy after birth |
Loss of myocytes, heart failure and early death |
[28] |
| Mtor | Component of mTORC1 |
Cardiac-specific Mtor-deficient mice |
Reduced myocyte proliferation; significant loss of myocytes |
>90% embryonic lethality; surviving mice have cardiac hypertrophy and severely impaired cardiac function |
[29] |
| Raptor | Component of mTORC1 |
Cardiac-specific Raptor-deficient mice |
Reduced protein synthesis; alterations in metabolism; increased apoptosis and autophagy |
Deletion in adult heart leads to heart failure; lack of adaptive hypertrophy after aortic banding |
[30] |
| Bcn1 | Required for the formation of autophagosomes |
Global Beclin1 heterozygous (+/−) knockout mice |
Reduced autophagy |
Reduced diabetes- induced cardiac damage; Reduced chronic myocardial I/R injury |
[3,9] |
| Bcn1 | Required for the formation of autophagosomes |
Cardiac-specific inducible transgenic |
Increased autophagy |
Enhanced diabetes- induced cardiac damage |
[9] |
| Atg5 | Important in autophagosome elongation and maturation |
Cardiac-specific Atg5-deficient mice |
Impaired autophagy |
Cardiac dysfunction by 10 months of age; death before 12 months of age |
[47] |
| Vps34 | Required for the formation of autophagosomes |
Conditional cardiac- specific Vps34- deficient mice |
Reduced autophagy |
Development of cardiac dysfunction and hypertrophy by 6 weeks of age |
[59] |
| FoxO1 | Transcription factor; regulates expression of genes involved in cell growth, metabolism, and autophagy |
Conditional cardiac- specific FoxO1- deficient mice |
Impaired activation of gene targets |
Reduced HFD- induced cardiac dysfunction |
[65] |
|
FoxO1
and FoxO3 |
Transcription factors; regulate expression of genes involved in cell growth, metabolism, and autophagy |
Conditional cardiac- specific FoxO1/FoxO3- deficient mice |
Impaired activation of gene targets |
Reduced HFD- induced cardiac dysfunction |
[65] |
| Prkaa2 | Energy sensor that activates autophagy during stress |
Cardiac-specific transgenic mice overexpressing a dominant negative AMPK |
Reduced autophagy |
Increased injury after acute myocardial ischemia |
[3] |
Acknowledgements
Å.B. Gustafsson is supported by National Institutes of Health grants P01HL085577, R01HL087023, R01HL101217, and R01HL092136.
Glossary
- Akt
a serine/threonine kinase that regulates numerous cellular survival processes including cell proliferation, apoptosis, and autophagy. An important survival kinase in the heart, Akt is activated via phosphorylation by PI3K and participates in activation of mTOR in response to insulin and growth factors.
- AMP-activated protein kinase (AMPK)
a serine/threonine kinase that is important for sensing changes in energy levels in the cell. AMPK responds to low cellular energy levels by upregulating autophagy through activation of ULK1, disinhibition of Beclin 1, or activation of FoxO transcription factors. Activation of AMPK occurs in response to a decline in ATP:AMP ratio and results in decreasing fatty acid and protein synthesis, and increased glucose transport.
- Autophagy
a catabolic cellular process that removes macromolecular structures by lysosomal degradation. Autophagy is important for the constitutive turnover of long-lived proteins and dysfunctional organelles under baseline conditions, and is rapidly upregulated for stress adaptation. Impaired autophagy leads to rapid heart failure.
- Beclin 1
an autophagy-promoting protein that forms a complex with VPS34 and AMBRA1 to initiate phagophore formation. Beclin 1 is a BCL-2 homology domain 3 (BH3)-containing protein that is maintained in an inactive state by binding to BCL-2.
- Forkhead box protein O (FoxO)
a family of transcription factors that regulate the transcription of genes associated with cell proliferation and homeostasis. FoxO1 and FoxO3 proteins are activated by AMPK and activate transcription of autophagy-promoting genes.
- Metformin
an anti-diabetic drug that activates AMPK to decrease hepatic glucose production, increase insulin sensitivity, and reduce circulating triglyceride and LDL cholesterol levels. Metformin activates autophagy in several cell types.
- mTOR/mTORC1
Mammalian target of rapamycin (mTOR) is part of the mTOR complex 1 (mTORC1), a master regulator of autophagy. The mTORC1 complex is activated by signaling of insulin/growth factors, amino acids, and nutrients, to inhibit autophagy. Conversely, deficiency of nutrients leads to induction of autophagy by mTORC1 deactivation.
- Phagophore
A double membrane that encloses and isolates the cytoplasmic components during macroautophagy.
- RAG GTPases
form a complex and respond to changes in intracellular amino acid levels. In the presence of amino acids, the active (RAG-GTP) complex promotes mTORC1 localization to the lysosome, where it can interact with RHEB to suppress autophagy. Amino acid deficiency deactivates the RAG complex and promotes mTORC1 deactivation and movement to the cytosol.
- Ras homolog enriched in brain (RHEB)
A GTP-binding protein that activates mTORC1 in response to both growth factor/insulin signaling and amino acid sensing. GTP-bound RHEB activates mTORC1, thereby inhibiting autophagy. RHEB is inactivated by TSC1/2, and inactive GDP-bound RHEB promotes autophagy via mTORC1 deactivation.
- Tuberous sclerosis (TSC)1/2
membrane proteins that form a complex that is inhibited by Akt. Inhibition of TSC1/2 by Akt promotes RHEB activation, activation of mTORC1 and autophagy suppression.
- ULK1
a kinase that regulates autophagy in response to signals from mTORC1 and AMPK. AMPK phosphorylates and activates ULK1 to promote autophagy, while phosphorylation by mTORC1 inhibits ULK1 activation by AMPK.
- Vacuolar protein sorting (VPS)34
Class III phosphatidylinositol 3-kinase that forms a complex with Beclin 1 and AMBRA1 to promote autophagosome formation. Vps34 converts phosphatidylinositol (PI) to phosphatidylinositol 3-phosphate (PI3P).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nakai A, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 2.Nishino I, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 3.Matsui Y, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circulation Research. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 4.Hamacher-Brady A, et al. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 2007;14:146–157. doi: 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- 5.Ruderman NB, et al. AMPK, insulin resistance, and the metabolic syndrome. J. Clin. Invest. 2013;123:2764–2772. doi: 10.1172/JCI67227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanamori H, et al. Autophagy limits acute myocardial infarction induced by permanent coronary artery occlusion. Am J Physiol Heart Circ Physiol. 2011;300:H2261–71. doi: 10.1152/ajpheart.01056.2010. [DOI] [PubMed] [Google Scholar]
- 7.Kubli DA, et al. Parkin Protein Deficiency Exacerbates Cardiac Injury and Reduces Survival following Myocardial Infarction. J Biol Chem. 2013;288:915–926. doi: 10.1074/jbc.M112.411363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X, et al. Akt2 knockout preserves cardiac function in high-fat diet-induced obesity by rescuing cardiac autophagosome maturation. J Mol Cell Biol. 2013;5:61–63. doi: 10.1093/jmcb/mjs055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu X, et al. Diminished Autophagy Limits Cardiac Injury in Mouse Models of Type 1 Diabetes. J Biol Chem. 2013;288:18077–18092. doi: 10.1074/jbc.M113.474650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giricz Z, et al. Autophagy, Myocardial Protection, and the Metabolic Syndrome. Journal of Cardiovascular Pharmacology. 2012;60:125–132. doi: 10.1097/FJC.0b013e318256ce10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang R, et al. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan L, et al. Autophagy in chronically ischemic myocardium. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13807–13812. doi: 10.1073/pnas.0506843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narendra D, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. The Journal of Cell Biology. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim D-H, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 16.Kim JE, Chen J. Regulation of peroxisome proliferator-activated receptor-gamma activity by mammalian target of rapamycin and amino acids in adipogenesis. Diabetes. 2004;53:2748–2756. doi: 10.2337/diabetes.53.11.2748. [DOI] [PubMed] [Google Scholar]
- 17.Hudson CC, et al. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Molecular and Cellular Biology. 2002;22:7004–7014. doi: 10.1128/MCB.22.20.7004-7014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cunningham JT, et al. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 19.Inoki K, et al. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, et al. PRAS40 Regulates mTORC1 Kinase Activity by Functioning as a Direct Inhibitor of Substrate Binding. J Biol Chem. 2007;282:20036–20044. doi: 10.1074/jbc.M702376200. [DOI] [PubMed] [Google Scholar]
- 21.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jewell JL, et al. Amino acid signalling upstream of mTOR. Nat. Rev. Mol. Cell Biol. 2013;14:133–139. doi: 10.1038/nrm3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith EM, et al. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- 24.Sancak Y, et al. Ragulator-Rag Complex Targets mTORC1 to the Lysosomal Surface and Is Necessary for Its Activation by Amino Acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long X, et al. Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J Biol Chem. 2005;280:23433–23436. doi: 10.1074/jbc.C500169200. [DOI] [PubMed] [Google Scholar]
- 26.Efeyan A, et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493:679–683. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 28.Riehle C, et al. Insulin receptor substrate signaling suppresses neonatal autophagy in the heart. J. Clin. Invest. 2013 doi: 10.1172/JCI71171. DOI: 10.1172/JCI71171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu Y, et al. Mechanistic Target of Rapamycin (Mtor) Is Essential for Murine Embryonic Heart Development and Growth. PLoS ONE. 2013;8:e54221. doi: 10.1371/journal.pone.0054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shende P, et al. Cardiac Raptor Ablation Impairs Adaptive Hypertrophy, Alters Metabolic Gene Expression, and Causes Heart Failure in Mice. Circulation. 2011;123:1073–1082. doi: 10.1161/CIRCULATIONAHA.110.977066. [DOI] [PubMed] [Google Scholar]
- 31.Kannel WB, et al. Role of diabetes in congestive heart failure: the Framingham study. Am. J. Cardiol. 1974;34:29–34. doi: 10.1016/0002-9149(74)90089-7. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, et al. Causes and characteristics of diabetic cardiomyopathy. Rev Diabet Stud. 2006;3:108–117. doi: 10.1900/RDS.2006.3.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abbott RD, et al. The impact of diabetes on survival following myocardial infarction in men vs women. The Framingham Study. JAMA. 1988;260:3456–3460. [PubMed] [Google Scholar]
- 34.Haffner SM, et al. Mortality from coronary heart disease in subjects with type 2 diabetes and in nondiabetic subjects with and without prior myocardial infarction. N. Engl. J. Med. 1998;339:229–234. doi: 10.1056/NEJM199807233390404. [DOI] [PubMed] [Google Scholar]
- 35.Xie Z, et al. Improvement of cardiac functions by chronic metformin treatment is associated with enhanced cardiac autophagy in diabetic OVE26 mice. Diabetes. 2011;60:1770–1778. doi: 10.2337/db10-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frustaci A, et al. Myocardial cell death in human diabetes. Circulation Research. 2000;87:1123–1132. doi: 10.1161/01.res.87.12.1123. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi S, et al. Suppression of autophagy is protective in high glucose-induced cardiomyocyte injury. Autophagy. 2012;8:577–592. doi: 10.4161/auto.18980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez CD, et al. The emerging role of autophagy in the pathophysiology of diabetes mellitus. Autophagy. 2011;7:2–11. doi: 10.4161/auto.7.1.13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mellor KM, et al. Myocardial autophagy activation and suppressed survival signaling is associated with insulin resistance in fructose-fed mice. Journal of Molecular and Cellular Cardiology. 2011;50:1035–1043. doi: 10.1016/j.yjmcc.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Resnick HE. Relation of weight gain and weight loss on subsequent diabetes risk in overweight adults. Journal of Epidemiology & Community Health. 2000;54:596–602. doi: 10.1136/jech.54.8.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dandona P, et al. Metabolic syndrome: a comprehensive perspective based on interactions between obesity, diabetes, and inflammation. Circulation. 2005;111:1448–1454. doi: 10.1161/01.CIR.0000158483.13093.9D. [DOI] [PubMed] [Google Scholar]
- 42.Li Z-L, et al. Transition from obesity to metabolic syndrome is associated with altered myocardial autophagy and apoptosis. Arterioscler. Thromb. Vasc. Biol. 2012;32:1132–1141. doi: 10.1161/ATVBAHA.111.244061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang L, et al. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sciarretta S, et al. Rheb is a critical regulator of autophagy during myocardial ischemia: pathophysiological implications in obesity and metabolic syndrome. Circulation. 2012;125:1134–1146. doi: 10.1161/CIRCULATIONAHA.111.078212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wohlgemuth SE, et al. Autophagy in the Heart and Liver During Normal Aging and Calorie Restriction. Rejuvenation Research. 2007;10:281–292. doi: 10.1089/rej.2006.0535. [DOI] [PubMed] [Google Scholar]
- 46.Wohlgemuth SE, et al. Skeletal muscle autophagy and apoptosis during aging: effects of calorie restriction and life-long exercise. Experimental Gerontology. 2010;45:138–148. doi: 10.1016/j.exger.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taneike M, et al. Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy. 2010;6:304–306. doi: 10.4161/auto.6.5.11947. [DOI] [PubMed] [Google Scholar]
- 48.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 49.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang HT, et al. Autophagy impairment induces premature senescence in primary human fibroblasts. PLoS ONE. 2011;6:e23367. doi: 10.1371/journal.pone.0023367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morselli E, et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 2010;1:e10. doi: 10.1038/cddis.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colman RJ, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willcox DC, et al. Caloric restriction and human longevity: what can we learn from the Okinawans? Biogerontology. 2006;7:173–177. doi: 10.1007/s10522-006-9008-z. [DOI] [PubMed] [Google Scholar]
- 55.Marzetti E, et al. Cellular mechanisms of cardioprotection by calorie restriction: state of the science and future perspectives. Clin. Geriatr. Med. 2009;25:715–32–ix. doi: 10.1016/j.cger.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hardie DG, et al. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He C, et al. Dissociation of Bcl-2-Beclin1 Complex by Activated AMPK Enhances Cardiac Autophagy and Protects Against Cardiomyocyte Apoptosis in Diabetes. Diabetes. 2013;62:1270–1281. doi: 10.2337/db12-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jaber N, et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proceedings of the National Academy of Sciences. 2012;109:2003–2008. doi: 10.1073/pnas.1112848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mammucari C, et al. FoxO3 Controls Autophagy in Skeletal Muscle In Vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 61.Sengupta A, et al. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009;284:28319–28331. doi: 10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hariharan N, et al. Deacetylation of FoxO by Sirt1 Plays an Essential Role in Mediating Starvation-Induced Autophagy in Cardiac Myocytes. Circulation Research. 2010;107:1470–1482. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cantó C, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 65.Battiprolu PK, et al. Metabolic stress-induced activation of FoxO1 triggers diabetic cardiomyopathy in mice. J. Clin. Invest. 2012;122:1109–1118. doi: 10.1172/JCI60329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Egan DF, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim J, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Inoki K, et al. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 69.Kim J, et al. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hosokawa N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Russell RC, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mack HID, et al. AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy. 2012;8:1197–1214. doi: 10.4161/auto.20586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sattelmair JR, et al. Effects of physical activity on cardiovascular and noncardiovascular outcomes in older adults. Clin. Geriatr. Med. 2009;25:677–702. viii–ix. doi: 10.1016/j.cger.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 75.Critchley J, et al. Explaining the increase in coronary heart disease mortality in Beijing between 1984 and 1999. Circulation. 2004;110:1236–1244. doi: 10.1161/01.CIR.0000140668.91896.AE. [DOI] [PubMed] [Google Scholar]
- 76.He C, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taylor F, et al. Statins for the primary prevention of cardiovascular disease. Cochrane Database Syst Rev. 2013;1:CD004816. doi: 10.1002/14651858.CD004816.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Andres AM, et al. Mitophagy is required for acute cardioprotection by simvastatin. Antioxid Redox Signal. 2013 doi: 10.1089/ars.2013.5416. DOI: 10.1089/ars.2013.5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou G, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Invest. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu H, et al. Cardiac autophagy is a maladaptive response to hemodynamic stress. J. Clin. Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jung HS, et al. Loss of Autophagy Diminishes Pancreatic β Cell Mass and Function with Resultant Hyperglycemia. Cell Metab. 2008;8:318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 82.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Joseph A-M, et al. Mitochondrial Dysregulation in the Pathogenesis of Diabetes: Potential for Mitochondrial Biogenesis-Mediated Interventions. Experimental Diabetes Research. 2012;2012:1–16. doi: 10.1155/2012/642038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Masini M, et al. Autophagy in human type 2 diabetes pancreatic beta cells. Diabetologia. 2009;52:1083–1086. doi: 10.1007/s00125-009-1347-2. [DOI] [PubMed] [Google Scholar]
- 85.Tamai T, et al. Rheb (Ras homologue enriched in brain)-dependent mammalian target of rapamycin complex 1 (mTORC1) activation becomes indispensable for cardiac hypertrophic growth after early postnatal period. J Biol Chem. 2013;288:10176–10187. doi: 10.1074/jbc.M112.423640. [DOI] [PMC free article] [PubMed] [Google Scholar]