Abstract
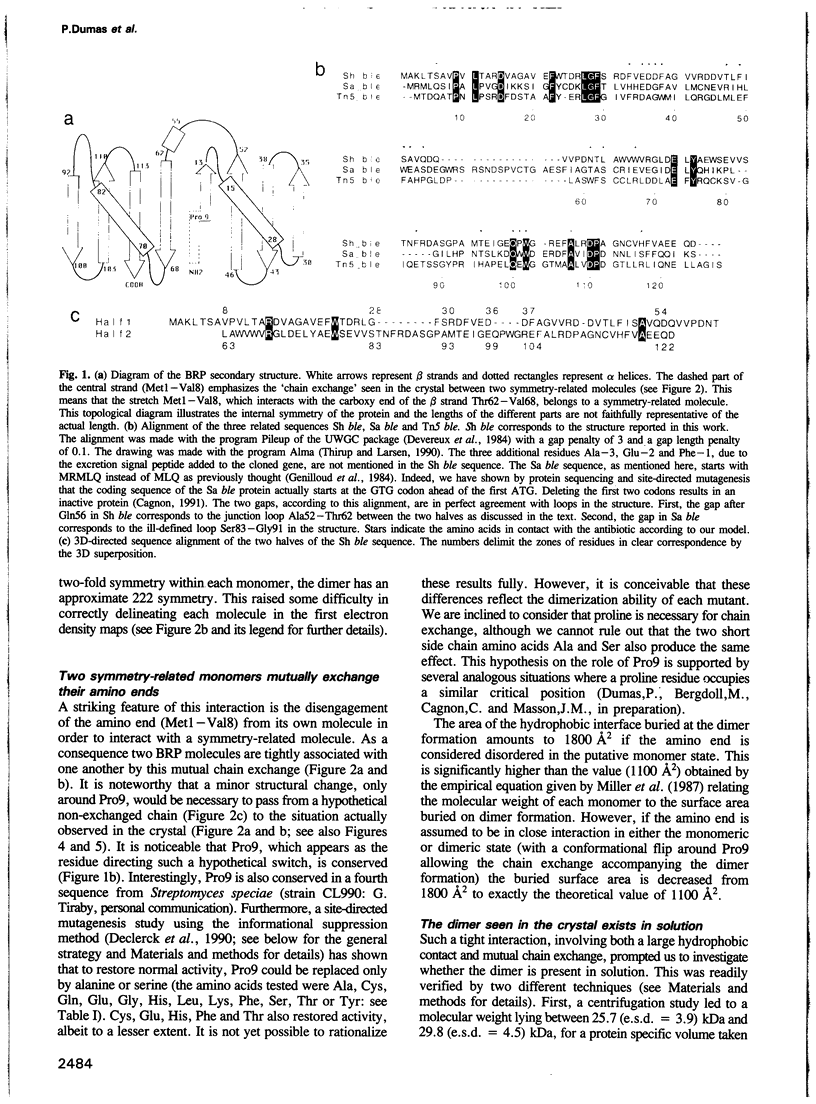
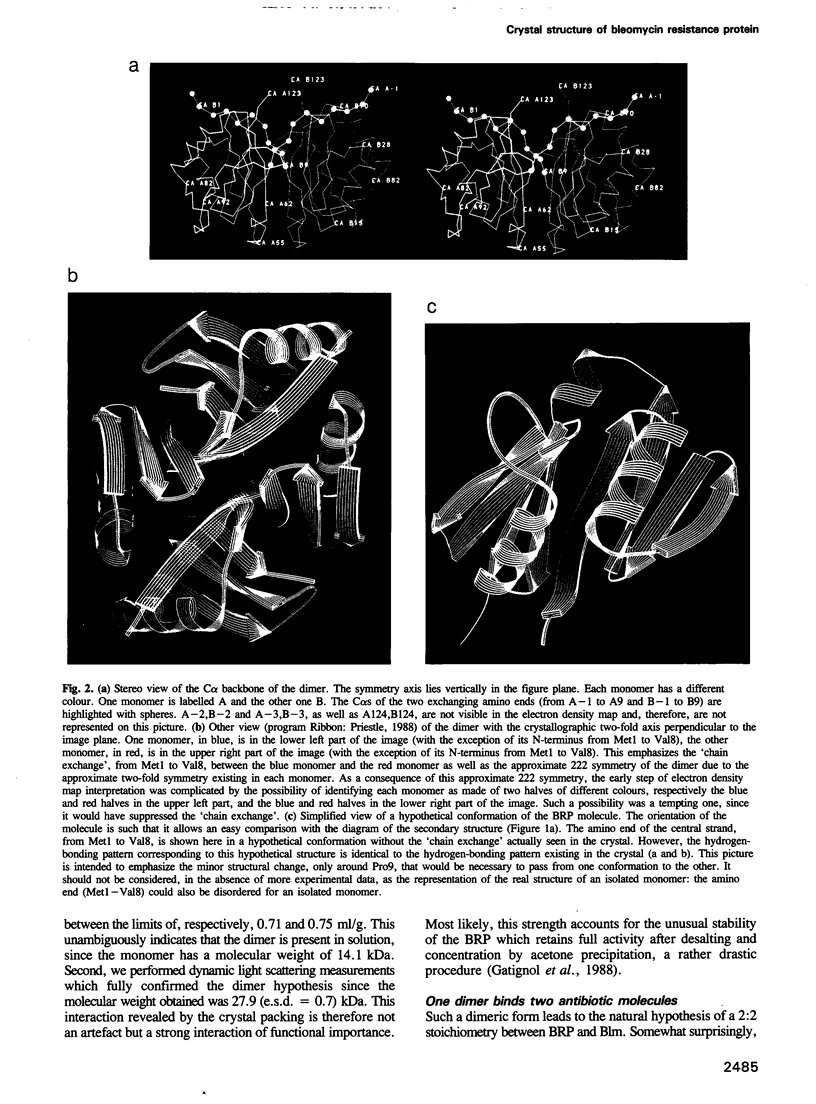
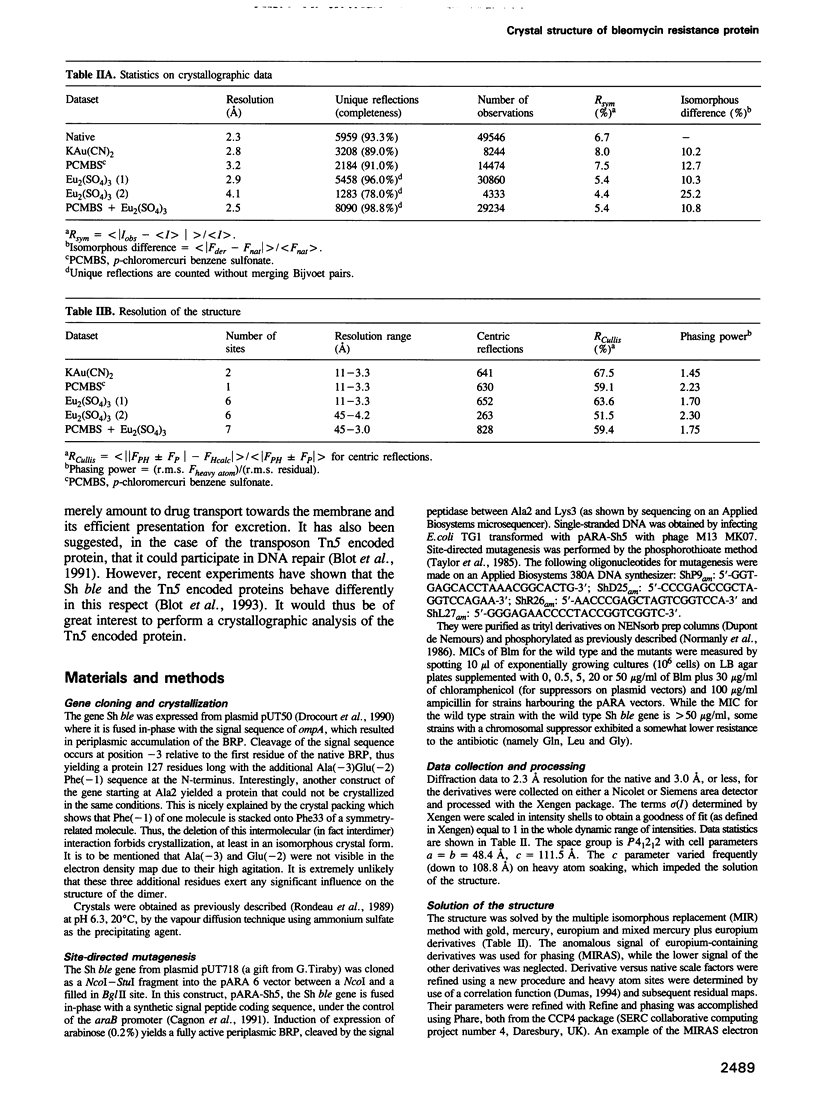
The antibiotic bleomycin, a strong DNA cutting agent, is naturally produced by actinomycetes which have developed a resistance mechanism against such a lethal compound. The crystal structure, at 2.3 A resolution, of a bleomycin resistance protein of 14 kDa reveals a structure in two halves with the same alpha/beta fold despite no sequence similarity. The crystal packing shows compact dimers with a hydrophobic interface and involved in mutual chain exchange. Two independent solution studies (analytical centrifugation and light scattering) showed that this dimeric form is not a packing artefact but is indeed the functional one. Furthermore, light scattering also showed that one dimer binds two antibiotic molecules as expected. A crevice located at the dimer interface, as well as the results of a site-directed mutagenesis study, led to a model wherein two bleomycin molecules are completely sequestered by one dimer. This provides a novel insight into antibiotic resistance due to drug sequestering, and probably also into drug transport and excretion.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birktoft J. J., Blow D. M. Structure of crystalline -chymotrypsin. V. The atomic structure of tosyl- -chymotrypsin at 2 A resolution. J Mol Biol. 1972 Jul 21;68(2):187–240. doi: 10.1016/0022-2836(72)90210-0. [DOI] [PubMed] [Google Scholar]
- Blot M., Heitman J., Arber W. Tn5-mediated bleomycin resistance in Escherichia coli requires the expression of host genes. Mol Microbiol. 1993 Jun;8(6):1017–1024. doi: 10.1111/j.1365-2958.1993.tb01646.x. [DOI] [PubMed] [Google Scholar]
- Blot M., Meyer J., Arber W. Bleomycin-resistance gene derived from the transposon Tn5 confers selective advantage to Escherichia coli K-12. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9112–9116. doi: 10.1073/pnas.88.20.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger R. M., Horwitz S. B., Peisach J., Wittenberg J. B. Oxygenated iron bleomycin. A short-lived intermediate in the reaction of ferrous bleomycin with O2. J Biol Chem. 1979 Dec 25;254(24):12999–12302. [PubMed] [Google Scholar]
- Cagnon C., Valverde V., Masson J. M. A new family of sugar-inducible expression vectors for Escherichia coli. Protein Eng. 1991 Oct;4(7):843–847. doi: 10.1093/protein/4.7.843. [DOI] [PubMed] [Google Scholar]
- Carter B. J., Murty V. S., Reddy K. S., Wang S. N., Hecht S. M. A role for the metal binding domain in determining the DNA sequence selectivity of Fe-bleomycin. J Biol Chem. 1990 Mar 15;265(8):4193–4196. [PubMed] [Google Scholar]
- Chien M., Grollman A. P., Horwitz S. B. Bleomycin-DNA interactions: fluorescence and proton magnetic resonance studies. Biochemistry. 1977 Aug 9;16(16):2641–2647. doi: 10.1021/bi00635a021. [DOI] [PubMed] [Google Scholar]
- Declerck N., Joyet P., Gaillardin C., Masson J. M. Use of amber suppressors to investigate the thermostability of Bacillus licheniformis alpha-amylase. Amino acid replacements at 6 histidine residues reveal a critical position at His-133. J Biol Chem. 1990 Sep 15;265(26):15481–15488. [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drocourt D., Calmels T., Reynes J. P., Baron M., Tiraby G. Cassettes of the Streptoalloteichus hindustanus ble gene for transformation of lower and higher eukaryotes to phleomycin resistance. Nucleic Acids Res. 1990 Jul 11;18(13):4009–4009. doi: 10.1093/nar/18.13.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatignol A., Durand H., Tiraby G. Bleomycin resistance conferred by a drug-binding protein. FEBS Lett. 1988 Mar 28;230(1-2):171–175. doi: 10.1016/0014-5793(88)80665-3. [DOI] [PubMed] [Google Scholar]
- Genilloud O., Garrido M. C., Moreno F. The transposon Tn5 carries a bleomycin-resistance determinant. Gene. 1984 Dec;32(1-2):225–233. doi: 10.1016/0378-1119(84)90050-7. [DOI] [PubMed] [Google Scholar]
- Hayes J. D., Wolf C. R. Molecular mechanisms of drug resistance. Biochem J. 1990 Dec 1;272(2):281–295. doi: 10.1042/bj2720281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroaki H., Uesugi S., Fujii S. The molecular recognition mechanism of DNA by bleomycin. Nucleic Acids Symp Ser. 1991;(25):147–148. [PubMed] [Google Scholar]
- Hénichart J. P., Bernier J. L., Helbecque N., Houssin R. Is the bithiazole moiety of bleomycin a classical intercalator? Nucleic Acids Res. 1985 Sep 25;13(18):6703–6717. doi: 10.1093/nar/13.18.6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iitaka Y., Nakamura H., Nakatani T., Muraoka Y., Fujii A., Takita T., Umezawa H. Chemistry of bleomycin. XX. The X-ray structure determination of P-3A Cu(II)-complex a biosynthetic intermediate of bleomycin. J Antibiot (Tokyo) 1978 Oct;31(10):1070–1072. doi: 10.7164/antibiotics.31.1070. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kleina L. G., Masson J. M., Normanly J., Abelson J., Miller J. H. Construction of Escherichia coli amber suppressor tRNA genes. II. Synthesis of additional tRNA genes and improvement of suppressor efficiency. J Mol Biol. 1990 Jun 20;213(4):705–717. doi: 10.1016/S0022-2836(05)80257-8. [DOI] [PubMed] [Google Scholar]
- Kuwahara J., Sugiura Y. Sequence-specific recognition and cleavage of DNA by metallobleomycin: minor groove binding and possible interaction mode. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2459–2463. doi: 10.1073/pnas.85.8.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenfant F., Labia R., Masson J. M. Probing the active site of beta-lactamase R-TEM1 by informational suppression. Biochimie. 1990 Jun-Jul;72(6-7):495–503. doi: 10.1016/0300-9084(90)90073-p. [DOI] [PubMed] [Google Scholar]
- Levy M. J., Hecht S. M. Copper(II) facilitates bleomycin-mediated unwinding of plasmid DNA. Biochemistry. 1988 Apr 19;27(8):2647–2650. doi: 10.1021/bi00408a002. [DOI] [PubMed] [Google Scholar]
- Miller S., Lesk A. M., Janin J., Chothia C. The accessible surface area and stability of oligomeric proteins. 1987 Aug 27-Sep 2Nature. 328(6133):834–836. doi: 10.1038/328834a0. [DOI] [PubMed] [Google Scholar]
- Nikolov D. B., Hu S. H., Lin J., Gasch A., Hoffmann A., Horikoshi M., Chua N. H., Roeder R. G., Burley S. K. Crystal structure of TFIID TATA-box binding protein. Nature. 1992 Nov 5;360(6399):40–46. doi: 10.1038/360040a0. [DOI] [PubMed] [Google Scholar]
- Normanly J., Kleina L. G., Masson J. M., Abelson J., Miller J. H. Construction of Escherichia coli amber suppressor tRNA genes. III. Determination of tRNA specificity. J Mol Biol. 1990 Jun 20;213(4):719–726. doi: 10.1016/S0022-2836(05)80258-X. [DOI] [PubMed] [Google Scholar]
- Normanly J., Masson J. M., Kleina L. G., Abelson J., Miller J. H. Construction of two Escherichia coli amber suppressor genes: tRNAPheCUA and tRNACysCUA. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6548–6552. doi: 10.1073/pnas.83.17.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Oppenheimer N. J., Rodriguez L. O., Hecht S. M. Structural studies of of "active complex" of bleomycin: assignment of ligands to the ferrous ion in a ferrous-bleomycin-carbon monoxide complex. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5616–5620. doi: 10.1073/pnas.76.11.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploegman J. H., Drent G., Kalk K. H., Hol W. G. Structure of bovine liver rhodanese. I. Structure determination at 2.5 A resolution and a comparison of the conformation and sequence of its two domains. J Mol Biol. 1978 Aug 25;123(4):557–594. [PubMed] [Google Scholar]
- Povirk L. F., Hogan M., Dattagupta N. Binding of bleomycin to DNA: intercalation of the bithiazole rings. Biochemistry. 1979 Jan 9;18(1):96–101. doi: 10.1021/bi00568a015. [DOI] [PubMed] [Google Scholar]
- Rondeau J. M., Cagnon C., Moras D., Masson J. M. Crystallization and preliminary X-ray data of a phleomycin-binding protein from Streptoalloteichus hindustanus. J Mol Biol. 1989 Jun 5;207(3):645–646. doi: 10.1016/0022-2836(89)90476-2. [DOI] [PubMed] [Google Scholar]
- Sausville E. A., Peisach J., Horwitz S. B. A role for ferrous ion and oxygen in the degradation of DNA by bleomycin. Biochem Biophys Res Commun. 1976 Dec 6;73(3):814–822. doi: 10.1016/0006-291x(76)90882-2. [DOI] [PubMed] [Google Scholar]
- Semon D., Movva N. R., Smith T. F., el Alama M., Davies J. Plasmid-determined bleomycin resistance in Staphylococcus aureus. Plasmid. 1987 Jan;17(1):46–53. doi: 10.1016/0147-619x(87)90007-2. [DOI] [PubMed] [Google Scholar]
- Story R. M., Weber I. T., Steitz T. A. The structure of the E. coli recA protein monomer and polymer. Nature. 1992 Jan 23;355(6358):318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Nagai K., Yamaki H., Tanaka N., Umezawa H. On the mechanism of action of bleomycin: scission of DNA strands in vitro and in vivo. J Antibiot (Tokyo) 1969 Sep;22(9):446–448. doi: 10.7164/antibiotics.22.446. [DOI] [PubMed] [Google Scholar]
- Takita T., Muraoka Y., Nakatani T., Fujii A., Umezawa Y., Naganawa H. Chemistry of bleomycin. XIX Revised structures of bleomycin and phleomycin. J Antibiot (Tokyo) 1978 Aug;31(8):801–804. doi: 10.7164/antibiotics.31.801. [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirup S., Larsen N. E. ALMA, an editor for large sequence alignments. Proteins. 1990;7(3):291–295. doi: 10.1002/prot.340070310. [DOI] [PubMed] [Google Scholar]
- Umezawa H. Chemistry and mechanism of action of bleomycin. Fed Proc. 1974 Nov;33(11):2296–2302. [PubMed] [Google Scholar]
- Voorintholt R., Kosters M. T., Vegter G., Vriend G., Hol W. G. A very fast program for visualizing protein surfaces, channels and cavities. J Mol Graph. 1989 Dec;7(4):243–245. doi: 10.1016/0263-7855(89)80010-4. [DOI] [PubMed] [Google Scholar]
- Wang B. C. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- Wilmot C. M., Thornton J. M. Beta-turns and their distortions: a proposed new nomenclature. Protein Eng. 1990 May;3(6):479–493. doi: 10.1093/protein/3.6.479. [DOI] [PubMed] [Google Scholar]







