Abstract
Eosinophilic esophagitis (EoE) is a food allergy-associated inflammatory disease characterized by esophageal eosinophilia. EoE has become increasingly common, but current management strategies are nonspecific. Thus, there is an urgent need to identify specific immunological pathways that could be targeted to treat this disease. EoE is associated with polymorphisms in the gene that encodes thymic stromal lymphopoietin (TSLP), a cytokine that promotes allergic inflammation, but how TSLP might contribute to EoE disease pathogenesis remains unknown. Here, we describe a new mouse model of EoE-like disease that developed independently of IgE but was dependent on TSLP-elicited basophils. Therapeutic TSLP neutralization or basophil depletion also ameliorated established EoE-like disease. Critically, in human subjects with EoE, we observed elevated TSLP levels and exaggerated basophil responses in esophageal biopsies, and a gain-of-function TSLP polymorphism was associated with increased basophil responses. Together, these data suggest that the TSLP-basophil axis could be therapeutically targeted to treat EoE.
INTRODUCTION
EoE is a food allergy-associated inflammatory disease that affects children and adults1–3. In industrialized countries, the incidence of EoE has increased dramatically in the past 30 years, resulting in a significant public health and economic burden2,4,5. Hallmark features of EoE include esophageal eosinophilia and inflammation and histological changes in the esophagus associated with stricture, dysphagia, and food impaction1–3. Currently, treatment strategies for EoE are nonspecific and impose a significant burden on patients. Although swallowed topical steroids can be effective in limiting EoE-associated inflammation, there are concerns regarding the long-term use of steroids, particularly in children2,6. Adherence to an elemental diet that eliminates exposure to foods that trigger EoE results in resolution of symptoms in many patients; however, this approach requires disruptive changes in lifestyle and eating habits2,6,7. Thus, there is an urgent need to identify new drug targets and more specific therapies7. The observations that immune suppression or removal of dietary trigger foods can ameliorate EoE symptoms indicate that EoE is a food antigen-driven disease mediated by aberrant immune responses1,2,8. Therefore, targeting the dysregulated immunological pathways that underlie EoE could offer new treatment strategies for this disease.
Studies investigating the immunological mechanisms that mediate EoE have shown that various immune cell types, including eosinophils, mast cells, TH2 cells that produce interleukin (IL)-4, IL-5, and IL-13, and IgE-producing B cells, may contribute to esophageal inflammation during EoE1–3,9. Further, recent work has shown that there is a strong association between a gain-of-function polymorphism in the gene that encodes the predominantly epithelial cell-derived cytokine TSLP and the development of EoE in children10,11. TSLP is associated with multiple allergic disorders10–16 and is thought to promote allergic inflammation by activating dendritic cells, inducing TH2 cell responses, supporting IgE production, and eliciting the population expansion of phenotypically and functionally distinct basophils12,17–21. However, whether TSLP directly promotes inflammatory responses associated with EoE and the mechanisms by which polymorphisms in TSLP and increased TSLP expression may contribute to the pathogenesis of EoE in patients remain unknown.
RESULTS
A new mouse model of experimental EoE-like disease
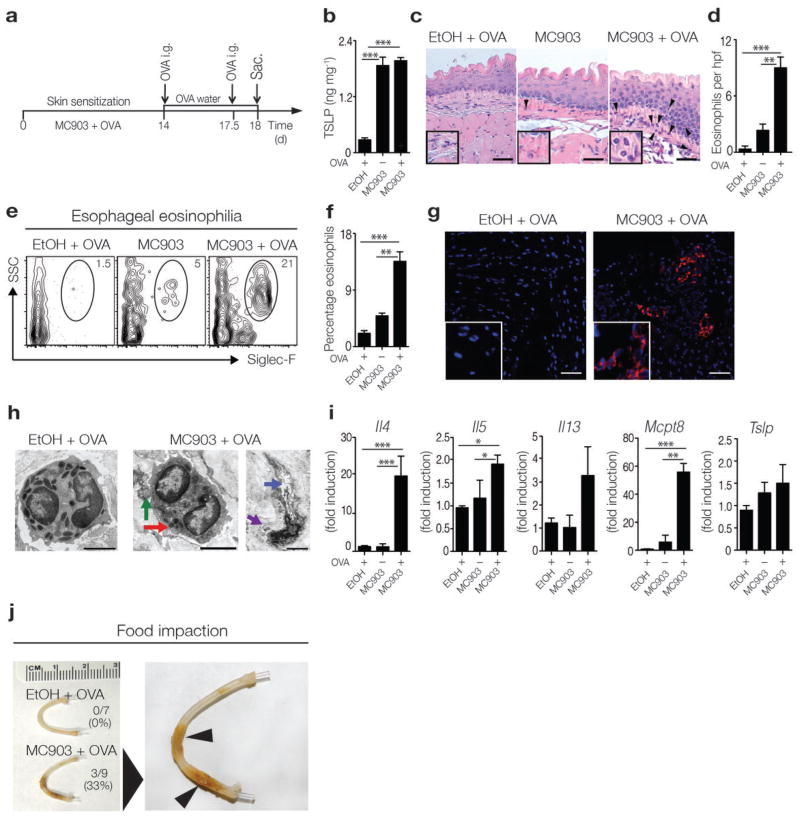
In order to investigate whether TSLP directly promotes EoE disease pathogenesis, we developed a novel mouse model of EoE-like disease that is associated with exaggerated TSLP production. Multiple studies in mouse models and humans suggest that sensitization to food allergens may occur at sites where the skin barrier is disrupted, such as atopic dermatitis lesions22–24. Thus, we employed a model of epicutaneous sensitization to a food antigen, ovalbumin (OVA), on an atopic dermatitis-like lesion that is associated with elevated TSLP production in the skin17,25–27 (Fig. 1a). Consistent with previous reports, wild-type (WT) BALB/c mice treated epicutaneously with the vitamin D analog MC903 exhibited increased TSLP expression in the skin (Fig. 1b). Epicutaneous sensitization to and subsequent oral challenge with OVA resulted in the development of experimental EoE-like disease that was characterized by inflammation, edema, and eosinophilia in the esophagus as measured histologically and quantified by enumeration of eosinophils per high-powered field (hpf) (Fig. 1c, d). Flow cytometric analysis (Fig. 1e, f) and immunofluorescent staining (Fig. 1g) also demonstrated that there was an accumulation of eosinophils in esophageal tissues of mice with EoE-like disease, and electron microscopic (EM) analysis revealed the presence of degranulated eosinophils in these tissues (Fig. 1h). EoE-like disease was also characterized by significantly increased expression of genes that encode TH2 cytokines and the basophil-specific protease Mcpt8 in esophageal tissues. Although not statistically significant, there was also an increase in Tslp expression (Fig. 1i). Further, a similar pattern of EoE-like disease was observed in mice that were epicutaneously sensitized to crude peanut extract (CPE) on an atopic dermatitis-like skin lesion (Supplementary Fig. 1a–c), confirming that sensitization to a natural food allergen in the presence of elevated levels of TSLP results in experimental EoE-like disease. Eosinophil accumulation in this model was not restricted to the esophagus, as mice with EoE-like disease also exhibited eosinophilia in the gastrointestinal tract (Supplementary Fig. 1d, e), associated with antigen-specific TH2 cytokine responses in the mesenteric lymph node and spleen (Supplementary Fig. 1f, g).
Figure 1.
Experimental mouse model of EoE-like disease. (a) Schematic of sensitization and intragastric (i.g.) challenge in WT BALB/c mice. (b) TSLP expression in supernatants of overnight-cultured skin (ears) measured by ELISA. Data depicted are from one experiment (EtOH + OVA, n = 3; MC903, n = 3; MC903 + OVA, n = 4), and are representative of three independent experiments. (c) Histological sections (H & E staining) from the esophagus. Arrows identify tissue-infiltrating eosinophils. Scale bar: 25 μm. (d) Number of eosinophils per hpf in the esophagus. (e) Representative flow cytometry plots showing frequencies of eosinophils in esophageal tissues. Data depicted in (c–e) are from one experiment (EtOH + OVA, n = 3; MC903, n = 3; MC903 + OVA, n = 4), and are representative of three or more independent experiments. (f) Frequencies of eosinophils in esophageal tissues as measured by flow cytometry. Data depicted are from three pooled experiments (EtOH + OVA, n = 7; MC903, n = 8; MC903 + OVA, n = 11). (g) Immunofluorescent staining for eosinophils (Siglec-F-specific mAb, red) in esophageal tissues. Counterstaining with DAPI (blue). Scale bar: 25 μm. Data are representative of two controls and three EoE-like disease samples. (h) Representative electron microscopy (EM) image of an eosinophil in the esophagus of control mice with intact granules with electron dense cores (left panel) or degranulating eosinophils in MC903 + OVA treated mice (right panel), showing loss of electron density in granule cores (red arrow), granule extrusion channels (blue), and loss of granule contents (purple). Scale bar: 2 μm. (i) mRNA expression levels of TH2 cytokines (Il4, Il5, Il13), the basophil-specific protease Mcpt8, and Tslp in the esophagus. Data depicted are from one experiment (EtOH + OVA, n = 3; MC903, n = 3; MC903 + OVA, n = 4), and are representative of three independent experiments. (j) Representative images of esophagi, with incidence of impaction. Arrows identify impacted food. Data depicted are from two pooled experiments (EtOH + OVA, n = 7, MC903 + OVA, n = 9). All parameters were assessed 12 h post-final oral antigen challenge. Data depicted in (a–i) are from mice challenged twice with OVA, and data depicted in (j) are from mice challenged repeatedly with OVA. Results are shown as mean ± sem, and a non-parametric, one-way Kruskal-Wallis ANOVA with Dunn’s post-hoc testing was used to determine significance. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
In addition to immunological parameters, human EoE is defined by physiological changes in esophageal tissue and signs of esophageal dysfunction, including food impaction, which occurs in approximately 40% of EoE patients1–3,28. To assess whether clinical manifestations of EoE were present in the model of experimental EoE-like disease, mice that had existing EoE-like disease were challenged repeatedly with OVA to induce prolonged esophageal inflammation. Analysis using optical coherence tomography (OCT), which allows for high-resolution imaging of live biological tissues based on optical scattering29,30, revealed that although not statistically significant, prolonged inflammation in EoE-like disease was characterized by increased thickness of the esophageal epithelium (Supplementary Fig. 2a, b). Critically, prolonged esophageal inflammation was also associated with food impaction in the esophagus of approximately 30% of fasted mice with EoE-like disease at the time of euthanasia, but food impaction was never observed in the esophagus of control mice (Fig. 1j). Collectively, these data indicate that this new model of EoE-like disease is characterized by a number of immunological and pathophysiological changes in esophageal tissues and signs of esophageal dysfunction similar to those observed in human EoE patients1–3,31–34.
EoE-like disease is dependent on TSLP but independent of IgE
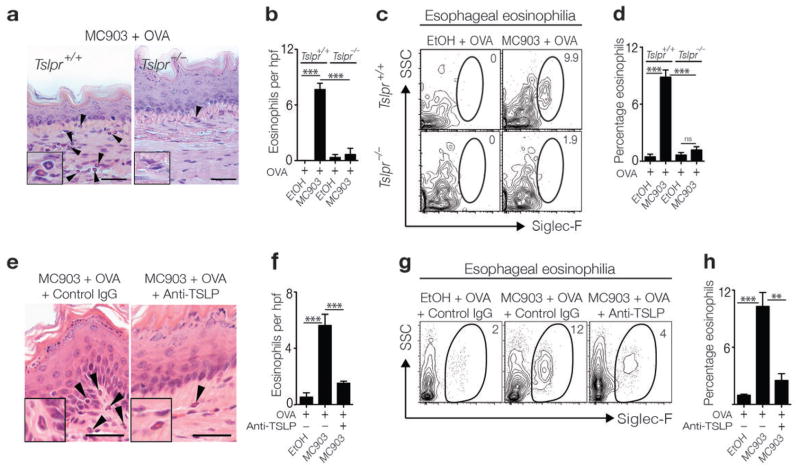
To interrogate whether TSLP directly promotes the pathogenesis of experimental EoE-like disease in mice, WT BALB/c (Tslpr+/+) mice or mice deficient in the TSLP receptor (TSLPR) (Tslpr−/−) were epicutaneously sensitized to OVA followed by oral antigen challenge. While sensitized and challenged Tslpr+/+ mice exhibited esophageal eosinophilia and associated inflammation, Tslpr−/− mice did not develop esophageal eosinophilia (Fig. 2a–d). Using an alternative approach to abrogate TSLP signaling, monoclonal antibody (mAb)-mediated neutralization of TSLP during epicutaneous sensitization with OVA in WT BALB/c mice also limited eosinophil infiltration in the esophagus following oral challenge (Fig. 2e–h).
Figure 2.
TSLP-TSLPR interactions are critical for the pathogenesis of EoE-like disease. (a) Histological sections (H & E staining) from the esophagus of BALB/c Tslpr+/+ or BALB/c Tslp−/− mice. Arrows identify tissue-infiltrating eosinophils. Scale bar: 25 μm. (b) Number of eosinophils per hpf in the esophagus. (c) Representative flow cytometry plots showing frequencies of eosinophils in esophageal tissues. Data depicted in (a–c) are from one experiment (Tslpr+/+ EtOH + OVA, n = 3; Tslpr+/+ MC903 + OVA, n = 5; Tslpr−/− EtOH + OVA, n = 3; Tslpr−/− MC903 + OVA, n = 5), and are representative of three independent experiments. (d) Frequencies of eosinophils in esophageal tissues as measured by flow cytometry. Data depicted are from three pooled experiments (Tslpr+/+ EtOH + OVA, n = 5; Tslpr+/+ MC903 + OVA, n = 11; Tslpr−/− EtOH + OVA, n = 5; Tslpr−/− MC903 + OVA, n = 12). (e) Histological sections (H & E staining) from the esophagus of WT BALB/c mice treated with an isotype control or TSLP-specific mAb. Arrows identify tissue-infiltrating eosinophils. Scale bar: 50 μm. (f) Number of eosinophils per hpf in the esophagus. (g) Representative flow cytometry plots showing frequencies of eosinophils in esophageal tissues. Data depicted in (e–g) are from one experiment (EtOH + OVA + IgG, n = 3; MC903 + OVA + IgG, n = 3; MC903 + OVA + anti-TSLP mAb, n = 3), and are representative of three independent experiments. (h) Frequencies of eosinophils in esophageal tissues as measured by flow cytometry. Data depicted are from three pooled experiments (EtOH + OVA +IgG, n = 5; MC903 + OVA + IgG, n = 9; MC903 + OVA + anti-TSLP mAb, n = 10). All parameters were assessed 12 h post-final oral antigen challenge. Data depicted are from mice challenged twice with OVA. Results are shown as mean ± sem, and a non–parametric, one-way Kruskal-Wallis ANOVA with Dunn’s post-hoc testing or a non-parametric, two-way ANOVA with Bonferroni post-hoc testing were used to determine significance. ns: not significant. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
To test whether TSLP was sufficient for the development of EoE-like disease during epicutaneous sensitization, mice were injected intradermally with exogenous recombinant TSLP (rTSLP) in the presence or absence of OVA and orally challenged (Supplementary Fig. 3a). Mice sensitized in the presence of rTSLP also exhibited esophageal eosinophilia following oral challenge (Supplementary Fig. 3b). In a complementary approach, Tslpr+/+ mice treated with control antibody or a TSLP-specific mAb and Tslpr−/− mice were sensitized with OVA on tape-stripped skin (Supplementary Fig. 3c). Tape-stripping was associated with elevated local TSLP production following physical perturbation of the skin barrier (Supplementary Fig. 3d and 35). While Tslpr+/+ mice treated with control antibody that were sensitized to OVA on tape-stripped skin exhibited esophageal eosinophilia following oral antigen challenge, Tslpr+/+ mice treated with a TSLP-specific mAb and Tslpr−/− mice did not develop esophageal eosinophilia (Supplementary Fig. 3e, f). Finally, the contribution of TSLP to the development of clinical signs of EoE-like disease was assessed. Although not statistically significant, repeated challenge with OVA following sensitization in the presence of MC903 was associated with increased thickness of the esophageal epithelium. Prolonged esophageal inflammation was also associated with an increased incidence of food impaction in the esophagus in Tslpr+/+ but not Tslpr−/− mice (Supplementary Fig. 4a, b). Collectively, these data indicate that TSLP-TSLPR interactions are necessary and sufficient for the development of experimental EoE-like disease in mice.
TSLP-TSLPR interactions are known to promote the production of IgE36,37, a critical mediator of allergic inflammation38, and class-switched B cells have been observed in the esophagus of EoE patients9,39,40. In addition, MC903-induced TSLP expression was associated with elevated systemic OVA-specific IgE levels (Fig. 3a), provoking the hypothesis that TSLP-dependent EoE-like disease in mice might be IgE-dependent. To directly test this, IgE-sufficient WT BALB/c (Igh-7+/+) mice and IgE-deficient (Igh-7−/−) mice were epicutaneously sensitized to OVA in the presence of MC903. Following oral challenge with antigen, both Igh-7+/+ and Igh-7−/− mice exhibited equivalent EoE-like disease, characterized by esophageal inflammation, elevated tissue eosinophilia (Fig. 3b–d), the presence of degranulated eosinophils in the esophagus (Fig. 3e), and significant increases in expression of genes that encode TH2 cytokines in esophageal tissues (Fig. 3f). These data demonstrate that EoE-like disease can occur in an IgE-independent manner and are consistent with recent findings from clinical studies suggesting that treatment with IgE-specific mAb does not ameliorate EoE in most patients41–44. Together, these data indicate that manipulation of the IgE pathway may not be an effective therapeutic approach for the treatment of EoE.
Figure 3.
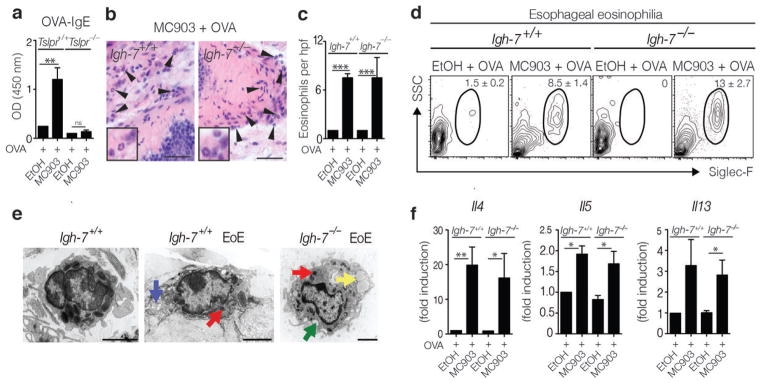
EoE-like disease development is independent of IgE. (a) OVA-specific serum IgE levels from BALB/c Tslpr+/+ and Tslpr−/− mice. Data depicted are from one experiment (EtOH + OVA Tslpr+/+, n = 3; MC903 + OVA Tslpr+/+, n = 4; EtOH + OVA Tslpr−/−, n = 3; MC903 + OVA Tslpr−/−, n = 4), and are representative of three or more independent experiments. (b) Histological sections (H & E staining) from the esophagus of BALB/c Igh-7+/+ and BALB/c Igh-7−/− mice. Arrows identify tissue-infiltrating eosinophils. Scale bar: 25 μm. (c) Number of eosinophils per hpf. (d) Representative flow cytometry plots showing frequencies of eosinophils in esophageal tissues. Data depicted in (b–d) are from one experiment (EtOH + OVA Igh-7+/+ n = 3; MC903 + OVA Igh-7+/+ n = 3; EtOH + OVA Igh-7−/− n = 3; MC903 + OVA Igh-7−/− n = 4), and are representative of three or more independent experiments. (f) Representative EM image of an eosinophil in the esophagus of control Igh-7+/+ mice with intact granules with electron dense cores (left panel) or degranulating eosinophils in MC903 + OVA treated Igh-7+/+ (middle panel) or Igh-7−/− (right panel) mice in various stages of degranulation, with loss of electron density in granule cores (red arrow), formation of granule extrusion channels (blue), complete loss of granule contents (green), and formation of lipid vesicles (yellow). Scale bar: 2 μm. (g) mRNA expression levels of TH2 cytokines in the esophagus. Data depicted are from one experiment (EtOH + OVA Igh-7+/+ n = 3; MC903 + OVA Igh-7+/+ n = 3; EtOH + OVA Igh-7−/− n = 3; MC903 + OVA Igh-7−/− n = 3), and are representative of two independent experiments. All parameters were assessed 12 h post-final oral antigen challenge. Data depicted are from mice challenged twice with OVA. Results are shown as mean ± sem, and a non-parametric, two-way ANOVA with Bonferroni post-hoc testing was used to determine significance. **, P ≤ 0.01; ***, P ≤ 0.001.
EoE-like disease is dependent on basophils
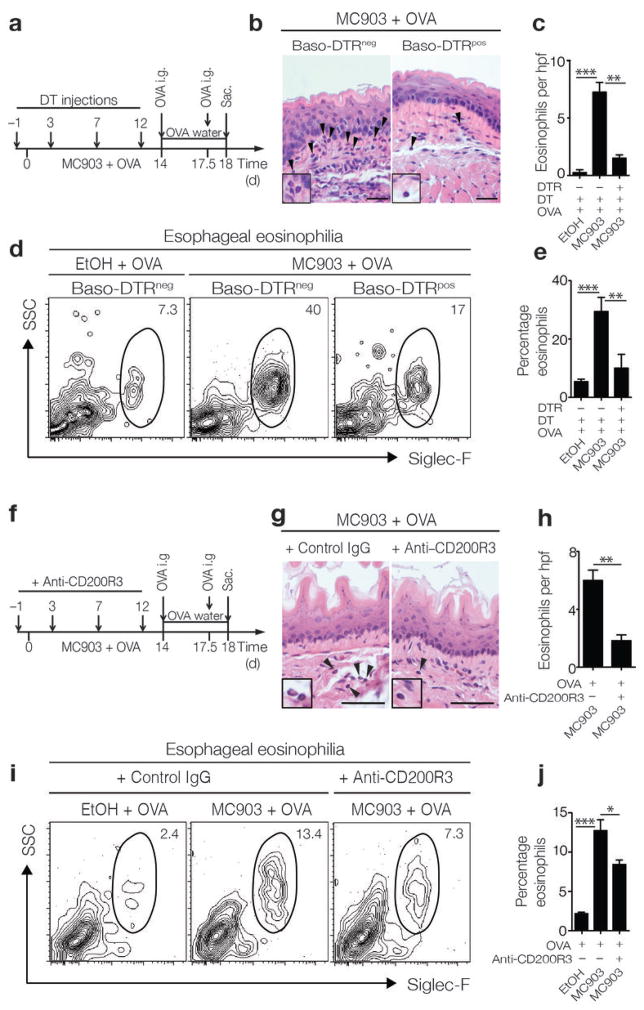
In addition to its role in promoting B cell and IgE responses, TSLP expression is associated with the selective expansion of a distinct population of basophils17,18. These data provoked the hypothesis that basophils may contribute to TSLP-dependent, IgE-independent EoE-like disease in mice. Consistent with this hypothesis, MC903-induced expression of TSLP in the skin was associated with TSLP-dependent, IgE-dependent systemic basophil responses (Supplementary Fig. 5a, b). To assess whether basophils contribute to the development of experimental EoE-like disease, we employed an established genetic approach to deplete basophils in vivo. C57BL/6 mice in which the diphtheria toxin receptor (DTR) is exclusively expressed by basophils (Baso-DTRpos)17,19,45 and DTR-negative littermate controls (Baso-DTRneg) were epicutaneously sensitized and orally challenged with OVA while being treated with diphtheria toxin (DT) (Fig. 4a). Consistent with results observed in BALB/c mice (Fig. 1b), increased TSLP production was observed in C57BL/6 Baso-DTRneg and Baso-DTRpos mice sensitized to OVA in the context of MC903 treatment (data not shown). Strikingly, while Baso-DTRneg mice that were epicutaneously sensitized and orally challenged with OVA exhibited increased frequencies of eosinophils in the esophagus, depletion of basophils in Baso-DTRpos mice (Supplementary Fig. 5c) led to a reduction in esophageal eosinophilia (Fig. 4b–e) and a reduction in expression of genes related to TH2 cytokine responses (Supplementary Fig. 6a–c).
Figure 4.
Basophils mediate the pathogenesis of EoE-like disease. (a) Schematic of in vivo basophil depletion strategy. C57BL/6 (Baso-DTRneg) or Baso-DTRpos mice in which the DTR is exclusively expressed on basophils were treated with DT during the course of epicutaneous sensitization. (b) Histological sections (H & E staining) from the esophagus. Arrows identify tissue-infiltrating eosinophils. Scale bar: 25 μm. (c) Number of eosinophils per hpf in the esophagus. (d) Representative flow cytometry plots showing frequencies of eosinophils in esophageal tissues. Data depicted in (b–d) are from one experiment (Baso-DTRneg EtOH + OVA, n = 3; Baso-DTRneg MC903 + OVA, n = 3; Baso-DTRpos MC903 + OVA, n = 4), and are representative of three independent experiments. (e) Frequencies of eosinophils in esophageal tissues as measured by flow cytometry. Data depicted are from three pooled experiments (Baso-DTRneg EtOH + OVA, n = 7; Baso-DTRneg MC903 + OVA, n = 10; Baso-DTRpos MC903 + OVA, n = 11). (f) Schematic of in vivo basophil depletion strategy using CD200R3-specific mAb in WT BALB/c mice. (g) Histological sections (H & E staining) from the esophagus. Arrows identify tissue-infiltrating eosinophils. Scale bar: 25 μm. (h) Number of eosinophils per hpf in the esophagus. (i) Representative flow cytometry plots showing frequencies of eosinophils in esophageal tissues. Data depicted in (g–i) are from one experiment (EtOH + OVA + IgG, n = 3; MC903 + OVA + IgG, n = 3; EtOH + OVA + anti-CD200R3 mAb, n = 3; MC903 + OVA + anti-CD200R3 mAb, n = 4), and are representative of three independent experiments. (j) Frequencies of eosinophils in esophageal tissues as measured by flow cytometry. Data depicted are from three pooled experiments (EtOH + OVA + IgG, n = 8; MC903 + OVA + IgG, n = 9; EtOH + OVA + anti-CD200R3 mAb, n = 8; MC903 + OVA + anti-CD200R3 mAb, n = 10). All parameters were assessed 12 h post-final oral antigen challenge. Data depicted are from mice challenged twice with OVA. Results are shown as mean ± sem, and a non-parametric, two-tailed Mann-Whitney t-test or a non-parametric, one-way Kruskal-Wallis ANOVA with Dunn’s post-hoc testing were used to determine significance. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Using an alternative approach, epicutaneously sensitized and orally challenged WT BALB/c mice were treated with a monoclonal antibody specific for CD200R3 (Ba103) to deplete basophils46 (Fig. 4f). Basophil depletion during sensitization (Supplementary Fig. 5d) reduced the accumulation of eosinophils in the esophagus following oral challenge with OVA (Fig. 4g–j). Collectively, these results indicate that basophils are significant contributors to the pathogenesis of experimental EoE-like disease in mice and may represent a novel therapeutic target to treat this disease in patients.
TSLP or basophils can be targeted to treat EoE-like disease
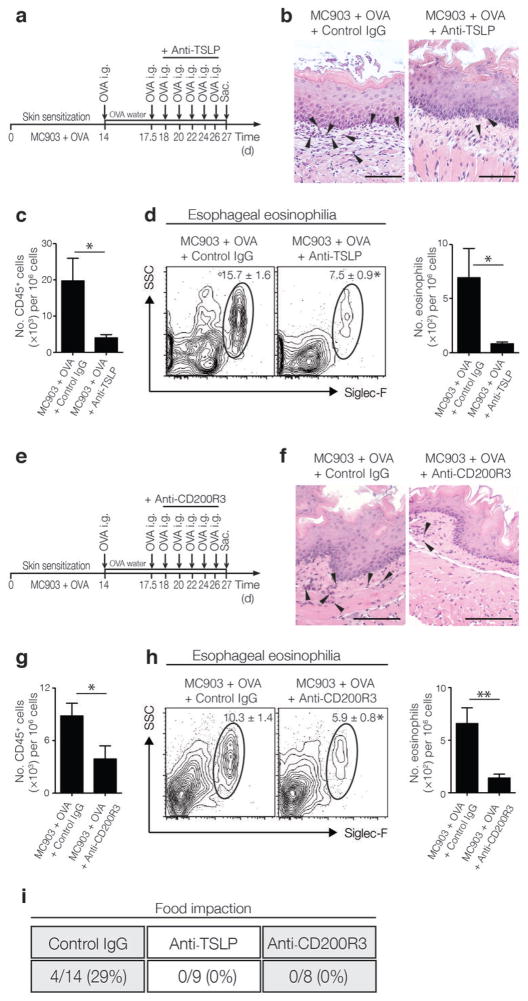
As TSLP and basophils were required during sensitization for the development of EoE-like disease in mice, we next tested whether the TSLP-basophil pathway could be therapeutically targeted to treat established EoE-like disease. First, mice were sensitized and challenged with OVA to establish EoE-like disease and were then treated with either an isotype control or a neutralizing TSLP-specific mAb during repeated antigen challenge (Fig. 5a). While mice with established EoE-like disease treated with a control antibody exhibited esophageal eosinophilia, mice that were treated with TSLP-specific mAb exhibited decreased esophageal eosinophilia as measured histologically (Fig. 5b). Flow cytometric analysis also revealed that the total immune cell infiltrate and esophageal eosinophilia were significantly reduced (Fig. 5c–d).
Figure 5.
Neutralization of TSLP or depletion of basophils ameliorates established EoE-like disease. (a) Schematic of treatment with TSLP-specific mAb in WT BALB/c mice in established EoE-like disease. (b) Histological sections (H & E staining) from the esophagus. Arrows identify tissue-infiltrating eosinophils. Scale bar: 25 μm. (c) Frequencies of CD45+ cells in esophageal tissues as measured by flow cytometry. (d) Representative flow cytometry plots showing frequencies and total numbers of eosinophils in esophageal tissues. Data depicted in (b–d) are from one experiment (MC903 + OVA + IgG, n = 5; MC903 + OVA + anti-TSLP mAb, n = 5), and are representative of three independent experiments. (e) Schematic of CD200R3-specific mAb basophil-depletion treatment in WT BALB/c mice in established EoE-like disease. (f) Histological sections (H & E staining) from the esophagus. Arrows identify tissue-infiltrating eosinophils. Scale bar: 25 μm. (g) Frequencies of CD45+ cells in esophageal tissues as measured by flow cytometry. (h) Representative flow cytometry plots showing frequencies and total numbers of eosinophils in esophageal tissues. Data depicted in (f–h) are from one experiment (MC903 + OVA + IgG, n = 4; MC903 + OVA + anti-CD200R3 mAb, n = 5), and are representative of three independent experiments. (i) Quantified incidence of food impaction. All parameters were assessed 12 h post-final oral antigen challenge. Data depicted are from mice challenged repeatedly with OVA. Results are shown as mean ± sem, and a non-parametric, two-tailed Mann-Whitney t-test was used to determine significance. *, P ≤ 0.05; **, P ≤ 0.01.
To test whether basophils contributed to the maintenance of EoE-like disease, mice with established EoE-like disease were treated with an isotype control or basophil-depleting CD200R3-specific mAb during repeated OVA challenge (Fig. 5e). Similar to the results observed following neutralization of TSLP, specific depletion of basophils resulted in decreased esophageal eosinophilia as measured histologically (Fig. 5f), and flow cytometric analysis also revealed a reduction in total immune cell infiltrate and eosinophil numbers in the esophagus (Fig. 5g–h). To test whether neutralization of TSLP or depletion of basophils were also associated with a resolution of signs of esophageal dysfunction, mice with established EoE-like disease treated with a control antibody, TSLP-specific mAb, or CD200R3-specific mAb were assessed for the incidence of food impaction. While food impaction was observed in about 30% of mice treated with a control antibody, impaction was not observed in mice in which TSLP or basophil responses were blocked (Fig. 5i). Taken together, these data demonstrate that TSLP neutralization or basophil depletion can be used to ameliorate inflammation and clinical symptoms of established experimental EoE-like disease in mice.
The TSLP-basophil axis is associated with EoE in humans
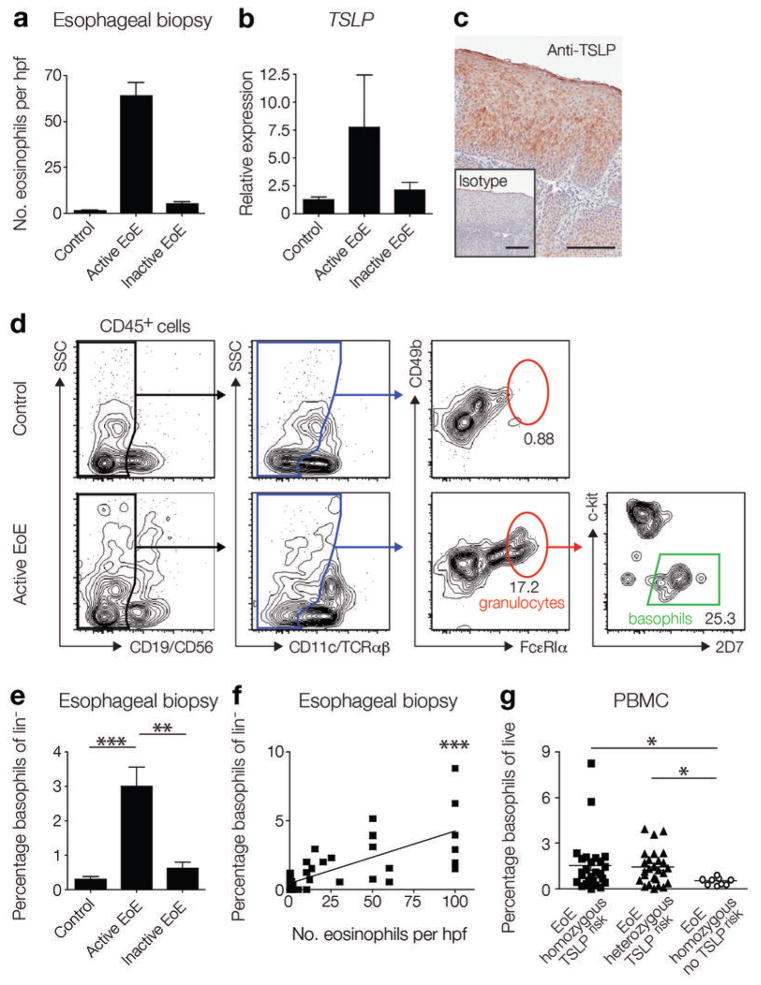
The roles of TSLP and basophils in experimental EoE-like disease in mice (Fig. 2 and 4) and the established association between gain-of-function polymorphisms in TSLP and EoE in human pediatric subjects10,11 provoked the hypothesis that the TSLP-basophil pathway may contribute to the pathogenesis of EoE in humans. To assess whether the TSLP-basophil axis is active in human subjects with EoE, TSLP and basophil responses were examined in esophageal biopsies from a cohort of pediatric subjects. This patient population was stratified based on the number of eosinophils counted in histologic sections from esophageal biopsies into the following groups: (i) control subjects without EoE, (ii) subjects with active EoE (≥15 eosinophils per hpf), or (iii) subjects with inactive EoE (<15 eosinophils per hpf and a prior clinical history of active EoE) (Fig. 6a). In agreement with previous studies10,11, TSLP expression in esophageal biopsies was increased in subjects with active EoE compared to control subjects or subjects with inactive EoE (Fig. 6b). Immunohistochemical staining revealed that stratified squamous epithelial cells exhibited positive staining for TSLP in esophageal biopsies from subjects with active EoE (Fig. 6c). Flow cytometric analysis was then employed to identify and quantify the inflammatory cell infiltrate in biopsies. Strikingly, increased frequencies of cells with a phenotype consistent with that of basophils (lin−, CD49b+, FcεRI+, c-kit−, 2D7+) were observed in esophageal biopsies from subjects with active EoE compared to those from control subjects or subjects with inactive EoE (Fig. 6d, e). Further, this basophil frequency positively correlated with the number of eosinophils counted per hpf in histological sections of esophageal biopsies (Fig. 6f). Additionally, a cohort of adult subjects was stratified based on the number of eosinophils counted in histologic sections (Supplementary Fig. 7a). Consistent with results observed in pediatric subjects, although not statistically significant, adult subjects with active EoE had an increased frequency of basophils in the esophageal biopsy as measured using flow cytometry that correlated positively with the number of eosinophils counted per hpf in histological sections (Supplementary Fig. 7b, c). Collectively, these data indicate for the first time that the TSLP-basophil axis is associated with active EoE in pediatric and adult subjects.
Figure 6.
The TSLP-basophil axis is active in human subjects with EoE. (a) Number of eosinophils per hpf in esophageal biopsy tissue sections were quantified for pediatric control subjects (n = 19) and subjects with active EoE (n = 16) or inactive EoE (n = 15). (b) Relative expression of TSLP in esophageal biopsies of pediatric control subjects (n = 8) and subjects with active EoE (n = 25) or inactive EoE (n = 10). (c) Immunohistochemical staining for TSLP (red) in an esophageal biopsy. Data are representative of 5 subjects with active EoE. Scale bar: 100 μm. (d) Basophils were identified by flow cytometry in esophageal biopsies from pediatric subjects with active EoE (plots are representative of 19 control subjects and 16 subjects with active EoE). (e) Frequencies of basophils in the lineage negative (lin−) compartment (see Methods) in esophageal biopsies from pediatric control subjects (n = 19) and subjects with active EoE (n = 16) or inactive EoE (n = 15). (f) Correlation of frequencies of basophils in pediatric esophageal biopsies and the number of eosinophils per hpf observed histologically (n = 50) (Spearman r = 0.6638). (g) Frequencies of basophils in the PBMCs of pediatric subjects with EoE that were homozygous (n = 26) or heterozygous for the TSLPrisk polymorphism (n = 26), or that lacked the TSLPrisk polymorphism (n = 9) were identified by flow cytometry. All data are shown as mean ± sem, and a non-parametric, two-tailed Mann-Whitney t-test or a non-parametric, one-way Kruskal-Wallis ANOVA with Dunn’s post-hoc testing were used to determine significance. Correlation analysis was performed using a non-parametric Spearman correlation (sensitivity analyses were performed), and a linear regression of the data is displayed. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
These findings, coupled with the association between the development of EoE and a previously identified gain-of-function polymorphism in TSLP associated with TSLP over-expression (TSLPrisk)10, provoked the hypothesis that there may be an association between the TSLPrisk polymorphism and enhanced basophil responses in human subjects with EoE. To directly test this, a new cohort of pediatric subjects with EoE genotyped for the presence of the TSLPrisk polymorphism was assessed for basophil frequencies in the peripheral blood mononuclear cells (PBMCs). Subjects that carried the TSLPrisk polymorphism exhibited significantly higher basophil frequencies in their PBMCs than subjects with EoE that did not carry the TSLPrisk polymorphism (Fig. 6g), establishing a genetic link between a gain-of-function TSLP polymorphism, increased peripheral basophil responses, and EoE. As with most human inflammatory diseases such as asthma, inflammatory bowel disease, and multiple sclerosis47–50, the development of EoE likely involves a complex interplay of genetic and environmental factors. However, these data suggest a model in which patients that carry the TSLPrisk polymorphism have a pre-disposition toward TSLP over-expression and associated peripheral basophilia that may increase the likelihood of developing EoE following encounter with trigger antigens (Supplementary Fig. 8).
DISCUSSION
Here we describe a new mouse model in which epicutaneous sensitization to a model food antigen followed by oral antigen challenge results in EoE-like disease. We demonstrate that TSLP and basophils, but not IgE, were required for the development of experimental EoE-like disease in mice and that antibody-mediated neutralization of TSLP or depletion of basophils was effective in preventing the development of experimental EoE-like disease. Critically, targeting TSLP or basophils was also effective in treating established EoE-like disease in mice. In addition, we identify for the first time the presence of increased basophil responses in the esophageal biopsy tissue of human subjects with EoE and a genetic link between a gain-of-function polymorphism in TSLP and increased peripheral basophil responses.
While all experimental model systems have limitations and do not recapitulate the diversity of symptoms reported in patients, the model of EoE-like disease we report here is associated with several characteristics of EoE in humans, including esophageal eosinophilia and associated esophageal dysfunction. In addition, this model is also characterized by gastrointestinal eosinophilia and systemic TH2 cytokine responses. EoE in patients is defined as a disease associated with eosinophilia in the esophagus. However, EoE patients often suffer from co-existing allergic disorders such as atopic dermatitis, allergic rhinitis, asthma, or intestinal food allergy2,7,51. These observations suggest that a subset of EoE patients with co-existing allergic diseases may present with manifestations of allergic disease at tissue sites outside of the esophagus52. Thus, the mouse model of EoE-like disease we describe may recapitulate a pan-allergic disease state present in some humans that have EoE and suffer from additional allergic diseases. While it is clear that EoE-like disease develops independently of IgE but is critically dependent on TSLP and basophils, further studies will be required to investigate whether the gastrointestinal eosinophilia in this model is dependent on IgE or TSLP-elicited basophils.
Previous studies in mouse models and humans have identified various immunological factors that are associated with EoE1–3,31–34,53–58. However, recent clinical trials that have targeted some of these factors, including IgE and IL-5, have failed to ameliorate symptoms of disease2,41,42,44,59,60, suggesting that these factors may not be critical for the pathogenesis of EoE. The demonstration that EoE-like disease in mice can develop independently of IgE but was critically dependent on TSLP and basophils may explain why previous clinical trials employing other candidate biologic therapies have not been successful. The identification of a role for TSLP and basophils in experimental EoE-like disease in mice, coupled with the association between TSLP and basophil responses and EoE in humans, indicate that targeting the TSLP-basophil axis may offer new opportunities for the clinical management of EoE in patients.
ONLINE METHODS
Mice
Male and female BALB/c and C57BL/6 mice were purchased from the Jackson Laboratories. BALB/c Tslpr+/+ and BALB/c Tslpr−/− mice were provided by Amgen, through Charles River Laboratories. BALB/c Igh–7−/− mice and C57BL/6 Baso–DTR mice were bred at the University of Pennsylvania. All mice were used at 8–12 weeks of age, and all experiments employed age-, gender- and genetic strain-matched controls, in order to account for any variations in data sets compared across experiments. Animals were bred and housed in specific pathogen–free conditions at the University of Pennsylvania. Animals requiring medical attention were provided with appropriate veterinary care by a licensed veterinarian and were excluded from the experiments described. No other exclusion criteria existed. All experiments were performed under Institutional Animal Care and Use Committee (IACUC) approved protocols and in accordance with the guidelines of the IACUC of the University of Pennsylvania.
Reagents and treatments
Mice were treated daily with 2 nmol MC903 (calcipotriol, Tocris Bioscience) in 20 μl of 100% EtOH on ears in the presence of 100 μg OVA for 14 d. As a vehicle control, the same volume of EtOH and OVA was applied. For tape-stripping, animals were shaved on the back, tape-stripped 6 times, and sensitized with 100 μg OVA or saline as control daily for 14 d. For TSLP injections, mice were subcutaneously injected with 5 μg rTSLP in the presence of 100 μg OVA on d 0, 3, 6, 9 and 12. For controls, mice were injected subcutaneously with PBS or rTSLP alone. For CPE sensitization, CPE was made from whole roasted peanuts (Sainsbury’s Ltd.) sterilized by gamma irradiation (Lillico Biotech) that were ground in an airflow cabinet using a pestle and mortar. The resulting paste was solubilized in pH 7.4 PBS (Gibco) and sonicated for two 20 min periods, with mixing in between. The solution was then filtered through a 75 μm tissue filter (BD Biosciences) to remove large particles of debris. LPS content was tested (Lonza) and reported less than 0.006 ng mL−1. Mice were treated daily with 2 nmol MC903 in 20 μl of 100% EtOH on ears in the presence of 100 μg CPE for 14 d. As a vehicle control, the same volume of EtOH and CPE was applied. Mice were challenged i.g with 50 mg OVA or 10 mg CPE on d 14 and 17.5 and euthanized on day 18. Upon first i.g. OVA or CPE challenge, mice were continuously fed water containing 1.5 g L−1 OVA or given continuous access to roasted peanut. Mice subjected to repeated challenge with OVA to induce prolonged inflammation in the esophagus were challenged i.g. with 50 mg OVA on d 14, 17.5, 18, 20, 22, 24, and 26 and euthanized on d 27. For depletion with TSLP-specific mAb17, mice were injected with 500 μg of control IgG or TSLP-specific mAb intraperitoneally every 3 d during the course of the experiment starting at d −1 or every other day starting at d 18. For basophil depletion by DT treatment, Baso-DTRpos or Baso-DTRneg littermate control mice were treated with 500 ng DT intraperitoneally on d −1, 3, 7 and 12. For depletion with CD200R3-specific mAb (Ba103)46, mice were injected with 100 μg of control IgG or CD200R3-specific mAb intravenously every 4 d during the course of the experiment starting at d −1 or every other day starting at d 18. To assess food impaction in the esophagus, mice exposed to prolonged esophageal inflammation were fasted for at least 30 min and up to 2 h. Mice were then euthanized, and their esophagi were examined for the presence of impacted food.
Cohort of human subjects with EoE
Pediatric participants from a cohort of control subjects or subjects with EoE at the University of Pennsylvania Penn-Children’s Hospital of Philadelphia (CHOP) Joint Center for Digestive, Liver and Pancreatic Medicine or the Center for Pediatric Eosinophilic Disorders at CHOP were analyzed and were provided under an IRB to M.-L.W. and A.J.B. Adult participants from a cohort of control subjects or subjects with EoE being treated at the Hospital of the University of Pennsylvania Division of Gastroenterology were also assessed and were provided under an IRB to G.W.F. and P.M.-K. Written consent was obtained from all participants or their parents or legal guardians, and for pediatric participants, verbal assent from the child was additionally obtained. Subjects defined as having EoE had no other chronic condition, except asthma, allergic rhinitis, food allergy, urticaria, or atopic dermatitis. Control subjects presented with epigastric abdominal pain but had normal endoscopic and microscopic results. Pediatric subjects with EoE were on proton pump inhibitor therapy, but subjects on systemic corticosteroid treatment or antibiotics were excluded. Subjects with active EoE had an esophageal eosinophil count of ≥15 per hpf after 8 weeks of treatment with a proton pump inhibitor. Subjects with inactive EoE had previously been diagnosed with active EoE but had an esophageal eosinophil count of < 15 per hpf at the time of sample collection. During routine endoscopy, three esophageal biopsies were collected for histological analysis of esophageal eosinophil counts. During the same procedure, two esophageal tissue biopsies were collected for research purposes, for either real-time PCR, immunohistochemistry, or flow cytometry. For flow cytometry, single cell suspensions were made by filtering the mechanically disrupted tissue through a 70 μm filter (BD Biosciences) for flow cytometry.
Peripheral blood from pediatric subjects from a cohort of control subjects or subjects with EoE that were genotyped for a gain-of-function TSLP polymorphism at the University of Pennsylvania Penn-CHOP Joint Center for Digestive, Liver and Pancreatic Medicine or the Center for Pediatric Eosinophilic Disorders at CHOP was analyzed and was provided under an IRB to J.M.S. and K.R.R. Peripheral blood was collected by venipuncture, and serum was isolated. PBMC were isolated by Ficoll gradient as previously described17, and cells were analyzed by flow cytometry. For genotyping of pediatric subjects with EoE, all samples were genotyped on either the Illumina HumanHap 550 or 610 BeadChips according to the manufacturer’s protocols. Data normalization and canonical genotype clustering were carried out using the Illumina Genome Studio package. Samples with call rate < 98% were excluded from further analysis.
Human real-time PCR and immunohistochemistry
For real-time PCR analysis of gene expression in human esophageal biopsies, human subject biopsy samples were collected and placed in RNAlater (Ambion). RNA was isolated using the mirVana miRNA Isolation Kit according to the manufacturer’s recommendations (Ambion) and reverse transcribed using a high-capacity cDNA reverse transcriptase kit (Applied Biosystems). Quantitative real-time PCR was performed using the Taqman Fast Universal PCR Master Mix kit and preformulated TaqMan Gene Expression Assays for TSLP (Applied Biosystems). Reactions were performed in triplicate using 96 well optical plates on a StepOnePlus Real-Time PCR System (Applied Biosystems). GAPDH was used as an endogenous control to normalize the samples using the CT method of relative quantification, where CT is the threshold cycle. For immunohistochemical staining for human TSLP, human esophageal biopsies were embedded in paraffin and sectioned. Sections were dewaxed and stained with a primary human TSLP-specific mAb or an isotype control antibody (validated by J.H. Yearley. and R. de Waal Malefyt), and positive staining was visualized using the DAB substrate kit (Vector Laboratories).
Flow cytometry
For mouse studies, esophageal tissues of 2–3 animals were pooled within each replicate experiment, opened longitudinally, digested in 1 mg mL−1 collagenase/DNase (Roche) for 30 min, and mashed through 70 μm nylon mesh filters. Single cell suspensions were incubated with Aqua Live/Dead Fixable Dye (Invitrogen) for dead cell exclusion and stained with fluorochrome-conjugated mAbs specific for mouse B220 (RA3-6B2), CD3ε (145-2C11), CD4 (GK1.5), CD5 (53-7.3), CD8 (53-6.7), CD11c (N418), NK1.1 (PK136), CD19 (eBio1D3), FcεR1 (MAR-1), IgE (23G3), CD45 (30-F11), CD49b (DX5), CD117 (c-kit) (2B8), or Siglec-F (E50-2440) (BD Bioscience, BioLegend, eBioscience), or fluorochrome-conjugated mAbs specific for human CD19 (HIB19), CD56 (B159), CD11c (B-ly6), TCRαβ (IP26), CD45 (HI30), CD49b (ebioY418), FcεR1(AER-37), CD123 (6H6), or c-kit (104D2) (BD Bioscience, BioLegend, eBioscience). For intracellular staining, surface stained cells were washed, fixed in 2% paraformaldehyde, permeabilized using eBioscience Permeabilization Buffer (eBioscience) according to manufacturer instructions, stained intracellularly with human 2D7-specific mAb (2D7) (eBioscience), washed, and resuspended in flow cytometry buffer. All cells were run on a 4-laser 14-color LSR II (BD Biosciences), and FlowJo 8.7.1 (Tree Star, Inc.) was used to analyze data. Mouse eosinophils were identified as live, lin− (CD3, CD5, CD19, CD11c, NK1.1), CD45+, Siglec-F+, side-scatter (SSC) high cells. Mouse basophils were identified as live, lin− (CD3, CD5, CD19, CD11c, NK1.1), c-kit−, CD49b+, IgE+ cells (or as FcεRI+ cells in Igh-7−/− mice). Human basophils in the esophageal biopsy were identified as live, lin− (CD19, CD56, CD11c, TCRαβ), CD49b+, FcεRI+, c-kit−, 2D7+ cells. Human basophils in the PBMC were identified as live, lin− (CD19, CD56, CD11c, TCRαβ), CD123+, FcεRI+ cells.
OCT
An OCT system operating at 1.3 μm center wavelength at 47 kHz axial scan rate (~30 frames per s) was developed and used for obtaining volumetric images of freshly excised mouse esophagus. The axial and transverse resolution was 6 μm and 10 μm in tissue, respectively, and the imaging depth was approximately 2 mm, sufficient to image through the entire thickness of the mouse esophagus. Prior to OCT imaging, the esophagus was removed from the mouse, and a plastic tube with 0.75 mm outer diameter was inserted, allowing for the lumenal surface to be clearly differentiated in cross-sectional images. The esophagus was immersed in saline solution to remove light reflection from the surface. Subsequently, 3D OCT images were obtained from multiple locations along the esophagus, with each data set covering 3×1.5×1.5 mm3. The thickness values of the squamous epithelial layer were measured from cross-sectional OCT images every 200 μm along the esophagus within each data set. Average squamous epithelial thickness values from the middle of the esophagus were calculated from each mouse by an investigator blinded to group allocations and were used for comparison between different groups.
Mouse cell cultures, ELISA, real-time PCR, histology, and EM
To measure skin TSLP levels, ears were incubated overnight in culture media, and cell-free supernatants were stored for a TSLP ELISA using a commercially available kit (eBioscience). For antigen re-stimulation, splenocytes or mesenteric LN cells were isolated, and single cell suspensions were stimulated with 200 μg OVA for 72 h. Cell-free supernatants were used for standard sandwich ELISA. Antigen-specific IgE responses were measured as described previously61. For histological analysis, at necroscopy, the esophagus was fixed in 4% paraformaldehyde and embedded in paraffin, and 5 μm sections were cut and stained with hematoxylin and eosin (H & E). For immunofluorescence, sections were dewaxed and stained with biotinylated SiglecF-specific mAb (R & D systems), followed by secondary staining with Cy3–conjugated streptavidin (Jackson Laboratory) and counterstaining with DAPI (Molecular Probes). For EM, esophageal tissues were fixed with 2.5% glutaraldehyde, 2.0% paraformaldehyde in 0.1M sodium cacodylate buffer, pH 7.4, overnight at 4°C. After buffer washes, the samples were post-fixed in 2.0% osmium tetroxide for 1 h at room temperature and rinsed in dH2O prior to en bloc staining with 2% uranyl acetate. After dehydration through a graded ethanol series, the tissue was infiltrated and embedded in EMbed–812 (Electron Microscopy Sciences). Thin sections were stained with uranyl acetate and lead citrate and examined with a JEOL 1010 electron microscope fitted with a Hamamatsu digital camera and AMT Advantage image capture software. For real-time PCR analysis, RNA was isolated from esophageal tissue using an RNeasy mini kit (QIAGEN) or the mirVana miRNA isolation kit (Ambion) according to the manufacturer’s instructions. cDNA was generated using a SuperscriptII reverse transcription kit (Invitrogen). Real-time quantitative PCR was performed on cDNA using SYBR green master mix (Applied Biosystems) and commercially available primer sets from Qiagen (Quantitect primer assays). Samples were run on a real-time PCR system (ABI 7500; Applied Biosystems), normalized to β-actin, and displayed as fold induction over controls.
Statistical analysis
Results are shown as mean ± sem. To determine group sizes necessary for adequate statistical power, power analysis was performed using preliminary data sets for all analyses presented. Mice of the indicated genotypes were assigned at random to treatment groups for all mouse studies. Mouse studies were not performed in a blinded fashion, except where indicated. Analyses of basophil responses in esophageal biopsy samples and peripheral blood were conducted in such a manner that the investigator was blinded to the disease state (number of eosinophils per hpf in the biopsy) and TSLP genotype until after flow cytometric analyses were completed. Analysis of TSLP expression levels in the biopsies of control subjects and those with EoE were not performed in a blinded fashion. All inclusion/exclusion criteria for mouse and human studies were pre-established. For mouse studies, statistical significance was determined using a non-parametric, two-tailed Mann-Whitney t-test, a non-parametric, one-way Kruskal-Wallis ANOVA test followed by Dunn’s post-hoc testing, or a non-parametric, two-way ANOVA followed by Bonferroni post-hoc testing. For human studies, a non-parametric, two-tailed Mann-Whitney t-test or a non-parametric, one-way Kruskal-Wallis ANOVA followed by Dunn’s post-hoc testing were used. Correlation analysis was performed using a non-parametric Spearman correlation (sensitivity analyses were performed), and a linear regression of the data is displayed. All data meet the assumptions of the statistical tests used. Within each group there is an estimate of variation, and the variance between groups is similar. For each statistical analysis, appropriate tests were selected based on whether the data was normally distributed and whether multiple comparisons were made. Results were considered significant at P ≤ 0.05. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. All statistical analyses were performed using Prism version 5.0a (GraphPad Software, Inc).
Supplementary Material
Acknowledgments
We thank members of the Artis laboratory for discussions and critical reading of the manuscript. Research in the Artis lab is supported by the US National Institutes of Health (AI061570, AI087990, AI074878, AI095776, AI102942, AI095466, AI095608, and AI097333 to D.A.), the Swiss National Science Foundation Prospective and Advanced Research Fellowships (PBBEP3_130438 and PA00P3_136468 to M.N.), T32-AI060516 and F32-AI098365 to E.D.T.W., T32-AR007465 and KL2-RR024132 to B.S.K., F32-AI085828 to M.C.S., the Australian National Health and Medical Research Council Overseas Biomedical Fellowship (613718 to P.R.G.), AI091759 to M.G.N., the State of Pennsylvania (SAP 4100042728 to P.M.S. and H.H.), K08-AI089982 to A.C., and the Burroughs Wellcome Fund Investigator in Pathogenesis of Infectious Disease Award to D.A. This work was supported by the US National Institutes of Health/US National Institute of Diabetes and Digestive and Kidney Diseases P30 Center for Molecular Studies in Digestive and Liver Diseases (P30-DK050306) and its pilot grant program and scientific core facilities (Molecular Pathology and Imaging, Molecular Biology, Cell Culture and Mouse), by the Joint Children’s Hospital of Philadelphia-Penn Center in Digestive, Liver and Pancreatic Medicine and its pilot grant program, and by the US National Institute of Allergy and Infectious Diseases Mucosal Immunology Studies Team (MIST) consortium (www.mucosal.org) (U01-AI095608). We also thank the Matthew J. Ryan Veterinary Hospital Pathology Lab and the Abramson Cancer Center Flow Cytometry and Cell Sorting Resource Laboratory (partially supported by NCI Comprehensive Cancer Center Support Grant (P30-CA016520)), the Skin Disease Research Center (supported by P30-AR057217), and the Electron Microscopy Resource Laboratory for technical advice and support. J.M.S. and K.R.R. acknowledge support from The Children’s Hospital of Philadelphia Institutional Development Fund, and J.M.S. also acknowledges support from the Department of Defense (A-16809.2). Human tissue samples were provided by M.-L.W. and A.J.B., funded by Abbot Nutrition (ANUS1013). Research in the Zhou lab is supported by the US National Institutes of Health (R00EB010071) and the Lehigh University start-up fund. The studies described here were supported in part by the Institute for Translational Medicine and Therapeutics Transdisciplinary Program in Translational Medicine and Therapeutics (UL1-RR024134 from the US National Center For Research Resources). The authors also wish to thank P. Just and N. Ruiz at eBioscience, Inc. for reagents, support, and invaluable technical advice.
Abbreviations
- CPE
crude peanut extract
- DT
diphtheria toxin
- DTR
diphtheria toxin receptor
- EM
electron microscopy
- EoE
eosinophilic esophagitis
- hpf
high-powered field
- IgE
immunoglobulin E
- IL
interleukin
- i.g
intragastric
- lin−
lineage negative
- mAb
monoclonal antibody
- OCT
optical coherence tomography
- OVA
ovalbumin
- PBMC
peripheral blood mononuclear cell
- rTSLP
recombinant thymic stromal lymphopoietin
- SSC
side-scatter
- TH2
T-helper type 2
- TSLP
thymic stromal lymphopoietin
- TSLPR
thymic stromal lymphopoietin receptor
- WT
wild-type
Footnotes
The content is solely the responsibility of the authors and does not represent the official views of the US National Center For Research Resources or the US National Institutes of Health.
AUTHOR CONTRIBUTIONS
M.N., E.D.T.W., B.S.K., M.C.S., P.R.G., M.G.N., A.B.M., A.A., C.Z., and D.A. designed and performed experiments. A.J.B., K.R.R., P.M.-K., A.C., G.W.F., M.-L.W., and J.M.S. provided human pediatric and adult esophageal biopsies and peripheral blood samples, K.R.C. analyzed pediatric esophageal biopsy histology, and D.A.H. and T.B.-W. coordinated patient care and clinical studies. A.E.M. and Q.J.S. provided CPE, M.K. provided Baso-DTR mice, K.O.-N. and H.K. provided anti-CD200R3 mAb, M.R.C. provided TSLPR-deficient mice and TSLP reagents, J.H.Y. and R.d.W.M. performed anti-human TSLP staining, and P.M.S. and H.H. provided genotype information on pediatric EoE patients. M.N., E.D.T.W., B.S.K., M.C.S., P.R.G., A.A., C.Z., M.-L.W., J.M.S., and D.A. analyzed the data. M.N., E.D.T.W., M.C.S., and D.A. wrote the manuscript, and all authors critically reviewed the manuscript.
COMPETING FINANCIAL INTERESTS
M.R.C. is an employee and shareholder of Amgen. The authors declare no competing financial interests.
References
- 1.Spergel JM. Eosinophilic esophagitis in adults and children: evidence for a food allergy component in many patients. Curr Opin Allergy Clin Immunol. 2007;7:274–278. doi: 10.1097/ACI.0b013e32813aee4a. [DOI] [PubMed] [Google Scholar]
- 2.Liacouras CA, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3–20. e26. doi: 10.1016/j.jaci.2011.02.040. quiz 21–22. [DOI] [PubMed] [Google Scholar]
- 3.Abonia JP, Rothenberg ME. Eosinophilic esophagitis: rapidly advancing insights. Annu Rev Med. 2012;63:421–434. doi: 10.1146/annurev-med-041610-134138. [DOI] [PubMed] [Google Scholar]
- 4.Straumann A, Simon HU. Eosinophilic esophagitis: escalating epidemiology? J Allergy Clin Immunol. 2005;115:418–419. doi: 10.1016/j.jaci.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Kapel RC, et al. Eosinophilic esophagitis: a prevalent disease in the United States that affects all age groups. Gastroenterology. 2008;134:1316–1321. doi: 10.1053/j.gastro.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 6.Straumann A, Schoepfer AM. Therapeutic concepts in adult and paediatric eosinophilic oesophagitis. Nat Rev Gastroenterol Hepatol. 2012 doi: 10.1038/nrgastro.2012.182. [DOI] [PubMed] [Google Scholar]
- 7.Arora AA, Weiler CR, Katzka DA. Eosinophilic esophagitis: allergic contribution, testing, and management. Curr Gastroenterol Rep. 2012;14:206–215. doi: 10.1007/s11894-012-0254-8. [DOI] [PubMed] [Google Scholar]
- 8.Markowitz JE, Spergel JM, Ruchelli E, Liacouras CA. Elemental diet is an effective treatment for eosinophilic esophagitis in children and adolescents. Am J Gastroenterol. 2003;98:777–782. doi: 10.1111/j.1572-0241.2003.07390.x. [DOI] [PubMed] [Google Scholar]
- 9.Mulder DJ, Justinich CJ. B cells, IgE and mechanisms of type I hypersensitivity in eosinophilic oesophagitis. Gut. 2010;59:6–7. doi: 10.1136/gut.2009.189316. [DOI] [PubMed] [Google Scholar]
- 10.Rothenberg ME, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. 2010;42:289–291. doi: 10.1038/ng.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sherrill JD, et al. Variants of thymic stromal lymphopoietin and its receptor associate with eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126:160–165. e163. doi: 10.1016/j.jaci.2010.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziegler SF. The role of thymic stromal lymphopoietin (TSLP) in allergic disorders. Curr Opin Immunol. 2010;22:795–799. doi: 10.1016/j.coi.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramasamy A, et al. A genome-wide meta-analysis of genetic variants associated with allergic rhinitis and grass sensitization and their interaction with birth order. J Allergy Clin Immunol. 2011;128:996–1005. doi: 10.1016/j.jaci.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 14.Liu M, et al. Genetic variants of TSLP and asthma in an admixed urban population. PloS one. 2011;6:e25099. doi: 10.1371/journal.pone.0025099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirota T, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet. 2011;43:893–896. doi: 10.1038/ng.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunninghake GM, et al. TSLP polymorphisms are associated with asthma in a sex-specific fashion. Allergy. 2010;65:1566–1575. doi: 10.1111/j.1398-9995.2010.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siracusa MC, et al. TSLP promotes interleukin-3-independent basophil haematopoiesis and type 2 inflammation. Nature. 2011;477:229–233. doi: 10.1038/nature10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siracusa MC, Wojno ED, Artis D. Functional heterogeneity in the basophil cell lineage. Adv Immunol. 2012;115:141–159. doi: 10.1016/B978-0-12-394299-9.00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giacomin PR, et al. Thymic Stromal Lymphopoietin-Dependent Basophils Promote Th2 Cytokine Responses following Intestinal Helminth Infection. J Immunol. 2012 doi: 10.4049/jimmunol.1200691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu YJ, et al. TSLP: an epithelial cell cytokine that regulates T cell differentiation by conditioning dendritic cell maturation. Annu Rev Immunol. 2007;25:193–219. doi: 10.1146/annurev.immunol.25.022106.141718. [DOI] [PubMed] [Google Scholar]
- 21.Soumelis V, et al. Human epithelial cells trigger dendritic cell mediated allergic inflammation by producing TSLP. Nat Immunol. 2002;3:673–680. doi: 10.1038/ni805. [DOI] [PubMed] [Google Scholar]
- 22.Hsieh KY, Tsai CC, Wu CH, Lin RH. Epicutaneous exposure to protein antigen and food allergy. Clin Exp Allergy. 2003;33:1067–1075. doi: 10.1046/j.1365-2222.2003.01724.x. [DOI] [PubMed] [Google Scholar]
- 23.Lack G. Update on risk factors for food allergy. J Allergy Clin Immunol. 2012;129:1187–1197. doi: 10.1016/j.jaci.2012.02.036. [DOI] [PubMed] [Google Scholar]
- 24.van den Oord RA, Sheikh A. Filaggrin gene defects and risk of developing allergic sensitisation and allergic disorders: systematic review and meta-analysis. BMJ. 2009;339:b2433. doi: 10.1136/bmj.b2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M, et al. Topical vitamin D3 and low-calcemic analogs induce thymic stromal lymphopoietin in mouse keratinocytes and trigger an atopic dermatitis. Proc Natl Acad Sci U S A. 2006;103:11736–11741. doi: 10.1073/pnas.0604575103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li M, et al. Induction of thymic stromal lymphopoietin expression in keratinocytes is necessary for generating an atopic dermatitis upon application of the active vitamin D3 analogue MC903 on mouse skin. J Invest Dermatol. 2009;129:498–502. doi: 10.1038/jid.2008.232. [DOI] [PubMed] [Google Scholar]
- 27.Leyva-Castillo JM, Hener P, Jiang H, Li M. TSLP Produced by Keratinocytes Promotes Allergen Sensitization through Skin and Thereby Triggers Atopic March in Mice. J Invest Dermatol. 2012;133:154–163. doi: 10.1038/jid.2012.239. [DOI] [PubMed] [Google Scholar]
- 28.DeBrosse SD, et al. Long-term outcomes in pediatric-onset esophageal eosinophilia. J Allergy Clin Immunol. 2011;128:132–138. doi: 10.1016/j.jaci.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang D, et al. Optical coherence tomography. Science. 1991;254:1178–1181. doi: 10.1126/science.1957169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou C, et al. Characterization of buried glands before and after radiofrequency ablation by using 3-dimensional optical coherence tomography (with videos) Gastrointest Endosc. 2012;76:32–40. doi: 10.1016/j.gie.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhattacharya B, et al. Increased expression of eotaxin-3 distinguishes between eosinophilic esophagitis and gastroesophageal reflux disease. Human Pathol. 2007;38:1744–1753. doi: 10.1016/j.humpath.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Blanchard C, et al. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J Allergy Clin Immunol. 2011;127:208–217. 217 e201–207. doi: 10.1016/j.jaci.2010.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu Blatman KS, Gonsalves N, Hirano I, Bryce PJ. Expression of mast cell-associated genes is upregulated in adult eosinophilic esophagitis and responds to steroid or dietary therapy. J Allergy Clin Immunol. 2011;127:1307–1308. e1303. doi: 10.1016/j.jaci.2010.12.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Justinich CJ, et al. Activated eosinophils in esophagitis in children: a transmission electron microscopic study. J Pediatr Gastroenterol Nutr. 1997;25:194–198. doi: 10.1097/00005176-199708000-00011. [DOI] [PubMed] [Google Scholar]
- 35.Oyoshi MK, Larson RP, Ziegler SF, Geha RS. Mechanical injury polarizes skin dendritic cells to elicit a T(H)2 response by inducing cutaneous thymic stromal lymphopoietin expression. J Allergy Clin Immunol. 2010;126:976–984. 984 e971–975. doi: 10.1016/j.jaci.2010.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoo J, et al. Spontaneous atopic dermatitis in mice expressing an inducible thymic stromal lymphopoietin transgene specifically in the skin. J Exp Med. 2005;202:541–549. doi: 10.1084/jem.20041503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jessup HK, et al. Intradermal administration of thymic stromal lymphopoietin induces a T cell- and eosinophil-dependent systemic Th2 inflammatory response. J Immunol. 2008;181:4311–4319. doi: 10.4049/jimmunol.181.6.4311. [DOI] [PubMed] [Google Scholar]
- 38.Finkelman FD. Anaphylaxis: lessons from mouse models. J Allergy Clin Immunol. 2007;120:506–515. doi: 10.1016/j.jaci.2007.07.033. quiz 516–507. [DOI] [PubMed] [Google Scholar]
- 39.Lucendo AJ, et al. Immunophenotypic characterization and quantification of the epithelial inflammatory infiltrate in eosinophilic esophagitis through stereology: an analysis of the cellular mechanisms of the disease and the immunologic capacity of the esophagus. Am J Surg Pathol. 2007;31:598–606. doi: 10.1097/01.pas.0000213392.49698.8c. [DOI] [PubMed] [Google Scholar]
- 40.Vicario M, et al. Local B cells and IgE production in the oesophageal mucosa in eosinophilic oesophagitis. Gut. 2010;59:12–20. doi: 10.1136/gut.2009.178020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rocha R, et al. Omalizumab in the treatment of eosinophilic esophagitis and food allergy. Eur J Pediatr. 2011;170:1471–1474. doi: 10.1007/s00431-011-1540-4. [DOI] [PubMed] [Google Scholar]
- 42.Foroughi S, et al. Anti-IgE treatment of eosinophil-associated gastrointestinal disorders. J Allergy Clin Immunol. 2007;120:594–601. doi: 10.1016/j.jaci.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stone KD, Prussin C. Immunomodulatory therapy of eosinophil-associated gastrointestinal diseases. Clin Exp Immunol. 2008;38:1858–1865. doi: 10.1111/j.1365-2222.2008.03122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sampson HA, et al. A phase II, randomized, doubleblind, parallelgroup, placebocontrolled oral food challenge trial of Xolair (omalizumab) in peanut allergy. J Allergy Clin Immunol. 2011;127:1309–1310. e1301. doi: 10.1016/j.jaci.2011.01.051. [DOI] [PubMed] [Google Scholar]
- 45.Sawaguchi M, et al. Role of mast cells and basophils in IgE responses and in allergic airway hyperresponsiveness. J Immunol. 2012;188:1809–1818. doi: 10.4049/jimmunol.1101746. [DOI] [PubMed] [Google Scholar]
- 46.Obata K, et al. Basophils are essential initiators of a novel type of chronic allergic inflammation. Blood. 2007;110:913–920. doi: 10.1182/blood-2007-01-068718. [DOI] [PubMed] [Google Scholar]
- 47.Mukherjee AB, Zhang Z. Allergic asthma: influence of genetic and environmental factors. J Biol Chem. 2011;286:32883–32889. doi: 10.1074/jbc.R110.197046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun L, Nava GM, Stappenbeck TS. Host genetic susceptibility, dysbiosis, and viral triggers in inflammatory bowel disease. Curr Opin Gastroenterol. 2011;27:321–327. doi: 10.1097/MOG.0b013e32834661b4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Renz H, et al. Gene-environment interactions in chronic inflammatory disease. Nat Immunol. 2011;12:273–277. doi: 10.1038/ni0411-273. [DOI] [PubMed] [Google Scholar]
- 50.Gourraud PA, Harbo HF, Hauser SL, Baranzini SE. The genetics of multiple sclerosis: an up-to-date review. Immunol Rev. 2012;248:87–103. doi: 10.1111/j.1600-065X.2012.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown-Whitehorn TF, Spergel JM. The link between allergies and eosinophilic esophagitis: implications for management strategies. Exp Rev Clin Immunol. 2010;6:101–109. doi: 10.1586/eci.09.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Straumann A, et al. Cytokine expression in healthy and inflamed mucosa: probing the role of eosinophils in the digestive tract. Inflam Bowel Dis. 2005;11:720–726. doi: 10.1097/01.mib.0000172557.39767.53. [DOI] [PubMed] [Google Scholar]
- 53.Akei HS, Mishra A, Blanchard C, Rothenberg ME. Epicutaneous antigen exposure primes for experimental eosinophilic esophagitis in mice. Gastroenterology. 2005;129:985–994. doi: 10.1053/j.gastro.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 54.Mavi P, Rajavelu P, Rayapudi M, Paul RJ, Mishra A. Esophageal functional impairments in experimental eosinophilic esophagitis. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1347–1355. doi: 10.1152/ajpgi.00013.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rajavelu P, Rayapudi M, Moffitt M, Mishra A. Significance of para-esophageal lymph nodes in food or aeroallergen-induced iNKT cell-mediated experimental eosinophilic esophagitis. Am J Physiol Gastrointest Liver Physiol. 2012;302:G645–654. doi: 10.1152/ajpgi.00223.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mishra A, Schlotman J, Wang M, Rothenberg ME. Critical role for adaptive T cell immunity in experimental eosinophilic esophagitis in mice. J Leukoc Biol. 2007;81:916–924. doi: 10.1189/jlb.1106653. [DOI] [PubMed] [Google Scholar]
- 57.Mishra A, Rothenberg ME. Intratracheal IL-13 induces eosinophilic esophagitis by an IL-5, eotaxin-1, and STAT6-dependent mechanism. Gastroenterology. 2003;125:1419–1427. doi: 10.1016/j.gastro.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 58.Mishra A, Hogan SP, Brandt EB, Rothenberg ME. IL-5 promotes eosinophil trafficking to the esophagus. J Immunol. 2002;168:2464–2469. doi: 10.4049/jimmunol.168.5.2464. [DOI] [PubMed] [Google Scholar]
- 59.Spergel JM, et al. Reslizumab in children and adolescents with eosinophilic esophagitis: results of a double-blind, randomized, placebo-controlled trial. J Allergy Clin Immunol. 2012;129:456–463. 463 e451–453. doi: 10.1016/j.jaci.2011.11.044. [DOI] [PubMed] [Google Scholar]
- 60.Castro M, et al. Reslizumab for poorly controlled, eosinophilic asthma: a randomized, placebo-controlled study. Am J Respir Crit Care Med. 2011;184:1125–1132. doi: 10.1164/rccm.201103-0396OC. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Z, et al. Thymic stromal lymphopoietin overproduced by keratinocytes in mouse skin aggravates experimental asthma. Proc Natl Acad Sci U S A. 2009;106:1536–1541. doi: 10.1073/pnas.0812668106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.