Abstract
Prognostic genes are key molecules informative for cancer prognosis and treatment. Previous studies have focused on the properties of individual prognostic genes, but have lacked a global view of their system-level properties. Here we examined their properties in gene co-expression networks for four cancer types using data from The Cancer Genome Atlas. We found that prognostic mRNA genes tend not to be hub genes (genes with an extremely high connectivity), and this pattern is unique to the corresponding cancer-type specific network. In contrast, the prognostic genes are enriched in modules (., a group of highly interconnected genes), especially in module genes conserved across different cancer co-expression networks. The target genes of prognostic miRNA genes show similar patterns. We identified the modules enriched in various prognostic genes, some of which show cross-tumor conservation. Given the cancer types surveyed, our study presents a view of emergent properties of prognostic genes.
Prognostic genes have properties (such as expression level or mutation status) that are informative regarding clinical outcomes. These genes are of particular biomedical interest in cancer research because of their potential as biomarkers, to help predict patients' survival, and to provide insights into the molecular mechanisms of tumor progression1-5. Over the past decades, tremendous efforts have been made to identify prognostic genes and build more effective models for stratifying individuals with cancer6-11. However, such studies have focused on individual prognostic genes and their clinical utilities, without investigating the emergent properties and behaviors of prognostic genes at the systems level.
Biological networks represent valuable platforms for understanding systems-level properties12-14. The commonly used biological networks include protein-protein interaction networks, signaling networks, metabolic networks, gene regulatory networks, and gene co-expression networks. Compared with other types of biological networks, using gene co-expression networks has several advantages15: nearly complete coverage of human genes, little bias due to the knowledge obtained from the published literature, and the ability to construct cancer-type–specific networks.
Using recently available cancer genomic data from The Cancer Genome Atlas (TCGA), we investigated the properties of prognostic genes in the gene co-expression networks of four representative cancer types (glioblastoma multiforme [GBM], ovarian serous cystadenocarcinoma [OV], breast invasive carcinoma [BRCA], and kidney renal clear cell carcinoma [KIRC])16-19. Here we focused on three primary questions about expression-based prognostic genes. First, are there network properties that distinguish prognostic genes from other genes in the co-expression networks? Second, do different types of prognostic genes show similar network properties? Third, do those patterns hold true across different cancer types? We performed a comparative analysis of prognostic genes in terms of key network properties ( e.g., whether they tend to be hub genes and enriched in modules) across the four cancer types. Our results reveal some common and distinct patterns of prognostic genes and identify modules associated with prognostic signatures. This study contributes to a comprehensive understanding of the informative behaviors of prognostic genes from the point of view of systems biology.
Results
Prognostic mRNA genes tend not to be hub genes
In this study, we focused on the four TCGA cancer types with adequate follow-up/survival data and sufficient sample size. The power of detecting prognostic genes varies from one cancer to another, which mainly depends on the sample size and the number of survival events (i.e., death). Here, we defined prognostic genes as those whose mRNA expression levels are significantly correlated with overall patient survival in two alternative ways: first, different numbers of prognostic mRNAs were identified based on the signal-to-noise ratio within each sample cohort; and second, the top 1000 mRNA genes most correlated with patient survival were identified per cancer type. We obtained very similar results using these two strategies, and throughout the text, we will mainly present the results based on the first method. With the first method, we identified 1,706, 728, 974 and 2,050 prognostic mRNA genes in GBM, OV, BRCA and KIRC, respectively (Fig. 1a, Methods). These prognostic genes showed great robustness through the assessment of subset samplings (Methods, Supplementary Figure 1a); and the four cancer types shared only a small portion (3%∼12%) of these prognostic genes (Supplementary Figure 1b). For each cancer type, we constructed a gene co-expression network from Agilent microarray data using weighted gene correlation network analysis (WGCNA)20,21. WGCNA is a well-established method designed for constructing co-expression networks from microarray-based expression data, and considers not only the co-expression patterns between two genes, but also the overlap of neighboring genes. As a result, we obtained four cancer-type–specific co-expression networks, each containing the same set of 17,813 genes (nodes). These co-expression networks are weighted networks in which any two nodes are connected with an edge weight (from 0 to 1, where 0 indicates no interaction, and 1 a strong interaction). Previous studies have indicated that a weighted network retains more information and is more robust and accurate than an unweighted one in network analysis15,21.
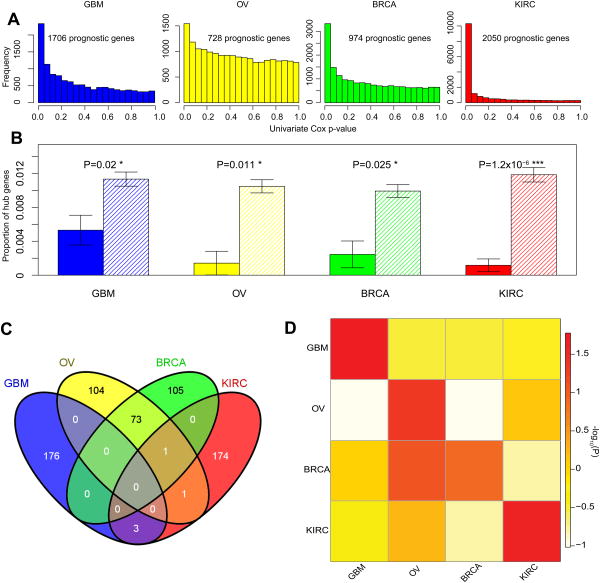
Figure 1. The depletion of prognostic mRNA genes in hubs.
(A) The P-value distributions of the correlations of mRNA expression with overall survival based on the univariate Cox model in the four cancer types. Based on the signal-to-noise ratio, prognostic mRNA genes were identified. (B) Prognostic mRNA genes are depleted in the hubs. Solid bars represent the proportions of hub genes among prognostic mRNA genes; striped bars represent the proportions of hub genes among non-prognostic mRNA genes. Error bars indicate ± 1 s.e.m., and P-values were calculated based on Fisher's exact tests. (C) The Venn diagram of hub genes across the four cancer types. (D) The heatmap showing the cancer-type–specific pattern of hub depletion. The color of each cell represents the depletion score of prognostic mRNA genes of a cancer type (column) in hub genes of another cancer type (row); row-wise scaled –log10(P-value) is plotted with red indicating significant, white indicating not significant. P-values were calculated based on Fisher's exact tests.
One key property for a gene in a biological network is connectivity, which reflects how frequently a node interacts with other nodes (for a weighted network, connectivity is defined as the sum of the weights across all edges of a node). According to the node connectivity, genes can be further classified into hub genes (with an extremely high level of connectivity) and non-hub genes. Hub genes are very important nodes, and in the protein interaction networks of various organisms, hub proteins tend to encode essential genes22-24. In the gene co-expression network, hub genes represent a small proportion of nodes with maximal information exchange with other nodes. For example, one prognostic hub gene in GBM is KLKL1, which is a serine protease with diverse physiological functions. We first examined the properties of connectivity and enrichment for the prognostic genes. We found that, on average, prognostic genes have higher connectivity, but this association does not follow a simple monotonic increasing trend. The prognostic genes appeared to be depleted in nodes with either extremely low or high connectivity (Supplementary Figure 2). To formally test whether prognostic genes are less likely to be hub nodes in the co-expression networks, we examined the connectivity distributions and defined the 1% (or 5%) of nodes with the highest connectivity as hub genes, according to the literature25-27. We found that prognostic genes are significantly depleted in the hubs across all four cancer types (Fig. 1b, Supplementary Figure 3 includes the bar plots of a random same-size set of non-prognostic genes for comparison; Supplementary Figure 4). In addition, we observed the same patterns when the top 1000 prognostic gene sets were used (Supplementary Figure 5).
In general, the hub genes defined in the different cancer networks are highly specific and show only a little overlap, except that OV and BRAC share some hub genes due to their pathophysiological similarity (Fig. 1c). To further examine whether the observed depletion of prognostic genes in hubs is unique to the cancer-specific network, we performed similar analyses using prognostic genes and hub genes defined in different co-expression networks. Fig. 1d shows the results in a heatmap format: significant depletions were primarily observed along the diagonal line, demonstrating that prognostic genes in one cancer type show a significant depletion in only the hubs of the corresponding co-expression network.
Prognostic mRNA genes are enriched in modules
Another important aspect of a gene co-expression network is modularity: genes that are highly interconnected within the network are usually involved in the same biological modules or pathways28,29. Using WGCNA, we defined the modules in each cancer co-expression network (Fig. 2a, Methods), and detected 85, 98, 55 and 81 modules in GBM, OV, BRCA and KIRC, respectively. For example, one common module across tumor types is enriched with genes related to the regulation of cell death and apoptosis. The corresponding proportions of module genes are 36.9%, 34.7%, 26.5%, and 29.6% in these cancer types. Strikingly, except for BRCA, prognostic mRNA genes show a significant enrichment in modules (Fig. 2b; Supplementary Figure 6 includes the bar plots of a random same-size of non-prognostic genes for comparison). Moreover, we obtained the same patterns using the top 1000 prognostic gene sets (Supplementary Figure 5d). In contrast to hub genes, there is substantial overlap of module genes across tumor types (Fig. 2c). This is reasonable since modules largely reflect the underlying biological processes.
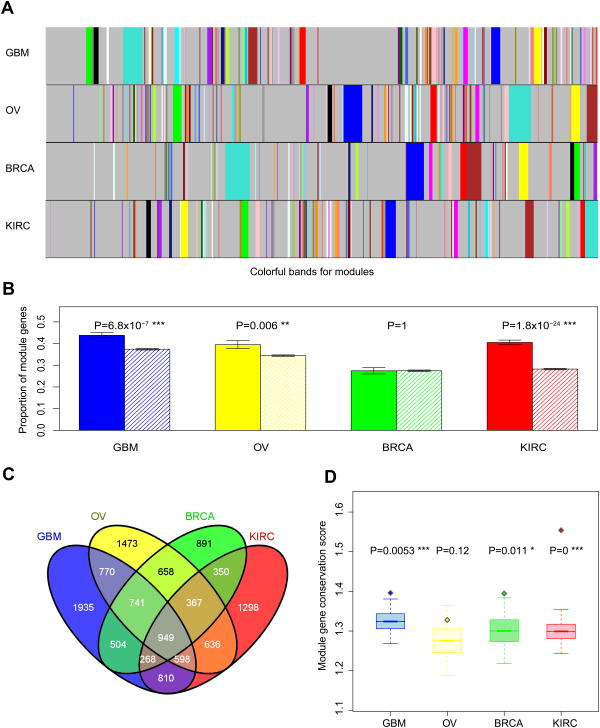
Figure 2. The enrichment of prognostic mRNA genes in modules.
(A) Modules defined from the weighted gene co-expression networks. Colorful bands represent modules in the network, with the biggest module in turquoise, second largest in blue, then brown, green, yellow and so on. (B) Prognostic genes are enriched in the modules. Solid bars represent the proportions of module genes among prognostic mRNA genes; striped bars represent the proportions of module genes among non-prognostic mRNA genes. Error bars indicate ± 1 s.e.m., and P-values were calculated based on Fisher's exact tests. (C) The Venn diagram of module genes across the four cancer types. (D) Boxplots (median ± 1 quartile) showing that prognostic genes tend to be more conserved module genes. Y-axis represents the module-gene conservation score, which ranges from 0 to 4, with 0 indicating not a module gene in any of the four cancer types and 4 indicating a module gene in all four cancer types. Each boxplot represents the mean-conservation-score distribution of 20,000 randomly sampled same-size gene sets; the diamond dot represents the mean conservation score of prognostic genes. P-values were calculated based on Wilcoxon rank sum tests.
To further examine the relationships between prognostic genes and modules in the co-expression networks, for each gene, we calculated a module–gene conservation score (range 0∼4), which indicates how frequently a gene is classified in a module among the four cancer types. We found that the conservation scores of prognostic genes are significantly higher than those of other genes in GBM, BRCA and KIRC; the same trend holds true in OV, but with a marginal significance (Fig. 2d). These results indicate that prognostic mRNA genes are enriched in module genes, especially in conserved module genes.
Target genes of prognostic microRNAs show similar patterns
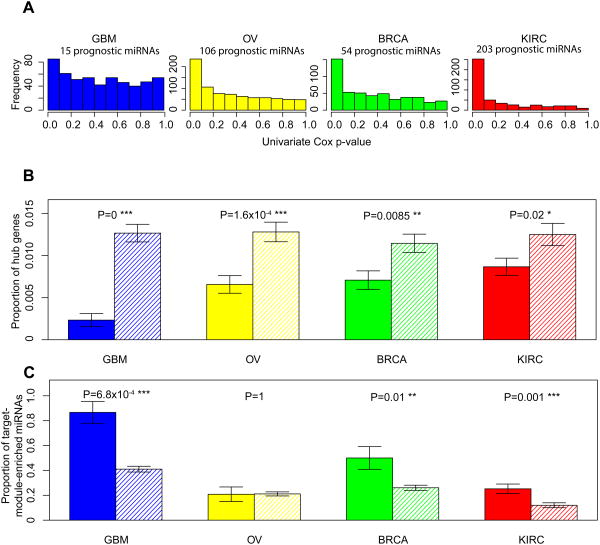
In addition to prognostic mRNA genes, we examined the network properties of prognostic miRNA genes, an important class of non-coding regulatory genes30,31. Similar to prognostic mRNAs, we defined prognostic miRNA genes based on the correlations of their expression levels with overall patient survival in two ways: first, different numbers of prognostic miRNAs were identified based on the signal-to-noise ratio within each sample cohort; and second, the top 50 miRNA genes most correlated with patient survival were identified per cancer type. We obtained very similar results using these two methods, and will mainly focus on the results from the first method. We identified 15, 106, 54 and 203 prognostic miRNAs in GBM, OV, BRCA and KIRC, respectively (Fig. 3a, Methods). Since the gene co-expression networks consist of mRNA genes only, we investigated the properties of the target genes of the prognostic miRNAs identified by a leading miRNA-target prediction program, TargetScan32. Strikingly, we observed the same patterns: the target genes of prognostic miRNAs are depleted in hub genes (Fig. 3b; Supplementary Figure 7 shows the results when the 5% of nodes with the highest connectivity were defined as hub genes). Considering the noise in miRNA target prediction, we further examined the predicted targets of prognostic miRNAs with different stringent criteria and obtained concordant results (Supplementary Figure 8, Methods). We also obtained the same results using the top 50 prognostic miRNA sets (Supplementary Figure 9). Moreover, the target genes of prognostic miRNAs are more likely to be enriched in modules than those of non-prognostic miRNAs (Fig. 3c; Supplementary Figure 9d shows the results for the top 50 prognostic miRNA sets). These results suggest that prognostic mRNA and miRNA genes share similar network properties in the cancer gene co-expression networks.
Figure 3. Target genes of prognostic miRNA genes show the same patterns.
(A) The P-value distributions of the correlations of miRNA expression with overall survival based on the univariate Cox model in the four cancer types. Based on the signal-to-noise ratio, prognostic miRNA genes were identified. (B) Target genes of prognostic miRNA genes are depleted in the hubs. Solid bars represent the proportions of hub genes among target genes of prognostic miRNAs; striped bars represent the proportions of hub genes among non-target genes of prognostic miRNAs. (C) Target genes of prognostic miRNAs are enriched in the modules. Solid bars represent the proportions of target-module–enriched miRNAs among prognostic miRNAs; striped bars represent the proportions of target-module–enriched miRNAs among non-prognostic miRNAs. Error bars indicate ± 1 s.e.m., and P-values were calculated based on Fisher's exact tests.
Some prognostic modules are conserved across tumor types
Since both prognostic mRNA genes and the targets of prognostic miRNAs are enriched in the modules of co-expression networks, we were also interested in identifying individual prognostic modules and determining their relationships across cancer types. For this purpose, we first identified modules enriched with prognostic mRNA genes (FDR < 0.1), and found 47 prognostic modules across the four cancers (GBM: 13, OV: 8, BRCA: 8, and KIRC: 18), with module sizes (number of genes within a module) ranging from 21 to 793 (Fig. 4a, Supplementary Data 1). For each cancer type, tumor subtypes classified by the gene expression of these prognostic modules showed distinct survival curves, highlighting their potential clinical relevance (Methods, Supplementary Figure 10). We further annotated their biological themes through gene ontology (GO) terms33. The common themes across the four cancers included “response to wounding and inflammation,” “regulation of cell death/apoptosis,” “RNA biosynthetic processing,” “translational elongation and termination,” “signaling pathway regulation,” “regulation of kinase cascade,” “cellular response to hormone/chemical stimulus,” and “multiple lipids metabolic and biosynthesis process.” Besides the common themes, there were specific module themes for each cancer type, such as “nervous system development,” “negative regulation of gliogenesis,” “type I interferon-mediated signaling pathway,” and “response to metal ion” in GBM; “negative regulation of histone modification” and “tRNA aminoacylation for protein translation” in OV; “chromatin remodeling” and “smooth muscle tissue development” in BRCA; and “positive regulate CREB transcription factor activity,” “interleukin-1-mediated signaling pathway,” “negative regulation of WNT receptor signaling pathway,” and “stress-activated MAPK cascade” in KIRC.
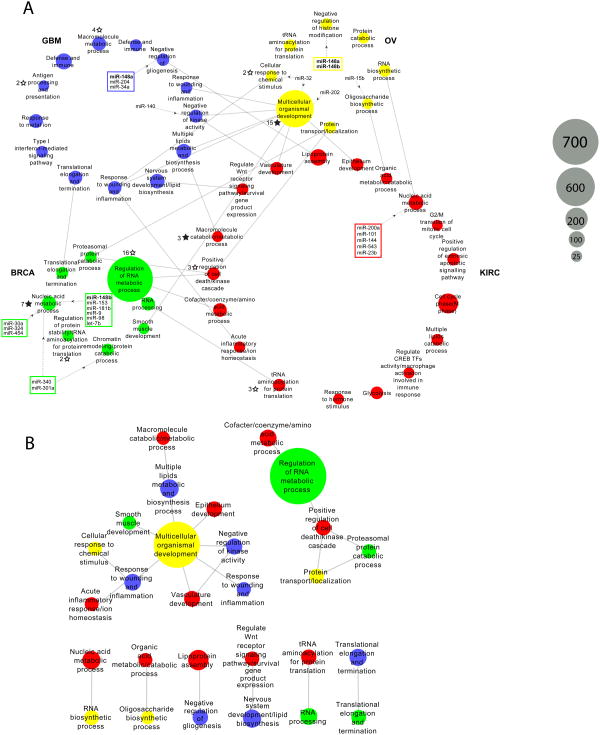
Figure 4. Integrative analysis of prognostic modules.
(A) The 47 prognostic modules are plotted in 4 circles, each representing one cancer type. Grey solid lines represent the conservation correspondence between two modules from two different cancer types. Dashed grey lines with black arrow represent the connections of miRNAs whose target genes are enriched in the module. Two or more miRNAs targeting the same module(s) are enclosed within a rectangle; miRNAs as module regulators in more than one cancer type are shown in boldface. Solid black stars mark the modules enriched with significantly mutated pan-cancer genes, and the associated number indicates the number of mutated genes; unfilled black stars mark enrichment that is significant only before multiple testing correction. (B) Plot showing a zoomed-in view of the 22 modules with cross-tumor conservation correspondence in (A).
Interestingly, among the 47 prognostic modules, 9 prognostic modules were also enriched with the targets of 23 prognostic miRNAs (FDR < 0.1, Fig. 4a). Among them, three miRNAs (miR-32, miR-301a and miR-340) show target enrichment in at least two modules, and two miRNAs (miR-148a and miR-148b) are associated with prognostic modules in two cancer types. In addition, 3 prognostic modules are enriched with significantly mutated genes identified in the TCGA Pan-Cancer project (FDR < 0.1, Fig. 4a).
To investigate the prognostic modules conserved across tumor types, we determined the module correspondence by considering the members that overlap between the two prognostic modules. This analysis revealed 22 pairwise correspondences among the 47 prognostic modules across the 4 cancers (Fig. 4b). Remarkably, one OV module annotated with “multicellular organismal development” acted like a module “hub,” directly linking 7 other modules of different cancer types with a wide range of biological themes.
Discussion
In this study, we performed a systematic analysis of the properties of prognostic genes in the context of biological networks across multiple cancer types. Importantly, we used the gene co-expression networks constructed from a single type of microarray as the investigation platform. This practice reduces various confounding factors in the data analysis, such as prior knowledge bias, which occurs from using other types of biological networks or batch effects when constructing the networks from expression data from multiple profiling techniques. Strikingly, although both prognostic genes and hub/module membership in the networks varied greatly from one cancer type to another, our study revealed some distinct properties of prognostic genes. The consistent nature of the patterns we found across multiple cancer types and both prognostic mRNA and miRNA genes (through their target genes) highlights the robustness of the observed pattern. This study provides the first systems-level understanding of the “informer” behaviors of cancer prognostic genes, thereby laying a foundation for how to incorporate the co-expression network information into prognostic modeling. For example, since both prognostic mRNAs and miRNAs appear to be preferably associated with some biological modules, on a practical level, further efforts are warranted for building module-based prognostic models, and the models thereby obtained will be more biologically interpretable. Furthermore, the analysis on prognostic modules across tumor types provides a unique perspective to elucidate the common/distinct biological processes involved in different cancer types, which may facilitate novel subtype classifications.
Hubs are topologically central in the co-expression network, having maximal informational connections with other genes. Despite prognostic genes having higher connectivity overall, we found them to be depleted in the hub nodes of the cancer-specific co-expression networks. One possible explanation for this finding is that the hubs act more like modulators, coordinating gene expression over many functional components (pathways). As hubs are associated with a very high level of activity of receiving and sending signals, their expression levels are often complicated by too many factors to correlate with the phenotype directly. This observation is compatible with the results from a genomic analysis of the hierarchical structure of gene regulatory networks34. Another explanation is that hubs may have more backup and feedback mechanisms to ensure their robust behavior, so their status may not sensitively reflect the properties of the whole system. In contrast, prognostic genes are enriched in modules, especially in module genes that are conserved across tumor types. Modules in the co-expression network represent groups of functionally related genes dedicated to specific biological processes, which perform essential functions from baseline housekeeping to activities related to tumor growth and invasion. Compared with genes that mainly work alone, the status of a module gene more frequently reflects the “group” behavior, and is therefore more informative regarding tumor progression.
MicroRNAs are an important class of regulatory genes that largely lead to gene silencing through either mRNA degradation or translational inhibition30,31. Recently, miRNAs have been widely implicated in tumor biology35,36. The target genes of prognostic miRNAs show patterns similar to those of prognostic mRNA genes in terms of hub and module properties. In particular, we found that some prognostic miRNAs have their target genes enriched in prognostic modules. It is likely that these miRNAs act as a master regulator, coordinating the behavior of the whole module through their targeting. For example, in GBM, such modules involve functions related to “negative regulation of gliogenesis,” and “negative regulation of kinase activity,” and “nervous system development and functions,” all of which are connected with the normal function and development of the brain.
Compared with single prognostic genes, biological modules enriched with prognostic genes may represent more robust prognostic signatures, and deserve further investigation. Our integrative analysis reveals several such modules of particular interest. In GBM, the module of “negative regulation of gliogenesis” (Fig. 4a) show an enrichment of target genes of 3 prognostic miRNAs (miR-148a, miR-204 and miR-34a). In terms of the Ingenuity Pathway Analysis (IPA) disease annotation, “tumorigenesis of neuroendocrine carcinoma,” “recurrence of carcinoma” and “squamous-cell carcinoma” are the top three terms. Interestingly, the upstream transcription regulators of this module (ZNF217, ASCL1, DNAJB6, DEK and E2F5) are also the predicted targets of the three prognostic miRNAs. In BRCA, there are two prognostic modules regulated by prognostic miRNAs. One is annotated as “nucleic acid metabolic process” (Fig. 4a). This module is enriched with the target genes of several prognostic miRNAs (miR-30a, miR-324, miR-454, miR-340, miR-301a, miR-148b, miR-153, miR-181b, miR-9, miR-98 and let-7b) associated with IPA terms “cell death and survival, cell cycle, cancer” and the estrogen receptor signaling pathway. It also shows an enrichment of significantly mutated pan-cancer genes (KIAA1109, NCOA3, TAF1, EP300, SETD2, STAG2 and TAF1L). Thus, this module appears to integrate multiple types of aberration signatures and likely plays an important role in the progression of breast cancer. Another BRCA module associated with prognostic miRNAs is annotated as “chromatin remodeling” and “protein catabolic process” (Fig. 4a), suggesting a key role of epigenetic modification in breast cancer. This module is associated with the AMPK signaling pathway, a known oncogenic signaling pathway in breast cancer37,38. In KIRC, one module annotated as “nucleic acid metabolic process” (Fig. 4a) has connections with 5 prognostic miRNAs. In OV, the module of particular interest is annotated as “multicellular organismal development.” The top IPA disease terms include “endometrial cancer,” “uterine serous papillary cancer,” “epithelia neoplasia,” ”metastatic colorectal cancer,” and “solid tumor,” and the enrichment of “ovarian adenocarcinoma” is also significant (P < 1×10-5). This module is also enriched with significantly mutated pan-cancer genes (APC, NAV3, COL11A1, TSHZ3, CDH1, RUNX1, AR, SYNE1, MN1, DCHS1, PDGFRA, TGFBR2, CDKN1A, ABCA9 and TNFAIP6). Interestingly, this module shows correspondence to quite a few prognostic modules in other cancer types (Fig. 4b). Thus, we speculate that this module may reflect a theme that is common across many cancer types. Further investigation is required to evaluate the clinical utility of these modules, such as in patient prognosis stratification and tumor subtype classification.
While our study provides some insight into the emergent properties of prognostic genes, more efforts are required to validate and extend our findings. We examined the properties of prognostic genes in the context of gene co-expression networks. A logical extension of this work would be to determine whether the observed patterns hold true in other biological networks, such as protein interaction networks or gene regulatory networks. A major challenge for that direction is that those networks are usually highly biased toward well-studied genes. A second logical extension would be to determine whether the same patterns hold true in the co-expression networks of other tumor types or diseases. Our findings are based on TCGA data, so a third and critical extension of this work would be to learn whether the patterns can be recapitulated by other independent sample cohorts with adequate survival data. Lastly, future efforts should be made to incorporate this systems-level understanding of prognostic genes into the practice of building effective prognostic models.
Methods
Identification of prognostic mRNA and miRNA genes
We obtained TCGA gene expression and overall survival data from Firehose (https://confluence.broadinstitute.org/display/GDAC/Home) and TCGA Pan-Cancer website (http://pancancer.soe.ucsc.edu/). All data are from pan-cancer 4.0 Freeze version. We detected prognostic mRNA or miRNA genes based on the raw Wald P-values generated from the univariate Cox model. We used two methods to define prognostic genes. First, depending on the signal-to-noise ratio, different false discovery rates (FDRs) were applied: for mRNA genes, 1,706 genes in GBM (P < 0.028, FDR < 0.2), 728 genes in OV (P < 0.019, FDR < 0.5), 974 genes in BRCA (P < 0.0053, FDR < 0.1), and 2,050 genes in KIRC (P < 2.24×10-6, FDR < 2×10-5); for miRNA genes, 15 miRNAs in GBM (P < 0.0053 , FDR < 0.2), 106 miRNAs in OV (P < 0.026 , FDR < 0.2), 54 miRNAs in BRCA (P < 0.01, FDR < 0.1), and 203 miRNAs in KIRC (P < 0.043, FDR < 0.1). We assessed the robustness of these prognostic genes through randomly sampled subsets: based on 100 subsets (75% of the original sample size), on average, 75.3%, 75.0%, 92.0% and 99.9% of the prognostic mRNA genes showed significant survival correlations in GBM, OV, BRCA and KIRC, respectively (Supplementary Figure 1). Second, the top 1000 mRNAs or top 50 miRNAs most correlated with overall survival were identified as prognostic genes. When gene or miRNA expression data were available from more than one platform (Agilent 244K microarray and Illumina HiSeq RNA-seq), we compared the Wald P-value distributions between the platforms and chose the one with a better signal-to-noise ratio (Table 1 and Supplementary Figure 11). We did not include other TCGA cancer types in this study because they did not have either the microarray data for co-expression network construction or sufficient prognostic genes identified (due to a limited sample size or inadequate follow-up time).
Table 1. Summary of TCGA genomic datasets used in this study.
| Cancer type | Co-expression network construction | Identification of prognostic mRNAs* | Identification of prognostic miRNAs* |
|---|---|---|---|
| GBM | Agilent 244K microarray >500 samples | Agilent 244K microarray >500 samples | Agilent 8×15K miRNA microarray >480 samples |
| OV | Agilent 244K microarray >560 samples | Agilent 244K microarray >560 samples | Agilent 8×15K miRNA microarray >560 samples |
| BRCA | Agilent 244K microarray >520 samples | Illumina HiSeq RNA-Seq >830 samples | Illumina RNA-Seq >830 samples |
| KIRC | Agilent 244K microarray >70 samples | Illumina HiSeq RNA-Seq >460 samples | Illumina RNA-Seq >480 samples |
When mRNA gene or miRNA expression data were available from more than one platform, we chose the one with a better signal-to-noise ratio (Methods).
Co-expression network construction
Given Agilent 244K microarray data for each cancer type, we used the WGCNA package20,21 to build a weighted gene co-expression network that contains 17813 nodes (genes). The key parameter, β, for weighted network construction was optimized to maintain both the scale-free topology and sufficient node connectivity as recommended in the manual. In such a network, any two genes were connected and the edge weight was determined by the topology overlap measure (TOM) provided in WGCNA. This measure considered not only the expression correlation between two partner genes, but also how many “friends” the two genes shared. The weights ranged from 0 to 1, and reflect the strength of the communication between the two genes. Given a network, we then obtained several key network properties such as the edge weight, node connectivity and modularity. Connectivity was defined as the sum of the weights across all the edges of a node, and the top 1% (or 5%) of the genes with the highest connectivity in the network were defined as hub genes. According to the connectivity distributions, this definition well covered the highly connected nodes in the power-law tails (Supplementary Figure 12). We obtained the same results when the hub genes were defined based on the adjacency matrix,20,21 in which the edge weight between two gene nodes depended on only their co-expression correlation (Supplementary Figure 13). We identified the modules using the advanced dynamic tree cut technique, built with the default value of SplitDepth for robust module detection in WGCNA39. We obtained the same results when the modules were defined with the SplitDepth value for more sensitive module detection (Supplementary Figure 14). The co-expression network has been deposited in Synapse (syn1445557).
Hub and module analysis of prognostic mRNA genes
For each cancer type, we used a Fisher's exact test to examine the enrichment or depletion of prognostic mRNA or miRNA genes in hub nodes. To examine the cancer-type specificity of the results, we performed the same analysis using hub and prognostic genes from any two cancer types. We used a Fisher's exact test to evaluate the enrichment of mRNA prognostic genes in modules. To study the conservation of module genes, we defined a score (from 0 to 4) for each gene, with 0 indicating the gene was not a module gene in any of the four cancers, and 4 indicating the gene was a conserved module gene in all four cancer types. We used a Wilcoxon rank sum test with continuity correction to test whether the conservation score was different between prognostic genes and other genes. We considered P < 0.05 to be statistically significant.
Analysis of prognostic miRNAs and their target genes
We first annotated the miRNAs to the corresponding miRNA families and then obtained the predicted conserved targets from TargetScan (Release 6.2)32,40. For hub analysis, the target genes of all prognostic miRNAs in a cancer type were combined to test the relations with hub genes, as described above. To examine the robustness of the results, target genes with different stringency criteria were used: high-confidence targets were identified as being predicted targets of more than one prognostic miRNA (2∼4). For module analysis, we first identified target-module-enriched miRNAs based on whether the target genes of a given miRNA were significantly enriched in at least one module in the co-expression network (FDR < 0.1). We then used a Fisher's exact test to examine whether prognostic miRNAs tended to be target-module enriched in a given cancer type.
Integrative module analysis
We used a hypergeometric test to identify prognostic modules as those that are significantly enriched with prognostic mRNAs (FDR < 0.1). To assess the clinical relevance of these prognostic mRNA modules, for each cancer type, we classified tumor samples into subtypes (or clusters) based on the expression of module genes (including both prognostic and non-prognostic genes) using non-negative matrix factorization41, and then tested the correlations of sample clusters with patient survival using log-rank tests. Similarly, we identified prognostic modules enriched with the target genes of a specific prognostic miRNA (FDR < 0.1). We obtained 224 high-confidence significantly mutated genes from TCGA Pan-Cancer Project (syn1750331) and tested their module enrichment in the same way. We used R package “topGO”42 to annotate modules with GO terms and used IPA (Ingenuity® Systems, www.ingenuity.com) to annotate modules with IPA knowledge-based terms. To identify module correspondence across tumor types, we used a hypergeometric test to examine the pairwise correspondence between any two cross-tumor prognostic modules (FDR < 0.1). The results showed great agreement with the cross-tumor module conservation score provided in WGCNA.
Supplementary Material
Acknowledgments
We gratefully acknowledge contributions from the TCGA Research Network and its TCGA Pan-Cancer Analysis Working Group (contributing consortium members are listed in Supplementary Note 1). The TCGA Pan-Cancer Analysis Working Group is coordinated by J.M. Stuart, C. Sander and I. Shmulevich. This study was supported by the National Institutes of Health (CA143883 and CA016672 to H.L.); UTMDACC – G.S. Hogan Gastrointestinal Research Fund, NIH/NCI Uterine SPORE Career Development Award and the Lorraine Dell Program in Bioinformatics for Personalization of Cancer Medicine to H.L. We thank LeeAnn Chastain for editorial assistance.
Footnotes
Author Contributions: H.L. conceived of and supervised the project. Y. Yang, L.H., Y. Yuan, J.L. and N.H. performed data analysis. Y. Yang and H.L. wrote the manuscript with input from all other authors.
Conflict of interest: The authors declare that they have no conflict of interest.
References
- 1.Sotiriou C, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–72. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 2.Bullinger L, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. New England Journal of Medicine. 2004;350:1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 3.Spentzos D, et al. Gene expression signature with independent prognostic significance in epithelial ovarian cancer. J Clin Oncol. 2004;22:4700–10. doi: 10.1200/JCO.2004.04.070. [DOI] [PubMed] [Google Scholar]
- 4.Zhao H, et al. Gene expression profiling predicts survival in conventional renal cell carcinoma. PLoS Med. 2006;3:e13. doi: 10.1371/journal.pmed.0030013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adler AS, Chang HY. From description to causality: mechanisms of gene expression signatures in cancer. Cell Cycle. 2006;5:1148–51. doi: 10.4161/cc.5.11.2798. [DOI] [PubMed] [Google Scholar]
- 6.Abba MC, Lacunza E, Butti M, Aldaz CM. Breast cancer biomarker discovery in the functional genomic age: a systematic review of 42 gene expression signatures. Biomark Insights. 2010;5:103–18. doi: 10.4137/BMI.S5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buyse M, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98:1183–92. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 8.Chakravarti A, et al. The prognostic significance of phosphatidylinositol 3-kinase pathway activation in human gliomas. J Clin Oncol. 2004;22:1926–33. doi: 10.1200/JCO.2004.07.193. [DOI] [PubMed] [Google Scholar]
- 9.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. New England Journal of Medicine. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 10.Langer C, et al. Prognostic importance of MN1 transcript levels, and biologic insights from MN1-associated gene and microRNA expression signatures in cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. J Clin Oncol. 2009;27:3198–204. doi: 10.1200/JCO.2008.20.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oscier DG, et al. Multivariate analysis of prognostic factors in CLL: clinical stage, IGVH gene mutational status, and loss or mutation of the p53 gene are independent prognostic factors. Blood. 2002;100:1177–84. [PubMed] [Google Scholar]
- 12.Furlong LI. Human diseases through the lens of network biology. Trends in Genetics. 2013;29:150–159. doi: 10.1016/j.tig.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–13. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 14.Cai JJ, Borenstein E, Petrov DA. Broker Genes in Human Disease. Genome Biology and Evolution. 2010;2:815–825. doi: 10.1093/gbe/evq064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao W, et al. Weighted gene coexpression network analysis: state of the art. J Biopharm Stat. 2010;20:281–300. doi: 10.1080/10543400903572753. [DOI] [PubMed] [Google Scholar]
- 16.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The Cancer Genome Atlas Research Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The Cancer Genome Atlas Research Network. Integrative analysis of genomic and molecular alterations in clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 22.Goh KI, et al. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–90. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 24.Liang H, Li WH. Gene essentiality, gene duplicability and protein connectivity in human and mouse. Trends in Genetics. 2007;23:375–378. doi: 10.1016/j.tig.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein M. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput Biol. 2007;3:e59. doi: 10.1371/journal.pcbi.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu H, Greenbaum D, Xin Lu H, Zhu X, Gerstein M. Genomic analysis of essentiality within protein networks. Trends in Genetics. 2004;20:227–231. doi: 10.1016/j.tig.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 27.Sun J, Zhao Z. A comparative study of cancer proteins in the human protein-protein interaction network. BMC Genomics. 2010;11(Suppl 3):S5. doi: 10.1186/1471-2164-11-S3-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han JD, et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 29.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–55. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 30.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 32.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 33.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu H, Gerstein M. Genomic analysis of the hierarchical structure of regulatory networks. Proc Natl Acad Sci U S A. 2006;103:14724–31. doi: 10.1073/pnas.0508637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 36.Calin GA, Croce CM. MicroRNA-cancer connection: The beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 37.Hadad SM, et al. Histological evaluation of AMPK signalling in primary breast cancer. BMC Cancer. 2009;9:307. doi: 10.1186/1471-2407-9-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang JT, et al. Resveratrol induces apoptosis in chemoresistant cancer cells via modulation of AMPK signaling pathway. Ann N Y Acad Sci. 2007;1095:441–8. doi: 10.1196/annals.1397.047. [DOI] [PubMed] [Google Scholar]
- 39.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–20. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 40.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci USA. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–7. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.