Abstract
The kidney, urine, and plasma proteomes are intimately related: proteins and metabolic waste products are filtered from the plasma by the kidney and excreted via the urine, while kidney proteins may be secreted into the circulation or released into the urine. Shotgun proteomics datasets derived from human kidney, urine, and plasma samples were collated and processed using a uniform software pipeline, and relative protein abundances were estimated by spectral counting. The resulting PeptideAtlas builds yielded 4005, 2491, and 3553 nonredundant proteins at 1% FDR for the kidney, urine, and plasma proteomes, respectively—for kidney and plasma, the largest high-confidence protein sets to date. The same pipeline applied to all available human data yielded a 2013 Human PeptideAtlas build containing 12,644 nonredundant proteins and at least one peptide for each of ~14,000 Swiss-Prot entries, an increase over 2012 of ~7.5% of the predicted human proteome. We demonstrate that abundances are correlated between plasma and urine, examine the most abundant urine proteins not derived from either plasma or kidney, and consider the biomarker potential of proteins associated with renal decline. This analysis forms part of the Biology and Disease-driven Human Proteome Project (B/D-HPP) and a contribution to the Chromosome-centric Human Proteome Project (C-HPP) special issue.
Keywords: Human Proteome Project, PeptideAtlas, LC-MS/MS, database, kidney, plasma, urine, proteome comparison
INTRODUCTION
Shotgun proteomics via tandem mass spectrometry (LC-MS/MS) is the most widely used workflow for detecting proteins and measuring their abundances in biological samples. PeptideAtlas1, 2 has become an important resource for defining the MS-detectable human proteome3 by collecting and reprocessing a large number of publicly available shotgun proteomics datasets. As a crucial component of the Human Proteome Project4–6, PeptideAtlas is defining proteomes of important human tissues and biofluids. The Human Plasma PeptideAtlas has been evolving for many years7–9, and recently several tissue/fluid-specific builds have been created.
Concurrently, the Human Proteome Organization (HUPO) has organized several initiatives to study in depth many human tissue/biofluid-based proteomes, among them the Human Kidney & Urine Proteome Project (HKUPP, www.hkupp.org) and the Human Plasma Proteome Project (HPPP, www.peptideatlas.org/hupo/hppp). The kidney, urine, and plasma proteomes are intimately related: proteins and metabolic waste products are filtered from the plasma by the kidney and excreted via the urine. Further, some kidney proteins pass directly into the urine, others are secreted or released into the extracellular fluid and the circulation, eventually making their way to plasma, and still others remain expressed only in the kidney. For the discovery of urine biomarkers for kidney diseases, knowledge of plasma and kidney proteomes is important because in most kidney diseases plasma proteins larger than 40–60 kDa pass through the impaired glomerular filter and appear in urine. With current methods, typically only albumin or total protein (or immunoglobulin chains) is assayed in urine.
Last year Cui, et al. published an analysis of the kidney glomerulus proteome10, represented by a non-redundant dataset of 1817 proteins produced via a stringent re-analysis of an earlier shotgun proteomics experiment11. These two publications comprise the only comprehensive proteomics survey of human kidney.12
In contrast, surveys of both urine and plasma have been performed by many laboratories over the years due to the perceived value of these body fluids as non-invasive specimens for biomarkers. A comprehensive survey of the urine proteome was completed by the Mann laboratory in 200613, identifying 1543 proteins. In 2009, the Steen laboratory reported 2362 proteins at <1% FDR14, and in 2011 the Pandey laboratory15 reported a list of 1823 proteins at <1% FDR. In 2010, Li, et al. identified 1310 proteins and added a focus of phosphoproteins and phosphorylation sites16. In 2011, Nagaraj and Mann investigated what can be done in a high-throughput, unfractionated manner17. By studying 7 individuals over 3 days they identified a total of 808 proteins, 587 of which were found in all analyses and that they called the “core urinary proteome”. In 2012, Zerefos, et al.18 reported 558 proteins, with emphasis on experimental estimation of molecular weight and recognition of isoforms. Urine, of course, contains many non-protein molecules; a survey of the human urine metabolome was recently completed19 that mined the literature for 445 compounds detected by a variety of methods including MS. (The same group had similarly compiled a list of 4229 serum compounds20.) Multiple web resources exist to support the study of the molecular composition of urine, including urineproteomics.org (Kentsis and Steen), the Urine Proteomic Website (UroProt, www3.niddk.nih.gov/intramural/uroprot) focused on urinary biomarkers, and The Kidney and Urinary Pathway Knowledge Base (KUPKB, www.kupkb.org) at the University of Manchester, implementing a multi-omics approach to biomarker discovery and pathway modeling using Semantic Web technologies21.
For human plasma, the most prominent surveys have been those by the HUPO PPP22 (Omenn et al, Proteomics 2005; States et al, Nature Biotech 2006), yielding a set of 3020 protein identifiers, condensed to a nonredundant, high-confidence list of 889 after a more sophisticated bioinformatics analysis23 including Bonferroni-type adjustment for multiple comparisons; by the Mann lab in 200824, with a high confidence list, excluding immunoglobulins, of 697 proteins; and by the PeptideAtlas team, combining 91 experiments from various labs to produce a high confidence list of 1929 proteins8. See Table 1 for a summary of all of the above studies.
Table 1.
Published surveys of urine, kidney, and plasma proteomes.
| reference | lab | sample & methods | proteins ID’d |
confidence & redundancy | in current work |
|---|---|---|---|---|---|
| Kidney | |||||
|
Miyamoto, et al., JPR 200711 |
Yamamoto | Glomerulus only. | 6686 | Peptide FDR per sample 1.9%. High redundancy. |
* |
|
Cui, et al.,Proteome Science 201310 |
Yamamoto | Re-analysis of Miyamoto, et al. |
1817 | Peptide FDR < 1%. Low redundancy. |
|
|
KidneyPA 2013 (current work) |
PeptideAtlas | Meta-analysis of 13 experiments |
4005 | Protein FDR 1%. Low redundancy. |
|
| Urine | |||||
|
Pisitkun, et al., PNAS 200428 |
Knepper | Urinary exosomes | 295 | unspecified | |
|
Adachi, et al. Genome Biology 200613 |
Mann | 10 healthy individuals, 9 pooled |
1543 | 99% significance level for one peptide, 95% for second. Stringent criteria for single peptide ID |
|
|
Gonzales, et al., J Am Soc Nephrology 200929 |
Knepper | Urinary exosomes | 1132a | Peptide FDR 1.9%. Only retain proteins with MW in range expected by 1D SDS-PAGE. |
|
|
Kentsis, et al., Proteomics Clin Applications 200914 |
Steen | 12 individuals | 2362 | Protein FDR < 1%. Mascot score threshold. No single hit identifications. |
* |
|
Li, et al., Rapid Commun. Mass Spectrom. 201016 |
Zeng | Non- prefractionation; focus on identifying phosphorylation sites |
1310 | Protein FDR < 1%. Low redundancy. |
|
|
Marimuthu, et al., JPR 201115 |
Pandey | 24 healthy individuals, pooled. Lectin-enriched and non-enriched. |
1823 | Protein FDR 1%. Single peptide hits included only if they mapped uniquely to a protein. |
* |
|
Nagaraj & Mann, JPR 201117 |
Mann | 5 male, 2 female, high-throughput single run assays, 3 days each. |
808b | Protein FDR 1%. ≥ 2 peptides per identification. Requires one peptide unique to protein group. |
|
|
Zerefos, et al., Proteomics 201218 |
Vlahou | "standard" EuroKUP COST Action sample. Multiple separation technologies. |
553 | 95% confidence, ≥ 2 peptides per identification. |
|
|
UrinePA 2013 (current work) |
PeptideAtlas | Meta-analysis of 15 experiments |
2491 | Protein FDR < 1%. Low redundancy. |
|
| Plasma | |||||
|
Anderson & Anderson, MCP 200230 |
Anderson | variety of methods | 289 | Various | |
|
Chan, et al., Clinical Proteomics 200431 |
Conrads | Pooled standard human serum |
1444 | SEQUEST DelCN <= 0.08 and Xcorr ≥ threshold dependent on charge, cleavage |
|
|
Omenn, et al., Proteomics 200522 |
HUPO | 18 laboratories analyzed a common set of samples |
3020 | ≥ 2 peptides per identification. High redundancy. |
* |
|
States, et al., Nat Biotechnol. 200623 |
Re-analysis of HUPO 3020 proteins |
889 | High confidence. Low redundancy. |
||
|
Schenk, et al., BMC Med Genomics 200824 |
Mann | Single pooled sample | 697c | High confidence. Low redundancy. |
|
|
Farrah, et al. MCP 20118 |
PeptideAtlas | Meta-analysis of 91 experiments |
1928 | Protein FDR 1%. Low redundancy. |
* |
|
PlasmaPA 2013 (current work) |
PeptideAtlas | Meta-analysis of 127 experiments |
3553 | Protein FDR 1%. Low redundancy. |
Unambiguous identifications
587 in all individuals, all days = "core urinary proteome"
Excludes immunoglobulins
In 2009, Jia and co-workers25 compared the urine protein list of the Mann lab with the HUPO PPP plasma protein list22, 23 to learn about kidney protein processing. Proteins known to be secreted by the prostate were subtracted from both lists. Three biofluid-based proteomes were subjected to Gene Ontology and molecular weight analyses: urine-only, urine and plasma, plasma-only. Cui, et al., in their 2013 glomerulus study, compared the kidney glomerulus proteome with previously published urine and plasma protein lists10. This comparison suggested the extent and characteristics of proteins present in kidney via plasma contamination and of those excreted into urine.
Here, we extend the previous comparative proteomics studies in several ways. First, we include not only glomerulus data but data from other parts of the kidney, and include urine and plasma datasets collected from many different laboratories for more complete tissue/biofluid-based proteome coverage; each resulting PeptideAtlas build has as many or more high confidence protein identifications than any report published to date for that tissue/biofluid-based proteome. Second, we use a standardized bioinformatics pipeline for all collated MS/MS data based on the Trans-Proteomic Pipeline26, 27 and at the core of the PeptideAtlas build process8. Third, for each tissue/biofluid-based proteome we estimate relative protein abundances using spectral counting, and normalize the abundances for comparison between proteomes.
Finally, we implement a bioinformatics tool to perform pair-wise and multi-way protein identification and abundance comparisons and perform Gene Ontology analysis on the results. We apply the tool to kidney, urine, and plasma, and provide the results in an online resource for examining proteome commonalities and differences, accessible at www.peptideatlas.org/hupo/hkup. We complete our report by using this resource to explore several biological questions.
EXPERIMENTAL PROCEDURES
Datasets from diverse experiments were collected for human kidney, urine, and plasma. See Table 2 and Supporting Information Table S1 for details. Thirteen kidney experiments on normal samples from cancerous nephrectomy were provided by T. Yamamoto; two from glomerulus were previously published10, 11. Fifteen urine experiments on samples from normal individuals were provided by five investigators. Finally, 127 experiments on primarily normal plasma samples were provided by many investigators; 69 of these had been included in the 2010 Human Plasma PeptideAtlas8. The 22 glycocapture enrichment experiments used in the previous study were excluded from the current study in order to obtain more accurate relative abundance estimations. For those interested in glycoproteins and glycopeptides, please refer to the previous study8.
Table 2.
Sample information for the Human Kidney, Urine, and Plasma PeptideAtlas builds used in the present study. In row Experiments, references are provided for data previously published (in whole or in part). See Supporting Information Table S1 for further detail
| Kidney | Urine | Plasma | |
|---|---|---|---|
| Laboratories | T. Yamamoto, Niigata U. | T. Yamamoto, Niigata U. Y.A. Goo, U. Washington A. Pandey, Johns Hopkins H. Steen, Boston Children's W. Qian, PNNL |
Hoffmann-La Roche S. Hanash, MD Anderson M. Snyder, Stanford D. Smith, PNNL HUPO PPP-I and many others |
| Samples | Normal samples from cancerous nephrectomy |
Samples from normal volunteers |
Primarily normal, plus 5 from graft-vs.-host disease and 10 from other disease samples. |
| Experiments | 9 glomerulus11 1 renal cortex 1 renal medulla 2 combined glomeruli, collecting ducts, distal tubules, and proximal tubules |
8 Yamamoto 4 Goo 1 Pandey15 1 Steen14 1 Qian32 |
127 various8 |
A PeptideAtlas build was constructed for each of the three sample types following a workflow described previously8. Briefly, most datasets were searched twice: (1) with X!Tandem33 + k-score34 against a target-decoy sequence database consisting of an Extended Complete Proteome (UniProt Complete Proteome (Swiss-Prot plus Trembl) release 2012_10 including Swiss-Prot varsplic entries and with appended peptides representing SNPs and other Swiss-Prot annotated variants, manuscript in preparation), about 500 sequences from the International Protein Index database35 (IPI version 3.71) that contain peptides putatively seen in previous PeptideAtlas builds yet not presently found in the Extended Complete Proteome, and cRAP common contaminants (www.thegpm.org/crap), plus one decoy sequence for each target entry, and (2) with SpectraST36 against NIST human ion trap spectral library v.05-30-2012 (http://peptide.nist.gov) with one decoy spectrum added for each library spectrum, or against a custom target/decoy spectrum library to accommodate special modifications. A few very large plasma datasets had been previously searched against IPI 3.71 with X!Tandem + K-score and also against NIST 2.0 with SpectraST; these previous search results were used in the present study because there was not enough compute time to search against the same database as the others. X!Tandem search parameters were set to detect N-terminal acetylation and pyroglutamic acid, plus any additional modifications expected according to the method of sample preparation for each specific sample. SpectraST searches detected the modifications included in the spectral library searched; for the NIST library this is primarily carbamidomethylation and oxidation. See Supporting Information, “Search parameters and modifications”, for further detail. Results were processed using the Trans-Proteomic Pipeline26. Identified peptides were mapped to a reference protein sequence database that included Extended Complete Proteome, the complete IPI v3.71, Ensembl v67.3737, cRAP, and all searched decoys. Redundancy was then removed from the resulting list of protein sequence identifiers as described previously8, except that Swiss-Prot identifiers were preferred when selecting among multiple similar sequences for canonical, NTT-subsumed, covering set, and protein group representative, superseding other criteria (protein probability, PSM count, number of distinct peptides, number of enzymatic termini). For each atlas build, this process was attempted with various PSM FDR filter thresholds until a threshold was found that produced a final list of canonical (nonredundant) proteins with a Mayu 38 decoy-estimated FDR between 0.008 and 0.015. For builds where this PSM FDR threshold admitted PSMs of probability < 0.9, a PSM probability threshold of ≥0.9 was applied to exclude low-probability PSMs; in these cases the final protein FDR was less than 0.008. The distinct peptides corresponding to the PSMs passing threshold comprise the final peptide list for the atlas build, and Mayu was used to estimate the PSM and peptide FDR as well. Because the total distinct peptide content of both the search database and the reference database is nearly equal, and the ProteinProphet algorithm39 of the Trans-Proteomic Pipeline reduces the data to nearly the same protein group count with both databases, the protein-level FDR can be accurately estimated after mapping to the reference database. Please refer to our 2011 Human Plasma PeptideAtlas publication8 for greater detail on the PeptideAtlas build process.
Additionally, using this same procedure, we created a fourth PeptideAtlas build containing nearly all the publicly released human data available to us, including much of the urine and all of the plasma data described above (the kidney data, and the Pandey, Steen, and Qian urine data, were not included due to time constraints but will be included in the next release) plus data from many other sample types totaling 515 experiments. This build is an extension of, and includes all the data in, the human build we described last year.3
So that we could conduct our comparative study using only the concise Swiss-Prot database, we then created two sets of Swiss-Prot identifiers for each atlas: complete mapping and nonredundant. The complete mapping included all Swiss-Prot identifiers containing any peptide in the final list of peptide identifications. Each PeptideAtlas build, following the Cedar protein inference method8, already contains a nonredundant set of protein identifiers called the canonical set. However, because this set contains some non-Swiss-Prot identifiers, and we wanted in the present study to consider only Swiss-Prot identifiers, we created a Swiss-Prot-only nonredundant set by starting with the complete mapping and removing (a) all identifiers that were subsumed by another Swiss-Prot identifier (i.e. whose peptides formed a proper subset of the peptides for another Swiss-Prot identifier), and (b) for each set of Swiss-Prot identifiers subsumed by the same non-Swiss-Prot identifier, all but the one with the most distinct peptides. This results in a set of Swiss-Prot identifiers nearly all of which contain peptide evidence to distinguish them from all others in the set. Like the PeptideAtlas canonical set, the Swiss-Prot non-redundant set is not to be considered a list of definitively identified proteins, but rather a parsimonious set of Swiss-Prot identifiers that explains all the peptide evidence.
When comparing atlas builds, we face the problem of how to decide which identifiers are shared in common between two builds. This is a problem throughout the field of proteomics, where multiple versions of multiple sequence databases make it very challenging to compare protein lists resulting from diverse experiments with diverse search and protein inference protocols. We take care of a large portion of this problem by applying a uniform bioinformatics pipeline to all three tissue/biofluid-based proteomes, resulting in protein lists from the same version of the same database (Swiss-Prot October 16, 2012). However, there is still the issue of peptides mapping to multiple sequences. When two nonredundant protein lists are compared, they may seem to have few proteins in common when they do in fact share identified peptides mapping to the same protein (see Supporting Information, “Finding commonalities between two proteomics protein sets,” for illustration). For this reason, we use the nonredundant set for the first proteome of any comparison and the complete mappings for the other(s).
The human proteins in the Global Proteome Machine Database (GPMDB)40, another repository of diverse proteomics datasets reprocessed through a uniform bioinformatics pipeline, were mapped to Swiss-Prot to facilitate comparison against PeptideAtlas. Protein identifiers were taken from the October 2013 GPMDB Guide to the Human Proteome (http://www.thegpm.org/lists/index.html#201008121), a complete mapping of peptides identified in GPMDB’s human datasets against the Ensembl37 database. The 69943 identifiers with Evidence Code = 4 (highest confidence) were submitted to PICR41 for mapping against Swiss-Prot. 35821 of these were found to map identically to 14841 distinct Swiss-Prot entries; these constitute a GPMDB EC=4 Swiss-Prot complete mapping.
A normalized spectral count (NSC) was computed for each Swiss-Prot identifier in each atlas according to the following formula, a simplification of the APEX method described by Lu and coworkers42:
NSCib: normalized spectral count for protein i in atlas build b
100,000=scaling factor to make NSC values fall into a convenient range of about 10−4 to 104 and to scale the numbers to a common size for a single dataset that identifies 100,000 PSMs with high confidence
n′ib: PSM count for protein i in atlas build b, adjusted for number of observable tryptic peptides in protein
Nb: total PSMs in atlas build b
nib: number of PSMs for peptides mapping to protein i in atlas build b
pi: total observable tryptic peptides in protein i
25: mean potential tryptic peptides per protein across human proteome (rough estimate)
Each Swiss-Prot entry was assigned the maximum NSC value for all splice variants observed (including the canonical form). NSC is a measure of the relative abundance of a protein within a (sub)proteome. It is an estimated answer to the question, “for every 100,000 observed protein molecules in the sample, how many are protein X?” For low-redundancy protein identification lists (the PeptideAtlas canonical lists and the nonredundant Swiss-Prot lists), the sum of the NSC values will approximate 100,000.
To gain insight into the relationships among the tissue/biofluid-based proteomes, 34 identifier sets were then compiled using NSC comparisons and set operations between sets of identifiers. See Table S4 in Supporting Information.
To determine which Gene Ontology (GO) terms are enriched among various sets of proteins, we employed the GOstats package43 (Bioconductor) running under the R statistical software. UniProt accessions (Swiss-Prot is a subset of UniProt) were mapped to Entrez gene IDs, and then the map was reversed and multiple mappings were resolved using the org.Hs.eg.db annotation package. 1612 (8%) of the Swiss-Prot IDs were missing from the map and thus were not included in this analysis. The analysis hyperGTest was run on each protein set with a P-value cutoff of 0.05 and parameters conditional=TRUE and testDirection=rep for all three GO ontologies. Enrichment for each protein set was measured by comparison against a custom universe as listed in Table S4 (Supporting Information). For each protein set, the (at most) 12 terms with the lowest P-values were output. See Figure 1 for a summary of the complete software pipeline.
Figure 1.
Software analysis pipeline used. See Experimental Procedures for details.
RESULTS AND DISCUSSION
By combining LC-MS/MS data from diverse laboratories worldwide, we have created PeptideAtlas builds containing high confidence peptide and protein identifications for each of three important human tissue/biofluid-based proteomes: kidney, urine, and plasma. These will be henceforth referred to as KidneyPA, UrinePA, and PlasmaPA. As seen in Table 3, for kidney and plasma we have approximately doubled the number of high-confidence, nonredundant protein identifications reported in any previous publication, while for urine we have approximately equaled the previous number. Protein identifier lists are provided in Supporting Information Table S2.
Table 3.
Tallies for PeptideAtlas builds for the three tissue/biofluid-basedsubprotomes, plus a build containing all available human data. The HumanAllPA numbers are an update over the values presented last year in the C-HPP special issue5 and are broken down by chromosome in Hancock, 2014 (this issue). The total of 14,133 identifiers in the Swiss-Prot complete mapping differs slightly from the total given for the neXtProt complete mapping in Marko-Varga, et al., 2014 (this issue) because the two databases, while containing nearly the same protein sequences, are not identical. (Each neXtProt release synchronizes with the most recent Swiss-Prot release and contains the same identifiers excluding 132 entries for immunoglobulin and T-cell receptor variable regions and the Ig mu heavy chain disease proteins (private communication with neXtProt); the Marko-Varga, et al. report in this issue uses a neXtProt release several months newer than the Swiss-Prot release used here and so contains further differences beyond the 132.)
| KidneyPA | UrinePA | PlasmaPA | HumanAllPA | |
|---|---|---|---|---|
| PSMs | 9.4 × 105 | 4.3 × 105 | 3.1 × 107 | 6.1 × 107 |
| Distinct peptides | 3.9 × 104 | 2.4 × 104 | 5.2 × 104 | 3.3×105 |
| Nonredundant proteins (PeptideAtlas canonical) |
4005 | 2491 | 3553 | 12,644 |
| Swiss-Prot | 3782 (94%) | 2325 (93%) | 3228 (91%) | 11,481 (91%) |
| Other | 223 | 166 | 325 | 1163 |
| PSM filter threshold | P<=0.9 | P<=0.9 | FDR=0.00005 | FDR=0.0002 |
| Decoy-estimated protein FDR |
0.2% | 0.08% | 1.5% | 1.1% |
| Swiss-Prot entries with independent peptide evidence (nonredundant) |
4287 | 2598 | 3677 | 12,934 |
| Swiss-Prot complete mapping |
5115 | 3175 | 4098 | 14,132 |
| Dynamic range measured | 5 × 105 | 5 × 105 | 8 × 107 | N/A |
| Largest NSC | 4647 | 7046 | 7646 | N/A |
| Smallest NSC | 0.010 | 0.013 | 0.00009 | N/A |
| Maximum non-redundant proteins identified in previous studies at FDR <= 1% |
181710 | 236214, 182315 |
19298 | not evaluated |
Additionally, we constructed an extension of the Human PeptideAtlas build we reported last year3 by adding more data. This new build, HumanAllPA, incorporates all of the data in KidneyPA and PlasmaPA, most of the data in UrinePA, plus data from many other diverse sample types—515 experiments in all. In addition to the 52 sample types listed in Figure 2 of last year’s report 3, we included breast cancer and colorectal cancer data, both of which yielded many thousands of protein identifications, plus four cell line sample types: LAPC4, hESC-NSC, HCT 116, and SW480+SW620, the latter two of which are both colorectal cancer cell lines. Of the 338,013 distinct peptides identified, 7.6% of them were found by SpectraST but not by X!Tandem, illustrating the utility of combining spectral library searching with sequence database searching44. The 2012 Human PeptideAtlas build contained peptides mapping to 12,629 Swiss-Prot entries, 11,868 of them with unique peptide evidence. This year, these numbers increase by about 1500 and 1000 respectively, or 7.5% and 5% of the predicted human proteome. About 1% of the identified peptides did not map to Swiss-Prot and are being investigated for possible inclusion therein. The Swiss-Prot complete mapping for HumanAllPA is provided in Supporting Information Table S3.
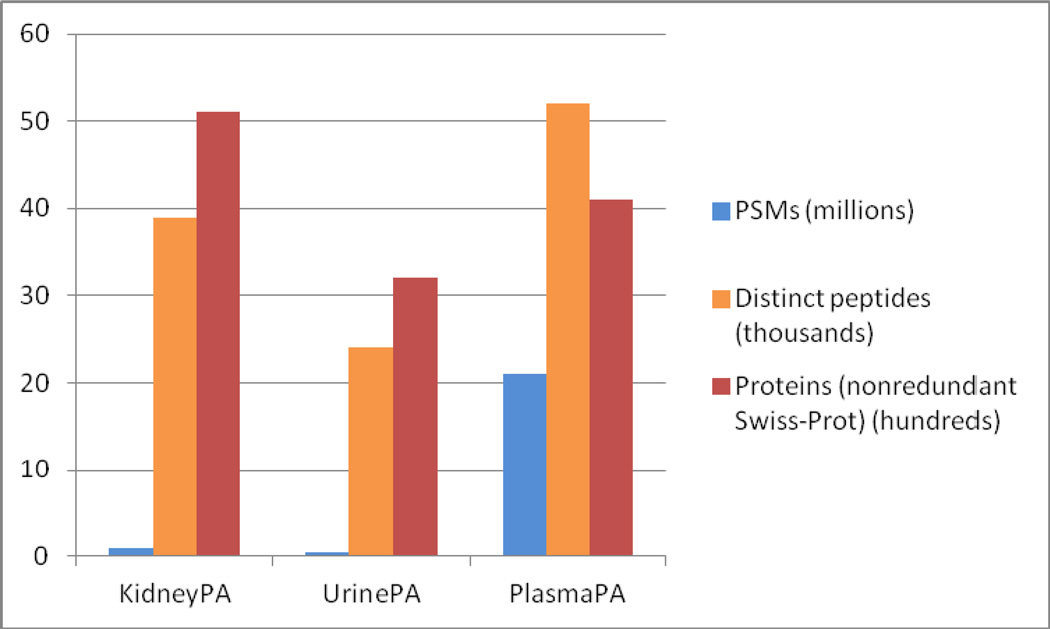
Figure 2.
PSM, peptide, and protein counts for each of the three tissue/biofluid-based proteome PeptideAtlas builds.
The Global Proteome Machine Database (GPMDB)40 is another repository of diverse proteomics datasets reprocessed through a uniform bioinformatics pipeline. Whereas PeptideAtlas contains only those peptides that support a nonredundant protein identification list of 1% FDR, GPMDB contains all peptide and protein identifications output by its pipeline, computing for each peptide a confidence (log(e)) value, and assigning to each protein identification the highest confidence value of all peptides mapped to that protein. Protein identifications are then assigned an evidence code (EC) of 1 (black), 2 (red), 3 (yellow), and 4 (green), with 4 being the highest confidence. We computed a Swiss-Prot complete mapping for the EC=4 human proteins in GPMDB (see Experimental Procedures). Of the 12,934 nonredundant Swiss-Prot entries in 2013 HumanAllPA, only 5% are missing from the GPMDB EC=4 Swiss-Prot complete mapping. Because GPMDB does not attempt to remove redundancy from their protein lists, we cannot compare a GPMDB nonredundant list against the PeptideAtlas complete mapping. However, only 12% of the 14,841 identifiers in the GPMDB EC=4 Swiss-Prot complete mapping are missing from HumanAllPA Swiss-Prot complete mapping. Thus, the sets of proteins observed in the two repositories are highly overlapping. Combined, their Swiss-Prot complete mappings cover 15,912 identifiers, or about 79% of the predicted human proteome as defined by Swiss-Prot, leaving 21% with no reliable peptide identifications in either repository.
As seen in Figure 2, the numbers of PSMs, distinct identified peptides, and nonredundant Swiss-Prot identifiers for each of the kidney, urine, and plasma atlases vary widely. Because of the large amount of plasma data collected over the past decade, beginning with the inception of the Human Plasma Proteome Project (HPPP)22 and subsequently from many other sources, PlasmaPA has by far the most PSMs. It also has the most identified peptides. KidneyPA has the second most PSMs, but leads with the largest number of proteins identified, presumably because it is derived from tissue samples containing cellular-level concentrations of proteins common to all cells. UrinePA has the fewest PSMs because it was constructed from the smallest amount of data. It also has the fewest identified peptides and proteins, partly because it has the fewest PSMs, and partly because the protein diversity at higher concentrations is simply smaller for urine.
The nonredundant Swiss-Prot list for KidneyPA included 4287 identifiers (73 of them immunoglobulin chains) with high enrichment (P-value < 1e-10) of many terms having to do with fundamental cellular processes (nucleic acid metabolic processes, translational elongation, small molecule catabolic process) relative to the entire set of proteins identified in any of the three sample types (see http://www.peptideatlas.org/hupo/hkup). Viral process, viral infectious cycle, and viral transcription are also enriched. In KidneyPA, 113 proteins were identified as transporters, exchangers, carriers or their related proteins, which mediate reabsorption or excretion of various molecules from or into urine, and 45 were mitochondria-related proteins. Proteomics, at least without special enrichment for membrane-embedded proteins, misses many proteins whose transcripts are highly expressed in kidney.
Of the 5115 Swiss-Prot identifiers in the KidneyPA complete mapping, 4880 (95%) were found in glomerulus data, either that of Miyamoto, et al.11 (reanalyzed by Cui, et al10) or later experiments on samples containing glomerulus only. This includes nearly all (99%) of the 1427 Cui, et al.10 proteins that could be mapped to Swiss-Prot (out of a total of 1817 nonredundant IPI identifiers appearing in 1478 genes). An additional 1206 Swiss-Prot identifiers in the KidneyPA Swiss-Prot complete mapping were identified from the Miyamoto data beyond what was identified by Cui, et al.; this can be attributed at least partly to the redundancy in the complete mapping and to the application of multiple search engines44. It is also possible that their protein FDR (not reported explicitly) is lower than ours.
The nonredundant Swiss-Prot list for UrinePA includes 2598 identifiers, 107 of them annotated in Swiss-Prot as immunoglobulin chains. The complete mapping (3175 identifiers) includes at least one identifier in each of 528 (90%) of the 587 protein groups in the “core urinary proteome” reported by Nagaraj and Mann17 as detected in seven individuals over three consecutive days. Seventeen of the groups we missed did not include a current Swiss-Prot identifier and thus could not be matched to our list, leaving only 42 Swiss-Prot “core urinary proteome” identifiers missing from UrinePA (see Supporting Information Table S5). Of the 1543 urine proteins found by Adachi, et al. in 200613, the 843 that were listed with Swiss-Prot identifiers mapped to 697 distinct Swiss-Prot entries, 614 (88%) of which are in UrinePA.
A GO analysis of UrinePA shows moderate enrichment (P-value < 1e-5) of several terms (see http://www.peptideatlas.org/hupo/hkup). In the molecular function ontology, enriched terms include receptor activity, extracellular matrix structural constituent, serine-type endopeptidase inhibitor activity, and child terms to binding and catalytic activity. This reflects intensive enzymatic digestion of many complexed molecules engulfed in lysosomes of the tubular cells. In the biological process ontology, enriched terms are associated with wound healing, immune response, protein activation, cell motility, blood vessel development, and negative regulation of endopeptidase activity, platelet degranulation, and cellular iron ion homeostasis. In the cellular component category, enriched terms include those relating to the extracellular region, the plasma membrane, the nucleosome, melanosome, and lysosome. Serum albumin is the protein with the second highest NSC in urine (3087, second to Ig kappa chain C region at 7479) at molecular weight 67 kDa. The kidney glomeruli allow plasma proteins to pass to the glomerular filtrate; the cutoff is thought to be around 40–60 kDa45. Only less than 1% of serum albumin is estimated to leak in the glomerular filtrate and most of serum albumin is reabsorbed at proximal tubules in the kidney. However, serum albumin is still a predominant protein in urine of healthy volunteers when examined by gel electrophoresis.
The average calculated MW of all identifiers observed in urine is 62 kDa, just a bit lower than the average of 68 kDa for all identifiers seen in any KUP atlas, and 476 (28%) of observed urine identifiers exceed 60 kDa compared to 35% in all of Swiss-Prot and 37% of all identifiers seen in any KUP atlas. In our study we use the MW reported by Swiss-Prot in the SQ line, which is simply the sum of the molecular weights of its amino acid residues. However, this can be a gross misestimate of the true molecular weight of the protein or protein fragment that is actually present in the sample. Many proteins undergo post-translational modifications; they can be glycosylated, phosphorylated, or have numerous other molecules covalently attached to the amino acid side chains, increasing their molecular weight. Proteins from a single gene may have sequence differences due to nonsynonymous single nucleotide polymorphisms or ORF variants at the DNA level, alternative splicing at the heterogeneous nuclear RNA level, and RNA editing before translation. Further, a protein can be proteolytically cleaved to generate functional fragments, or can also be cleaved into smaller fragments—after cell death, for example— and multiple fragments detected, giving the appearance that the whole protein has been detected. Comparing the abundance of peptides from different regions of the protein sequence can provide clues to presence of the cleavage. Cui, et al.10 in their Figure 2 showed that the calculated MW of most glomerulus proteins is significantly different from the experimental (actual) MW based on electrophoretic gel mobility, and that except for large (>75–95 kDa) proteins the calculated MW is usually an overestimate. N-terminal cleavage involving about 25 amino acids occurs routinely in proteins with a secretion signal and many other variant N-termini are generated46.
Finally, the peptides identified in plasma mapped to 3553 nonredundant Swiss-Prot protein identifications (117 of them immunoglobulin chains)—by far the largest nonredundant list of confidently identified plasma proteins to date. [See Experimental Procedures for the expansion of the PlasmaPA since our previous report8.] Highly enriched (P-value < 10−7) terms in the biological process ontology (see http://www.peptideatlas.org/hupo/hkup) include humoral immune response, endocrine pancreas development, platelet degranulation, translational elongation and termination, cellular component disassembly (and child terms macromolecular complex disassembly, protein activation cascade, and lymphocyte mediated immunity). The most highly enriched term (1.6e-85) is the cellular compartment term extracellular region – not surprising given that plasma is a collector of proteins that have been secreted by or have escaped from cells.
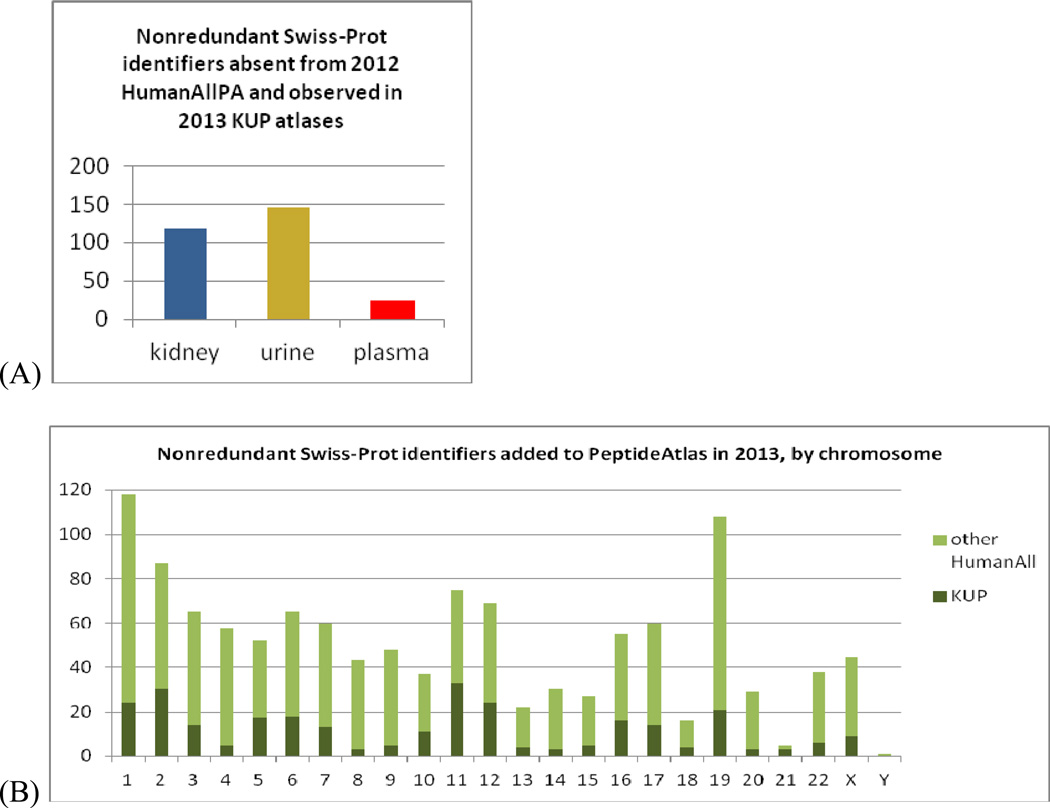
Among the three nonredundant HKUP Swiss-Prot identifier lists, 289 identifiers are found that were counted as “unseen” or “missing”5 (had no identified peptides) in our JPR 2013 report3 (Figure 3). The largest number of these is from urine. A total of 1216 identifiers from the nonredundant Swiss-Prot list for HumanAllPA had likewise been counted as “unseen” in 2012, representing about 6% of the estimated human proteome. A disproportionate number of these are from chromosome 19 (figure 3B); a Gene Ontology analysis shows these to be enriched in the P-value range 10−5 – 10−10 in terms related to biological regulation: DNA-dependent regulation of transcription, regulation of macromolecule biosynthetic process, regulation of cellular biosynthetic process, and regulation of nucleobase-containing compound metabolic process; also in DNA binding and metal ion binding (in particular, zinc ion binding). Also enriched among newly-detected chromosome 19 identifiers is the term RNA biosynthetic process (P-value 10−8).
Figure 3.
Nonredundant Swiss-Prot identifiers that were counted as “unseen” or “missing”5 (had no identified peptides) in our JPR 2013 report3. (A) From each of the three KUP atlas builds. Note that some are seen in multiple KUP atlas builds. (B) From KUP, HumanAllPA, by chromosome.
Comparability of result sets
Before comparing the proteins seen in each of these tissue/biofluid PeptideAtlas builds, let us consider the criteria that proteomics result sets ideally should possess in order for such comparisons to be meaningful.
Sample sources: Experiments should be conducted on samples collected from the same individual or pool of individuals. Ideally, all samples collected from an individual are collected at the same time.
Sample handling and LC-MS/MS technology: Experiments should be conducted using the same sample preparation (enzymatic digestion, protein extraction) and LC-MS/MS technology, ideally in the same laboratory.
Depth: Experiments should be conducted to comparable depth (comparable numbers of LC-MS/MS runs; comparable technical sophistication), or comparison informatics must account for variations in depth.
Search libraries/databases: Data should be searched against the same libraries and/or databases so that, in each case, the universe of possible peptide identifications is the same.
Search algorithms: Data should be searched using the same search algorithms. Different search algorithms will identify different peptides44. However, the biases do not appear to lead to inclusion or exclusion of specific proteins or classes of proteins, so this criterion is not so important.
Modifications searched: Searches for amino acid modifications should be conducted in a consistent manner. Otherwise, bias with regard to particular proteins may result. For example, if N-terminal acetylation is searched for in one data collection but not another, proteins with N-terminal acetylation will have higher likelihood of being identified in the former vs. the latter.
Mapping to proteins: Most importantly, results must be mapped to the same protein sequence databases using the same sequence identifiers.
Protein inference: The same protein inference method (i.e. method for removing redundancy from the list of proteins with peptide evidence) should be used.
Error rate: Protein result sets should have well-defined and low (<= 1%) false discovery rates.
The present work fulfills criteria 3, 6, 7, 8, and 9.
Because each of our tissue/biofluid-based subprotome atlas builds is created from a different pool of individuals, criterion 1 is not fulfilled and it is likely that certain proteins seen in one build but not another are due to differences among phenotypes (including health states and other temporary physiological conditions) rather than differences among the sets of proteins commonly found in tissues/biofluids. Of course, for any atlas build, the more samples in which a particular protein is observed, the smaller the likelihood that the protein is specific to certain phenotypes.
It is likely that some small additional bias was introduced because a variety of sample preparation and LC-MS/MS technologies were used, in a variety of laboratories (criterion 2).
For criterion 3, we adjusted for varying depths by estimating relative abundances using spectral counting, then performing abundance-based comparisons, using psuedo-counts in place of zero abundance values.
We fulfilled criteria 4 and 5 (search algorithms and libraries/databases) to the extent possible, but, as described in Experimental Procedures, some few plasma datasets were searched differently from the rest (SpectraST not used, SpectraST searched against in-house library, SpectraST searched against an older NIST library, X!Tandem searched against an older database). This likely resulted in fewer identified spectra from those datasets, and thus fewer identified peptides and proteins for the final plasma atlas build, with a small bias toward peptides discoverable in the older library or database. Because the proportion of plasma data differently searched is small, and because the peptides covered by the different releases of the library/database are largely the same, this bias is likely slight.
Cellular localization
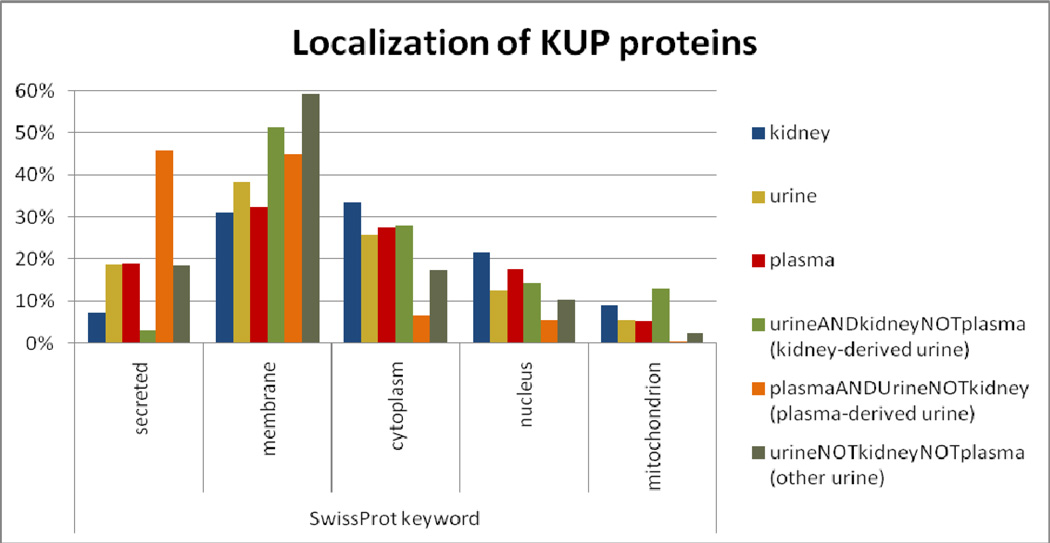
Figure 4 shows the cellular localization of proteins in KidneyPA, UrinePA, and PlasmaPA, as indicated by Swiss-Prot keywords. Plasma and urine have about three times the proportion of secreted proteins as kidney, whereas kidney has more cytoplasmic, nuclear, and mitochondrial proteins. More striking are the results for the various subsets of urine proteins: “kidney-derived”, “plasma-derived”, and “neither” (note that the 57% of UrinePA proteins seen in both KidneyPA and PlasmaPA—those that could derive from either kidney or plasma—are not in any of these three subsets). A full 46% of the UrinePA proteins in the “plasma-derived” set are annotated as secreted, compared to only 3% of “kidney-derived”. Conversely, the proportions of plasma-derived urine proteins that are annotated cytoplasmic, nuclear, or mitochondrial are much lower than those of kidney-derived urine proteins, with an extreme low of only 2 plasma-derived urine proteins (0.3%) annotated mitochondrial. Curiously, nearly 60% of UrinePA identifiers seen in neither PlasmaPA nor KidneyPA are annotated as membrane proteins.
Figure 4.
For six identifier sets, the proportion of identifiers with various Swiss-Prot cellular localization keywords. Some identifiers have multiple keywords.
Distributions of relative abundances
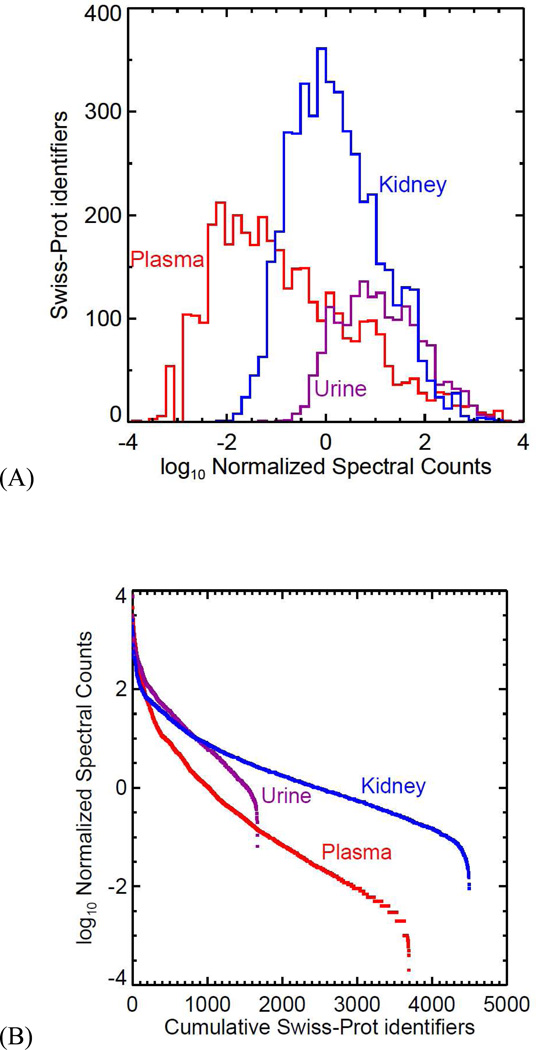
The range of NSC values observed for each atlas is illustrated in Figure 5. The extent of the left terminus of each curve in (A) is determined by the amount of data collected because the lowest possible NSC value is proportional to 1 divided by the total number of PSMs in the atlas. The extents of the right termini of the three curves in (A) are approximately the same: the largest NSC values for the three atlases are, on a log scale, nearly identical. Had not most of the plasma samples been subjected to routine depletion of the most abundant plasma proteins, albumin would constitute about half the total protein concentration and would thus have an NSC value of about 50,000, extending the plasma curve farther right to a value of about 4.7 (=log10(50,000)), and also causing the non-depleted proteins to have lower NSC values, extending the plasma curve farther to the left as well. See Figure S1 (Supporting Information) for a comparison of plasma curves over the history of the Human Plasma PeptideAtlas.
Figure 5.
(A) The normalized spectral counts (NSC) for each HKUP atlas, binned on a log scale. (B) The same data are plotted with cumulative protein counts on the X-axis and log(NSC) on the Y-axis to produce familiar dynamic range curves, showing more directly the varying numbers of proteins identified in each atlas.
Comparisons among tissue/biofluid-based proteomes
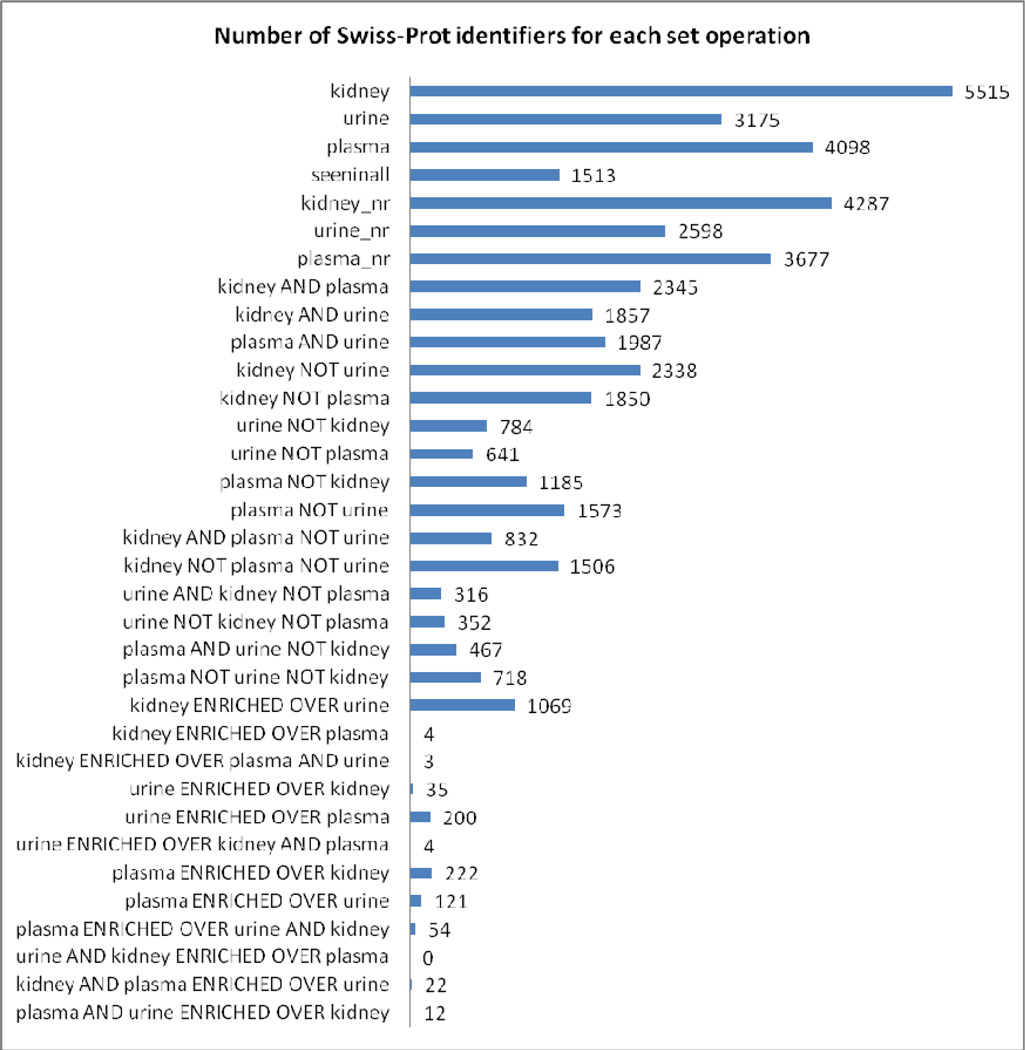
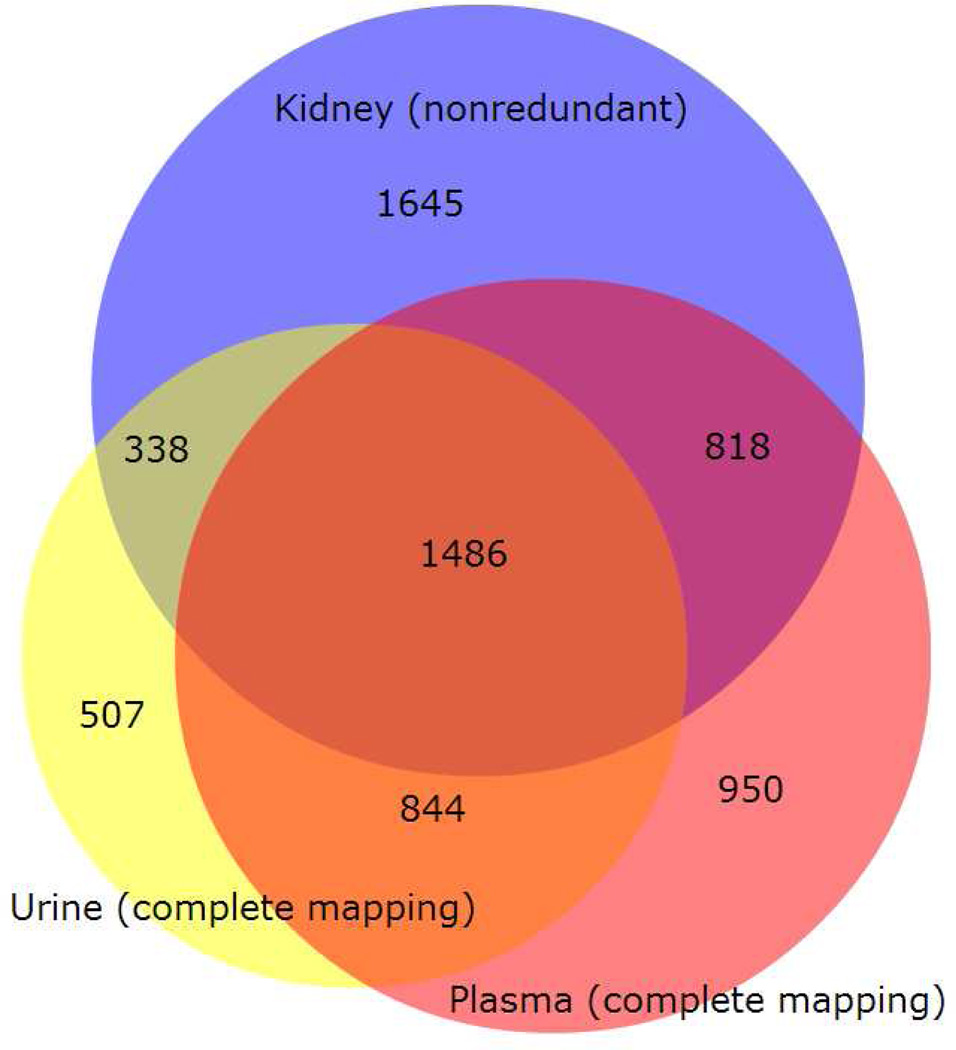
Our atlas comparison tool produces, for any three atlas builds, 34 sets of protein identifiers that facilitate study of the similarities and differences among the corresponding proteomes. Those for the current study are listed in Figure 6 and fully disclosed in Supporting Information Tables S2 and S4. Figure 7 illustrates graphically the formation of the set, “seen in all”. The identifier lists for each set, along with links to UniProt, can be browsed at www.peptideatlas.org/hupo/hkup. Further, an analysis to determine enrichment of Gene Ontology terms for each set was performed, and the 12 most enriched terms with p-value < 0.05 in each ontology (Biological Process, Cellular Component, Molecular Function) can be viewed in either graphical or tabular format at www.peptideatlas.org/hupo/hkup. Together, these resources provide a convenient way to investigate the relationships among the three tissue/biofluid-based proteomes.
Figure 6.
Thirty-four protein sets were derived from the original three using the redundancy reducing method, NSC comparisons, and set operations described in Experimental Procedures and in Table S4 (Supporting Information). All these sets, along with their GO analyses, can be browsed at www.peptideatlas.org/hupo/hkup.
Figure 7.
Nonredundant Swiss-Prot identifier set for KidneyPA intersected with the complete mappings for UrinePA and PlasmaPA. Diagram created using BioVenn47
An identifier is considered enriched in one atlas compared to another if its NSC in the first is at least 2σ times its NSC in the second, where σ is the standard deviation of the distribution of the logarithms of the ratios between NSC values for the two atlases for all identifiers. Any identifier not observed in a particular atlas is assumed to be present at a very low concentration, so for the purpose of calculating enrichment we replace each zero NSC value with a pseudo-count of half the smallest observed NSC value for that atlas. Note that, therefore, any identifier that appears in atlas A but not in atlas B will be listed under A NOT B no matter how small its NSC value in A, but it will only appear in A ENRICHED OVER B if its NSC value in A is greater than 2σ times half the smallest NSC value in B. See Table S4 (Supporting Information) for set definitions using set notation.
We made use of the analyses available at www.peptideatlas.org/hupo/hkup to investigate three questions: to what extent is urine dilute plasma? What is the nature of four proteins enriched at > 2σ in UrinePA relative to KidneyPA and PlasmaPA? What is the biomarker potential for genetic loci associated with renal decline?
Urine as dilute plasma
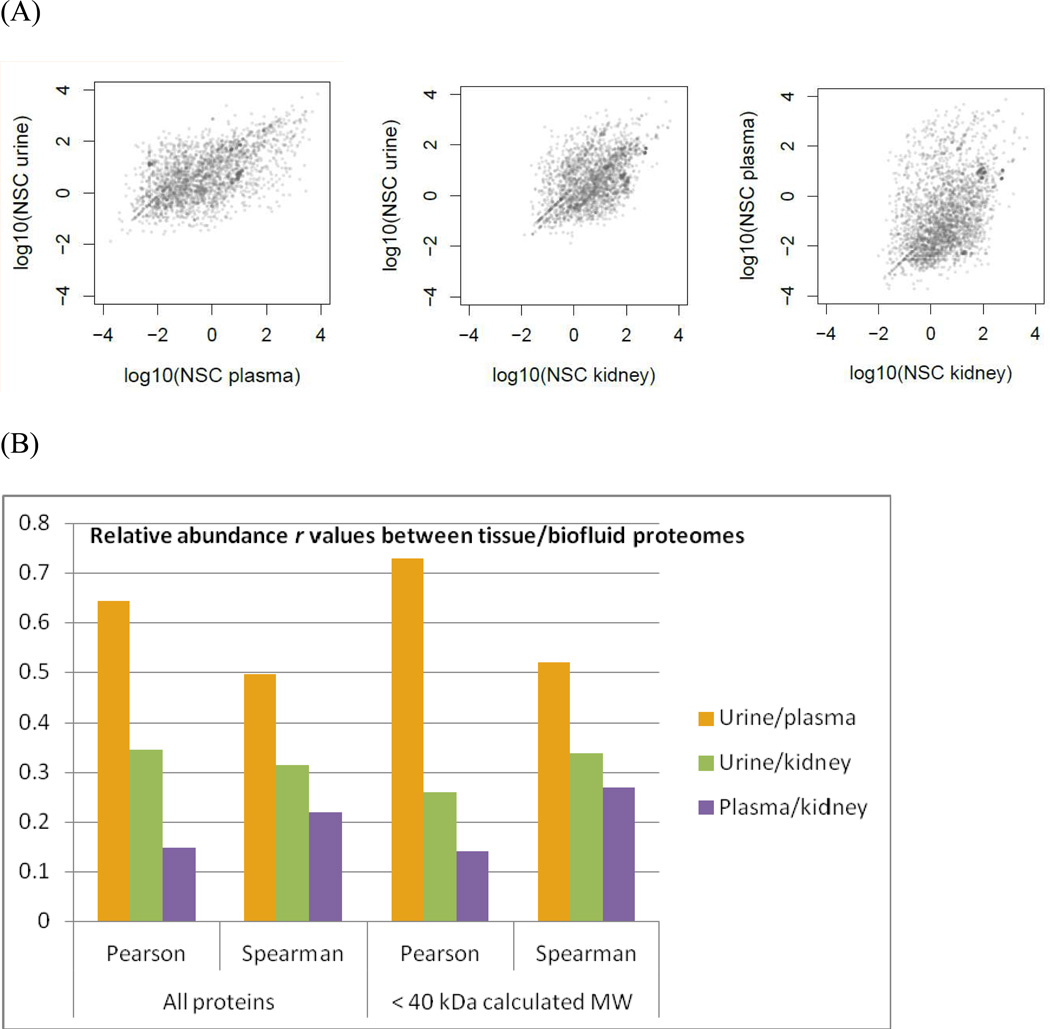
The 61 highest abundance PlasmaPA identifiers (NSC>486) are all seen in UrinePA. The calculated MWs of these proteins range between 9 and 187 kDa, with only nine above 60 kDa, suggesting that most if not all of them can pass through the glomerulus. For the 2330 identifiers in both PlasmaPA and UrinePA, the correlation coefficients for their abundances are higher than for urine/kidney and plasma/kidney, reflecting the reality that urine is primarily a filtrate of plasma wherein proteins smaller than 40–60kDa pass to the urine and larger ones are retained in the plasma. (see Figure 8). When proteins with a calculated MW larger than 40kDa are removed, the correlations increase somewhat. This may be the first study to show statistically what can be inferred from the biological relationship between these two body fluids: that their protein concentrations are proportional.
Figure 8.
(A) NSC values for proteins shared between two tissue/biofluid-based proteomes are plotted on a log/log scale. (B) Correlation coefficients (r) for (A). Urine/plasma is more strongly correlated than the other two pairs. For urine/plasma, the Pearson correlation is stronger than the Spearman, indicating that the relationship is fairly linear. In contrast, for plasma/kidney, the Spearman correlation is stronger, indicating that the (weak) correlation is monotonic but not linear. Restricting the analysis to small (<40kDa calculated MW) proteins strengthens the Pearson correlation for urine/plasma and weakens it for urine/kidney.
Urine proteins highly enriched relative to kidney and plasma
Three hundred ninety-two identifiers are seen in UrinePA but not in KidneyPA or PlasmaPA. When we conservatively assume that these are actually present in both kidney and plasma, just below the levels of detection, then only four are found to be enriched at >2σ in UrinePA relative to the other two (Table 4). All four are glycoproteins and we examine them below.
Table 4.
Four proteins significantly enriched in urine relative to plasma and kidney. (These proteins are absent from both KidneyPA and PlasmaPA, but for the purpose of calculating enrichment, we conservatively assume that they are actually present in each atlas build at an NSC equal to half the smallest NSC for that build.)
| Swiss- Prot |
Description | glycoprot | calc. MW (kDa) | chromosome | NSC in urine | PSMs | peptides | samples (of 15) | max mRNA expression in BioGPS |
HumanAllPA sample types (in addition to urine) |
HPA tissues with strong, reliable staining |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P01133 | Epidermal growth factor |
yes | 134 | 4 | 361 | 6454 | 99 | 15 | Kidney | seminal plasma | none |
| Q16769 | Glutaminyl- peptide cyclotransferase |
yes | 41 | 2 | 218 | 982 | 31 | 15 | Whole blood | plasma breast cancer vitreous humor neutrophil prostatic secretion seminal plasma |
none |
| P19835 | Bile salt-activated lipase |
yes | 79 | 9 | 98 | 590 | 37 | 13 | Pancreatic islet | colorectal cancer | lymph node germinal center cells, pancreas exocrine glandular cells, stomach lower glandular cells, tonsil germinal center cells |
| Q6UX06 | Olfactomedin-4 | yes | 57 | 13 | 93 | 567 | 23 | 11 | not in BioGPS | B-cell platelets colorectal cancer monocyte neutrophil protatic secretion seminal plasma |
not yet analyzed by HPA |
Pro-epidermal growth factor (P01133, EGF) is seen in all 15 urine experiments. Whereas the active form comprises only 53 residues, the pro-form is a membrane-spanning protein of 1185 residues, and the observed peptides cover the entire 1010 residue extracellular region. EGF was discovered in salivary gland and is a major growth factor for wound healing tied to inate responses such as licking of wounds 49. It is surprising that EGF is not seen in KidneyPA, since EGF mRNA expression is highest in kidney of the 84 different tissues and cell types in BioGPS50 —even higher than in salivary gland—or in PlasmaPA, given that EGF was detected in platelet-rich plasma at least as early as 198351. The portions of the kidney where EGF is produced are unclear; according to Harris 199152, the mRNA is localized to the thick ascending limb of Henle and distal convoluted tubule. However, EGF has been localized to the proximal tubules by immunohistochemistry 53. In HumanAllPA, EGF is seen exclusively in urine, with the exception of a single observation in seminal plasma. As EGF receptor was detected in glomeruli and tubules of the kidney by immunohistochemistry 54, EGF may play an important role in regeneration of renal epithelial cells.
Glutaminyl-peptide cyclotransferase (Q16769), a secreted enzyme, catalyzes the formation of N-terminal pyroglutamic acid on peptides, and its mRNA is present most abundantly in whole blood according to BioGPS. Although absent from PlasmaPA, it is found in HumanAllPA in a single glycocapture-enriched plasma sample. (Enriched samples were omitted from PlasmaPA to provide more accurate abundance estimations.)
Bile salt-activated lipase (BAL) (P19835) is an enzyme secreted by the pancreas and mammary glands that aids digestion of fats. In the Human All PeptideAtlas it is seen only in urine samples and in a very rich colorectal cancer sample. According to UniProt, mutations are known to result in an autosomal dominant inheritance of early onset diabetes (by age 25). This enzyme was also detected in the urine of healthy subjects in a 2006 study55 which cites previous work that demonstrated BAL reaches the blood from the pancreas via a transcytosis motion through enterocytes. Leaving aside the question of how this protein of ~80 kDa can be filtered through the glomerulus, it appears that this enzyme should exist in plasma, yet does not appear in PlasmaPA. Notably, BAL is absent from the 558 urine proteins identified in Zerefos, et al18 and also from the urine survey of Li, et al.16
Olfactomedin-4 (Q6UX06), a 510 residue secreted protein, is known to play a role in cell adhesion via interactions with cadherin and extracellular lectins, according to UniProt. However, while known to be secreted, UniProt citations show it is also known to be present in intracellular compartments, in particular in mitochondria. Interestingly, olfactomedin has high mRNA expression in pancreas, small intestine and colon, and has been shown to be a marker for both pancreatic and gastrointenstinal cancers56, 57. A recent glycocapture proteomics study of >40 human tissues performed at the Institute for Systems Biology (Watts, et al. and Harris, et al., in preparation) identifies olfactomedin in >20 of the tissues, including bladder, kidney, and prostate, any or all of which could explain its appearance in urine via secretion pathways.
Tissue/biofluid-based proteome analysis elucidates results of GWAS study and points to potential biomarkers
Kottgen, et al.58 reported confirmation of five loci and discovery of 16 new loci statistically associated with decline in kidney function (glomerular filtration rate, measured with creatinine or cystatin C); they also found 7 loci associated with creatinine production and secretion. We examined each of the 11 loci for which the SNP variant was actually in the named gene, rather than somewhere nearby (their Table 2), as well as three of the creatinine-associated loci; these 14 included all three that produced non-synonymous amino acid substitutions in the corresponding protein. The results are summarized in Table 5 and detailed in Supporting Information (“Integrated Analysis of Genomic Variation and Protein Detection in Kidney, Urine, and Plasma: Seeking Clues for New Biomarker Candidates”). In brief, we found that of the 14 loci, one emerges as a strong candidate for biomarker studies: DAB2, which, together with MYH9 and megalin, form a trio of loci whose protein products are known to interact physically and have each been detected in KidneyPA as well as UrinePA and/or PlasmaPA. Five additional loci have evidence in KidneyPA, but the evidence for one (PRKAG2) is subsumed by a related protein, and none of the five is present in PlasmaPA or UrinePA, the two biofluids most commonly used for biomarker detection.
Table 5.
Fourteen loci found by Kottgen, et al. 58 to be associated with chronic kidney disease, plus three related loci. All are discussed in detail in Supporting Information. Among these, the trio DAB2/MYH9/megalin emerge as promising biomarker candidates, as there is evidence that the protein products for all three exist in kidney (are all seen in KidneyPA) and additionally in plasma and/or urine.
| Gene ID | SwissProt accession |
Description | KidneyPA NSC |
UrinePA NSC |
PlasmaPA NSC |
kidney PSMs |
kidney peps |
kidney sample types |
|---|---|---|---|---|---|---|---|---|
| SHROOM3 | Q8TF72 | Protein Shroom3 | 0.15 | 0 | 0 | 11 | 5 | glomerulus |
| GCKR | Q14397 | Glucokinase regulatory protein |
0 | 0 | 0 | |||
| NAT8 | Q9UHE5 | Probable N- acetyltransferase 8 |
1.78 | 0 | 0 | 144 | 5 | cortex & medulla |
| ALMS1 | Q8TCU4 | Alstrom syndrome protein 1 |
0 | 0 | 0 | |||
| DAB2 | P98082 | Disabled homolog 2 | 1.78 | 0 | 0.007 | 36 | 8 | cortex, glom, mixed, medulla |
| MYH9 | P35579 | Myosin-9 | 194 | 6.1 | 0.29 | all | ||
| Megalin |
P98164 (LRP-2) |
LDL receptor-related protein 2 |
6.67 | 67 | 0 | 1369 | 73 | all |
| SLC34A1 | Q06495 | Sodium-dependent phosphate transport protein 2A |
0 | 0 | 0 | |||
| PRKAG2 | Q9UGJ0 | 5'-AMP-activated protein kinase subunit gamma-2 |
0.081 | 0 | 0 | 3 | 1 | glomerulus |
| PIP5KIB | O14985 | Mucin-5B | 0 | 0 | 0 | |||
| ATXN2 | Q99700 | Ataxin-2 | 0 | 0 | 0 | |||
| DACH1 | Q9UI36 | Dachshund homolog 1 | 0.32 | 0 | 0 | 4 | 2 | glomerulus |
| UBE2Q2 | Q8WVN8 | Ubiquitin-conjugating enzyme E2 Q2 |
0 | 0 | 0 | |||
| SLC7A9 | P82251 | B(0,+)-type amino acid transporter 1 |
0 | 0 | 0 | |||
| CPS1 | P31327 | Carbamoyl-phosphate synthase [ammonia], mitochondrial |
0 | 0 | 0.012 | |||
| SLC22A2 | O15244 | Solute carrier family 22 member 2 |
3.1 | 0 | 0 | 44 | 2 | cortex, medulla |
| SLC6A13 | Q9NSD5 | Sodium- and chloride- dependent GABA transporter 2 |
0 | 0 | 0 | 14 | 2 |
CONCLUSION
We provide a year 2013 update to the Human All PeptideAtlas and present PeptideAtlas builds containing high-confidence, nonredundant protein identifications for three important human proteomes, kidney, urine, and plasma, for the ultimate goal of defining the human proteome. By employing the standardized bioinformatics pipeline of PeptideAtlas to re-analyze datasets from diverse sources, we produced protein lists which are easily comparable. The resulting peptide and protein identifications can be mined at www.peptideatlas.org using the Browse Proteins and Browse Peptides features that have always been available at PeptideAtlas. In addition, a master spreadsheet of the Swiss-Prot protein identification lists is provided (Supporting Information Tables S2 and S3), including relative abundances (NSC values) for each proteome, proteome comparison results and a wealth of associated information on each protein such as molecular weight, cellular localization, HPA observations, BioGPS transcript localization, and PeptideAtlas samples and sample types. Finally, we provide a Gene Ontology analysis for each tissue/biofluid-based proteome comparison at www.peptideatlas.org/hupo/hkup, enabling investigation into the nature of proteins shared or not shared among these proteomes.
We have made use of these resources to address three questions: What are the abundance correlations between each pair of tissue/biofluid-based proteomes? What is the nature of proteins highly enriched in urine over both kidney and plasma? Of genetic loci previously associated with declining kidney function, which do we observe in kidney, urine, and plasma, and at what relative abundances? These studies only scratch the surface of what can be done with this PeptideAtlas tissue/biofluid-based proteome comparison. This work will greatly benefit the HPP, supporting all groups performing research under the auspices of the HPP including both the C-HPP (chromosome-centric) and B/D-HPP (biology-disease-centric).
The results from each of the three PeptideAtlas builds presented here, along with the human brain build, liver build, and “other” build have been merged into neXtProt59, and can be explored in the context of many other annotations there. The “other” build contains all samples that are not included in the kidney, urine, plasma, brain, and liver builds. PeptideAtlas will continue to expand as more data are collected through ProteomeXchange60 and collaborations, including the introduction of builds for other human proteomes for other tissue types and biofluids.
Supplementary Material
ACKNOWLEDGEMENTS
We extend our sincere thanks to David Campbell for discussion and PeptideAtlas support, and to Akhilesh Pandey, Hanno Steen, and Weijun Qian for their recent dataset contributions. This work was supported in part with federal funds from the National Science Foundation MRI grant No. 0923536, by the American Recovery and Reinvestment Act (ARRA) funds through National Institutes of Health; from the National Human Genome Research Institute grant No. RC2 HG005805, the National Institute of General Medical Sciences under grant No. R01 GM087221 and 2P50 GM076547/Center for Systems Biology] grant No.s RM-08-029, P30U54ES017885, U54DA021519, and UL1RR24986 (GSO), the US Department of Defense (Award # W81XWH-11-1-0487), the Japan Society for the Promotion of Science KAKENHI Grants 21390262 and 228071, the European Union FP7 grant 'ProteomeXchange' grant No. 260558, and by the Luxembourg Centre for Systems Biomedicine and the University of Luxembourg.
Abbreviations
- HUPO
Human Proteome Organization
- HKUPP
Human Kidney & Urine Proteome Project
- HPPP
Human Plasma Proteome Project
- KUP
Kidney, Urine, Plasma
- PSM
peptide-spectrum match
- NSC
normalized spectral count
- GO
Gene Ontology
- MW
molecular weight
- B/D-HPP
Biology and Disease-driven Human Proteome Project
- C-HPP
Chromosome-centric Human Proteome Project
- HPA
Human Protein Atlas
- MS
mass spectometry
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, Chen S, Eddes J, Loevenich SN, Aebersold R. The PeptideAtlas project. Nucleic Acids Res. 2006;34(Database issue):D655–D658. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 3.Farrah T, Deutsch EW, Hoopmann MR, Hallows JL, Sun Z, Huang CY, Moritz RL. The state of the human proteome in 2012 as viewed through PeptideAtlas. J Proteome Res. 2013;12(1):162–171. doi: 10.1021/pr301012j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Legrain P, Aebersold R, Archakov A, Bairoch A, Bala K, Beretta L, Bergeron J, Borchers CH, Corthals GL, Costello CE, Deutsch EW, Domon B, Hancock W, He F, Hochstrasser D, Marko-Varga G, Salekdeh GH, Sechi S, Snyder M, Srivastava S, Uhlen M, Wu CH, Yamamoto T, Paik YK, Omenn GS. The human proteome project: current state and future direction. Mol Cell Proteomics. 2011;10(7) doi: 10.1074/mcp.M111.009993. M111 009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marko-Varga G, Omenn GS, Paik YK, Hancock WS. A first step toward completion of a genome-wide characterization of the human proteome. J Proteome Res. 2013;12(1):1–5. doi: 10.1021/pr301183a. [DOI] [PubMed] [Google Scholar]
- 6.Aebersold R, Bader GD, Edwards AM, van Eyk JE, Kussmann M, Qin J, Omenn GS. The biology/disease-driven human proteome project (B/D-HPP): enabling protein research for the life sciences community. J Proteome Res. 2013;12(1):23–27. doi: 10.1021/pr301151m. [DOI] [PubMed] [Google Scholar]
- 7.Deutsch EW, Eng JK, Zhang H, King NL, Nesvizhskii AI, Lin B, Lee H, Yi EC, Ossola R, Aebersold R. Human Plasma PeptideAtlas. Proteomics. 2005;5(13):3497–3500. doi: 10.1002/pmic.200500160. [DOI] [PubMed] [Google Scholar]
- 8.Farrah T, Deutsch EW, Omenn GS, Campbell DS, Sun Z, Bletz JA, Mallick P, Katz JE, Malmstrom J, Ossola R, Watts JD, Lin B, Zhang H, Moritz RL, Aebersold R. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol Cell Proteomics. 2011;10(9) doi: 10.1074/mcp.M110.006353. M110 006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrah T, Deutsch EW, Aebersold R. Using the Human Plasma PeptideAtlas to study human plasma proteins. Methods Mol Biol. 2011;728:349–374. doi: 10.1007/978-1-61779-068-3_23. [DOI] [PubMed] [Google Scholar]
- 10.Cui Z, Yoshida Y, Xu B, Zhang Y, Nameta M, Magdeldin S, Makiguchi T, Ikoma T, Fujinaka H, Yaoita E, Yamamoto T. Profiling and annotation of human kidney glomerulus proteome. Proteome Sci. 2013;11(1):13. doi: 10.1186/1477-5956-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyamoto M, Yoshida Y, Taguchi I, Nagasaka Y, Tasaki M, Zhang Y, Xu B, Nameta M, Sezaki H, Cuellar LM, Osawa T, Morishita H, Sekiyama S, Yaoita E, Kimura K, Yamamoto T. In-depth proteomic profiling of the normal human kidney glomerulus using two-dimensional protein prefractionation in combination with liquid chromatography-tandem mass spectrometry. J Proteome Res. 2007;6(9):3680–3690. doi: 10.1021/pr070203n. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto T. Proteomics database in chronic kidney disease. Adv Chronic Kidney Dis. 2010;17(6):487–492. doi: 10.1053/j.ackd.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7(9):R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kentsis A, Monigatti F, Dorff K, Campagne F, Bachur R, Steen H. Urine proteomics for profiling of human disease using high accuracy mass spectrometry. Proteomics Clin Appl. 2009;3(9):1052–1061. doi: 10.1002/prca.200900008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marimuthu A, O'Meally RN, Chaerkady R, Subbannayya Y, Nanjappa V, Kumar P, Kelkar DS, Pinto SM, Sharma R, Renuse S, Goel R, Christopher R, Delanghe B, Cole RN, Harsha HC, Pandey A. A comprehensive map of the human urinary proteome. J Proteome Res. 2011;10(6):2734–2743. doi: 10.1021/pr2003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li QR, Fan KX, Li RX, Dai J, Wu CC, Zhao SL, Wu JR, Shieh CH, Zeng R. A comprehensive and non-prefractionation on the protein level approach for the human urinary proteome: touching phosphorylation in urine. Rapid Commun Mass Spectrom. 2010;24(6):823–832. doi: 10.1002/rcm.4441. [DOI] [PubMed] [Google Scholar]
- 17.Nagaraj N, Mann M. Quantitative analysis of the intra- and inter-individual variability of the normal urinary proteome. J Proteome Res. 2011;10(2):637–645. doi: 10.1021/pr100835s. [DOI] [PubMed] [Google Scholar]
- 18.Zerefos PG, Aivaliotis M, Baumann M, Vlahou A. Analysis of the urine proteome via a combination of multi-dimensional approaches. Proteomics. 2012;12(3):391–400. doi: 10.1002/pmic.201100212. [DOI] [PubMed] [Google Scholar]
- 19.Bouatra S, Aziat F, Mandal R, Guo AC, Wilson MR, Knox C, Bjorndahl TC, Krishnamurthy R, Saleem F, Liu P, Dame ZT, Poelzer J, Huynh J, Yallou FS, Psychogios N, Dong E, Bogumil R, Roehring C, Wishart DS. The human urine metabolome. PLoS One. 2013;8(9):e73076. doi: 10.1371/journal.pone.0073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R, Gautam B, Young N, Xia J, Knox C, Dong E, Huang P, Hollander Z, Pedersen TL, Smith SR, Bamforth F, Greiner R, McManus B, Newman JW, Goodfriend T, Wishart DS. The human serum metabolome. PLoS One. 2011;6(2):e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jupp S, Klein J, Schanstra J, Stevens R. Developing a kidney and urinary pathway knowledge base. J Biomed Semantics. 2011;2(Suppl 2):S7. doi: 10.1186/2041-1480-2-S2-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Omenn GS, States DJ, Adamski M, Blackwell TW, Menon R, Hermjakob H, Apweiler R, Haab BB, Simpson RJ, Eddes JS, Kapp EA, Moritz RL, Chan DW, Rai AJ, Admon A, Aebersold R, Eng J, Hancock WS, Hefta SA, Meyer H, Paik YK, Yoo JS, Ping P, Pounds J, Adkins J, Qian X, Wang R, Wasinger V, Wu CY, Zhao X, Zeng R, Archakov A, Tsugita A, Beer I, Pandey A, Pisano M, Andrews P, Tammen H, Speicher DW, Hanash SM. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005;5(13):3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 23.States DJ, Omenn GS, Blackwell TW, Fermin D, Eng J, Speicher DW, Hanash SM. Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nat Biotechnol. 2006;24(3):333–338. doi: 10.1038/nbt1183. [DOI] [PubMed] [Google Scholar]
- 24.Schenk S, Schoenhals GJ, de Souza G, Mann M. A high confidence, manually validated human blood plasma protein reference set. BMC Med Genomics. 2008;1:41. doi: 10.1186/1755-8794-1-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia L, Zhang L, Shao C, Song E, Sun W, Li M, Gao Y. An attempt to understand kidney's protein handling function by comparing plasma and urine proteomes. PLoS One. 2009;4(4):e5146. doi: 10.1371/journal.pone.0005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, Sun Z, Nilsson E, Pratt B, Prazen B, Eng JK, Martin DB, Nesvizhskii AI, Aebersold R. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10(6):1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedrioli PG. Trans-proteomic pipeline: a pipeline for proteomic analysis. Methods Mol Biol. 2010;604:213–238. doi: 10.1007/978-1-60761-444-9_15. [DOI] [PubMed] [Google Scholar]
- 28.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101(36):13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, Star RA, Kleta R, Wang NS, Knepper MA. Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol. 2009;20(2):363–379. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 31.Chan KC, Lucas DA, Hise D, et al. Serum/Plasma Proteome. Clinical Proteomics. 2004;1(1):101–225. [Google Scholar]
- 32.Sigdel TK, Kaushal A, Gritsenko M, Norbeck AD, Qian WJ, Xiao W, Camp DG, 2nd, Smith RD, Sarwal MM. Shotgun proteomics identifies proteins specific for acute renal transplant rejection. Proteomics Clin Appl. 2010;4(1):32–47. doi: 10.1002/prca.200900124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20(9):1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 34.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol. 2005;1 doi: 10.1038/msb4100024. 2005.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4(7):1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 36.Lam H, Deutsch EW, Eddes JS, Eng JK, King N, Stein SE, Aebersold R. Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics. 2007;7(5):655–667. doi: 10.1002/pmic.200600625. [DOI] [PubMed] [Google Scholar]
- 37.Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho- Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, Garcia-Giron C, Gordon L, Hourlier T, Hunt S, Juettemann T, Kahari AK, Keenan S, Komorowska M, Kulesha E, Longden I, Maurel T, McLaren WM, Muffato M, Nag R, Overduin B, Pignatelli M, Pritchard B, Pritchard E, Riat HS, Ritchie GR, Ruffier M, Schuster M, Sheppard D, Sobral D, Taylor K, Thormann A, Trevanion S, White S, Wilder SP, Aken BL, Birney E, Cunningham F, Dunham I, Harrow J, Herrero J, Hubbard TJ, Johnson N, Kinsella R, Parker A, Spudich G, Yates A, Zadissa A, Searle SM. Ensembl 2013. Nucleic Acids Res. 2013;41(Database issue):D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reiter L, Claassen M, Schrimpf SP, Jovanovic M, Schmidt A, Buhmann JM, Hengartner MO, Aebersold R. Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol Cell Proteomics. 2009;8(11):2405–2417. doi: 10.1074/mcp.M900317-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 40.Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J Proteome Res. 2004;3(6):1234–1242. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 41.Cote RG, Jones P, Martens L, Kerrien S, Reisinger F, Lin Q, Leinonen R, Apweiler R, Hermjakob H. The Protein Identifier Cross-Referencing (PICR) service: reconciling protein identifiers across multiple source databases. BMC Bioinformatics. 2007;8:401. doi: 10.1186/1471-2105-8-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25(1):117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 43.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23(2):257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 44.Shteynberg D, Nesvizhskii AI, Moritz RL, Deutsch EW. Combining results of multiple search engines in proteomics. Mol Cell Proteomics. 2013;12(9):2383–2393. doi: 10.1074/mcp.R113.027797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boron WF, Boulpaep EL. Medical Physiology: A Cellular And Molecular Approach. Elsevier/Saunders; 2011. [Google Scholar]
- 46.Fahlman RP, Chen W, Overall CM. Absolute proteomic quantification of the activity state of proteases and proteolytic cleavages using proteolytic signature peptides and isobaric tags. J Proteomics. 2013 doi: 10.1016/j.jprot.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 47.Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25(2):288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sorensen OE, Thapa DR, Roupe KM, Valore EV, Sjobring U, Roberts AA, Schmidtchen A, Ganz T. Injury-induced innate immune response in human skin mediated by transactivation of the epidermal growth factor receptor. J Clin Invest. 2006;116(7):1878–1885. doi: 10.1172/JCI28422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, Su AI. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10(11):R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oka Y, Orth DN. Human plasma epidermal growth factor/beta-urogastrone is associated with blood platelets. J Clin Invest. 1983;72(1):249–259. doi: 10.1172/JCI110964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harris RC. Potential physiologic roles for epidermal growth factor in the kidney. Am J Kidney Dis. 1991;17(6):627–630. doi: 10.1016/s0272-6386(12)80336-2. [DOI] [PubMed] [Google Scholar]
- 53.Bernardini N, Bianchi F, Lupetti M, Dolfi A. Immunohistochemical localization of the epidermal growth factor, transforming growth factor alpha, and their receptor in the human mesonephros and metanephros. Dev Dyn. 1996;206(3):231–238. doi: 10.1002/(SICI)1097-0177(199607)206:3<231::AID-AJA1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 54.Nakanishi K, Sweeney W, Jr, Avner ED. Segment-specific c-ErbB2 expression in human autosomal recessive polycystic kidney disease. J Am Soc Nephrol. 2001;12(2):379–384. doi: 10.1681/ASN.V122379. [DOI] [PubMed] [Google Scholar]
- 55.Comte B, Franceschi C, Sadoulet MO, Silvy F, Lafitte D, Benkoel L, Nganga A, Daniel L, Bernard JP, Lombardo D, Mas E. Detection of bile saltdependent lipase, a 110 kDa pancreatic protein, in urines of healthy subjects. Kidney Int. 2006;69(6):1048–1055. doi: 10.1038/sj.ki.5000133. [DOI] [PubMed] [Google Scholar]
- 56.Kobayashi D, Koshida S, Moriai R, Tsuji N, Watanabe N. Olfactomedin 4 promotes S-phase transition in proliferation of pancreatic cancer cells. Cancer Sci. 2007;98(3):334–440. doi: 10.1111/j.1349-7006.2007.00397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu L, Wang L, Chen S. Olfactomedin 4, a novel marker for the differentiation and progression of gastrointestinal cancers. Neoplasma. 2011;58(1):9–13. doi: 10.4149/neo_2011_01_9. [DOI] [PubMed] [Google Scholar]
- 58.Kottgen A, Pattaro C, Boger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV, O'Connell JR, Li M, Schmidt H, Tanaka T, Isaacs A, Ketkar S, Hwang SJ, Johnson AD, Dehghan A, Teumer A, Pare G, Atkinson EJ, Zeller T, Lohman K, Cornelis MC, Probst-Hensch NM, Kronenberg F, Tonjes A, Hayward C, Aspelund T, Eiriksdottir G, Launer LJ, Harris TB, Rampersaud E, Mitchell BD, Arking DE, Boerwinkle E, Struchalin M, Cavalieri M, Singleton A, Giallauria F, Metter J, de Boer IH, Haritunians T, Lumley T, Siscovick D, Psaty BM, Zillikens MC, Oostra BA, Feitosa M, Province M, de Andrade M, Turner ST, Schillert A, Ziegler A, Wild PS, Schnabel RB, Wilde S, Munzel TF, Leak TS, Illig T, Klopp N, Meisinger C, Wichmann HE, Koenig W, Zgaga L, Zemunik T, Kolcic I, Minelli C, Hu FB, Johansson A, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Schreiber S, Aulchenko YS, Felix JF, Rivadeneira F, Uitterlinden AG, Hofman A, Imboden M, Nitsch D, Brandstatter A, Kollerits B, Kedenko L, Magi R, Stumvoll M, Kovacs P, Boban M, Campbell S, Endlich K, Volzke H, Kroemer HK, Nauck M, Volker U, Polasek O, Vitart V, Badola S, Parker AN, Ridker PM, Kardia SL, Blankenberg S, Liu Y, Curhan GC, Franke A, Rochat T, Paulweber B, Prokopenko I, Wang W, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Shlipak MG, van Duijn CM, Borecki I, Kramer BK, Rudan I, Gyllensten U, Wilson JF, Witteman JC, Pramstaller PP, Rettig R, Hastie N, Chasman DI, Kao WH, Heid IM, Fox CS. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42(5):376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaudet P, Argoud-Puy G, Cusin I, Duek P, Evalet O, Gateau A, Gleizes A, Pereira M, Zahn-Zabal M, Zwahlen C, Bairoch A, Lane L. neXtProt: organizing protein knowledge in the context of human proteome projects. J Proteome Res. 2013;12(1):293–298. doi: 10.1021/pr300830v. [DOI] [PubMed] [Google Scholar]
- 60.Vizcaino JADEW, Wang R, Csordas A, Reisinger F, Rios D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz P-A, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley RJ, Kraus H-J, Albar JP, Martinez-Bartolome S, Apweiler R, Omenn GS, Martens L, Jones AR. H. Henning ProteomeXchange: globally co-ordinated proteomics data submission and dissemination. Nat Biotechnol. 2014 doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.