Abstract
Candida albicans public proteomic data sets, though growing steadily in the last few years, still have a very limited presence in online repositories. We report here the creation of a C. albicans PeptideAtlas comprising near 22000 distinct peptides at a 0.24 % False Discovery Rate (FDR) that account for over 2500 canonical proteins at a 1.2% FDR. Based on data from 16 experiments, we attained coverage of 41% of the C.albicans open reading frame sequences (ORFs) in the database used for the searches. This PeptideAtlas provides several useful features, including comprehensive protein and peptide-centered search capabilities and visualization tools that establish a solid basis for the study of basic biological mechanisms key to virulence and pathogenesis such as dimorphism, adherence, and apoptosis. Further, it is a valuable resource for the selection of candidate proteotypic peptides for targeted proteomic experiments via selected reaction monitoring (SRM) or SWATH-MS.
Keywords: Candida albicans, PeptideAtlas, Proteotypic peptides
Introduction
Candida albicans is a fungus of great clinical importance. In addition to asymptomatically colonizing mucous membranes as a commensal in a large percentage of the population, it may cause severe opportunistic infections in specific cases such as patients with weakened immune defenses, a common circumstance in cancer and AIDS patients. C. albicans infections are also a threat to patients in post-surgical situations and intensive care unit stays. In this respect, invasive candidiasis remains nowadays one of the major types of nosocomial infections and a challenge in terms of economical and health costs [1–3]. From the perspective of proteomics, recent studies have provided new insights into the C. albicans biology and suggested new clinical biomarker candidates for diagnosis and prognosis of invasive candidiasis [4–7].
However, the clinical relevance of this organism is not reflected in the number of large-scale publicly available proteomics resources. Up to the current date, the PRIDE [8] database includes only 15 experiments accounting for 1786 identified proteins. The more C. albicans-focused Proteopathogen database [9] comprises several hundred protein identifications including data from gel based proteomics, and other major proteomics online resources such as the Global Proteome Machine Database (GPMDB [10]) or Tranche [11] contain no C. albicans data whatsoever.
As for the genomic data, according to Candida Genome Database (CGD), currently the most comprehensively annotated C. albicans sequence repository [12], the C. albicans genome contains 6215 ORFs (as of May 28, 2013), out of which 1497 are annotated as verified, i.e. representing genes for which there is empirical evidence that the ORF actually encodes a functionally characterized protein. In contrast, 4566 ORFs are termed uncharacterized, indicating there exists no conclusive evidence for the existence of a protein product. This data implies that most part of the predicted proteome, over 70% of the ORFs, is still unknown or has not been properly annotated yet. An extensive characterization of the C. albicans proteome will therefore be of great value to increase our knowledge in proteins involved in mechanisms of virulence and infection and, thus serve as a basis to design strategies for diagnosis, vaccination and treatment of invasive candidiasis.
Since its inception, the PeptideAtlas project [13] has encouraged mass spectrometry data submission by the community and has thus grown to a large compilation of atlases of different species including human tissue and body fluid specific builds (brain, plasma [14] and urine), microbial builds (Halobacterium [15], Mycobacterium tuberculosis [16], Streptococcus [17], Leptospira, Plasmodium [18], Saccharomyces [19] and Schizosaccharomyces [20]); invertebrate (C.elegans, Drosophila [21] and Apis Mellifera [22]) builds; and a pig and a bovine milk [23] build. The PeptideAtlas project, as a multi-species compendium of proteomes, is continuously increasing its biological diversity. The recent S. pombe atlas [23] attains a large coverage of its proteome by ad hoc extensive fractionation and high-resolution LC-MS/MS, and contributes in the sense that some of the fission yeast biological processes have a high degree of conservation with the corresponding pathways in mammalian cells. The incorporation of C. albicans resolves the previous absence of fungal pathogens in the PeptideAtlas and their under representation in any public proteomic data repository.
Furthermore, the proven utility of PeptideAtlas as a resource for selecting proteotypic peptides for Selected Reaction Monitoring (SRM) [24] or SWATH-MS [25] will enable a starting point for future targeted proteomics workflows in C. albicans.
Material and Methods
Empirical data compilation
Large amounts of mass spectrometry data corresponding to many and diverse measurements of the C. albicans proteome initially intended for different purposes were assembled in order to build the PeptideAtlas. A range of proteomic methods, protocols and different biological conditions were used to generate the data as shown in Table 1. These include membrane protein extractions [26], morphological yeast to hypha transition experiments [27] and phosphoprotein enrichment treatments. The combination of these diverse datasets resulted in an unprecedented overall coverage of the C. albicans proteome. Protein samples were obtained as previously described in [27]. Briefly, cells of the clinical isolate SC5314 were grown in YPD medium for standard growth, whereas hyphal form growth was induced using either Lee medium pH 6.7 or heat-inactivated fetal bovine serum. Protein extracts were then obtained by mechanical cell disruption using either glass beads in the MSK cell homogenizer or the Fast-Prep cell breaker. Protein digests were obtained by trypsinization and separated via HPLC. All spectra acquisition runs were performed by LC-MS/MS in a data-dependent manner in different instruments and set-ups. Table 1 provides an overview of the experiments along with the instrument used for the mass spectrometry and the corresponding number of raw spectra data files that were acquired.
Table 1.
List of experiments collected to construct the C albicans PeptideAtlas.
| # experiment | sample (as named in the web interface) | labeling/treatment | instrument type | #raw files |
|---|---|---|---|---|
| 1 | Calb_acidic_subproteome | - | LTQ | 3 |
|
| ||||
| 2 | Calb_memb | - | LTQ | 8 |
|
| ||||
| 3 | SILAC_phos_OrbitrapVelos_1 | SILAC. IMAC+TiO2 | Orbitrap Velos | 3 |
| 4 | SILAC_phos_OrbitrapVelos_2 | SILAC. IMAC+TiO2 | Orbitrap Velos | 3 |
| 5 | SILAC_phos_OrbitrapVelos_3 | SILAC. IMAC+TiO2 | Orbitrap Velos | 3 |
| 6 | SILAC_phos_OrbitrapVelos_4 | SILAC. IMAC+TiO2 | Orbitrap Velos | 3 |
| 7 | SILAC_phos_OrbitrapXL_1A | SILAC. IMAC | Orbitrap XL | 11 |
| 8 | SILAC_phos_OrbitrapXL_1A_TiO2 | SILAC. IMAC+TiO2 | Orbitrap XL | 5 |
| 9 | SILAC_phos_OrbitrapXL_1B | SILAC. IMAC | Orbitrap XL | 6 |
| 10 | SILAC_phos_OrbitrapXL_1B_TiO2 | SILAC. IMAC+TiO2 | Orbitrap XL | 6 |
| 11 | SILAC_phos_OrbitrapXL_2 | SILAC. IMAC | Orbitrap XL | 6 |
| 12 | SILAC_phos_OrbitrapXL_3 | SILAC. IMAC | Orbitrap XL | 6 |
| 13 | SILAC_phos_OrbitrapXL_4 | SILAC. IMAC | Orbitrap XL | 5 |
|
| ||||
| 14 | Calb_extract_3TOF | - | Triple TOF | 2 |
|
| ||||
| 15 | Hyphal_extract_OrbitrapVelos | - | Orbitrap Velos | 4 |
| 16 | Yeast_extract_OrbitrapVelos | - | Orbitrap Velos | 4 |
In addition, raw MS data from unpublished, SILAC labeled and phosphoprotein enriched samples generated from studies focused on Candida interaction with host immune cells and from experiments studying the hyphal and yeast-form proteomes, were added to the collection.
Peptide and protein identification
PeptideAtlas ensures consistency and quality of the stored data by processing the raw spectra sets by the Trans-Proteomic Pipeline (TPP) [28], a suite of software tools for processing shotgun proteomics datasets. The TPP tools are run in a well-established sequential pipeline spanning steps from creating appropriate standard files to be used as input by the search engine to statistical validation of protein inference and calculation of the False Discovery Rate (FDR).
The collected raw spectra files in different proprietary file formats were converted to the standard format for mass spectrometry output data mzML [29], searched using X!Tandem [30] with the K-score algorithm plug-in [31] and the output search results were converted to the search engine-independent pep1XML format [32]
The target fasta sequence file used for the search was obtained from the Candida Genome Database (CGD) [12] at: http://www.candidagenome.org/download/sequence/C_albicans_SC5314/Assembly21/ Common contaminants from the common Repository of Adventitious Proteins (cRAP) were appended. Then for each of these sequences, counterpart reversed decoy sequences were appended.
PeptideProphet [33] was then run on the search results to model the distributions of correctly and incorrectly assigned peptide-to-spectrum matches (PSMs). It then assigns probabilities of being correct for each PSM, yielding a sensitive, flexible approach to report results in a comparable manner. Next, iProphet [34] was used to combine additional sources of evidence including multiple identifications of the same peptide across spectra, experiments, and charge and modification states, allowing a more precise integration of evidence supporting the identification of each unique peptide sequence. ProteinProphet [35] was then run to refine iProphet probabilities by adding the information at the protein level, like the number of sibling peptides within a protein and to compute final protein level probabilities. The prophet tools together combine multiple layers of evidence and refine the model iteratively to achieve an optimal analysis of the data. Finally MAYU [36] estimated FDR at different levels for each contributing experiment and for the entire dataset based on the PSMs to decoy proteins.
This process followed the pipeline first implemented in the construction of the human plasma PeptideAtlas described in [14] and successfully applied to other builds such as the bovine milk and mammary gland PeptideAtlas [23]
Construction of the PeptideAtlas
The PeptideAtlas building process calculates the cumulative number of identified peptide and proteins across the experiments, gathers information on protein to genome location mappings and estimates the peptides Empirical and Predicted Suitability scores (ESS, PSS). The genomic mappings, since C. albicans is not present in the Ensembl database, which is the default PeptideAtlas uses to that purpose, were extracted from a generic feature file located at the following url: http://www.candidagenome.org/download/gff/C_albicans_SC5314/archive/C_albicans_SC5314_version_A21-s02-m05-r10_features.gff
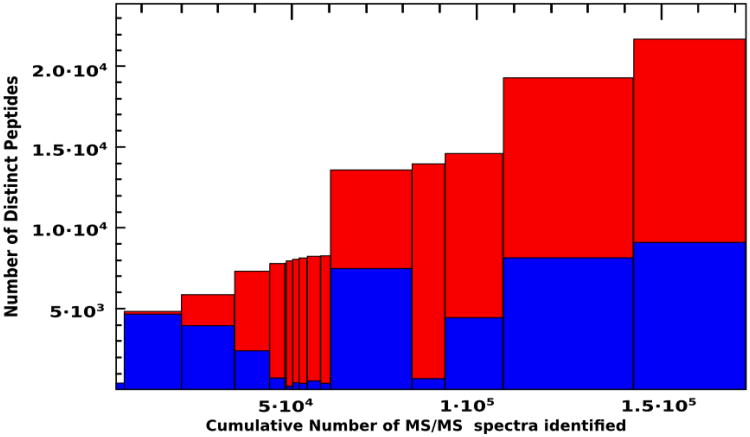
An overview of how the different experiments contribute, in terms of number of identified spectra and peptides, to the atlas build is depicted in Figure 1.
Figure 1.
Histogram showing the cumulative number of distinct peptides in the C. albicans PeptideAtlas. Each bar represents a different experiment that has contributed to the build. Bar width is proportional to number of high confidence PSMs. Height of the blue section of the bar represents the number of distinct peptides in each experiment and total height of the bar (red plus blue sections) indicates the cumulative number of peptides. The order of experiments is the same as in Table 1.
Besides, and due to the particularly rich number of identifications in experiments aimed at detection of phosphorylated proteins (experiments #3 to #13), a similarly processed version of the PeptideAtlas was created including in this case PTMProphet results which provide, alongside each modified residue, the probability that the post-translational modification is truly detected at that site.
Results and Discussion
Assessment of Proteome coverage and Functional enrichment analysis
The assembled proteomic datasets (Table 1) were subject to uniform data processing in order to build the C. albicans PeptideAtlas. The PSM assignment and protein inference processes were conducted by means of the consistent and robust pipeline TPP. The prophet tools integrate various levels of information and report identification results in statistical terms so that spectrum assignments, peptide to protein mappings and protein groups are statistically validated, leading to an overall improved sensitivity for a defined FDR level. As a result the generated C. albicans PeptideAtlas comprises 21938 peptides identified at a 0.24% FDR allocated to 2562 proteins at a 1.2% FDR, that is, a coverage of 41.3% of the 6209 C. albicans translated ORF sequences from the fasta database used for searches. While the presented instance of the C. albicans PeptideAtlas has reached unprecedented coverage, it does not represent a final representation of the respective proteome. Like other PeptideAtlas instances for other species, the C. albicans atlas will be expanded upon submission and processing of new MS data generated in ongoing projects.
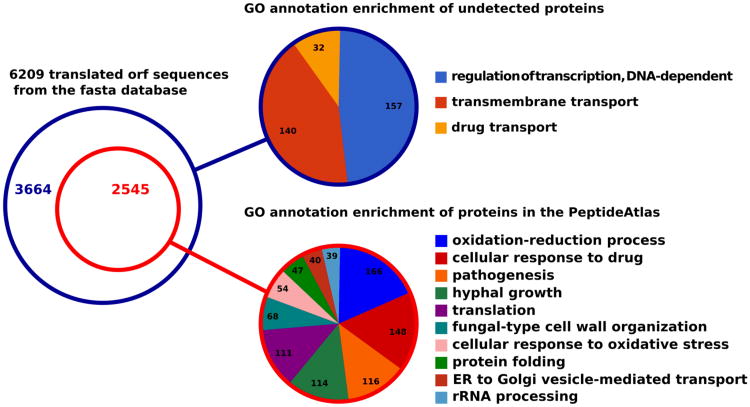
To determine the biological functions encompassed by the covered part of the proteome in this PeptideAtlas a Gene Ontology (GO) annotation enrichment analysis was carried out for the list of all detected C. albicans canonical proteins, excluding decoy hits, using the biological process ontology and Genecodis software [37]. Predictably, it generated a diverse array of clusters heterogeneously annotated, among which the largest in number of proteins are associated with the GO terms oxidation-reduction process, cellular response to drug, pathogenesis and hyphal growth respectively (Figure 2). The enrichment in some very generic GO terms such as oxidation-reduction process, cellular response to drug and translation supports the hypothesis that the diversity of experiments assembled to build the atlas provides a representative, unbiased subset of the C. albicans proteome. In contrast, the more precise groups resulting from the analysis related to pathogenesis, hyphal growth and fungal-type cell wall organization are consistent with the large contribution to the atlas by the experiment aimed at identifying proteins from cells in hyphal form and by the profusion of these sort of annotations in the source database.
Figure 2.
Gene Ontology annotation enrichment analysis for both the covered and undetected proteome subsets. All shown GO annotations correspond to the biological process ontology and were found significant for a p-value cut-off below 0.01.
As for the set of proteins present in the fasta database used for the searches that are not covered in the PeptideAtlas, they were subject to a similar analysis and were found to be enriched in annotations related to the transmembrane transport GO term (Figure 2). These proteins are not easily observed by LC-MSMS techniques as previously reported [20]. Also, we observed an enrichment in regulation of transcription, DNA-dependent in the undetected part of the proteome. Given the short life span and low abundance of many transcription factors it is plausible that they were not detected in the collected dasets and their under representation in proteomic data has also been reported in other proteomic studies and in PeptideAtlas instances from other species [20, 38, 39]. The low number of protein groups significantly associated with GO annotations in the undiscovered set is understandably due to the fact that 2460 out of 3665 of the undetected protein sequences, roughly two thirds, correspond to unnamed ORFs, meaning, that little is known about their biological function.
In addition to the groups of functionally characterized proteins, this PeptideAtlas offers solid empirical evidence for the existence of 1564 proteins, showing a ProteinProphet probability score greater than 0.9, corresponding to uncharacterized ORFs in the CGD database (i.e., one-third of all 4566 uncharacterized ORFs).
Proteins of interest. Case of use
From the clinical angle, the characterization of the C albicans proteome is focused on particular subproteomes, including cell surface constituents, and the set of proteins involved in the yeast-to-hypha transition. The cell wall, as the outermost cell structure represents the contact surface with host cells and therefore gathers many antigens, virulence factors and Pathogen Associated Molecular Patterns (PAMPs) [40]. Proteins involved in hyphal growth are also relevant in pathogenesis, in the sense that hyphae have been proven key for invasiveness whereas the switch back to yeast form plays a role in dissemination [41].
Within these groups, a selected set of proteins of interest present in the atlas, are the adhesins from the ALS family with a role in invasiveness Als2p and Als3p; the required for cell wall biogenesis and organization glycosidases Phr1p, Phr2p and Utr2p; mannosyltransferases Pmt1p, Pmt4 and Pmt6; the involved in the cell-wall glucan metabolism Mp65p and Ecm33p, and the hyphal cell wall constituents Hwp1, Csp37p and Rbt1p.
Other relevant proteins in the atlas are the ones related to apoptosis, since those would make an ideal target for the treatment of invasive candidiasis. Among those, the atlas contains Mca1p, Bcy1p, Ras1p and three unnamed ORFs with orthologous in other species showing roles in the apoptotic process (orf19.713, orf19.967 and orf19.7365).
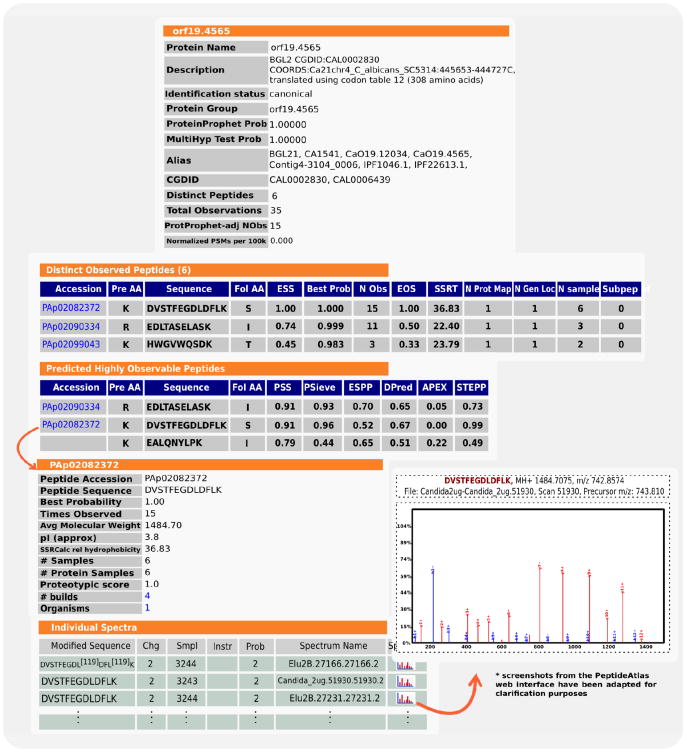
For any particular protein of interest, the PeptideAtlas web interface provides tools to explore the data. A user can browse through a set of protein and peptide-centric views as illustrated in Figure 3 for the specific case of Bgl2p, a cell wall glucosyltransferase. Its corresponding observed peptides are highlighted in the protein sequence and sorted by the Empirical Suitability Score (ESS), which represents the proportion of the number of samples in which the peptide is observed with regard to the number of samples in which the original protein is observed. This parameter, in combination with others, such as number of protein mappings, genome location and amino acid composition will help the user to select candidate proteotypic peptides for a targeted proteomics (SRM, Selected Reaction Monitoring) experiment.
Figure 3.
Protein- and peptide-centric views for Bgl2p are depicted. Distinct observed peptides are ranked by the BestProb parameter (representing the PeptideProphet probability). Of those, most probably, some will also be present in the following Predicted Highly Observable Peptides table were peptides are ranked by PSS, a combination of different prediction algorithms. For all observed peptides, spectra from the different experiments are also available.
Concerning those cases where a selected protein of interest is not observed in the selected build, the PeptideAtlas also provides the Predicted Suitability Score (PSS), a value resulting from the combination of different observability prediction algorithms based upon physico-chemical properties derived from the amino acid composition and previous training data sets as described in [42].
The build that assembles the phosphoprotein enrichment experiments may be of great potential interest biological processes such as signal transduction, since it encompasses a number of kinases and phosphatases. A total of 421 different phosphopeptides were detected and allocated to 210 phosphoproteins. The largest number of phosphorylation sites occur in S, 410 phosphopeptides contain, at least, one phosphorylation in S; 79 phosphopeptides contain, at least, one phosphorylation in T; and 10 phosphopeptides contain one phosphorylation in Y.
Conclusions
This C albicans PeptideAtlas build provides empirical identification evidence for 21938 unique peptides including 421 phosphopeptides at a 0.24% peptide-level FDR that account for a high-confidence set (as defined in [14]) of 2562 canonical proteins at a 1.2% protein-level FDR representing thus a significant advance in the proteomic characterization of C. albicans.
Through the web interface, an important set of tools are made available to the scientific community, enabling a solid foundation to study different basic biological processes like dimorphism, signal transduction, apoptosis and the interaction with the human host. Furthermore, its value as a resource for proteotypic peptide selection is of great potential interest for future SRM experiments.
The current version of the PeptideAtlas can be found at https://db.systemsbiology.net/sbeams/cgi/PeptideAtlas/buildDetails?atlas_build_id=323 and the version including PTM results at: https://db.systemsbiology.net/sbeams/cgi/PeptideAtlas/buildDetails?atlas_build_id=324
Significance.
This C. albicans PeptideAtlas resolves the previous absence of fungal pathogens in the PeptideAtlas project. It respresents the most extensive characterization of the proteome of this fungus that exists up to the current date, including evidence for uncharacterized ORFs. Through its web interface, PeptideAtlas supports the study of interesting proteins related to basic biological mechanisms key to virulence such as apoptosis, dimorphism and adherence. It also provides a valuable resource to select candidate proteotypic peptides for future (SRM) targeted proteomic experiments.
Acknowledgments
The Proteomics Unit UCM-Parque Científico de Madrid is a member of the ProteoRed-Spanish National Institute for Proteomics.
We are thankful to María Luisa Hernáez and Jose Antonio Reales for helping in sample obtention from the hyphal and yeast form protein extracts and to Antonio Serna for providing the tandem mass spectra from the triple-TOF instrument. Also Aida Pitarch helped in the preparation of the manuscript.
This work was supported by BIO 2009-07654 and BIO 2012-31767 from the Ministerio de Economía y Competitividad, PROMPT (S2010/BMD-2414) from the Comunidad de Madrid, and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD12/0015) -co-financed by European Development Regional Fund “A way to achieve Europe” ERDF.
EWD, ZS, and RLM are supported in part by the National Institute of General Medical Sciences, under grant No. R01 GM087221, 2P50 GM076547/Center for Systems Biology, the National Science Foundation MRI [grant No. 0923536], the EU FP7 grant ‘ProteomeXchange’ [grant number 260558], and by the Luxembourg Centre for Systems Biomedicine and the University of Luxembourg.
RA is supported in part by ERC advanced grant ‘Proteomics v3.0’ (grant no. 233226) of the European Union
Abbreviations
- SRM
Selected Reaction Monitoring
- CGD
Candida Genome Database
- FDR
False Discovery Rate
- PSM
Peptide-Spectrum Match
- PRIDE
Protein Identifications Database
- PSS
Predicted Suitability Score
- ESS
Empirical Suitability score
References
- 1.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39:309–17. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 2.Moran C, Grussemeyer CA, Spalding JR, Benjamin DK, Reed SD. Comparison of costs, length of stay, and mortality associated with Candida glabrata and Candida albicans bloodstream infections. Am J Infect Control. 2010;38:78–80. doi: 10.1016/j.ajic.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tong KB, Murtagh KN, Lau C, Seifeldin R. The impact of esophageal candidiasis on hospital charges and costs across patient subgroups. Curr Med Res Opin. 2008;24:167–74. doi: 10.1185/030079908x253401. [DOI] [PubMed] [Google Scholar]
- 4.Fernández-Arenas E, Cabezón V. Integrated proteomics and genomics strategies bring new insight into Candida albicans response upon macrophage interaction. Mol Cell Proteomics. 2007;6:460–78. doi: 10.1074/mcp.M600210-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Pitarch A, Nombela C, Gil C. Prediction of the clinical outcome in invasive candidiasis patients based on molecular fingerprints of five anti-Candida antibodies in serum. Mol Cell Proteomics. 2011;10:M110.004010. doi: 10.1074/mcp.M110.004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitarch A, Nombela C, Gil C. Candida albicans biology and pathogenicity: insights from proteomics. Methods Biochem Anal. 2006;49:285–330. [PubMed] [Google Scholar]
- 7.Pitarch A, Nombela C, Gil C. Contributions of proteomics to diagnosis, treatment, and prevention of candidiasis. Methods Biochem Anal. 2006;49:331–61. doi: 10.1002/0471973165.ch18. [DOI] [PubMed] [Google Scholar]
- 8.Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–9. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vialás V, Nogales-Cadenas R, Nombela C, Pascual-Montano A, Gil C. Proteopathogen, a protein database for studying Candida albicans--host interaction. Proteomics. 2009;9:4664–8. doi: 10.1002/pmic.200900023. [DOI] [PubMed] [Google Scholar]
- 10.Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J Proteome Res. 2004;3:1234–42. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 11.Smith BE, Hill JA, Gjukich MA, Andrews PC. Tranche distributed repository and ProteomeCommons. org Methods Mol Biol. 2011;696:123–45. doi: 10.1007/978-1-60761-987-1_8. [DOI] [PubMed] [Google Scholar]
- 12.Costanzo MC, Arnaud MB, Skrzypek MS, Binkley G, Lane C, Miyasato SR, et al. The Candida Genome Database: facilitating research on Candida albicans molecular biology. FEMS Yeast Res. 2006;6:671–84. doi: 10.1111/j.1567-1364.2006.00074.x. [DOI] [PubMed] [Google Scholar]
- 13.Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, et al. The PeptideAtlas project. Nucleic Acids Res. 2006;34:D655–8. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrah T, Deutsch EW, Omenn GS, Campbell DS, Sun Z, Bletz JA, et al. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol Cell Proteomics. 2011;10:M110.006353. doi: 10.1074/mcp.M110.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van PT, Schmid AK, King NL, Kaur A, Pan M, Whitehead K, et al. Halobacterium salinarum NRC-1 PeptideAtlas: toward strategies for targeted proteomics and improved proteome coverage. J Proteome Res. 2008;7:3755–64. doi: 10.1021/pr800031f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schubert OT, Mouritsen J, Ludwig C, Röst HL, Rosenberger G, Arthur PK, et al. The Mtb Proteome Library: A Resource of Assays to Quantify the Complete Proteome of Mycobacterium tuberculosis. Cell Host Microbe. 2013;13:602–12. doi: 10.1016/j.chom.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lange V, Malmström JA, Didion J, King NL, Johansson BP, Schäfer J, et al. Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol Cell Proteomics. 2008;7:1489–500. doi: 10.1074/mcp.M800032-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindner SE, Swearingen KE, Harupa A, Vaughan AM, Sinnis P, Moritz RL, et al. Total and putative surface proteomics of malaria parasite salivary gland sporozoites. Mol Cell Proteomics. 2013;12:M110.024505. doi: 10.1074/mcp.M112.024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King NL, Deutsch EW, Ranish JA, Nesvizhskii AI, Eddes JS, Mallick P, et al. Analysis of the Saccharomyces cerevisiae proteome with PeptideAtlas. Genome Biol. 2006;7:R106. doi: 10.1186/gb-2006-7-11-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunaratne J, Schmidt A, Quandt A, Neo SP, Sarac OS, Gracia T, et al. Extensive Mass Spectrometry-Based Analysis of the Fission Yeast Proteome: The S. pombe PeptideAtlas. Mol Cell Proteomics. 2013;12:M112.023754. doi: 10.1074/mcp.M112.023754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loevenich SN, Brunner E, King NL, Deutsch EW, Stein SE, Aebersold R, et al. The Drosophila melanogaster PeptideAtlas facilitates the use of peptide data for improved fly proteomics and genome annotation. BMC Bioinformatics. 2009;10:59. doi: 10.1186/1471-2105-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan QWT, Parker R, Sun Z, Deutsch EW, Foster LJ. A honey bee (Apis mellifera L.) PeptideAtlas crossing castes and tissues. BMC Genomics. 2011;12:290. doi: 10.1186/1471-2164-12-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bislev S, Deutsch E, Sun Z. A Bovine PeptideAtlas of milk and mammary gland proteomes. Proteomics. 2012;12:2895–9. doi: 10.1002/pmic.201200057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deutsch E, Lam H, Aebersold R. PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Reports. 2008;9:429–34. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11:O111.016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cabezón V, Llama-Palacios A, Nombela C, Monteoliva L, Gil C. Analysis of Candida albicans plasma membrane proteome. Proteomics. 2009;9:4770–86. doi: 10.1002/pmic.200800988. [DOI] [PubMed] [Google Scholar]
- 27.Monteoliva L, Martinez-Lopez R. Quantitative proteome and acidic subproteome profiling of Candida albicans yeast-to-hypha transition. J Proteome Res. 2010;10:502–17. doi: 10.1021/pr100710g. [DOI] [PubMed] [Google Scholar]
- 28.Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, et al. A guided tour of the Trans Proteomic Pipeline. Proteomics. 2010;10:1150–9. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martens L, Chambers M, Sturm M. mzML—a community standard for mass spectrometry data. Mol Cell Proteomics. 2011;10:R110.000133. doi: 10.1074/mcp.R110.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–7. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 31.MacLean B, Eng J, Beavis R, McIntosh M. General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics. 2006;22:2830–2. doi: 10.1093/bioinformatics/btl379. [DOI] [PubMed] [Google Scholar]
- 32.Keller A, Eng J, Zhang N. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol. 2005;1:2005.0017. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 34.Shteynberg D, Deutsch EW, Lam H, Eng JK, Sun Z, Tasman N, et al. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol Cell Proteomics. 2011;10:M111.007690. doi: 10.1074/mcp.M111.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 36.Reiter L, Claassen M, Schrimpf S. Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol Cell Proteomics. 2009;8:2405–17. doi: 10.1074/mcp.M900317-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012;40:W478–83. doi: 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding C, Chan DW, Liu W, Liu M, Li D, Song L, et al. Proteome-wide profiling of activated transcription factors with a concatenated tandem array of transcription factor response elements. Proc Natl Acad Sci U S A. 2013;110:6771–6. doi: 10.1073/pnas.1217657110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simicevic J, Schmid AW, Gilardoni PA, Zoller B, Raghav SK, Krier I, et al. Absolute quantification of transcription factors during cellular differentiation using multiplexed targeted proteomics. Nat Methods. 2013;10:570–6. doi: 10.1038/nmeth.2441. [DOI] [PubMed] [Google Scholar]
- 40.Vialás V, Perumal P, Gutierrez D, Ximénez-Embún P, Nombela C, Gil C, et al. Cell surface shaving of Candida albicans biofilms, hyphae and yeast form cells. Proteomics. 2012;8:2331–9. doi: 10.1002/pmic.201100588. [DOI] [PubMed] [Google Scholar]
- 41.Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell. 2003;2:1053–60. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol. 2007;25:125–31. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]