Abstract
NMR has matured into a technique routinely employed for studying proteins in near physiological conditions. However, applications to larger proteins are impeded by the complexity of the various correlation maps necessary to assign NMR signals. This article reviews the data analysis techniques traditionally employed for resonance assignment and describes alternative protocols necessary for overcoming challenges in large protein spectra. In particular, simultaneous analysis of multiple spectra may help overcome ambiguities or may reveal correlations in an indirect manner. Similarly, visualization of orthogonal planes in a multidimensional spectrum can provide alternative assignment procedures. We describe examples of such strategies for assignment of backbone, methyl, and nOe resonances. We describe experimental aspects of data acquisition for the related experiments and provide guidelines for preliminary studies. Focus is placed on large folded monomeric proteins and examples are provided for 37, 48, 53, and 81 kDa proteins.
Keywords: Large protein, Spectral overlap, Resonance assignment, Spectra analysis, Nuclear Magnetic Resonance (NMR)
1. Introduction
Over the years, nuclear magnetic resonance has evolved into a popular technique for studying biological systems in near physiological conditions. This popularity results in a large part from the versatility of the technique: NMR allows for studies of kinetics, thermodynamics, dynamics and structural features of molecules at atomic resolution. In turn, this versatility results from the ability to manipulate spins in a predictable manner and generate a multitude of correlation maps in which NMR signals, displayed as correlation cross-peaks, report on molecular properties. Thus, a multitude of experiments have been designed to probe molecular features with NMR parameters such as signal line-widths, scalar couplings, residual dipolar couplings, and nuclear Overhauser effects. However, to translate these spectroscopic features into molecular information, the investigator must first assign the observed NMR signals to the corresponding nuclei in the molecule under study. Consequently, a correspondingly large number of experiments have been developed to assign NMR signals, many of which will be discussed here. The chances of succeeding are intimately tied to the spectroscopic properties of the biological sample, which may present challenges such as spectral crowding, line-broadening, signal losses or frequency degeneracies. Here, we focus on folded monomeric proteins, i.e. a single polypeptidic chain consisting of a large number of residues (300–1000), rather than macromolecular assemblies, oligomeric proteins, and large unfolded proteins. Arguably, folded monomeric proteins provide a good framework to discuss all challenges. This article describes how analyzing various combinations of NMR spectra can help overcome such obstacles. Thus, the majority of the discussion presented centers on the analysis of NMR data rather than on the acquisition of the data. We do not review an exhaustive list of NMR experiments, but instead we introduce a series of protocols employing a select number of NMR experiments.
It is our hope that these protocols may be used as an introduction for new investigators or as a source of inspiration for spectros-copists meeting an impasse while assigning NMR spectra of challenging proteins.
The data visualization and protocols are described in a manner that should facilitate their implementation in popular assignment software packages. Many of the spectra layouts are readily available when using the program CARA [1], but can easily be reproduced with CCPN [2], SPARKY [3] or NMR View [4].
2. Terminology and notation
Frequency degeneracy: refers to signals that have overlapping frequencies in one dimension, whereas spectral overlap refers to signals that are partially or completely superimposed in all dimensions of a given multidimensional spectrum. We often refer to frequency degeneracy in contrast to spectral overlap to emphasize that signals may be resolved (free from overlap) and yet still be subject to degeneracies in frequencies along one or more dimensions in the spectrum; in general, the term refers to the frequency degeneracy of one type of nucleus (e.g. 13Cα). Signals suffering from spectral overlap necessarily feature frequency degeneracies along multiple dimensions of a spectrum.
Spectral crowding: confinement of a multitude of NMR signals within a given spectral region. Spectral crowding occurs for proteins with a larger number of residues, but also for intrinsically unfolded proteins and often for alpha-helical proteins. Even when the signals are only subject to partial overlap, the abundance of correlations within a small spectral region greatly increases the likelihood of erroneous assignments, in particular with limited resolution.
Autocorrelated relaxation originating from a particular mechanism is denoted by the label of this mechanism (e.g. DD for a general dipolar relaxation, or NH for the contribution of the HN–N dipole/dipole interaction); cross-correlated relaxation uses the labels of each interaction involved (e.g. CSA/DD for the interference between a chemical shift anisotropy (CSA) and a dipolar interaction or N/NH for the interference between nitrogen CSA and the HN–N dipole/dipole interaction).
TROSY (transverse relaxation optimized spectroscopy): The TROSY principle is the exploitation of relaxation interferences, or cross-correlated relaxation, to reduce transverse relaxation rates. Usage of the term TROSY can be somewhat confusing since the literature employs the same acronym to refer to either the TROSY principle, or to a 2D pulse sequence using this method, or specifically to using N/NH and H/NH cross-correlated relaxation (excluding other relaxation mechanisms). In this article, the term will most often be used in an adjectival sense, i.e. as a qualifier within the name of a pulse sequence to indicate that some part of it incorporates use of the TROSY principle. To make it explicitly clear that this is not meant to imply concatenation of a TROSY pulse sequence block with other pulse sequence blocks specified within the same name, it will be written as a prefix in lower case italics “trosy-”. The notation HN-trosy- will be used (instead of just trosy-) when it is specifically N/NH and H/NH cross-correlated relaxation that is being used. Thus, for instance, 2D-HN-trosy-HSQC here means a 2D experiment correlating 15N and 1H signals through single-quantum coherences and employing N/NH and H/NH cross-correlated relaxation. trosy-HNCA is a pulse sequence with magnetization transfers and evolution periods designed to minimize transverse relaxation. trosy-HNCA not only incorporates the TROSY pulse sequence block but features additional modifications when compared to HNCA (e.g. no composite pulse decoupling of protons).
Backbone experiments, backbone triple resonance experiments: ensemble of all triple resonance experiments used for assigning backbone resonances. For small molecules, these consist of HNCA, HN(CO)CA, HN(CA)CO, HNCO, HNCACB, and HN(CO)CACB, sometimes with additional experiments not discussed in this article. The nuclei enclosed in brackets participate in magnetization transfers but are not frequency labeled during the pulse sequence. For large proteins, the minimal set comprises trosy-HNCA, trosy-HN(CA)CO, trosy-HNCO, and trosy-HN(CA)CB. See Sections 4 and 5.2.
Spin system: for a given residue type, the corresponding set of spins that can be observed by NMR. In larger proteins, this is often limited to H, N, CA, CB, CO and the methyls of selected residues (A, I, L, V, etc.). We will use the notation {H, N} to refer to a system comprising a pair of amide proton and nitrogen, {H, N, CO} if we need to consider the carbonyl carbon, etc.
Anchor correlations: correlations common to a set of experiments that can be used to relate the information provided by each experiment. For example, HN-anchors in trosy-HNCA and trosy-HN(CA)CO allow for pairing C’ and Cα chemical shifts, or HC-anchors in HMCMCBCA and HCCH-TOCSY can group Cα and Cβ together with Cδ1 and Cδ2 chemical shifts.
Sequential fragment: group of spin systems for which sequential correlations have been unambiguously identified. The signals are not necessarily assigned.
Fragment/sequence alignment: statistical comparison between the chemical shifts observed in a sequential fragment with those predicted from the protein primary sequence and the BMRB chemical shift database.
Labeling scheme: isotopic composition of a given protein according to residues, chemical groups, isotope types, etc. We use the following convention: the isotope is mentioned first, followed by the chemical group, and finally the residue types; e.g. 13C-CO-P denotes a sample in which the carbonyl carbon of prolines is enriched in 13C. If a combination of isotopes applies to chemical groups and/ or residues, they are listed together, e.g. 1H,13C-Me-ILV. Absence of group or residue label indicates that the labeling applies to the entire residue and to all residues in the protein, e.g. 13C-Ala-15N, denotes a protein with uniform 15N labeling and with all alanines uniformly enriched in 13C. For clarity, we use the prefix U to emphasize that an isotope is used uniformly in cases where another isotope is used uniformly with some exceptions. For example, 1H–13C–Me–2H–ILV–2H–U–15N denotes a protein uniformly deuterated with the exception of isoleucine, leucine, and valine (which have protonated methyls), uniformly enriched in 15N (with no exception) and enriched in 13C for the methyls of isoleucine, leucine, and valine only. 1H–Me–2H-ILV–2H–U–15N–U–13C denotes a protein uniformly enriched in 15N and 13C and deuterated with the exception of Ile, Leu, and Val methyls.
3. Preliminaries: sample design and quality control
The success of assigning NMR resonances depends critically on the design of a suitable sample. Thus, much time and effort should be dedicated to screening various constructs (e.g. proteins with different boundaries in their primary sequences, fusion proteins, co-expression with binding partners, etc.) and optimizing buffers for protein expression, protein extraction and for NMR measurements. The methods available for performing such screenings have been reviewed elsewhere [5,6], and, here, we will merely discuss preliminary NMR experiments that should be performed before recording spectra for assigning resonances.
NMR is exquisitely sensitive to measuring conditions and will differentiate samples that behave identically when probed with other analytical methods. For example, size exclusion chromatography or gel electrophoresis (native or non-native) may indicate that a protein of interest is monomeric whereas the protein dimerizes in the high concentrations found in the NMR tube. Many such undetected sample properties may have adverse effects on the quality of the spectra. Examples include slow aggregation, which results in signal losses over the course of the measurements, oligomerization, which results in line broadening, or partial unfolding, which results in a large dynamic range in signal intensities. Because these adverse effects may not be detected prior to NMR investigations, it is important to carefully characterize the sample in situ.
Proper folding of the protein can be probed with a two-dimensional H/N correlation map (e.g. HN-HSQC [7] or HN-trosy-HSQC [8]). Partial unfolding, or long unstructured regions in the molecule, results in spectral crowding at the center of the amide proton spectral region since the corresponding residues are subject to poorly differentiated chemical environments. In addition, lack of structural constraints leads to fast internal dynamics (ps–ns) resulting in much slower relaxation and hence in sharp and intense lines. If the sample conditions are not modified, it will be impossible to record spectra that are optimal for both the structured (well-folded) and unstructured (unfolded) regions. Evolution times required for encoding indirect dimensions will either be too short for the flexible regions, resulting in a truncation of the free induction decay and hence sinc wiggles that degrade the spectrum in all areas surrounding the corresponding sharp signals, or too long for the structured region, increasing the amount of noise per signal and hence decreasing the signal-to-noise ratio for these residues. Thus, it is preferable to design a new construct (reduce the length of the polypeptide chain to reduce the number of unfolded residues) and/or screen for a new NMR buffer (stabilize the folded state), as exemplified in Fig. 1a.
Fig. 1.
Optimization of NMR buffer (a) and concentration test (b). (a) Top, in phosphate buffer, 150 mM NaCl, pH 6.7, this 12 kDa alpha-helical protein displays signs of improper folding and aggregation. Precipitation was observed within hours. Bottom, in PIPES, 150 mM NaCl, 2 mM MgCl2, the more uniform signal intensity indicates that the protein is now well folded and aggregation was not observed for several days. (b) HN-trosy-HSQC ID spectra of a 48 kDa protein at 1.5 mM (red), diluted to 260 µM (purple), and subsequently re-concentrated to 390 µM (black). All measurement conditions are identical. Concentrations were determined by UV spectroscopy, and the one of the diluted sample was verified by calculation using the final volume after dilution. The agreement between the concentrations estimated with the two methods indicates that no solute has been lost through precipitation. See text for details of interpretation.
The purity of the sample can easily be probed and monitored. One dimensional (1D) proton detected experiments, such as WATERGATE [9], display not only the protein signals but also those of additives present in the buffer. Comparison of 1D spectra will reveal inconsistencies between buffer preparations and also possible contaminants. During the course of an investigation, all samples should result in near identical 1D proton spectra.
Aggregation and oligomerization depend on the protein concentration; dilution tests should be performed to identify the optimal concentration for NMR measurements. These tests are achieved by recording a series of 1D experiments (WATERGATE or 1D trace of HN-HSQC) and monitoring the signal intensities as a function of the protein concentration, although recording 2D experiments as well is recommended, time permitting (see below). A non-linear relationship between the protein concentration and the signal amplitudes observed indicates (transient) oligomerization/aggregation at higher concentration or an increase in the solution’s viscosity (see 4). To prevent systematic losses during sample manipulation, it is preferable to perform successive dilutions or use a combination of dilutions and concentrations rather than iterative concentrations. Fig. 1b provides an example in which concentrations higher than 900 µM are subject to some form of oligomerization. The sample was initially concentrated to 1.5 mM and subsequently diluted six fold to 260 µM. For this protein, the result is quite startling: the signal intensities in 1D-traces of HN-trosy-HSQC are stronger at the lower of these two concentrations. Inspection of 2D HN-trosy-HSQC signals at high concentration displays line-broadening indicative of aggregation (not shown). The sample at 260 µM was then concentrated to 390 µM, where the signals are still almost proportional to the protein concentrations (260/390 = 2/3~0.7; signal(260)/signal(390) = 0.76). To identify the optimal condition, the same sample was concentrated to 1.3 mM and diluted first to 1.1 mM, then 900 µM (not shown). Concentrations were determined by UV spectroscopy; for samples obtained by dilution, the concentration was also calculated using the final volume after dilution. The agreement between the concentrations estimated from UV and those calculated using volume changes demonstrate that no solute is lost through precipitation. In the end, 1.1 mM gave the strongest signals, but intensities were proportional to concentrations up to 900 µM only. Moreover, some signals were broadened beyond detection in 2D correlation maps at 1.1 mM. Consequently, the remainder of the investigation was performed at concentrations below 900 µM.
The dilution tests performed above establish the optimal concentration for measurements, but do not demonstrate that the protein is monomeric. The size of the molecular assembly can be determined from spectroscopic techniques such as dynamic light scattering or fluorescence anisotropy. In particular, the fluorescence anisotropy of the dye ANS (l-Anilinonaphthalene-8-Sulfonic Acid) can be used to report on the size of arbitrary proteins, without requiring tryptophan or fluorescent probes. These methods are powerful tools to help optimize buffers for NMR measurements. However, it is best to also use NMR methods to directly probe the sample in situ since reversible transient binding events may affect NMR signals but may be invisible to other methods. Rotational and translational diffusion can both be measured by NMR to analyze the oligomeric state of the protein. Translational diffusion is estimated by proton-detected 1D pulse-field gradient experiments [10–12]. Rotational diffusion can be assessed by the 1D TRACT experiment [13]. These parameters can then be used to evaluate the molecular size. To probe for exchange phenomena (conformational fluctuations or transient self-association), the result of the TRACT experiment can be compared to a global estimation of nitrogen transverse relaxation times 15N-T2’s (again from 1D experiments); in contrast to 15N-T2, the TRACT experiment is insensitive to relaxation by chemical shift modulation and hence, to a large extent, to exchange. In the end, estimation of the protein’s oligomeric state in the NMR tube and preliminary assessment of dynamics will dictate the choice of NMR experiments to be conducted and possibly indicate that further sample optimization must be performed.
4. Challenges and solutions for optimal data collection
This section describes experimental challenges encountered in the assignment of large proteins and introduces concepts used to resolve these challenges. More detailed descriptions are provided when discussing each experiment in Section 5. In large monomeric proteins, the high number of resonances leads to spectral crowding and overlap. The long sequence of amino acids increases the likelihood of repetitions in the primary sequence, which in turn increases the likelihood of frequency degeneracies. Because the protein has a large volume there is a higher risk that large regions may not be accessible to the solvent. Thus, a number of amide protons may exchange very slowly with the solvent, preventing their detection in deuterated samples. The large volume occupied by the protein also limits the highest concentration attainable or degrades spectroscopic measurements at high concentrations (see 3). For example, a 2 mM sample of a 50 kDa protein corresponds to a 10% w/v solution and the viscosity is likely to be substantially increased, resulting in increased relaxation. Finally, the spins are subject to larger relaxation rates even under ideal measurement conditions. In summary, large monomeric proteins display crowded spectra with broad signals, in which many residues may not be detected.
Relaxation represents the return of the nuclear spins polarization to their values at equilibrium. Longitudinal relaxation causes the return of the spin state populations to values corresponding to this equilibrium polarization. Longitudinal relaxation affects longitudinal components of the magnetization vector, in the Bloch formalism, and its rate is denoted R1 = 1/T1, where T1 is the longitudinal relaxation time. A short longitudinal relaxation time is often considered beneficial since it allows for short recycling time and, hence, for the accumulation of a large number of transients per unit time. To reach the polarization at equilibrium, the coherences present during detection and during spin manipulations must disappear over time. This decoherence phenomenon is dubbed transverse relaxation since it affects transverse components of the magnetization vector in the Bloch formalism. The transverse relaxation rate is denoted R2 = 1/T2, where T2 is the transverse relaxation time. A long transverse relaxation time is favorable to NMR experiments since it allows for spin manipulation without losses in signal intensity. In summary, transverse relaxation affects coherences (single- or multiple-quantum), whereas longitudinal relaxation affects magnetization and longitudinal n-spin orders.
Both transverse and longitudinal relaxation have important consequences for NMR of large proteins. Larger molecules have a slower tumbling rate, which provides a more efficient relaxation mechanism for transverse relaxation, and, hence, coherences relax faster than in smaller proteins. This increased relaxation has two major impacts on NMR spectra. (i) Interferograms during detection and signal encoding periods decay faster and lead to broader Lorentzians after Fourier transformation (the line-width at half-height is given by R2/π). This line broadening degrades the maximal resolution that can be obtained in a given spectrum and, together with spectral crowding, further increases the occurrence of signal overlap. (ii) The coherences decay rapidly during the spin manipulation periods necessary for correlation maps, resulting in lower correlation efficiency, and, hence, lower signal-to-noise ratios. Many signals may not be detected at all after spin manipulations, leading to incomplete information. In addition, many proteins are subject to exchange phenomena that further enhance transverse relaxation. While this contribution is independent of the molecular size, the resulting deterioration in spectral quality has more dramatic consequences for larger proteins, which already suffer from large relaxation rates. Thus, studies of large proteins subject to structural fluctuations are particularly challenging. Overall, transverse relaxation has adverse effects on NMR spectra of larger proteins and must be combated.
Whereas the slower tumbling of larger proteins enhances transverse relaxation, it reduces longitudinal relaxation. This results in longer T1, and, hence, longer recovery times are needed between transient recordings. This reduces the sensitivity of NMR experiments, i.e. the signal-to-noise obtained in a given time. Protons are an exception to this behavior [14]. Long-range dipolar interactions between protons provide a mechanism behaving like transverse relaxation, i.e. with increased efficiency for slower tumbling. This mechanism becomes favorable for supramolecular assemblies larger than 350–400 kDa. This exception notwithstanding, relaxation affects larger proteins in a negative manner, whether through a reduction in T2 or because of an increase in T1.
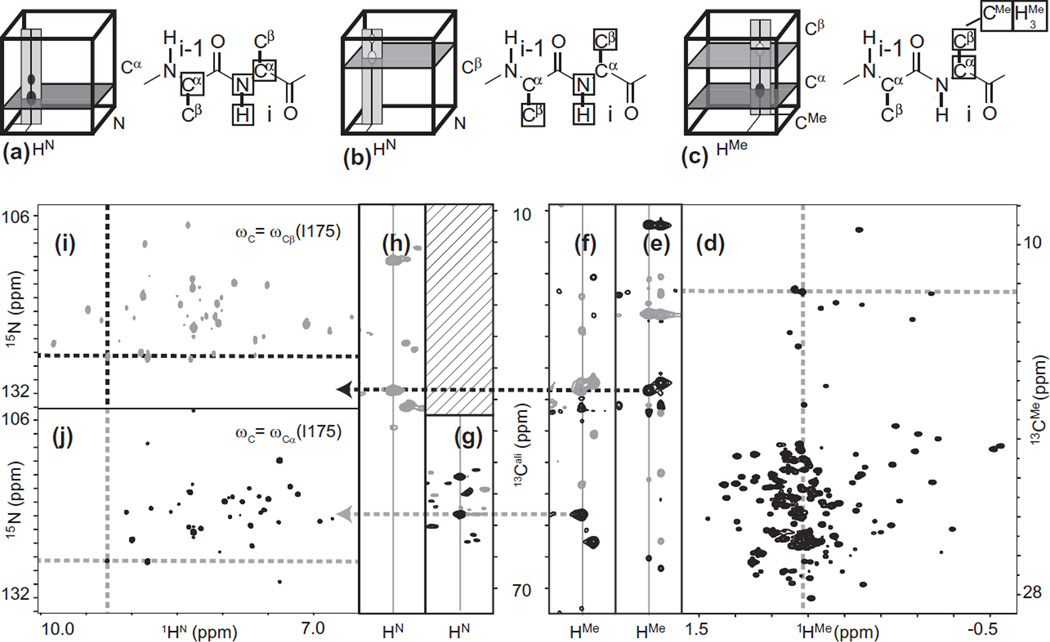
Both sample manipulation and spin manipulation are used to overcome adverse effects of spin relaxation. Most sample modifications consist of changing the isotopic composition of the protein (the so-called protein labeling scheme) to minimize transverse relaxation. Deuteration of carbon moieties dramatically reduces dipolar relaxation (by a factor of about 36 for autocorrelated mechanisms) and is a necessity for molecules larger than about 20–25 kDa [15]. For proteins in the range 20–35 kDa partial deuteration may be sufficient to reduce relaxation and has the advantage of preserving a high number of NMR signals, which can be used as atomic probes for functional and structural studies [16]. For higher molecular weights, perdeuteration (>98% 2H incorporation) is required and will be assumed for the majority of experiments described in this article. Spectroscopic probes can be re-introduced in deuterated proteins by selective protonation of target chemical moieties. The most popular approach was introduced by Gardner and Kay [17,18] and consists in preparing a protein featuring protons on methyl groups of Val, Leu and Ile (delta position) in a protein otherwise deuterated on carbons. Because these methyl groups are prevalent in hydrophobic cores, they have proven to be particularly efficient probes for structural investigations. Subsequent advances involved diastereoscopic selective protonation of Leu and Val methyls [19,20], as well as protocols for producing samples with Met [21–24], Ala [25,26], Ile (γ2) [27], and Thr methyls [28–30].
Transverse relaxation is further minimized by careful pairing of the labeling schemes with appropriate NMR pulse sequences. In the simple example of a deuterated protein, carbon coherences must be maintained in-phase with respect to deuterium, and composite pulse deuterium decoupling needs to be applied during related evolution periods. Absence of decoupling would result in a dramatic loss in sensitivity due to quadrupolar relaxation of deuterons, which affects carbon coherences antiphase with respect to deuterium. As a consequence deuterium pulses must be carefully calibrated to ensure efficient decoupling. More involved pulse schemes have been designed to manipulate relaxation effects. One of the major breakthroughs in the past 20 years was the exploitation of relaxation interferences, or cross-correlated relaxation, to reduce transverse relaxation rates. The method, developed by Pervushin and Wüthrich, was named TROSY for transverse relaxation optimized spectroscopy. The first implementation used the relaxation interference between the HN–N dipolar interaction and either the nitrogen CSA or the proton CSA, to reduce nitrogen or proton relaxation, respectively [8,31]. Other examples of TROSY-based techniques involve aromatic (using the CAr/CArHAr CSA/DD interference) [32] and methylene groups (using CH/CH DD/DD interferences) [33].
Besides the HN-TROSY effect, the most important TROSY effect involves coherences of methyl groups. Tugarinov and Kay observed that, for large proteins, HMQC pulse sequences give much improved signal-to-noise for methyl signals compared to HSQC [34]. This improvement originates from CH/HH dipolar interferences, which enhance the signal-to-noise of the central line in a CMeHMe multiple-quantum spectrum. Other advantages of methyl moieties include further reduction in relaxation by fast internal motion averaging and threefold advantage in proton concentration due to the degeneracy of methyl proton frequencies. When combined with deuteration, as in methyl protonated proteins in which all non-methyl non-exchangable protons are replaced by deuterons, these effects have rendered methyl groups the most significant spectroscopic probes of very large proteins. Methods for assigning their resonances, and related distance constraints, are discussed in Sections 6 and 7.2, respectively.
Regardless of the labeling scheme and TROSY methods, transverse relaxation must be accounted for during the various magnetization transfer delays present in pulse sequences. In particular, INEPT and COSY transfer delays that are calculated from scalar couplings must be reduced to account for transverse relaxation. Thus, before recording a multidimensional experiment it is customary to array the various delays of the experiment to maximize signal-to-noise. One should however keep in mind that multiple scalar couplings may be utilized for a given transfer and signal optimization must be interpreted with caution.
Sensitivity losses due to reduced longitudinal relaxation rates, R1, have proven to be more difficult to overcome. For smaller proteins, less than 15–20 kDa, it is possible to accelerate longitudinal relaxation by increasing the efficiency of proton HNH DD relaxation, as proposed by Brutscher and co-workers [35,36]. The recycling delay can then be reduced to a few milliseconds, thus permitting the acquisition of more transients per unit time and increasing sensitivity. This improvement requires selective manipulation of detected protons (e.g. HN), while leaving other protons at equilibrium. Unfortunately, an efficient acceleration necessitates samples with protonated carbons, which are subject to faster transverse relaxation than deuterated samples. For larger proteins, the resulting loss in signal-to-noise cannot be overcome by a fast repetition rate and amount to a reduction rather than an improvement in sensitivity. A similar amplification of R1 can be achieved by sample manipulation rather than spin manipulation. By adding relaxation agents which increase R1 more substantially than R2, such as the complex Ni(II)D02A, Chen and co-workers managed to reduce the recycling time by 70% [37]. However, experiments employing flip-back pulses benefit less from this method and residues in the core of the protein are expected to have a lesser improvement. Perhaps for these reasons, this method has not yet found a broad range of applications. Thus, in general, longer recycle delays are used for larger proteins than for smaller proteins, to account for relaxation. Just as effects of transverse relaxation are sometimes accounted for by optimizing (reducing) delays in pulse sequences, so it is customary to experimentally verify that an optimal recycling delay is used. This value is estimated by a simple array of the recycling delay and monitoring signal intensities. However, it is sensitivity (i.e. signal-to-noise in a given time) rather than the signal-to-noise ratio itself that must be probed. Thus, while a fully relaxed signal may require 5 s, 2 s may already provide 98% of signal intensity. In fact, sensitivity may be further improved by probing the cumulative effect of relaxation and signal averaging; while a recycle time of 3 s may optimize the signal-to-noise when recording 8 transients, an even better result may be obtained with 1.5 s and 16 scans. The size of potential phase cycling procedures may also play a role in deciding which acquisition set-up is employed.
This section has shown that many of the factors impeding studies of larger proteins can be overcome to various extents. Nonetheless, one indisputable fact remains: large monomelic proteins have a large number of atoms and assigning their signals is dramatically more challenging than for their smaller counterparts. The remainder of this article focuses essentially on this aspect.
5. Sequential assignment of backbone resonances
Assigning the signals observed in NMR spectra to the nuclei of a protein can be described in three steps. (i) So-called spin systems are populated with the chemical shifts of amino-acid nuclei detected in a given series of experiments; the chemical shifts typically comprise those of amide proton and nitrogen, as well as those of carbonyl, alpha and beta carbons. (ii) Correlations between signals of systems belonging to sequential residues enable one to link these systems into sequential fragments. (iii) Residue-specific chemical shifts are used to assign residue types (Ala, Gly, Ser, etc.) to spin-systems and each fragment is mapped to the amino acid sequence of the protein.
5.1. Identification of spin systems
In the assignment procedure, H–N correlations play a central role because all residues, with the exception of prolines and the N-terminal residue, feature a single pair of amide proton and nitrogen. Their 2D-correlation map is used as a starting point for the assignment and is used to monitor progress. Backbone assignment is often considered completed when all signals in an H–N correlation map have been assigned. For small proteins, with little overlap, the first step in defining spin systems consists in peak picking all H/N signals in an HSQC spectrum. The corresponding peaks, or {H, N} systems, will then be used as anchors when seeking sequential correlations. For larger proteins, however, it is highly preferable to peak pick HN-anchors by inspecting the 3D–HNCO experiment summarized in Section 5.2. The experiment is the most sensitive of all triple-resonance experiments and can be acquired in little time (6 h 30 in the example discussed in 5.2, i.e. for a 48 kDa protein at 800 µM). In addition, the spectrum provides only one correlation per residue, making it the 3D counterpart of a 2D-HN-HSQC. While automated peak picking procedures are available in most software packages, it is preferable to inspect the spectrum manually and identify partial overlaps that may be ignored by peak picking algorithms. A rapid and efficient procedure consists in inspecting H/C and N/C strips simultaneously for each HN-anchor. Thus, the HN-anchors are first identified in a 2D HN-correlation map, while the carbonyl carbon shifts of residues i – 1 are determined along the carbon dimension of the H/CO and N/CO strips. Simultaneous inspection of both strips allows for revealing potential overlap in the 2D-HN spectrum, whilst pairing the carbon shift with the appropriate HN-anchor. For example, two systems may have complete degeneracy in proton and near degeneracy in nitrogen. Resolving the signals in the 3D spectrum of HNCO, when the carbonyl carbon shifts are different, will allow for determining each nitrogen frequency and hence differentiate the two systems. Clearly, such a distinction is impossible if only H/CO strips are inspected. An extreme example of a quadruple overlap in H/N is discussed in Section 5.4 and Fig. 5. The same precaution is followed when the signals of other carbons must be identified (e.g. Cα or Cβ). For instance, H/Cα and N/Cα strips of each {H, N, CO} system are examined simultaneously to assign alpha carbon resonances and extend the system to {H, N, CO, CA}. This procedure minimizes mispairing resonances of alpha carbons to HN-anchors and thus to carbonyl carbons as well: the carbonyl carbon of a residue might otherwise be otherwise paired with the alpha carbon of another residue, which would result in incorrect sequential (Section 5.2) and residue type (Section 5.3) assignments. Such mispairing can have dramatic consequences, as discussed in Section 5.2. Overall, it is critical to thoroughly inspect all resonances while peak picking systems in larger proteins and, in case of ambiguity, it is preferable to not to assign carbon signals to specific {H, N} systems. Section 5.4 will show how unpicked signals can still be utilized for sequential assignment.
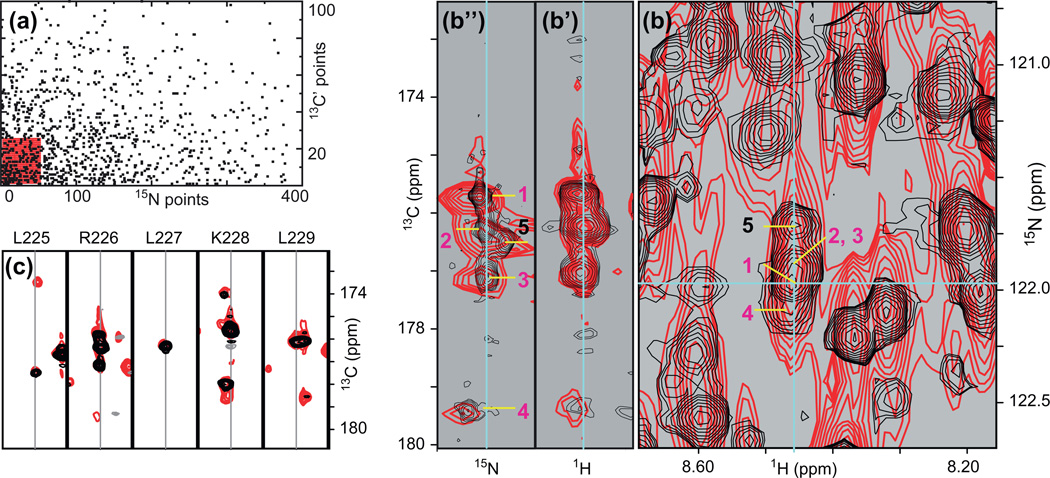
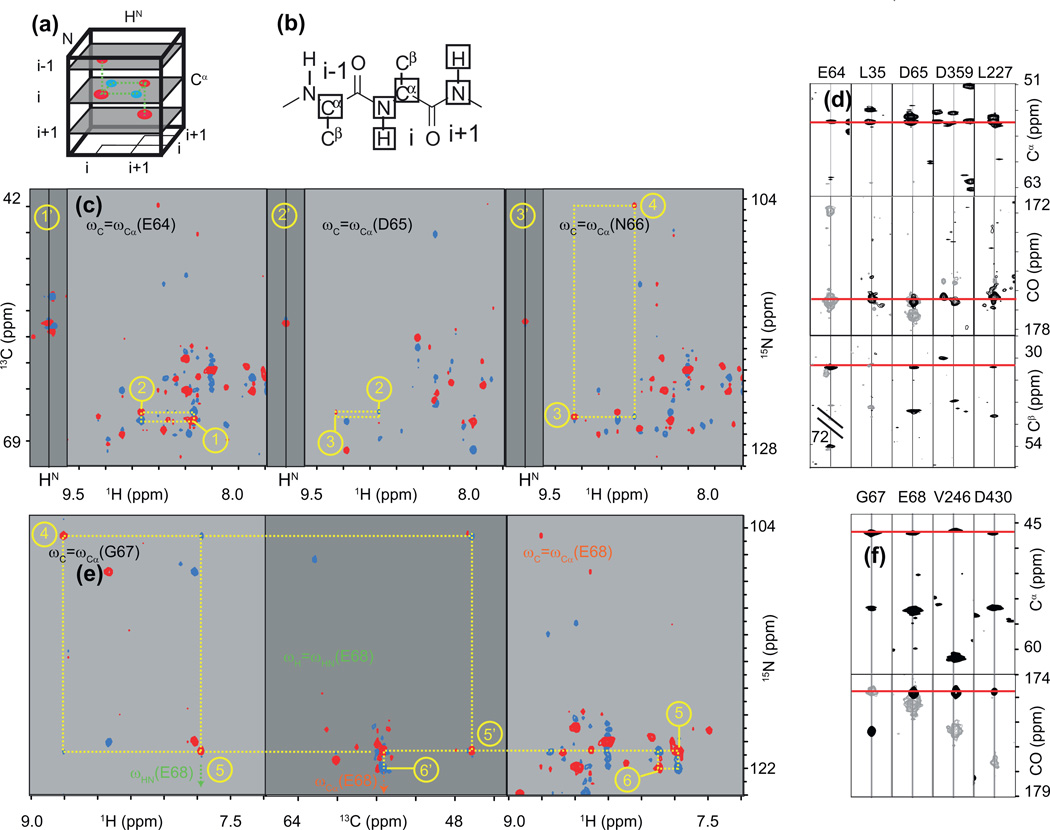
Fig. 5.
Application of non-uniform sampling (NUS) to the assignment of resonances of larger proteins. (a) Comparison of acquisition schemes for uniform (red) and nonuniform sampling (black). In both cases, 1025 complex points are being acquired and the experimental time is the same. The NUS schedule has been optimized for a trosy-HNCO recorded on a 48 kDa protein (Table 1). (b) Comparison of H/N projections of the 3D-HNCO experiment recorded with the schedules shown in (a). With a linear acquisition (red) only one (H, N) correlation is observed near the resonance of R226 (blue crosshair). With NUS (black), four (H, N) correlations can be distinguished. (b’) H/CO strip and (b”) N/CO strip at the shifts of R226. The 13C dimension shows four correlations in the linear acquisition (two of which cannot be assigned in the uniformly sampled spectrum because of simultaneous degeneracies in 1H and 15N frequencies). NUS reveals a fifth correlation and due to the higher resolution all five signals can now be assigned to different (H, N) correlations. (c) trosy-HNCO H/CO strips of a sequential fragment encompassing R226 showing both uniform (red) and non-uniform sampling (black). See text for details of interpretation.
5.2. Identification of sequential residues
The principles of the conventional approach for assigning protein backbone resonances were developed in the early 1990s following cost-effective production of 15N–13C (and later also 2H) labeled proteins. A series of 3D triple resonance experiments is used to identify signals belonging to sequential residues. For small proteins, the canonical list of backbone triple resonance experiments consists of HNCA, HN(CO)CA, HN(CA)CO, HNCO, HNCACB, and HN(CO)CACB or CBCA(CO)NH. The many aspects that must be considered for optimal data acquisition have been reviewed by Sattler et al. [38]. For larger proteins, high magnetic fields are needed to increase spectral resolution. Unfortunately, at high fields the transverse relaxation of carbonyl carbons is dramatically increased and magnetization transfers involving carbonyl carbons (in HN(CO)CA and HN(COCA)CB) result in mediocre signal-to-noise. Thus, only a subset of experiments can be used for assignment, resulting in a reduction in the information available for sequential assignment. In other words, while larger proteins require additional information to overcome the complexity of the task, less experimental data is available. This provides an added motivation for ensuring that the data from the subset of experiments used for larger proteins is exploited in an optimal manner during analysis, e.g. by following the protocols described in this article. The subset consists of trosy-HNCO, trosy-HNCA, trosy-HN(CA)CO and trosy-HN(CA)CB, which each provide both sequential and intra-residue correlations with the exception of trosy-HNCO, which only provides sequential correlations. These experiments are modified to minimize relaxation losses via the HN-TROSY method. Multiple HN-TROSY schemes have been developed over the years. At the time of writing, two implementations of HN-TROSY have distinct benefits when applied to backbone triple resonance experiments. Loria et al., and simultaneously Salzman et al., noted that ST2-PT derived HN-TROSY schemes can be concatenated with the last 13C → 15N INEPT of these experiments, thus reducing the length of the pulse sequence by 5.5 ms and improving sensitivity [39,40]. In contrast, Yang and Kay, and later Nietlispach, use a selection scheme involving longitudinal components of the density operator, hence with slower relaxation [41,42]. Which scheme is most appropriate cannot be predicted in a general manner since the outcome will depend on the sample properties, such as the labeling scheme that may vary substantially (e.g. percentage 2H) or the presence of ms or µs dynamics. Fortunately, a single H/N plane recorded on an HNCO is sufficient to compare both strategies and make a decision.
Sequential assignment exploits the similarity of 1JNCα and 2JNCα couplings within and between residues. Thus, a nitrogen single-quantum coherence evolving under JNCα can become antiphase with respect to both the alpha carbon of the same residue and that of the preceding residue. These antiphase coherences are then used either directly to correlate protons and nitrogens with alpha carbons, or indirectly to make transfers to other carbons. 2JNCα is most often slightly smaller than 1JNCα and intra-residue signals are therefore usually stronger than those of inter-residue correlations; this property is used for a tentative assignment of sequential versus intra-residue correlation.
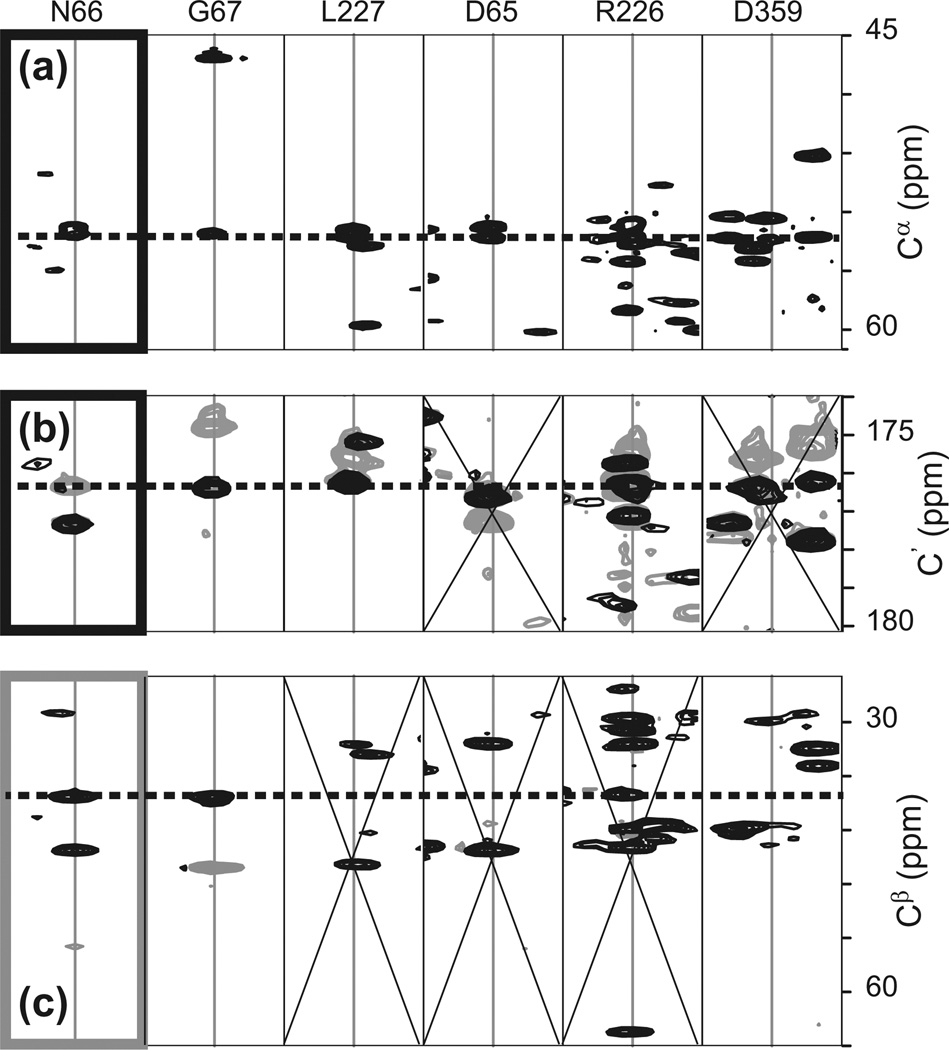
The simplest three dimensional experiment that uses 1JNCα and 2JNCα is the trosy-HNCA experiment, with a magnetization transfer summarized as H(i) → N(i) → Cα (i), Cα (i – 1)(t1) → N(i)(t2) → H(i)(t3). HNCA is the most sensitive of all sequential triple-resonance experiments and is often recorded first to assess the feasibility of the assignment process. It provides correlations of the form [ωH(i), ωN(i), ωcα (i)] and [ωH(i), ωN(i), ωCα (i – 1)], Fig. 2a). Thus each {H, N} system is correlated both to its own Cα carbon and also to that of the preceding residue. To identify signals of sequential residues, H/Cα and N/Cα strips are extracted for all {H, N} spin systems. Strips of sequential residues will feature a common correlation along the Cα dimension and the strips can be compared side by side to identify this feature. Most software packages provide an automated procedure, which will be described for a case in which the succeeding residue is being sought. Fig. 3 shows an example where the successor of residue N66 needs to be identified. First, the reference strip with correlations [ωH(i), ωN(i), ωCα (i)] and [ωH(i), ωN(i), ωcα (i–1)] is selected (labeled N66). The strips of all other {H, N, Cα} systems are then sorted according to the difference in chemical shifts between picked alpha carbons and the frequency ωCα (i); if picking was accurate, the first strip should feature the correlation [ωH(i+1), ωN(i+1), ωCα (i)] as well as [ωH(i+1), ωN(i+1), ωCα (i+1)]. As seen in Fig. 3a, this procedure is usually unsuccessful in identifying signals of sequential residues, because of degeneracies in carbon frequencies. Thus, here, five systems can potentially be the successor of N66. To overcome this ambiguity, the same strategy is repeated with spectra containing sequential correlations with other carbon moieties. The next experiment containing sequential correlations is the HN(CA)CO experiment, with a magnetization pathway H(i) → N(i) → Cα(i), Cα(i–l)→ CO(i), CO(i–1)(t1)→Cα(i), Cα(i–1)→N(i)(t2)→H(i)(t3), thus providing correlations [ωH(i), ωN(i) ωco(i)] and [ωH(i), ωN(i), ωco(i–1)], see Fig. 2c. The intra-residue and sequential correlations are provided by carbonyl carbons, while the two remaining frequencies correspond to the same HN-anchor as used in HNCA. Carbonyl carbons play a central role for assigning large proteins because the complementary experiment HNCO, that only features sequential correlations, is very sensitive. Thus the combination of HNCO and HN(CA)CO may often be the only means of distinguishing intra-residue and sequential resonances. The identification of sequential systems can then be performed with the joint information of trosy-HNCA and the trosy-HN(CA)CO/trosy-HNCO pair of experiments. Depending on the software, one or more of several different approaches can be used. The carbonyl carbon strips can be sorted independently from the alpha carbon strips and the best matches can be compared. Alternatively, the strips of the carbonyl carbons can be displayed by using the alpha carbons sorting, thus revealing mismatched carbonyl carbons for systems correlated by alpha carbons only (this is the strategy depicted in Fig. 3). Finally, both alpha and carbonyl carbons can be used simultaneously to sort the systems. All methods are equivalent in their effect: Cα and CO sequential correlations are combined to help identify the appropriate neighboring residue. Fig. 3b shows how the combination of trosy-HNCO and trosy-HN(CA)CO data can greatly reduce ambiguities coming from simple Cα strip matching. Two out of the five candidate sequential systems, D65 and D359, can now be discarded. Unfortunately, this advantage is often impaired by the fast relaxation of carbonyl carbons, which results in broad signals that are in addition often weak in trosy-HN(CA)CO. Consequently, in our example, three spin-systems provide sequential correlations to N66 both in trosy-HNCA and trosy-HNCO/trosy-HN(CA)CO. To identify the true sequential system, one must use the HN(CA)CB experiment. This experiment uses the transfer H(i) → N(i) → Cα(i), Cα(i – 1) → Cβ(i), Cβ(i – 1)(t1) → Cα(i), Cα(i – 1) → N(i)(t2) → H(i)(t3), to provide correlations [ωH(i), ωN(i), ωcβ(i)] and [ωH(i), ωN(i), ωcβ(i – 1)]. see Fig. 2c. We note that the transfer from Cα to Cβ has been set so that only Cβ coherences are encoded during t1. It is also possible to reduce the duration of this transfer to simultaneously provide Cα correlations, as typically done for small proteins (the experiment is then denoted trosy-HNCACB). Because a shorter transfer suffers less from transverse relaxation it may appear that this setting would be beneficial to larger proteins. However, in general, the reduction in relaxation losses does not compensate for dividing the signal between Cα and Cβ coherences. Since Cα coherences are independently provided by the very sensitive trosy-HNCA experiment, it is thus preferable to adjust the tro-sy-HN(CA)CB for optimal Cβ signal intensities. By using the Cβ correlations together with those of trosy-HNCA, trosy-HNCO and trosy-HN(CA)CO, it becomes possible to unequivocally assign the system of G67 as the successor of N66, Fig. 3c.
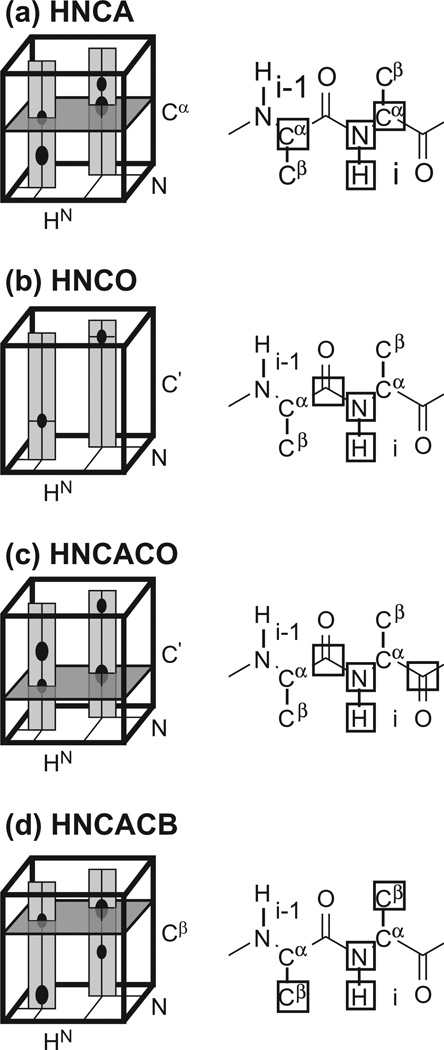
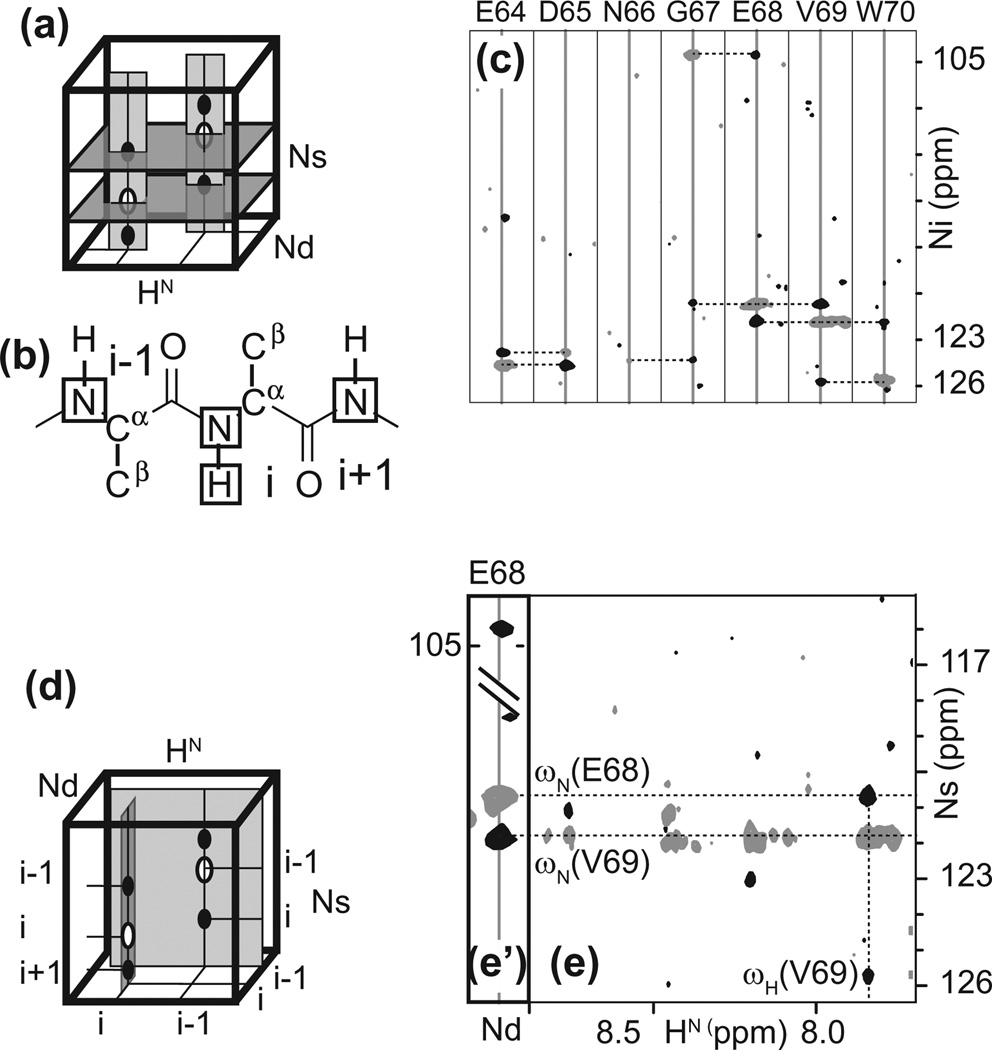
Fig. 2.
Correlations used for assigning backbone resonances with conventional experiments in large proteins. The cartoons on the left depict signals for two sequential residues. Strips that are extracted for comparisons are shown in light grey. The dark gray planes emphasize signals common to sequential residues. (a) HNCA. (b) HNCO. (c) HN(CA)CO. (d) HN(CA)CB. On the right, the rectangles indicate the set of nuclei that are correlated by each experiment for a residue i in a protein.
Fig. 3.
Strip matching for identifying sequential residues of a 48 kDa protein. The strips of the residue for which a successor is being sought (N66) are highlighted by the thick frames. (a) Comparison of Cα strips extracted from from trosy-HNCA. (b) C strips extracted from trosy-HN(CA)CO (grey) and from trosy-HNCO (black). (c) Cβ strips extracted from trosy-HN(CA)CB. Signals in grey have opposite phases and correspond to alpha carbons. The dotted lines indicate the carbon frequencies used for comparisons. Panels marked with an X denote sequential residue candidates that can be discarded. In this example, the combination of all three experiments identifies a unique sequential residue candidate. See text for details of interpretation.
The conventional approach for connecting sequential spin systems presented in this section is extremely successful and popular for small proteins. However, there are a number of setbacks that occur for large proteins. For example, differences in the magnitudes of 2JNCα and 1JNCα as well as relaxation effects may result in a sequential correlation (i – 1) of stronger amplitude than the intra-residue correlation (i) and the strongest signal in a given backbone triple resonance spectrum could then be erroneously assigned to an intra-residue correlation. In general, this situation disrupts the assignment procedure since no sequential spin system is identified. However, a spin system may accidentally match the misassigned correlations (the inter-residue is now an intra-residue signal) and corrupt assignments. Clearly, the likelihood of such a situation increases with the number of residues in the protein and large proteins are more prone to erroneous assignments of this form. In small proteins, experiments such as HN(CO)CA or HN(CO-CA)CB (or CBCA(CO)NH) provide only sequential correlations and resolve this ambiguity. In large proteins, at high field, only trosy-HNCO provides such inter-residue specific correlations with sufficient signal-to-noise, although partial information may be obtained from trosy-HN(CO)CA at fields lower or equal to 750 MHz. Thus, the assignment of sequential versus intra-residue correlations must be open to question until corroborated by multiple experiments.
In addition to ambiguities, the variation in scalar coupling magnitude and relaxation effects may lead to incomplete information: Small 2JNCα and large relaxation rates may preclude a sequential correlation so that only the signal of an intra-residue correlation is detected. Unfortunately, the absence of a second correlation cannot be distinguished from a degeneracy between the carbon frequencies of two sequential residues without experiments such as trosy-HN(CO)CA or trosy-HN(COCA)CB. Since these experiments have poor sensitivity at the high fields needed for large proteins, alternative methods have been designed to differentiate sequential and intra-residue correlations. In place of funneling the magnetization pathway through carbonyl carbons, these alternative methods rely on either editing or selecting one correlation only, as in intra-trosy-HNCA [43,44] and seq-trosy-HNCA [45,46], which provide intra- and inter-residue correlations, respectively. These experiments can be performed at high field but may not rescue systems featuring smaller 2JNCα Indeed, while the transfer periods can be optimized for a single value of JNCα, rather than a compromise between 1JNCα and 2JNCα. as in trosy-HNCA, this may not necessarily mean that all outliers are detected. In contrast, trosy-HN(CO)CA uses 1JNCO, which is homogenous for all residues in a protein. Thus, for a given sample at a given magnetic field, it is recommended to compare H/C planes of both strategies before choosing the experiment to be recorded. In summary, in larger proteins the relative assignment of sequential versus intra-residue correlations must often be open to doubt.
Partial overlap in H and N dimensions may also lead to erroneous assignments: the carbonyl carbon of a residue may be paired with the alpha carbon of another residue with H/N overlap. In small proteins, such a mispairing is usually identified because no sequential residue can be found. However, for larger systems, the abundance of degenerate carbon frequencies may provide accidental false positives, leading to incorrect sequential connectivities and resulting in wrong assignment. One way to help avoid such mispairing is to always examine H/Cα and N/Cα strips of each {H, N, C} system simultaneously and to record the data with as high a resolution as possible. Section 5.4 shows how maximal resolution can be obtained without resorting to prohibitively long data acquisition times. In general, it becomes critical to have multiple sets of sequential correlations to validate the various assumptions that need to be tested (sequential signal stronger than intra-residue, absence of sequential signal, degeneracy of carbon frequencies). Section 5.5 describes experiments that provide such additional correlations.
5.3. Primary sequence chemical shift mapping
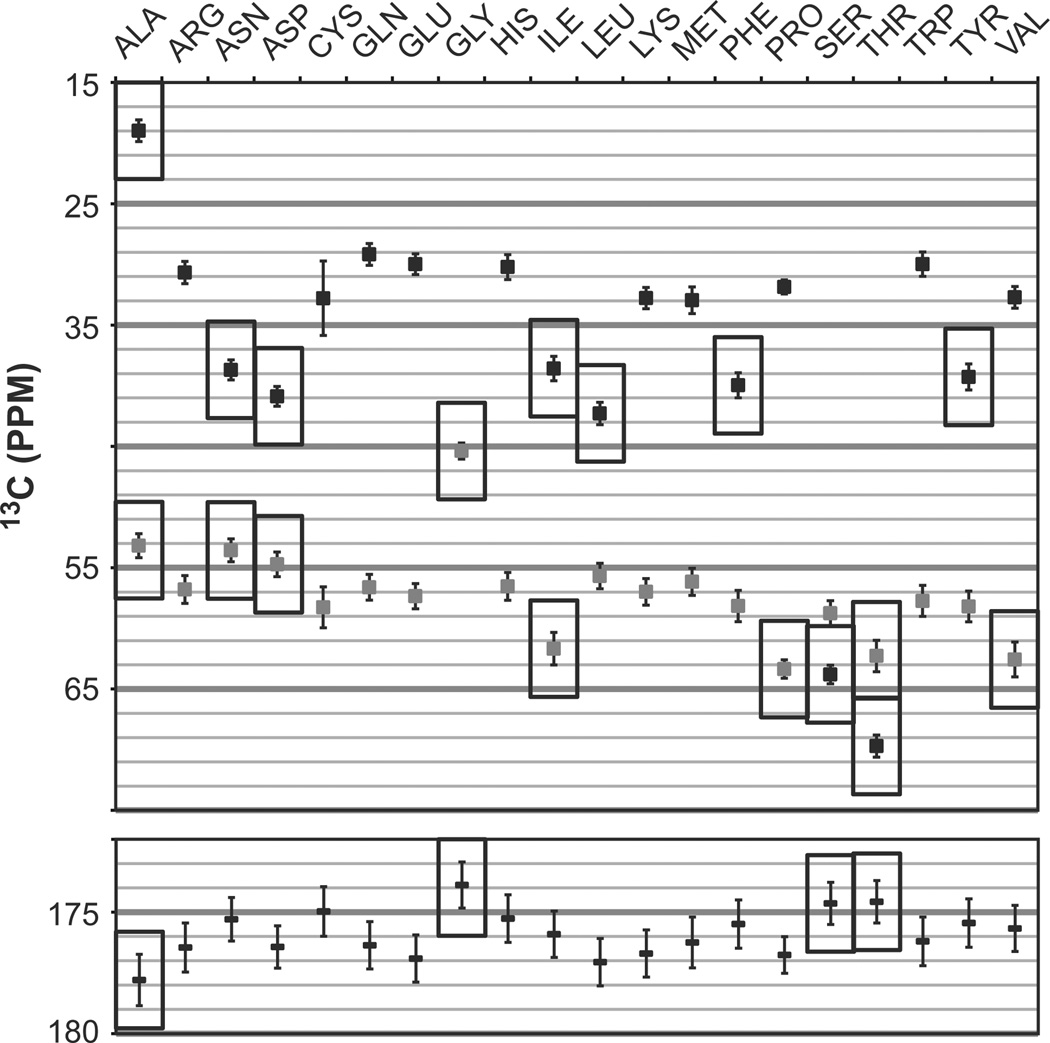
Once spin systems have been linked to form a so-called sequential fragment, i.e. a series of spins systems identified as being sequential, the residue type of each spin-system needs to be identified. To do so, we exploit the fact that NMR frequencies report on the nuclei’s chemical environments. The most useful chemical shifts for residue type assignment involve alpha carbons, beta carbons and carbonyl carbons. Fig. 4 displays the mean values of these chemical shifts calculated from the Biological Magnetic Resonance Data Bank. As can be seen, with a few exceptions, many amino acids have common chemical shifts and individual spin systems can rarely be assigned to a residue type. Instead, all chemical shifts identified in a sequential fragment are compared residue per residue to those predicted from the primary sequence and the BMRB statistics. In effect, this corresponds to a sequence alignment of the sequential fragment onto the sequence of amino acids using chemical shifts as the criterion for alignment. Many NMR assignment software packages feature automated routines for performing this so-called sequence alignment and provide a ranking of polypeptide fragments that match the spin systems. For larger proteins, many factors complicate this procedure. Redundancies in sequences of residues are more frequent (e.g. the sequence ADGS may be found several times) and redundancies in chemical shift patterns are even more frequent (e.g. DAES or NAQS give the same profile). Thus, large proteins require identification of longer fragments, relative to smaller proteins, before an unambiguous sequence alignment can be made. In addition, the larger size and greater complexity of the molecule increases the likelihood of encountering “outlier” residues, i.e. residues displaying chemical shifts departing from the BMRB statistics, for example when they are near aromatic rings or subject to unusual torsion angles. Thus, it becomes critical to add another level of scrutiny during sequence alignment to identify unambiguous chemical shift patterns and possible outliers. Indeed, some residues have unique spectroscopic signatures and can be used as check points. They include Alanine, with low Cβ (20 ppm) and to some extent Cα (53 ppm) and high CO chemical shifts (177 ppm), Glycine, with a very low Cα (45 ppm) chemical shift and a low carbonyl carbon shift (174 ppm), and Leucine with a unique Cβ at 42 ppm and a rather low Cα at 55 ppm. Serine residues have high Cα and Cβ at 59 and 64 ppm, respectively. Likewise Threonine has a Cα at 62 ppm and the Cβ with the highest shift, at 70 ppm. In addition, other residues have characteristic chemical shifts. Asparagine, Aspartate, Phenylalanine, and Tyrosine have beta carbons appearing at about 40 ppm, and the first two amino acids tend to have low Cα chemical shifts. Isoleucine and Valine have a high alpha carbon shifts, both around 62 ppm, and outlier Cβ of 38 and 33 ppm, respectively. Prolines are not detected but their sequential correlations can be identified with high Cα (63 ppm) and low Cβ (32 ppm). One drawback in using automated sequence alignment is that the chemical shifts of all systems are typically compared with the same degree of weighting, which does not account for residue-specific characteristic chemical shifts. However, the corresponding systems should be assigned to the relevant residue type in a manner that supersedes statistical analysis. This aspect of the assignment necessitates human intervention and, thus, it is critical to clearly identify all possible unambiguous residue types in a fragment and assign the corresponding residue types manually.
Fig. 4.
Mean chemical shifts and standard deviations used for residue type assignment. Data compiled from BRMB as updated on 02/14/2013. (Top) Alpha (grey) and beta (black) carbon shifts. (Bottom) Carbonyl carbon shifts. Rectangles highlight chemical shifts with values characteristic of a given residue type or small group of residue types. Often, chemical shifts of different carbons must be considered together for reside type identification.
In larger proteins it is frequent that backbone triple resonance spectra are insufficient to delineate residue type candidates or to identify outliers. A rather efficient means of accelerating the backbone assignment procedure and increasing its reliability simply consists in analyzing experiments providing side-chain chemical shifts while performing the sequential assignment. For example, while the Cα, Cβ and CO shifts of an outlier Isoleucine may suggest a Valine residue, the chemical shift of an Ile δ1 methyl would differentiate both residue types even under extreme perturbation of the residue’s magnetic environment. Such improvement in residue type determination is a major incentive to acquire all data at once. If spectroscopic methods are insufficient to delineate residue types, biochemical methods can be employed to produce samples with residue-specific isotope labeling [47,48].
The protein secondary structure, if it is known, can be used to facilitate sequential assignment. The statistical averages of Cα, Cβ and CO chemical shifts used in sequence alignment primarily report on side-chain substituents (e.g. hydroxyl, thiol, carboxylic groups, etc.). However, these carbons have been shown to be good reporters of secondary structures [49]. Cα and Cβ are affected by the dihedral angles φ and ψ, which have characteristic values in alpha-helices and beta-sheets, and carbonyl carbons are subject to different hydrogen bonds in each environment [50]. Cα and CO experience downfield shifts in alpha-helices and upfield shifts in beta-strands, while the opposite occurs for Cβ frequencies. These effects can be exploited during assignment with help from secondary structures predicted with bioinformatics tools [51], for instance to distinguish between two sequences of residues matching a sequential fragment but having different secondary structure environments. This property is best exploited when used in conjunction with NOESY spectra, which provide an orthogonal means of identifying secondary structures, i.e. without considering chemical shifts. NOESY spectra have characteristic patterns in alpha-helices, parallel beta-strands and antiparallel beta-strands. In larger proteins, alpha-helices provide multiple (HN, HN) sequential correlations, (i, i ± 1), (i, i ± 2), and, for long mixing times, (i, i ± 3), while beta-strands provide non-sequential through-space correlations and weak (i, i ± 1) correlations. While secondary structure is rarely employed when assigning small proteins, it can become an important tool for larger proteins, in particular to disambiguate repetitive sequence elements (e.g. QERAS in an alpha-helix and QERAS in a beta-sheet).
In this section, we have underlined the many challenges that occur during assignment of larger proteins using a conventional approach relying on strip matching of trosy-HNCA, trosy-HN(CA)CO, and trosy-HN(CA)CB data. Overall, they all can be summarized by the fact that limitations in resolution, signal detection, or ambiguities arising from spectral crowding, frequency degeneracies or redundancies in the amino acid sequence, require the formulation of multiple hypotheses throughout the assignment procedure: sequential connectivities may need to be tentatively assigned (i.e. one candidate arbitrarily selected), residue type determination may need to be assumed, secondary structure may be presumed, etc. Only once the assignment is completed, and possibly once a structure is available, can the user confirm these hypotheses. Clearly, this is a rather uncomfortable situation and the following sections of this review present strategies to avoid resorting to such hypotheses.
5.4. Maximizing resolution with non-uniform sampling
In the previous sections we have greatly emphasized the importance of resolving NMR signals during resonance assignment. Line-broadening effects in large proteins can be overcome to some extent by using deuterated samples and TROSY techniques. These strategies increase the maximal resolution that can be achieved for a given sample, i.e. a resolution defined by the so-called natural line width of the signal, l/πT2, where T2 (=1/R2) is the transverse relaxation time of the nucleus in the related dimension. However, this improvement can only be exploited if the digital resolution in the spectrum matches this line-width. Traditionally, the interfero-grams in indirect dimensions, e.g. t1, are obtained by monotonically sampling N evolution delays at increments Δt1 = 1/SW1 (where SW1 is the spectral width) up to a value t1max = N1 * Δt1, leading to a digital resolution Δv1 = l/t1max. Thus, the benefits of TROSY and deuteration come at the cost of the long acquisition times required to sample all N1 data points. For example, a 50 kDa perdeuterated protein requires the acquisition of about 400 complex points in 15N for optimal resolution at 900 MHz, amounting to 5 h of spectrometer time with 16 scans and a 1.5 s recycling delay. This acquisition time increases exponentially with the dimensionality of the experiment, which require N = N1 *N2 * … *Nn points for an n-dimensional experiment. Thus, only two-dimensional experiments can be recorded with optimal resolution within reasonable experimental time. This restriction may be bypassed by sampling only a subset Ns of evolution time points out of the N points that encompass the timax in each dimension, a process named non-uniform sampling (represented graphically in Fig. 5a) [52–54]. Ns is often referred to as the number of sampled points and dictates the time needed for acquisition. N is the size of the so-called sampling space and relates to the resolution in each indirect dimension as N = N1 * N2 * … * Nn, where Ni-is the number of points in dimension i covered by the sampling process. The acceleration provided by non-uniform sampling is described by the sampling factor, s = Ns/N (corresponding to an acceleration factor of 1/s). The matrix containing the coordinates of all Ns points sampled in the sampling space is called the sampling schedule. Because the interferogram is no longer sampled uniformly, Fourier transformation would result in major artifacts and alternative processing methods must be used to produce the spectrum. Maximum Entropy reconstruction [54–56], multiple-way decomposition [57,58], compressed sensing [59,60], and iterative soft threshold methods [61,62] can all provide faithful NMR spectra. A comparison of processing techniques is beyond the scope of this review article; all methods have been reviewed [63–66] and they all have undergone sufficient development to allow for straightforward processing. Other strategies pose constraints on the sampling schedule, i.e. on the choice of the Ns points selected from the N points encompassing the sampling space [67–73]. A comparison made by using Maximum Entropy reconstruction recently revealed that the original random sampling procedure, with exponential damping to match relaxation [52], provided the most faithful spectrum [74], although spectacular results were later obtained with density probabilities following Poisson distribution [62,75,76] or modified radial sampling [69]. Overall, non-uniform sampling of NMR spectra has now gained in popularity, and features for acquisition and processing of NUS data are readily available in the latest releases of the acquisition software of both major NMR spectrometer manufacturers.
Non-uniform sampling is particularly suited to providing maximal resolution and, hence, facilitating NMR studies of larger proteins. NUS was originally employed to rapidly provide spectra while maintaining a resolution comparable to those obtained with traditional acquisition. For instance, for small proteins, the complete set of six 3D backbone triple-resonance experiments requires about one week of acquisition with uniform sampling, but can be acquired with NUS within a day [77]. This acceleration was made possible by sampling about 22% of the points, as defined by the sampling factor s = Ns/N, in which N = N1*N2 was the same for non-uniform and uniform experiments. However, rather than reducing the time needed for acquisition (by reducing Ns), one may instead maintain a constant spectrometer time and increase the resolution (by increasing N). Such an application of nonuniform sampling allows for further reduction in the sampling factor. Indeed, a central condition for a faithful spectral reconstruction, regardless of the method used for reconstruction, is that enough points be sampled to reproduce the many NMR signals along indirect dimensions (personal communications from Sven Hyberts and Vladislav Orekhov). Clearly, the minimum number of sampled points (Ns) will depend on the method used for processing. However, once Ns is sufficiently large, the size of the sampling space (N) is only limited by signal-to-noise (see below for further information). In other words, the sampling factor can be reduced more dramatically by increasing N than by reducing Ns. Under those conditions, sampling factors in triple resonance 3D spectra can be as low as 2–4% (see [78] and below). Thus, optimal resolution for a 3D spectrum can be obtained within the same time required for conventional acquisition of a low resolution spectrum. Along the same lines, while the faithful reconstruction of a two-dimensional experiment may require sampling 20% of the points in the indirect dimension, only 2–4% are needed in a three dimensional experiment because fewer signals appear in each dimension. For the same reason only 1% of the points need be sampled to faithfully produce a 4D spectrum, as recently demonstrated [62]. As a rule of thumb, each additional indirect dimension allows for reducing the sampling factor by a factor of 5 (Sven Hyberts, personal communication). We note that the ability to optimize resolution becomes even more important when acquiring data at high fields. Indeed, the dwell time, Δti, for a given spectral width in ppm decreases as the field strength increases. Consequently, more points are needed to reach the same value of timax = NiΔti at high field than at low field. Thus, NUS acquisition is a necessity for acquisitions at high field. In summary, NUS is an ideal method for maximizing the resolution of multidimensional spectra and, thus, for helping alleviate spectral crowding in larger proteins.
The generation of a sampling schedule is a critical aspect of non-uniform acquisition since the quality of the reconstructed spectra ultimately depends on the data sampled. Fortunately, a number of software packages are now available to assist in designing such schedules [62,79,80]. We present here some general guidelines applicable to all these software packages, and which were used when setting up the experiments to record the spectra used as examples in the remainder of this article.
Predict or estimate relaxation rates affecting evolution periods. The rates will be used to define the targeted resolution and may be needed for defining the distribution of sampled points. Software for designing schedules (see point iii) usually provides estimates of relaxation rates, but it is advisable to account for labeling schemes such as the level of deuteration. Calculated rates typically underestimate relaxation and, when in doubt, it is recommended to increase these values. If possible, it is best to roughly estimate relaxation rates experimentally, e.g. by arraying a spin-echo in the pulse sequences used to collect the data.
Define the resolution targeted in each dimension. This step will define N, the size of the sampling space. In each dimension the number of points covered by non-uniform sampling is calculated from Ni = timax/Δti, Maximal resolution is achieved for timax = πT2. However, Rovnyak et al. have shown that sensitivity deteriorates when points are sampled after a value estimated at 1.2T2 [81]. Moreover, the resolution may need to be constrained to simultaneously satisfy restrictions on the sampling factor and the number of transients that need to be accumulated, together with the acquisition time.
Generate the schedule with software. The size of the schedule, Ns, and the sampling space, N, must be varied to restrain the sampling factor to values exceeding about 20%, 4% and 1% for 2D, 3D, and 4D experiments, respectively. The exact value of the minimal sampling factor is empirical since it depends on spectral crowding and on the method used for processing the data. The values provided come from the empirical rule of thumb that 20% of the points be sampled per dimension (see above). During this step, one must also choose the distribution used to randomly select sampled points. Data points collected during real-time evolution periods feature exponential damping by the appropriate relaxation rate, while constant time periods may use simple random distributions. For schedules sampling semi-constant time periods, one can account for the residual relaxation by using a relaxation rate scaled by (timax – Ti)/timax in the damped distribution, where Ti, is the duration of the constant time period. The programs used to process the data (MddNMR for MWD/compressed sensing [60,65], rnmrtk for MaxEnt [56], istHMS for FM, 11-norm, MaxEnt, iterative soft thresholding [62]) usually provide a module to generate sampling schedules. Other tools with user-friendly interfaces are now available on-line or as separate programs, e.g. Sample Scheduler (aka Schedule Tool, http://sbtools.uchc.edu/nmr/sample_scheduler/) or Poisson Gap Java Applet (http://gwagner.med.harvard.edu/intranet/istHMSv2/gensched_old.html). The latest versions of spectrometer acquisition software provided by the two major spectrometer manufacturers also now include schedule generators. Overall, non-uniform sampling is suitable for routine acquisition and should rapidly gain in popularity.
We illustrate the advantages of NUS acquisition during backbone resonance assignments with data collected on an 800 µM sample of a 48 kDa protein. Table 1 shows the parameters used for the acquisition of the triple resonance experiments trosy-HNCA, trosy-HNCO, trosy-HN(CA)CO, and trosy-HN(CA)CB. In this example, relaxation rates were estimated experimentally by determining the duration of spin echo that needs to be introduced into a pulse sequence to yield a decrease of 2/3 in signal-to-noise (exp(−R2T2) = 1/e = 0.36 ~ 1/3). The target resolution was set with for optimal resolution with little loss in sensitivity [81] (see legend to Table 1 for definition of ). For trosy-HNCA and trosy-HN(CA)CB, the resolution was limited to the magnitude of the 1Jcαcβ scalar coupling, ~35 Hz, under which 13C coherences evolve. This prevents resolving multiplet components, which would lead to sensitivity losses. Sampling factors were set to 2% for aliphatic carbons and 3% for the broader carbonyl carbons. For trosy-HN(CA)CB the number of points sampled, Ns, was further reduced and the number of scans was increased to compensate for sensitivity losses. All pulse sequences feature HN-TROSY with transfer concatenation [39,40] and have been modified to allow for semi-constant time evolution in 15N [82,83].
Table 1.
Acquisition parameters for the data presented in Figs. 3–7 and 11. : maximal evolution time for optimal resolution and sensitivity (1.2R2). N*: total number of points corresponding to with, in parentheses, corrections to account for scalar couplings. N: number of points adjusted to match time allocated. Ns: sampling schedule sizes. Number of scans: 4 (HNCO), 16(HNCA), 24(HN(CA)CO), and 48 (HN(CA)CB).
| Coherence | N* | N | Experiment | Sampling factor (%) | Ns | Acquisition time | ||
|---|---|---|---|---|---|---|---|---|
| 15N | 125 | 400 | 400 | |||||
| 13CO | 29 | 100 | 100 | HNCO/HN(CA)CO | 3 | 1250 | 6 h 30/1 d 15 h 00 | |
| 13Cα | 155 | 1120(206) | 256 | HNCA | 2 | 2000 | 2 d | |
| 13Cβ | 155 | 2456(412) | 256 | HN(CA)CB | 2 | 2000 | 6 d |
The resolution provided by non-uniform sampling greatly facilitates the identification of spin-systems in the HNCO spectrum. The high sensitivity of the trosy-HNCO experiment allows for recording both uniform and non-uniformly sampled spectra within 6 h 30 min (4 scans, 800 µM sample) allowing for a comparison between them. Fig. 5a compares the sampling space of uniform (red) and non-uniform schedules (black). The uniform schedule consists of a matrix of 50 (15N) × 25 (13C) = 1250 complex points, while the non-uniform schedule consists of 1250 points randomly sampled from a matrix of 400 (15N) × 100 (13C) = 40,000 complex points. This schedule provides an 8-fold increase in resolution for 15N and 4-fold for 13C when compared to uniform sampling of 1250 points, and provides a factor 50 in acceleration compared to sampling all 40,000 points. Thus the NUS spectrum recorded in 6 h 30 min would require more than 13 days to reach the same resolution with uniform sampling. Fig. 5b shows an overlay of the H/N projections of 3D-trosy-HNCO spectra recorded with conventional (red) and NUS (black) acquisition. We focus on the spectral region around the blue cross-hair. While four (H, N) correlations (labeled 1, 2, 4, and 5 in Fig. 5b) can be distinguished by their 1H and 15N shifts in the NUS H/N projection (black contours), all four correlations appear as a single very broad signal in the H/N projection of the conventionally acquired spectrum (red contours). In H/ N and N/C strips taken from the full 3D spectrum (panels b’ and b”), dispersion along the carbonyl carbon dimension indeed resolves four correlations in the uniform spectrum (labeled 1–4, in pink). However, the lack of resolution only permits a distinction between two sets of correlations; 1 and 4, on one hand, and 2 and 3, on the other hand, have seemingly identical proton and nitrogen chemical shifts. Thus, the carbonyl frequencies observed in trosy-HNCO cannot be assigned to distinct HN-anchors. The investigator may also notice that the signal labeled 2 displays some asymmetry (panels b’ and b”). However, the near frequency degeneracy in all three dimensions would preclude defining a distinct carbonyl carbon frequency. Inspection of the NUS spectrum (black) reveals that this asymmetry is indeed due to a fifth signal (labeled 5 in black in panel b”) and the high resolution allows for identification of 13C and 15N frequencies, thus unveiling a distinct {H, N, CO} spin system. Likewise, the increased resolution in the NUS spectrum resolves the signals of 1 and 4, and, in the end, three carbonyl carbon frequencies (1,4, and 5) can unambiguously be assigned to three distinct HN-anchors in the NUS spectrum. As mentioned in Section 5.2, it is critical to be able to distinguish between the HN-anchors of different spin-systems so that the carbon frequencies identified in trosy-HNCA, trosy-HN(CA)CO and trosy-HN(CA)CB are assigned accurately to each system. With NUS, {H, N, CO, Ca, Cb} systems will be accurate for 1, 4 and 5 because the three HN-anchors are resolved. With the conventional spectrum, carbonyl, alpha, and beta carbons would need to be tentatively assigned to each of the three {H, N} systems which have indistinguishable HN-anchors. We note that, even with non-uniform sampling, two systems have indistinguishable HN-anchors in this example (2 and 3). We will see below and in Fig. 7 how to bypass such a limitation. Panel c highlights other advantages of NUS when analyzing H/C strips. First, the increased resolution in 15N prevents so-called “leak-through”, i.e. the contamination of (H, C) correlations with (near) degeneracies in 15N frequencies. This improvement is best illustrated for L225, where two (H, C) correlations are seen at the same proton frequency in the uniform spectrum, thus preventing assignment, whereas only one (H, C) correlation is seen in the NUS spectrum. The advantages of high resolution in the 13C dimension can be appreciated in the strip of L229. Here, two systems have (partially) overlapping (H, N, C) correlations. While the proton frequencies are different enough to distinguish between both systems, the overlap in all three dimensions prevents the detection of two distinct carbonyl frequencies and, in the uniformly sampled spectrum, a common carbonyl carbon frequency must be assigned to both spin-systems. Thus, the 13C frequency of each system is inaccurate, and sequential systems may be identified erroneously by strip matching. When NUS has been used, the gain in resolution is sufficient to determine the frequency of every carbonyl carbon, thus increasing the accuracy in strip matching. The resolution achieved by using non-uniform sampling alleviates a number of ambiguities and, in the end, greatly facilitates and improves the reliability of spin-system determination.
Fig. 7.
Assignment by synchronization. Shown are H/N planes of trosy-HNCA (a), trosy-HN(CA)CB (b), and trosy-HN(CA)CO (c, gray) in overlay with trosy-HNCO (c, black). The planes are chosen at the carbon frequencies of D65 (intra-residue correlation). The dotted-line circle indicates the (H, N) coordinate of the residue for which a successor is being sought (D65). The dotted crosshairs indicate the position of the signal of the successor, N66, which displays sequential correlations with D65 that appear in all H/N planes. All spectra were recorded with non-uniform sampling. See text for details of interpretation.
Fig. 6 illustrates how the increased resolution benefits sequential assignment using strip matching. Three sequential residues, E64, D65, and N66, have similar Cα frequencies and the HNCA spectra could not have been used to confirm these sequential connectivities without the high resolution offered by the NUS spectra (Fig. 6a). An equally important advantage is shown in the insert of Fig. 6b, for the carbonyl frequencies of V69 and W70. The sequential and intra-residue correlations of V60 overlap to the extent that only one signal would be detected without NUS. In addition, the {H, N, C} spin-system of W70 partially overlaps with another system leading to inaccuracy in determining the carbonyl carbon frequency, as described above for L229. With the optimal resolution provided by NUS, the intra-residue and sequential signals of V69 are resolved and the frequency of the sequential correlation of W70 can be determined, thus confirming that the systems of V69 and W70 are sequential. Similarly, the resolution provided by NUS was necessary for distinguishing between inter- and intra-residue Cβ signals for E68, V69, and W70 in trosy-HN(CA)CB, as seen in panel c. Thus, optimal resolution was needed in all dimensions of all 3D spectra to determine the sequential fragment shown in Fig. 6.
Fig. 6.
H/C strips of a sequential fragment for spectra recorded with NUS acquisition. (a) trosy-HNCA of a 48 kDa protein. Non-uniform sampling resolves the signals of E64, D65 and N66 alpha carbons, which would otherwise produce overlapping sequential and intra-residue signals. (b) trosy-HN(CA)CO (gray) and trosy-HNCO (black). The insert shows that the carbonyl carbons signals of E68 and V69 can be distinguished in the H/CO strip of V69. The H/CO strip of E70 shows that the 13C resolution in trosy-HNCO is sufficient to resolve a partial overlap in the 1H and 13C dimensions. (c) trosy-HN(CA)CB. The lines (\\) indicate an interruption in the chemical shift scale for E64. The signals in gray have an opposite phase and belong to the alpha carbon of a glycine (G67). See text for details of interpretation.
We have described a number of setbacks when using strip-matching to identify sequential residues, such as misassigning sequential versus intra-residue correlations in a system or assigning the wrong carbon resonances in two systems with overlapping HN-anchors. The ultra-high resolution of NUS allows one to employ an alternative approach for finding sequential systems. So far we have analyzed the 3D spectra of backbone triple resonance experiments with an ensemble of strips along the H/C and N/C dimensions, each with sequential and intra-residues correlations along the carbon dimension, for example providing [ωH(i), ωN(i), ωcα(i)] and [ωH(i) ωN(i),ωcα(i–1)]. However, by inspecting the cartoons in Fig. 2 one can see that sequential correlations can be revealed by inspecting H/N planes across a carbon frequency of interest. Indeed, the carbon frequency of an intra-residue correlation is the same as the frequency of the sequential correlation of a successor spin-system. Thus, both correlations [ωN(i), ωcα(i)] and [ωH(i+1), ωH(i+1), ωcα(i)] appear in the H/N plane of trosy-HNCA at a frequency ωcα = ωcα(i). Thus, one can inspect H/N planes positioned at frequencies of intra-residue correlations to find the (H, N) coordinates of successor residues (Fig. 7). Since the spectra of trosy-HNCA, trosy-HN(CA)CO and trosy-HN(CA)CB all share the H and N dimensions, the simultaneous inspection of their H/N planes allows the true sequential spin system to be distinguished from those that accidentally share one carbon frequency. In effect, this is the same procedure as described in Fig. 3 for strip comparison, when the data of three spectra are analyzed simultaneously, although it is now applied to correlations in H/N planes in place of a series of H/C strips. In the absence of high resolution, such a comparison of H/N planes is however cumbersome and time-consuming, and in the end inferior to a more rapid strip matching. A limited resolution in 13C results in “leak through” of signals with similar frequencies and a multitude of (H, N) correlations need to be monitored. In contrast, strip comparison would rapidly reveal which systems do not have identical carbon frequencies since they will appear misaligned. However, a comparison of H/N planes does not depend on peak picking and hence, would bypass errors associated during spin-system definitions. With the high resolution provided by NUS, the number of correlations appearing in an H/N plane is greatly reduced and the procedure becomes tractable. Fig. 7 shows an example in which the successor of residue D65 is being sought, the (H, N) coordinates of which are denoted by the cross-cursor. Clearly, even with high resolution, many candidates are observed in each trosy-HNCA, trosy-HN(CA)CB, and trosy-HN(CA)CO. However, only one (H, N) coordinate is present in all three H/N planes, indicated by a dashed circle, and the successor of D65 is identified unambiguously. Because this correlation is identified by manual inspection of spectroscopic data, it will be revealed regardless of errors in defining the spin system of the successor and potential mistakes can be corrected. Unlike strip-matching, which relies on peak picking and assigning inter- and intra-residue correlations, the procedure we have described relies on spectral visualization only and will bypass peak-picking errors. Indeed, for N66 the sequential and intra-residue correlations had signals of similar amplitudes and were misassigned so that the successor of D65 could not be identified with strip matching. Thus, the procedure described in this paragraph, while more time-consuming than strip comparisons, may be a powerful tool to resolve mistakes associated with peak picking.
This section has demonstrated the critical need for obtaining high resolution in NMR spectra during studies of large proteins. We have shown how non-uniform sampling can provide such resolution and described many ways in which this improvement can be exploited. However, the challenges of assigning large proteins are such that, while optimal resolution is necessary, it is not sufficient for completing the task. Alternative methods to the conventional set of backbone triple resonance experiments must be employed, as described in the following two sections.
5.5. Experiments with sequential nitrogen and proton correlations
The previous sections have shown how methods traditionally applied to small proteins require adaptation and modification for studies of larger proteins. Unfortunately, even with all the improvements discussed, these experiments are usually insufficient for completing the assignment of NMR signals in such cases. Notably, the abundance of degeneracies for carbon frequencies prevents identification of many sequential systems. In particular, alpha carbons and beta carbons, while good reporters of residue types and secondary structure elements, are poorly sensitive to variations in local environment. Thus, two residues in redundant sequences of amino acids (e.g. EAS) and in the same secondary structure environment (e.g. an alpha helix) are likely to feature (nearly) identical Cα and Cβ chemical shifts. In contrast, amide nitrogens and protons have frequencies that reflect subtle differences in environment, resulting in the optimal dispersion observed in H and N dimensions. Indeed, the success of NMR studies of proteins is in large part due to the exquisite spectral dispersion of HN- HSQC spectra and their derived 3D experiments which enables identification of residue specific correlations. Consequently, a number of experiments have emerged since the early 1990s to provide sequential nitrogen or amide proton correlations. Weisemann, Bermel and co-workers [84] developed the H(NCA)NH and (H)N(CA)NH experiments, which provide sequential correlations to both successor and predecessor residues via amide proton and amide nitrogen resonances, respectively (see cartoon of (H)N(CA)NH in Fig. 8a and b). The magnetization flow is summarized by H(i) → N(i)(t1) → Cα(i), Cα(i–1) ⇒ N(i), N(i–1), N(i+ l)(t2) → H(i), H(i– 1), H(i+ l)(t3), where the double arrow denotes a transfer where two carbon single-quantum coherences each evolve under both 1JNCa and 2JNCa scalar couplings to simultaneously transfer magnetization from Cα to N and from one residue to another. This transfer is the only major distinction between (H)N(CA)NH and HNCA and will be referred to as a mixing period. The details of this pathway and the correlations provided by the experiment are discussed below (Eq. (1)). At about the same time, Grzesiek and Bax designed a 4D-HN(C0CA)NH experiment [85] providing both amide proton and nitrogen sequential connectivities for successor residues only. Many variants appeared during the end of the decade [86–90] leading to the so-called HNN and HN(C)N experiments, popular for studies of unfolded proteins [91]. Unfortunately, these experiments suffer from very poor sensitivity and their application to larger proteins seemed unlikely. In HN(CA)NH-type experiments sequential correlations are distributed between both successor and predecessor residues, amounting to a factor two in signal losses when compared to backbone experiments and increasing spectral crowding. On the other hand, the carbonyl carbon coherences used in HN(COCA)NH-type experiments to funnel magnetization through a single pathway are prohibited at the high fields necessary for studies of large proteins due to their fast relaxation. More importantly, HN-TROSY techniques were designed for coherences of nitrogens that are scalar coupled to detected protons and the sequential cross-peaks in HN(CA)NH would not benefit from HN-TROSY. Thus, a considerable delay occurred between the development of TROSY backbone experiments and TROSY HNCANH types of experiments. The resulting pulse sequence, a double trosy-(H)N(CA)NH, here referred to simply as trosy-(H)N(CA)NH, employs the IPAP technique to apply HN-TROSY to sequential nitrogen correlations, and a variant, s-trosy-(H)N(CA)NH, uses coupling network discrimination (as in seq- and intra-HNCA) in place of carbonyl coherences to select correlations with successors only and simplify the spectrum [92]. We emphasize that s-trosy-(H)N(CA)NH only simplifies the spectrum by editing and does not improve sensitivity by combining magnetization pathways. In addition to double-TROSY, non-uniform sampling was utilized to further increase the sensitivity by increasing the number of scans while maintaining high resolution. Application of NUS for sensitivity improvement was recently described in great detail [93–95]. Finally, a selective Cα shaped pulse allows for optimizing the mixing period in presence of relaxation. The magnetization flow is described by:
| (1) |
demonstrating that the magnetization originating from one proton is propagated to three different residues. This magnetization flow can be rearranged to reveal the sequential correlations associated with each HN-anchor:
| (2) |
Fig. 8.
(H)N(CA)NH and assignment by matching correlations in orthogonal planes. (a) Cartoon representation of the trosy-(H)N(CA)NH spectrum depicting signals for two sequential residues. Strips that are extracted for comparisons are shown in light grey. Nd: N direct (directly attached to H); Ns: N sequential. (b) The coherences correlated to the {H, N} system of residue i are depicted by squares on a peptide fragment. (c) Strips of a sequential fragment. Intra-residue correlation peaks are in gray while inter-residue correlation peaks have opposite phases and are in black. The lack of signals in the strip of N66 results in part from the overlap between the positive intra-residue signal of N66 and the negative sequential cross-peak with N65. All other strips show two sequential correlations. (d) Cartoon of trosy-(H)N(CA)NH highlighting correlations that can be compared in orthogonal planes, (e) H/Ns plane of trosy-(H)N(CA)NH at the frequency ωNd= ωN(V69). (e’) Nd/ Ns strip at ωH = ωH(E68) and ωNd = ωN(E68). The example demonstrates how both nitrogen and proton frequencies of V69 can be determined simultaneously with trosy-(H)N(CA)NH. See text for details of interpretation.
Thus, an HN-anchor is correlated to nitrogens of the same residue and to those of successor and predecessor residues, showing correlation signals with coordinates [ωH(i), ωN(i), ωN(i)], [ωH(i), ωN(i), ωN(i –1)] and [ωH(i), ωN(i), ωN(i+1)], as represented in the cartoons of Fig. 8a, b, and d. In addition, the phase of the in-tra-residue correlation is opposite to those of the two sequential cross-peaks. Fig. 8c illustrates the advantages of the method for strip-matching. Because there is a redundancy in sequential correlations, each strip can be matched to a sequential residue by two matches rather than only one as provided by trosy-HNCA, trosy-HN(CA)CO, or trosy-HN(CA)CB. This redundancy greatly improves the confidence of the sequential assignment since erroneous sequential assignment would now require two pairs of nitrogen frequencies to be degenerate in place of one pair of carbon frequencies. Moreover, the redundancy provides some robustness when sequential correlations are missing. For example, residue N66 is subject to two adverse effects: the intra-residue and sequential correlation to N65 overlap in a destructive manner and the Cβ of N66 is affected by the Cα selective pulse. Thus, no signals appear for this residue in the trosy-(H)N(CA)NH spectrum. Nevertheless, the sequential correlation between G67 and N66, as observed in the strip of G67, can be used to link G67 and N66 and extend the sequential fragment. The link between 65 and 66, however, had to be established with the procedure previously described in Section 5.4 and Fig. 7. Thus, the redundancy in sequential correlations provides either a proof-reading mechanism or a failsafe mechanism for identifying sequential residues.
The presence of intra- and sequential correlations with both predecessors and successors enables a very powerful protocol for identifying sequential systems. Because the HN-anchor of a given residue (i) provides a correlation to its successor, [ωH(i), ωN(i), ωN(i + 1)], and the HN-anchor of this successor (i+ 1) provides a correlation to its predecessor, [ωH(i +1), ωN(i +1), ωN(i)], it is possible to simply inspect the 3D spectrum for matching correlations and identify in a single step the {H, N} system of the successor. Fig. 8d depicts this property in a cartoon and Fig. 8e shows an example for linking residues E68 and V69. In this figure, the label Ns denotes a dimension featuring intra-and sequential correlations, whereas Nd features only direct correlations with HN and hence only intra-residue correlations. First the Nd/Ns strip of E68 is inspected revealing correlations to its predecessor and successor (panel e’). The latter provides the frequency coordinates [ωH(i), ωN(i), ωN(i+ 1)], and hence ωN(i+ 1). Next a 2D plane HN/Ns, selected at the position ωN(i + 1) in the dimension Nd, is juxtaposed to the Nd/Ns strip. This plane can be described as [ωH. ωN(i; + 1), ωN], where ωH and ωN denote continuous variables that feature the correlation [ωH(i+1), ωN(i+1), ωN(i)], thereby providing ωH(i + 1). These correlations can all easily be identified by drawing horizontal and vertical cursor lines as shown in panel e. A very strong advantage of this method is that it does not depend on previous HN-anchor definitions (peak picking). Thus, this procedure reveals sequential correlations to an {H, N} system whether or not the other systems have been accurately defined.
While very powerful, the procedure we described in the previous paragraph only provides sequential proton frequencies in an indirect manner and relies on nitrogen correlations only. For more difficult situations it is preferable to also record a complementary experiment with sequential proton correlations. These correlations can be obtained by recording trosy-H(NCA)NH in addition to trosy-(H)N(CA)NH or by substituting both experiments with a trosy-4D-HN(CA)NH. Both options come at the cost of a large increase in spectrometer time. Fortunately, these sequential correlations can also be obtained by a minor modification of the trosy-(H)N(CA)NH. Careful inspection of Eq. (2) reveals that sequential proton correlations can simply be obtained by separating the existing sequential nitrogen correlations along a carbon alpha dimension:
| (3) |
Eq. (3) shows that the following correlations have the same alpha carbon frequencies: [ωH(i), ωN(i), ωCα(i)], –[ωH(i), ωN(i+1), ωCα(i)], –[ωH(i+1), ωN(i). ωCα(i)], and [ωH(i+1), ωN(i+1), ωCα(i)]. The negative signs have been introduced to indicate that the two corresponding correlations have a phase opposite to the remaining two correlations. This set of correlations gives rise to a rectangular pattern with the (H, N) coordinates of two sequential residues at two diagonally opposite corners and sequential cross peaks, with opposite phases, at the remaining two corners (cartoon in Fig. 9a and b). The correlations can also be regrouped according to common proton and nitrogen frequencies, giving [ωH(i), ωN(i), ωCα(i)] and [ωH(i), ωN(i), ωcα(i–1),]. This pair of correlations corresponds to that observed in the trosy-HNCA experiment. Thus, the trosy-(H)NCA(N)H experiment provides a spectrum comparable to trosy-HNCA, but with additional proton and nitrogen sequential correlations that unambiguously identify signals of sequential residues in each H/N plane. Consequently, residues can simply be assigned by navigating between H/N planes and identifying rectangular patterns, with alpha carbon correlations identifying H/N planes of sequential residues. This sequential inspection of H/N planes is sometimes referred to as a stairway backbone walk. Fig. 9c shows an example for a difficult fragment of three residues subject to overlaps in a 48 kDa protein. The numbers circled in the figure are used to denote intra-residue correlations and will be written in parentheses in this paragraph. The first plane is chosen at the carbon frequency of E64 (defined in the H/C strip in dark gray, (1’))- This plane displays the (H, N) correlation of E64 (signal in red, labeled (1)) as well as two sequential cross-peaks (blue) that reveal the (H, N) coordinates of D65 (2). The corresponding alpha carbon frequency is determined by examination of the H/C strip (2’). The H/N plane of D65 does not provide a rectangular pattern as a consequence of the nitrogen overlap between D65 and N66, which was already detrimental to the trosy-(H)N(CA)NH spectrum. However, in trosy-(H)NCA(N)H, one of the sequential cross-peaks can be observed and the (H, N) coordinate of N66 can be found (3). Similarly, the H/N plane of N66 only displays three of the four expected correlations, which is sufficient for identifying the (H, N) coordinates of G67 (4). Thus, as in trosy-(H)N(CA)NH, the redundancy in sequential correlations provides robustness to the method. The advantage of the stairway backbone walk over strip matching of conventional triple resonance experiments is demonstrated in Fig. 9d. The successor of E64 cannot be identified by the combined information of trosy-HNCA, trosy-HN(CA)CO, and (trosy-HN(CA)CB. This lack of success comes in spite of the optimal resolution provided by non-uniform sampling; four residues have degenerate frequencies in Cα, CO, and Cβ In contrast, assignment with trosy-(H)NCA(N)H is relatively trivial as seen on points (1) and (2) in Fig. 9c.
Fig. 9.
trosy-(H)NCA(N)H and “backbone stairway” assignment. (a) Cartoon representation of the trosy-(H)NCA(N)H spectrum. Intra-residue (H, N) correlations appearing at a given carbon frequency are in red and sequential cross-peaks are in blue. (b) Coherences correlated to the {H, N} system of a residue i are depicted as squares on a peptide fragment. (c) Stairway assignment using H/C strips (dark gray) and H/N planes (light gray). The numbers in yellow denote the order in which the signals of sequential residues are identified, and those with a prime (‘) indicate dimensions orthogonal to those without a prime. See main text for further explanation. (d) Unsuccessful strip matching of E64 with D65, corresponding to points (1) and (2) in (c). Signals of trosy-HN(CA)CO are in gray and those of trosy-HNCO in black. The lines (\\) denote an interruption in the chemical shift scale of E64 in trosy-HN(CA)CB. (e) Stairway assignment using C/N planes (dark gray) and H/N planes (light gray). (f) Unsuccessful strip matching of G67 and E68, corresponding to (5) and (6) in (e). Signals of trosy-HN(CA)CO are in gray and those of trosy-HNCO in black. See text for details of interpretation.
The stairway backbone walk can also be performed by inspecting orthogonal planes of trosy-(H)NCA(N)H instead of navigating along H/N planes (Fig. 9e). First, an H/N plane is displayed and (H, N) coordinates of sequential {H, N} spin systems are readily identified. For example, the successor of G67 (4) can be determined by recognizing the rectangular pattern, which provides simultaneously ωH(E68) and ωN(E68). To provide further elongation of the sequential fragment, the C/N plane at ωH(E68) is displayed. Inspection of Eq. (3) shows that C/N planes feature the correlations [ωH(i), ωN(i–1), ωcα(i–1)]. [ωH(i), ωN(i). ωcα(i–1)]. [ωH(i). ωN(i), ωcα(i)]. and [ωH(i) ωN+1(i), ωcα(i)]. The first two correlations have already been observed in the previous H/N plane, while the last two provide not only the carbon frequency of the current residue, which needs to be used to display the next H/N plane, but also the nitrogen frequency of the new successor. Here, ωcα(E68) and ωN(V69) are the new frequencies provided by inspection of the C/N plane at ωH(E68). Next, the H/N plane at ωCα(E68) confirms ωN(V69) and reveals ωH(V69). This procedure is equivalent to that described above but has the advantage that the same sequential nitrogen correlations are monitored both in C/N planes and in H/N planes. We have emphasized in Section 5.2 the importance of inspecting sequential correlations along several dimensions (H/C strips and N/C strips). Here, the second stairway backbone walk procedure inherently ensures inspection of all dimensions, hence minimizing the risks of erroneous assignments and providing a second means of resolving overlap; sequential nitrogen correlations may be subject to crowding in H/N planes but not in N/C planes, and vice versa. Overall, the protocol provides a rapid and reliable means of performing sequential assignment.
While we have extensively described the advantages of using trosy-(H)N(CA)NH or trosy-(H)NCA(N)H, here referred to jointly as HNCANH, both methods suffer from at least two major drawbacks. First, they have the lowest sensitivity when compared to the popular backbone triple resonance experiments trosy-HNCA, trosy-HN(CA)CO, trosy-HNCO, and trosy-HN(CA)CB. Sensitivity losses due to relaxation are comparable to those of trosy-HN(CA)CB: the experiments have the same N → CA transfers, benefit from HN-TROSY, and the total duration of the CA → CB and CB → CA transfers in trosy-HN(CA)CB (~28 ms) is comparable to the mixing transfer (28–35 ms) in HNCANH. The slightly longer delay in HNCANH experiments is compensated for by constant time evolution of Cα single quantum coherences in place of real time evolution of Cβ single quantum coherences, i.e. subject to both relaxation and evolution under (multiple) scalar couplings. However, HNCANH provides sequential cross-peaks between two nitrogens in addition to carbons and hence suffers from an additional factor of two in signal-to-noise losses. The second drawback of HNCANH experiments is that the plethora of sequential correlations requires high resolution in all dimensions to combat the increase in spectral crowding. Thus, semi-constant time nitrogen evolution periods are needed and long t1max must be sampled. Sensitivity and resolution considerations render non-uniform sampling a necessity, so that a high number of transients may be accumulated to improve signal-to-noise while attaining a high resolution within a reasonable experimental time. Recent efforts in popularizing NUS may lessen the impact of this requirement, making HNCANH experiments more accessible. Even so, acquisition of spectra with sufficient signal-to-noise typically requires six to eight days of measurements. Nevertheless, the sacrifice in experimental times needed to overcome sensitivity limitations is justified by the advantages of obtaining three sets of sequential correlations in all three dimensions of the spectrum.
5.6. Resolving overlap with higher dimensions
Overcoming crowding and overlap challenges by adding dimensions to NMR correlation maps is a logical extension of past efforts. Indeed, detailed structural NMR studies of proteins only became possible with the advent of two-dimensional NMR and became popular with three-dimensional experiments. Consequently, four-dimensional spectra are commonly recorded during studies of larger molecules, as will be further discussed in Section 7. Notably, the group of Lewis Kay have developed a strategy for assigning NMR backbone resonances that relies primarily on four-dimensional experiments. The three 4D experiments all encode the frequencies of alpha and carbonyl carbons and amide protons and nitrogens. The 4D-trosy-HNCOCA experiment [96] has a transfer summarized as H(i) → N(i) → CO(i – 1)(t1) → CA(i – 1)(t2) → CO(i – 1)-N(i)(t3) → H(i)(t4) and provides the correlations [ωH(i), ωN(i), ωCO(i – 1), ωCα(i – 1)]. Thus, it relates the HN-anchor of a residue to the COCA-anchor of its predecessor. 4D-trosy-HNCACO [96] is the four dimensional counterpart of the trosy-HN(CA)CO experiment with a magnetization flow summarized by H(i) → N(i) → CA(i), CA(i – 1)(t1) → CO(i), CO(i – 1)(t2) → CA(i), CA(i – 1) → N(i)(t3) → H(i)(t4). It provides primarily the correlations [ωH(i), ωN(i), ωCO(i), ωCα(i)] and less frequently [ωH(i), ωN(i), ωCO(i – 1), ωCα(i – 1)]. This allows for identification of {H, N, CO, CA} spin systems. Together, these two 4D experiments enable the assignment of sequential {H, N, CO, CA} spin systems, as discussed below in more detail. Finally, 4D-trosy-HN COi–1CAi [97] is most often necessary to remove ambiguities. The transfers involved in this experiment can be summarized as H(i) → N(i) → CO(i → 1)(t1)/CA(i), CA(i – 1)(t2) → N(i)(t3) → H(i)(t4), where the “/” emphasizes that magnetization is transferred from nitrogen simultaneously to both CO and Cα. The major correlation is [ωH(i), ωN(i), ωCO(i – 1), ωCα(i)], which is complementary to the correlations provided by 4D-trosy-HNCOCA and 4D-trosy-HNCACO. 4D-trosy-HN COi–1CAi also provides occasionally the correlation [ωH(i), ωN(i), ωCO(i – 1), ωCα(i – 1)], which is redundant in all experiments and hence readily differentiated from the major correlation. Each 4D experiment provides a complete CO/Cα correlation plane to each HN-anchor. The most efficient way to utilize them is indeed to consider the 4D hypercube as a means of correlating H/N planes with CO/CA planes. Fig. 10 depicts part of the assignment of the 81 kDa protein Malate synthase G and highlights the need for combining all data [98]. The procedure relies mainly on inspecting CO(i)/CA(i) planes in 4D-trosy-HNCACO and H/N planes in 4D-trosy-HNCOCA. In addition, CO(i – 1)/CA(i) planes of 4D-trosy-HN COi–1CAi and H/N planes of 4D-NOESY are used to resolve ambiguities. We start by displaying the CO(i)/CA(i) plane of 4D-trosy-HNCACO at the coordinates [ωH(A33), ωN(A33)] (panel a). This reveals the frequencies of [ωCO(A33), ωCα(A33)], which can be used to position the H/N plane of 4D-trosy-HNCOCA (panel b). The H/N plane at [ωCO(A33), ωCα(A33)] provides the (H, N) coordinates of the succeeding residue, [ωH(A34), ωN(A34)]. Often this procedure can be repeated to elongate the sequential fragment. However, here, the CO(i)/CA(i) plane of 4D-trosy-HNCACO at [ωH(A34), ωN(A34)] shows many (CA, CO) correlations that prevent the assignment of the frequencies ωCO and ωCα to the spin system of A34 (panel e), so two additional 4D experiments must also be used. 4D-trosy-HN COi–1CAi reveals correlations between the Cα observed in trosy-HNCACO and the carbonyls of their predecessors, here A33 (panel d). These can then be related to the previously assigned correlations observed in the 4D-trosy-HNCOCA (panel c), in effect singling out the correlation of panel e in 4D-HNCACO that is paired with A33 in 4D-trosy-HNCOCA and hence must belong to A34. Displaying the H/N plane of the same 4D-trosy-HNCOCA spectrum at the newly found coordinates [ωCO(A34), ωCα(A34)] should provide the (H, N) coordinates of F35. Unfortunately, as for CO/CA planes, many signals are observed in the H/N plane (panel f). This, time the assignment is rescued by also inspecting the HN/HN plane of a 4D-HSQC-NOESY-HN-trosy-HSQC [99–101] at [ωH(A34), ωN(A34)] (panel g), which reveals a cross-peak to [ωH(F35), ωN(F35)] also observed in panel f. Use of NOESY data is contingent on the presence of sequential nOe’s and hence is most likely to be successful in alpha-helices. In the example of MSG, the suite of 4D experiments was combined with 3D backbone experiments for completing the assignment of this 723 residue protein.
Fig. 10.
Backbone assignment with 4D-trosy-HNCACO, 4D-trosy-HNCOCA and 4D-trosy-HNCOi–1CAi. The frequencies of the two invisible dimensions are indicated on each plane visualized. The frequencies determined following plane inspections are indicated near the arrows. (a) CO/CA plane of 4D-trosy-HNCACO revealing the signal of A33. (b) Corresponding H/N plane in 4D-trosy-HNCOCA revealing the (H, N) correlation of A34. (c, d and e) CO/CA planes of 4D-trosy-HNCACO, 4D-trosy-HNCOCA and 4D-trosy-HNCOi–1 CAi, used to identify the carbon shifts of A34. (f) H/N plane of 4D-trosy-HNCOCA and (g) H/N plane of 4D-HN-HSQC-NOESY-HN-trosy-HSQC used to identify the (H, N) correlation of F35. The colors are coded according to the residue for which the correlations belong. See text for details of interpretation. Adapted with permission from “V. Tugarinov, R. Muhandiram, A. Ayed, LE. Kay, Four-dimensional NMR spectroscopy of a 723-residue protein: chemical shift assignments and secondary structure of malate synthase g, J. Am. Chem. Soc. 124 (2002) 10025–10035” Copyright 2002 American Chemical Society [98].
The experiments described in this section provide a very powerful protocol for unambiguous assignment of resonances. However, four-dimensional experiments suffer from several drawbacks and have rarely been used in the 20 years since their development. One obstacle is simply the ease of use, since navigating across a four dimensional hypercube may seem intimidating and cumbersome. However, many software packages now allow for simultaneous visualization of 4D and 2D or 3D related spectra, thus providing points of references to the user. Similarly, restrictions in data storage and computer memory have been overcome with progress in hardware. Perhaps the most stringent limitation relates to the long spectrometer time necessary for encoding the three indirect dimensions. Typical acquisition times amount to about one week for modest resolution and with only a few scans accumulated. For relatively dilute proteins, the measurement time may become prohibitively long. Clearly, non-uniform sampling is ideally suitable for overcoming the limitation in resolution and methods have been developed to process spectra with more than three dimensions [69,102–104].
As an alternative to recording 4D (or higher dimension) spectra, one may consider measuring reduced dimensionality spectra. The principle of the technique relies on combining several dimensions of an N–D experiment by co-evolving coherences in different periods of the pulse sequence to produce an (N–m)–D spectrum, thereby considerably shortening the acquisition time [105]. The (N–m)–D spectrum may result in split signals, in which the magnitudes of the splitting and the central frequencies are used to provide correlated frequencies. A correlation set can be obtained by using the so-called GFT method [106]. However, this method does not resolve overlap issues in the (N–m)–D spectra; overlaps are identified but frequencies cannot be assigned to individual overlapping systems. Consequently, while it is a powerful method for rapid assignment of small proteins, it is not suitable for general application to larger systems. Alternatively, the acquisition scheme of the (N–m)–D experiment can be designed to produce a projection of the N–D experiment with an adjustable angle of projection. In this implementation, N–m = 2 in general. That is, two-dimensional spectra are used as read-out experiments. If sufficient projection angles are sampled, the N–D spectrum may be obtained by projection reconstruction [107]. Unfortunately, the reconstructed spectrum is prone to artifacts that render the analysis of crowded spectra impracticable, in particular when signal intensities have a large dynamic range. In fact, co-evolution of multiple dimensions can be viewed as a special case of non-uniform sampling and recent studies have shown that randomization is necessary for best removal of artifacts in a reconstructed spectrum [74,108]. However, the assignment of the frequencies in the reduced dimensions may not necessarily need a reconstructed spectrum. The position of each signal in the (N–m)–2D projection spectrum is a function of the frequencies of the related spin-system and, in principle, a limited number of projections is sufficient for determining these frequencies. This methodology is the basis of automated projection reconstruction spectroscopy (APSY), in which an increasing number of different 2D projection planes is measured until all expected signals, or a target subset thereof, are assigned [109,110]. A major drawback is that the limiting sensitivity of the overall method is dictated by the signal-to-noise of each individual 2D plane. In contrast, the sensitivity of a complete N–D experiment is a function of its entire acquisition time, not of an (N–m)-dimensional subset of the data matrix. In addition, the projection angles that need to be recorded are not known a priori so neither the time for convergence, i.e. the number of planes necessary for assignment, nor the total experimental time can be predicted. For these reasons APSY seems likely to be limited currently to proteins which provide a high signal-to-noise in their spectra and, hence, is not applicable in a straightforward manner to routine acquisitions for larger proteins. Thus, at the time of writing, recording higher dimensionality experiments is the most reliable solution to resolving spectral overlap.
6. Assignment of methyl side-chain resonances
A number of NMR applications can readily be carried out following the assignment of {H, N, CO, CA, CB} spin systems, e.g. chemical shift perturbation analysis, studies of backbone dynamics, prediction of domain orientation with residual dipolar couplings, etc. However, many studies will require assignment of side-chain signals for successful completion. Side-chains are better probes of binding surfaces, since most backbone atoms are shielded by side-chains; they are necessary probes for determining structures; they provide a complementary view of protein dynamics. As mentioned in Sections 3 and 4, of all side-chain moieties, methyls are predominantly used in larger proteins since they benefit from a TROSY effect and provide selective proton spin-probes in a deuterated environment, thus simultaneously minimizing relaxation and simplifying the spectra. In addition, methyl probes can be introduced selectively for some residue types (e.g. Ala, or Ile d1) or groups of residue types (e.g. Leu and Val together) depending on the biosynthesis pathway of the related amino acids. This targeted labeling strategy can provide further spectral simplification and a means to design labeling schemes tailored to a particular investigation. Consequently, pulse sequences have been developed specifically for assigning methyl resonances. In this section, we will briefly mention available methods and describe in more detail those applicable to proteins larger than 40 kDa.
For proteins up to 35–40 kDa, methyl resonances may be assigned by a conventional strategy used for assigning aliphatic side-chains in general. This method can also be used for partially deuterated samples of proteins in the 25–35 kDa range and we will first describe its application to this type of samples and later briefly emphasize differences for samples selectively protonated on methyls. In short, the frequencies of all side-chain aliphatic carbons and protons are paired with the already assigned {H, N, CO, CA, CB} spin systems. The most efficient and rapid means of obtaining appropriate correlations relies on the combination of trosy-H(CCO)NH and trosy-(H)C(CO)NH (these experiments are referred to with various denominations such as trosy-HCCONH and trosy-CCONH, or trosy-HCCCONH and trosy-CCCONH) [82]. trosy-H(CCO)NH has the magnetization flow H(t1) → C ↔ Cα → CO → N(t2) → H(t3) and provides the correlations [ωH(i), ωN(i), {ωHali(i – 1)}] in which {ωHali(i – 1)} denotes the ensemble of all aliphatic proton frequencies of residue i – 1. The double arrow, ↔, denotes a transfer mediated by TOCSY [111], which (ideally) spreads the magnetization from one carbon to all carbons belonging to a network of (pairwise) scalar coupled aliphatic carbons. All other arrows denote INEPT transfers. The trosy-(H)C(CO)NH experiment is obtained by encoding frequencies of aliphatic carbons in place of those of aliphatic protons, H → C(t1) ↔ Cα → CO → N(t2) → H(t3); it provides the correlations [ωH(i), ωN(i), {ωCali(i – 1)}]. trosy-H(CCO)NH and trosy-(H)C(CO)NH provide side-chain correlations for the preceding residue of the {H, N, CO, CA, CB} spin system featuring (H, N) correlations. Thus, the success of assigning side-chain signals with these experiments depends on prior assignment of backbone resonances, or at least on accurate identification of sequential residues. In addition, the method cannot be applied to spin systems for which a successor amide has not been identified. Together, the pair of experiments assigns aliphatic protons and carbon frequencies to {H, N, CO, CA, CB} spin systems but does not pair proton and carbon frequencies belonging to the same chemical group. Hence, (H, C) correlations remain unassigned at this stage. In many instances, pairing of aliphatic proton and carbon frequencies can simply be obtained by trial and error monitored on H/C correlation maps. Proton and carbon frequencies are tentatively assigned to a candidate chemical group of the spin system (e.g. Thr, Hγ and Cγ) and an H/C correlation map is inspected to verify the presence of a signal at these coordinates. Further confidence can be obtained by comparing the HC/H strip of a 3D 13C edited NOESY spectrum with the HN/H strip of H(CCO)NH. Successful pairing of Hali and Cali resonances into an {H, C} spin system may lead to the observation of common correlations in both strips. A much safer procedure relies on using HCCH-TOCSY [112] or HCCH-COSY [113] experiments, which feature magnetization pathways described by H(t1) → C(t2)↔ C → H(t3) and provide the correlations [ωHali(i), ωC(i), {ωHali}]. The first two correlations provide a map corresponding to an HC-HSQC for so-called source {H, C} systems. In HCCH-TOCSY, {ωHali} represents all protons which are attached to carbons within a network of scalar coupled carbons that includes the carbon in the source {H, C} system. In HCCH-COSY, only carbons directly scalar coupled to the source carbon lead to correlations with the corresponding proton. The redundancy in correlated frequencies can be exploited to provide an assignment procedure similar to those described in Sections 5.5 and 7, i.e. by comparing orthogonal planes in the spectra. Clearly, for larger proteins, HCCH-TOCSY suffers dramatically from spectral crowding and provides little useful information. In addition, both TOCSY and COSY derived experiments can be extremely insensitive and feature broad signals, and their application is most often limited to proteins below 25 kDa, even with partial deuteration. The assignment of methyl resonances in partially deuterated samples is a noteworthy exception to these limitations. Mulder and coworkers showed that, in a partially deuterated 35 kDa protein, direct excitation of carbon coherences in a 3D-CCH-TOCSY in place of polarization transfer from proton to carbons avoided losses during INEPT transfers, and was advantageous in spite of a much lower initial polarization [114]. Further, selection of CHD2 moieties by a simple sensitivity enhanced scheme [115,116] alleviated spectral crowding due to inhomogeneous isotope incorporation in methyl groups, which would otherwise produce additional signals for CH3 and CH2D. Finally, sensitivity enhancement was implemented in both carbon dimensions [117]. This method provides the assignment of Thr, Ala, Ile, Leu, Val methyls and is expected to be applicable to proteins up to 40 kDa.
For proteins larger than 40 kDa, selectively methyl protonated samples are required. Minor modifications to the original trosy-H(CCO)NH and trosy-(H)C(CO)NH experiments include implementation of 2H decoupling during periods involving aliphatic carbon coherences. Because the pair of experiments identifies the methyl resonances of the predecessor spin system and because only methyls of I, L, V are protonated, the assignment of methyl resonances also provides a virtually failure-proof validation of the related residues’ backbone assignments. Unfortunately, even with a perdeuterated background, relaxation losses throughout the pulse sequence result in very poor sensitivity and must be compensated for by using long acquisition times. For example, six days of acquisition for each experiment only provided very limited assignment for a 48 kDa protein (800 µM). The next paragraphs show how alternative strategies can provide methyl assignment in more challenging cases.
Tugarinov, Kay and coworkers have designed experiments sensitive enough for assigning methyls of proteins with sizes in the 100’s of kDa, and providing superior data to those of trosy-HCCONH experiments for proteins larger than 40 kDa. The underlying idea is to provide correlations between methyl groups and alpha and beta carbons. These correlations can then be paired with alpha and beta carbon frequencies of {H, N, CO, CA, CB} systems, thereby assigning methyl resonances. The HMCM(CG)CBCA experiment correlates Cβ and Cα frequencies to the corresponding methyl resonances of Ile (δ1), Leu, and Val [19,118]. The magnetization flow is different for each residue type. For Ile (δ1), and Leu, I,L-HMCM(CG)CBCA can be summarized as HMe → CMe → Cc → Cβ → Cβ, Cα(t1) → Cβ → Cγ → CMe(t2) → HMe(t3), where HMe = Hδ1 or Hδ2 for Leu and Hδ1 for Ile, likewise for CMe, and Cγ is Cγ1 for Ile. The experiment provides the correlations [ωHδ(i), ωCδ(i), ωCα(i)] and [ωHδ(i), ωCδ(i), ωCβ(i)]. For Val, the magnetization flow in the same pulse sequence is Hγ → Cγ → Cβ → Cα → Cβ, Cα(t1) → Cα → Cβ → Cγ(t2) → HMe(t3) and the correlations are [ωHγ(i), ωCγ(i), ωCα(i)] and [ωHγ(i), ωCγ(i), ωCβ(i)] (Fig. 11c). The experiment may then be denoted V-HMCM(CBCA)CBCA. Note that the magnetization flow is suboptimal for Val, in which an unnecessary excursion through Cα single quantum coherences occurs. In addition, in spite of minimal losses due to relaxation, the experiment often provides incomplete sets of correlations for Leu and Val. At some point in the experiment, the coherence of carbons at the methyl branch points (Cβ for Val and Cγ for Leu) should become antiphase with the carbon down the chain (Cα for Val and Cβ for Leu) while becoming in-phase with respect to the source methyl. To minimize the amount of coherence that inevitably becomes antiphase with the second methyl, the delay of this transfer must be set to a value that results in an incomplete transfer. This limitation is overcome when the experiment is paired with selective protonation and 13C enrichment of a single methyl moiety in Val and Leu, increasing the sensitivity by 2.5. The samples need not be (but can be) stereospecifically labeled for achieving such an improvement; the only condition is that the 13C nuclei form an unbranched network of coupled nuclei. Unfortunately, the production of such samples comes at increased monetary cost, in particular for stereospecific labels. For either stereospecific or non-stereospecific labeling, some of the missing correlations can be recovered by recording the more sensitive experiment I,L-HMCMCGCB/V-HMCMCBCA. The experiment provides the same correlations as V-HMCM(CBCA)CBCA for Val, but without unnecessary transfers, and hence with much increased sensitivity. In addition, the resulting spectrum provides [ωHδ(i), ωCδ(i), ωCγ(i)] and [ωHδ(i), ωCδ(i), ωCβ(i)] for Ile and Leu, which can rescue a number of correlations to Cβ carbons that are missing in the less sensitive I,L-HMCM(CG)CBCA. The correlations provided by both experiments, referred here together as HMCMCali experiments, can be paired to {H, N, CO, CA, CB} spin systems by comparing strips of each HMCMCali, trosy-HNCA and trosy-HN(CA)CB experiments (Fig. 11a – c). However, this procedure is tedious and subject to human error, and a more reliable method is described below.
Fig. 11.
Assignment of Ile, Leu, and Val methyl resonances. Cartoons of trosy-HNCA (a), trosy-HN(CA)CB (b), and HMCM(CG)CBCA (c) highlighting planes displayed in (d, i, and j) (darker grey) and strips shown in (e, f, g, and h). The squares on the peptide fragments indicate the nuclei that are correlated in the respective experiments. (d) HMeCMe-HMQC of a 53 kDa protein. The dashed lines indicate the position of the orthogonal strips in (b and c), at the coordinates of I175 (crosshair). (e) HMe/Cali strip of HMCMCGCB (V-HMCMCBCA for valines, not shown). Signals of Cγ and Cβ have opposite signs; Cγ appear in gray and Cβ in black. (f) HMe/Cali strip of HMCM(CG)CBCA (V-HMCM(CB)CACB for valines, not shown). Signals of Cβ and Cα have opposite signs; Cβ appear in gray and Cα in black. (g) HN/Cα strip of HNCA and (h) HN/Cβ strip of HN(CA)CB for I175. (i) HN(CA)CB and (j) HNCA H/N planes at the carbon frequencies of I175 Cβ and Cα, respectively. The crosshair denotes the (H, N) coordinates of I175. See text for details of interpretation.
To assign methyl resonances, the alpha and beta carbon frequencies provided by HMCMCali must be identified in trosy-HNCA and trosy-HN(CA)CB. Thus, an {HMe, CMe, CA, CB} spin system is compared to an {H, N, CA, CB} system and both are merged if they are found to belong to the same residue. In other words, the correlations of an unassigned HC-anchor with Cα and Cβ are matched to those of assigned HN-anchors. This can be achieved without resorting to peak lists, as shown in Fig. 11d – j for an isoleucine methyl. A target HC-spin system is selected in an H/C correlation map (panel d) and strips of HMCM(CG)CBCA(f) and HMCMCGCB(e) are displayed to reveal the associated Cα and Cβ frequencies. Next, the H/N planes of trosy-HNCA and trosy-HN(CA)CB are displayed simultaneously, each at the relevant carbon frequency identified with HMCMCali experiments (i, j). The cursor is synchronized for both experiments to reveal an (H, N) correlation that appears in both spectra. H/C strips in each experiment are also inspected to verify that none of the H/N correlations results from near degeneracies in carbon frequencies (“leak through” of off-plane correlations). This protocol is sufficient to assign the majority of the methyl resonances but it is not uncommon that several {H, N, CA, CB} systems match the target {HMe, CMe, CA, CB}. To resolve such ambiguities it may be necessary to record an HMCM(CGCBCA)CO experiment [19], which provides correlations between methyl groups and carbonyl carbons. Together with an HN(CA)CO spectrum, this experiment extends the sizes of the spin systems that are compared, which are now {H, N, CA, CB, CO} and {HMe, CMe, CA, CB, CO}, respectively. Alternatively, the correct matching of {H, N} and {H, C} spin systems may be verified by using NOESY experiments. For example, H/H strips of a NOESY-HN-trosy-HSQC can be compared with H/H strips of a NOESY-HC-HMQC to probe for common resonances. This comparison may reveal a correlation between the amide and methyl protons in each NOESY experiment, thus identifying the systems to be paired. However, the absence of such an intra-residue correlation cannot be used to rule out a system since the amide and methyl groups may be too remote to result in a NOESY cross-peak. At an advanced stage in resonance assignment, unambiguous nOe cross-peaks to residues near in space may be used to delineate two candidates; in such a situation the pattern of nOe cross peaks will be similar in the 15N and 13C edited spectra for the correct assignment, but in an ideal situation, no common nOes will be observed for the wrong candidate. As for intra-residue correlations, this method often fails due to different environments for methyl and amide protons. Fortunately, in general, the HMCMCali experiments will provide unambiguous assignment of methyl resonances. Thus, similar experiments have been designed to assign methyls of threonines (3D THR-HMCMCBCA [119]), alanines (4D ALA-HMCBCACO [120] or 3D ALA-HMCMCA and 3D ALA-HMCM(CA)CO [121]), and isoleucines at the γ2 position (3D ILE(γ2)-HMCM(CBCA)CO, 3D ILE(γ2)-HMCMCA, and 3D ILE(γ2)-HMCMCB [121]). Some of the pulse sequences for isoleucine and alanine methyls can be combined, and in samples containing protonated methyls for both Ala and Ile, the resonances may be assigned simultaneously [121]. Methyls in methionines are not scalar coupled to other side-chain carbons and cannot be assigned with such experiments; they are typically assigned using NOESY spectra and mutagenesis [22].
In samples that contain 13C and 1H in both methyls of Leu and Val, pairs of methyls that belong to the same residue can be identified by a simple inspection of the HMe/CMe plane at either Cα or Cβ frequencies in the third dimension. In other words, identification of {Hγ1, Cγ1, CA, CB} and {Hγ2, Cγ2, CA, CB} results in an {Hγ1, Cγ1, Hγ2, Cγ2, CA, CB} spin systems for Val, and similarly for methyls at the δ position for Leu. However, as mentioned throughout this article, Cα and Cβ display poor dispersion, which, when combined with spectral crowding in the HMe/CMe planes, may impair the assignment of one or both methyls due to signal overlap. Some assignments may nevertheless be rescued when using samples that are uniformly labeled in 13C. For such samples, the entire assignment procedure relying on HMCMCali experiments is facilitated and becomes more efficient with help from an HC(C)H-TOCSY experiment optimized for methyl groups. Because the pair of methyl carbons is only separated by two bonds, short TOCSY mixing times are sufficient to obtain correlations between methyl moieties, so that the experiment gains from having significantly lower relaxation losses than when seeking correlations between all aliphatic carbons using long mixing times. In addition, the methyl spectral widths are much smaller than those needed for correlating all aliphatic moieties and high resolution can be achieved rapidly. Together with the deuterated background in the sample, these effects promote this otherwise insensitive method to an experiment that can be recorded in very little time. The correlations provided by HC(C)H-TOCSY are (ωHδ1, ωCδ1, ωHδ2) and (ωHδ2, ωCδ2, ωHδ1) for Leu and (ωHγ1, ωCγ1, ωHγ2) and (ωHγ2, ωCγ2, ωHγ1) for Val. These correlations are best exploited by using the procedure described in Sections 5.5 and 7, i.e. by comparing orthogonal planes in the 3D spectrum. This identifies HC-anchors that belong to the same spin system, resulting in {Hδ1, Cδ1, Hδ2, Cδ2} spin systems for Leu and {Hγ1, Cγ1, Hγ2, Cγ2} for Val. This provides an obvious advantage for assigning both methyl signals with HMCMCali experiments: only one {HMe, CMe, CA, CB} system needs to be assigned to an {H, N, CA, CB} spin system and the second methyl may be assigned with the TOCSY experiment. In other words, even HC-anchors suffering from partial overlap in methyl proton and carbon dimensions and from overlap in alpha carbon and beta carbon dimensions may be assigned indirectly via the assignment of a paired HC-anchor that is resolved in the corresponding HMe/CMe plane. This provides some advantage over using samples with the single methyl labeling pattern, although the latter clearly benefit from increased sensitivity. Overall, the experiments described in this section have proven extremely successful and allowed for studies of systems as challenging as the 670 kDa proteasome [122].
7. Assignment of distance constraints
Many biomolecular NMR studies can be performed once backbone and methyl resonances are assigned. However, a protein structure must be available for a molecular interpretation. NMR structures consist of an ensemble of models generated by using structural constraints that are obtained through NMR parameters. Many such parameters do not require other assignments than those described in Sections 5 and 6 and their utilization in larger proteins is then only limited by the precision of their measurements, which in turn is most often limited by sensitivity. Examples of constraints used for large proteins include bond orientations provided by residual dipolar couplings [123,124], distances between a spin-labeled probe and other atoms [125], including transient contacts via paramagnetic relaxation enhancement [126], and solvent access provided by hydrogen/deuterium exchange measurements [127,128]. In particular, the chemical shifts assigned during backbone resonance assignment can readily be converted to torsion angle constraints [129–131] and, for smaller proteins, may even provide a structural model [132–135]. However, for larger monomeric proteins, a solution structure determined by NMR requires a set of distance constraints. This section presents procedures for assigning a maximal number of constraints with a limited number of probes, as typically available for large proteins when using selective labeling schemes.
The reliability of NMR structural models depends critically on the assignment of cross-peaks in NOESY spectra. The nOe signal intensities can then be converted in a parametric manner into distance constraints for structure calculations. Inaccuracies resulting from internal motions or relayed transfers of magnetization (spin diffusion) are compensated to some extent by the redundancy in information provided by the large number of constraints typically measured. This can be summarized by the adage “safety comes in numbers”: while each individual distance measurement may not be precise or accurate, their combined use nevertheless leads to a reliable description of the molecule. The result is an ensemble of structures that best represent the observed constraints while minimizing structural violations (clashes between atoms, forbidden angles in a Ramachandran plot, etc.). Regions with a low root mean square deviation (rmsd) are expected to be rigid, while those with a high rmsd may indicate flexible regions, insufficient constraints (because they are unassigned or labeled as ambiguous and discarded), or conflicting constraints (assigned constraints mutually inconsistent with a single mean structure). The latter two effects depend directly on the completeness and accuracy of the nOe cross-peak assignment. For larger proteins, the correlation maps consist of 3D or 4D NOESY spectra edited by 15N and/or 13C chemical shifts and measured on samples that are predominantly deuterated. The sample deuteration is inevitably accompanied by a reduction in the number of spectroscopic probes available and hence by a reduction in redundant distance constraints, so that “safety in numbers” is not necessarily guaranteed any more. In fact it is not uncommon for the fold of a large NMR structure, based on sparse constraints, to be substantially altered by modifying only a few constraints. Thus, the accuracy of the model critically depends on assigning unambiguously a maximal number of constraints, and the remainder of this section will discuss a few powerful strategies to resolve ambiguities and maximize the number of constraints that can be assigned.
7.1. Assignment of distance constraints between amide protons
The 3D-NOESY-HN-trosy-HSQC experiment is used during many steps of an NMR investigation of larger proteins and several methods can be used to exploit its information content. Indeed, we have already referred to this experiment during both backbone assignment (Section 5) and methyl assignment (Section 6). Its magnetization pathway can be summarized by Hall(t1) → HN → N(t2) → HN(t3), and provides correlations [ωHN(i), ωN(i), {ωHall(j)}], in which {ωHall(j)} represents the frequencies of all protons (amide, aliphatic, aromatic, hydroxyl, etc.) present in the sample’s labeling scheme. While a simple 1H–15N sample has been used to provide supplementary constraints for a 37 kDa protein [136], in general, for larger proteins, the sample is predominantly deuterated and this paragraph will focus on amide to amide nOe cross-peaks, [HN(i), HN(j)]. Departing from the previous two sections, the objective is no longer resonance assignment (assignment of NMR signals to corresponding atoms) but instead cross-peak assignment (assignment of nOe cross-peaks to corresponding pairs of protons). Software packages typically include a semi-automated routine that ranks all candidate proton assignments in a given dimension according to the difference between their shifts and the chemical shift of the correlation observed in the NOESY spectrum. This provides a rapid means of identifying cross peaks for correlations involving protons with unique chemical shifts. However, the latter case is rare in larger proteins and a multitude of protons often match a signal. Moreover, the ranking may be altered by inaccuracies in peak picking. Fortunately, the limitations of this approach, which basically relies on a list of numbers, can be overcome to some extent by using the strategy described in Fig. 12, analogous to that described for nitrogen sequential correlations in Section 5.5 and Fig. 8. Again the principle relies on redundancies in correlations throughout the spectrum: the identity of a residue j in a correlation [Hd(i), Hn(j), N(i)] is obtained by inspecting the 3D spectrum for the related correlation [Hd(j), Hn(i), N(j)] (Fig. 12a). Hd denotes the detected proton dimension and Hn denotes the indirect dimension that features nOe’s. Fig. 12 shows an example for a NOESY-HN-trosy-HSQC recorded on a 53 kDa U-2H,15N,13C protein. First, the cross-peak inspected (labeled with “?”) is monitored on an Hd/Hn strip (Fig. 12b). Next an orthogonal N/Hn plane, at frequency ωHd = ωHd(j) is displayed next to the strip. The horizontal dashed cursor allows for identification of diagonal signals at ωHn = ωHn(j) and the horizontal dotted cursor reveals nOe cross-peaks at ωHn = ωHn(i). The vertical dashed cursors highlight the nitrogen frequencies that are determined with this procedure. Thus, the cross-peak labeled “?” is assigned through both ωH and ωN, for instance by placing a cross-hair at these coordinates in an assigned HN-trosy-HSQC. In this example, the procedure reveals that the inspected cross-peak in fact consists of the superposition of two cross-peaks and readily assigns both residues involved by providing the chemical shifts of the nitrogens of both residues. Since this protocol does not rely on peak picking, it may also provide a means of correcting erroneous assignments and is often used during backbone assignment in the presence of ambiguous sequential correlations. We emphasize that, while the procedure can be used as a positive criterion for confirming cross-peak assignment, it cannot be used to rule out candidates. Indeed, the relaxation properties of residues i and j and the proton densities in their environments may be substantially different and only one of either the [HN(i), HN(j), N(i)] or [HN(j), HN(i), N(j)] correlation may be observed in the spectrum: the absence of a redundant cross-peak does not exclude a possible assignment.
Fig. 12.
HN → HN nOe assignment by matching correlations in orthogonal planes of 3D-NOESY-HN-trosy-HSQC. (a) Cartoon of NOESY-HN-trosy-HSQC (or HSQC-NOESY) highlighting correlations that can be compared in orthogonal planes. Hd: dimension with 1H directly connected to 15N. Hn: dimension with nOe cross-peaks. White ovals denote autocorrelation signals (diagonal peaks), while black ovals indicate correlation signals resulting from nOes (cross-peaks). (b) Hd/Hn strip of R182 in a 53 kDa protein. The cross-peak to be assigned is labeled with “?”. The horizontal dotted and dashed lines show the proton frequencies of R182 and of the unassigned signal, respectively. (c) N/Hn plane at the Hd frequency of the cross-peak with label “?”. The vertical dashed lines highlight the nitrogen chemical shifts of residues that feature a cross-peak with R182. Here two residues with degenerate proton frequencies, D180 and Y184, each contribute to the nOe cross-peak labeled “?”. See text for details of interpretation.
In larger proteins, correlations between amide protons and other protons (methyls are discussed in the next section) are traditionally assigned only if they are unambiguous and only once a preliminary model is available to verify the assignment. That is, in general, they are used as a tool to lower the rmsd of the structural ensemble at an intermediate stage of structure calculation, rather than a means to modify the initial fold of the model. One important exception relates to nOes between amide and alpha pro- tons, which greatly help in lowering the local rmsd for backbone atoms, in particular for beta-sheets. Hα can be assigned with the sensitive carbon detected 13C-MQ-HACACO experiment [137] and the assignment of their nOe cross-peaks is facilitated by the predictable patterns that are expected in secondary structured elements. Thus, while the 3D-NOESY-HN-trosy-HSQC experiment cannot be used with protonated samples as in smaller proteins, it provides a useful means to supplement constraints obtained with other samples.
7.2. Assignment of distance constraints involving methyl protons
The success of the structure determination of a large protein depends principally on the accurate assignment of distance constraints involving methyl and amide groups. We have strongly emphasized the advantages of using perdeuterated samples with selective protonations of particular types of methyl groups, which provides a solution to combat both relaxation losses and spectral crowding. The success of using Ile, Leu and Val as a set of structural probes [17,138], which is facilitated by the possibility of labeling their α-ketoacid precursors, results from their preponderance in the core of proteins. Together they represent about 20% of the amino acids found in proteins [139]. Subsequently, methods for introducing methyls have been designed for all remaining methyl-containing amino acid types. Inclusion of Alanine methyl groups [25,26] add another 8–10% to a protein’s composition [24,140,141]. Methionine methyls [21–24] are a good probe of binding surfaces since their distribution is biased towards protein/protein interfaces [142]. Finally, selective labeling of threo-nine methyls [28–30] further helps define protein interfaces [143] and provides a functional enzymatic probe. However, for all selectively methyl labeled samples, the strategy reduces the number of distance constraints that are available for the structure calculation, thus potentially jeopardizing the “safety comes in numbers” criterion for producing reliable structures. As a consequence it is critical to obtain multiple high-resolution NOESY correlation spectra to ensure a maximal number of unambiguous nOe cross-peak assignments.
The separation of proton frequencies in selectively methyl labeled samples allows for measuring high quality spectra within a short experimental time by using the so-called time-shared technique [144,145]. Time-shared acquisition enables simultaneous recording of correlation maps involving different nuclei. Its simplest implementation simultaneously provides 2D HN/N and HC/C correlation maps by performing the following transfers in parallel: HN → N(t1) → HN(t2) and HC → C(t1) → HC(t2). The method has found some limited application for backbone resonance assignments [146,147] but was mainly implemented and improved for measurements of NOESY correlation maps [148–152]. However, time-shared experiments have not become popular for small 15N–13C labeled proteins, largely because different solvents and pulse sequences are needed for recording optimal HN/N and HC/C spectra. The pulse sequence encoding HC/C correlation maps cannot contain water flip-backs (which would compromise the detection of Ha resonances) and the data is best recorded in D2O. In D2O, amide protons are exchanged to deuterons and hence are not detected. Likewise, water presaturation could be used to suppress water while minimizing the loss of alpha proton signals but would significantly degrade the spectra of amide groups due to saturation transfer [153]. In contrast, time-shared experiments are ideally applicable to selectively methyl labeled samples such as 1H–13C– Me–ILV–2H–U–15N since both methyl and amide proton resonances are remote from the water resonance and a water flip-back [153] strategy can be used. Likewise, NOESY-HSQC involving aromatic and amide protons can be edited simultaneously with 15N and 13C and, since HN-TROSY and HC-TROSY schemes are similar, a TS-NOESY-HN/HCAr-trosy-HSQC pulse sequence was developed shortly after the emergence of the HN-TROSY technique [154]. The first time-shared experiment for a methyl labeled sample employed a sensitivity enhanced gradient selection strategy [155] to provide a TS-NOESY-HN-trosy-HSQC/HC-PEP-HSQC. The method was later improved with the more sensitive TS-NOESY-HN-trosy-HSQC/HC-HSQC, which implemented the TROSY scheme of Niet-lispach and encodes 13C chemical shifts with a States-TPPI scheme [156]. Finally, a TS-NOESY-HN-trosy-HSQC/HC-HMQC [157], was designed, with optimal relaxation for both 15N and 13C chemical shift encoding. However, the spin manipulations necessary for the experiment come at the cost of a reduction in sensitivity. A time-shared TS-HC/HN-HSQC-NOESY was also developed to provide cross-peaks along the detected dimension, i.e. with optimal resolution, and was tested up to 37 kDa [158]. The optimal resolution offsets somewhat the disadvantages of the lower signal-to-noise of the spectrum. In practice, it may be necessary to rapidly estimate which implementation of the time-shared technique is most suitable for given experimental conditions.
Obtaining 15N and 13C edited NOESY spectra simultaneously has clear advantages for studies of larger proteins. Clearly the savings in spectrometer time can be used for optimizing resolution and sensitivity. In addition, this may allow for the recording of all 3D NOESY data for a protein with limited stability, thus saving time and money for the preparation of a second sample that would otherwise be required to acquire the second spectrum. However, a perhaps less obvious advantage comes from having 15N and 13C correlation maps recorded from a single sample and during the same experimental run, thus providing nOe cross-peaks with identical proton frequencies in NOESY-HN-trosy-HSQC and NOESY-HC-HMQC spectra for a given proton nucleus. This property was exploited early on after designing time-shared experiments (e.g. in [149]) and becomes a critical asset for assigning nOes in larger proteins. Here, we present a procedure that makes use of all dimensions obtained in both 15N and 13C dispersed spectra and allows for assigning correlations between amide and methyl protons. Fig. 13 depicts an example for a 53 kDa 1H–13C–Me–ILV–2H– U–15N protein. The procedure is similar to that depicted in Section 5.5 and Fig. 8, and in Section 7.1 and Fig.12:the redundancy of (H,H) correlations in the two spectra is used to provide nOe assignments by inspecting an H/H strip and an orthogonal plane in the 3D experiment. However, when assigning a correlation between protons attached to different heteronuclei (15N and 13C), the plane and the strip belong to different spectra (Fig. 13a and d). We start by assigning an nOe cross-peak between a methyl and amide proton as it appears in the NOESY-HN-trosy-HSQC spectrum. First, an HN/H strip of the NOESY-HN-trosy-HSQC is selected (here G294, panel b) and a targeted nOe cross-peak [ωHN(i), ωN(i), ωHMe(j)] to a yet to be determined methyl is inspected (the horizontal dotted line denotes the proton frequency of this cross-peak). Displaying the C/H plane at the 1H(Me) frequency of this cross-peak (panel c) reveals a correlation [ωHN(i), ωCMe(j), ωHMe(j)] with the original HN spin (cross-peak at the intersection of the horizontal dashed and vertical dotted lines in panel c), thus identifying the carbon frequency of the {H, C} spin system (vertical dotted line) and, thereby, enabling assignment of the nOe cross-peak. Clearly, the procedure can be performed in the opposite order, starting from an HMe/H strip (panele) and inspecting the N/H plane at the 1H(HN) frequency of this cross-peak (panel f) to identify the {H,N} system with a cross-peak at the intersection of the vertical dashed and horizontal dotted lines in panel f. This procedure allows for assignment of an [ωHN(i), ωCMe(j), ωHMe(j)] correlation in the original HMe/H strip (panel e). The advantages of this protocol over the more rapid semi-automated listing and selection of spin systems described in Section 7.1 become obvious when one remembers that methyl groups are confined to a spectral width of about 1–1.5 ppm. The list of systems with matching frequencies often comprises at least or so 10 candidates and, in the absence of other information, assignment depends on an educated guess that will need to be tested during the subsequent rounds of structure calculations, by monitoring distance violations. More importantly, the procedure overcomes inaccuracies in peak-picking. Like wise, the crucial importance of the simultaneous acquisition of NOESY-HC-HMQC and NOESY-HN-trosy-HSQC becomes apparent, since even minimal discrepancies in the proton frequencies of an (H, H) correlation observed in both 15N and 13C edited spectra would jeopardize an accurate assignment of nOe cross-peaks.
Fig. 13.
HN → HC nOe assignment by matching correlations in orthogonal planes of 3D-NOESY-HN-trosy-HSQC and 3D-NOESY-HC-HSQC. (a) Cartoon representation of the procedure used to assign nOe cross peaks to methyls in a 3D NOESY-HN-trosy-HSQC. (b) HN/H strip of G294 in 3D NOESY-HN-trosy-HSQC recorded on a 53 kDa protein. The horizontal dotted line denotes the proton frequency of the methyl that needs to be assigned and the horizontal dashed line denotes the frequency of HN G294. (c) C/H plane of 3D NOESY-HC-HSQC at the frequency ωHMe = ωHMe(V293) identified in (b). The vertical dotted line denotes the carbon frequency of V293, the only residue with a cross-peak at ωHN(G294). Note that other residues with apparent cross-peaks to G294 can be discarded after viewing expansions (not shown here) around the cross-peaks, which reveals subtle but unambiguous differences in proton frequencies. (d) Cartoon representation of the procedure used to assign nOe cross peaks to amides in a 3D NOESY-HC-HSQC. (e) HMe/H strip of V293 in 3D NOESY-HC-HSQC. The horizontal dashed line now denotes the amide proton that needs to be assigned. (f) N/H plane of 3D NOESY-HN-trosy-HSQC at the frequency ωHN = ωHN(G294) identified in e. See text for details of interpretation.
The methods described so far in this section fail when the frequencies of both protons involved in an (H, H) correlation are subject to degeneracies, for example, when (HN, HMe) cross-peaks appear multiple times at the same position in NOESY-HN-trosy-HSQC and NOESY-HC-HSQC spectra. The abundance of amide proton frequencies and the spectral confinement of methyl protons render such a situation rather frequent in larger proteins for (HN, HN), (HN, HMe), and (HMe, HMe) correlations. Thus, it is routinely necessary to record four-dimensional NOESY correlation maps to assign nOe cross peaks unambiguously in large proteins. Four different spectra are necessary for a complete and unambiguous assignment of nOes involving amide and methyl protons; they correspond to the four nOe transfers: HN → HN(I), HN → HMe(II), HMe → HN(III), and HMe → HMe(IV). Note that while transfers II and III may appear redundant, since the same correlations are encoded, the correlations that can be observed often differ owing to the difference in environment between the methyl and proton probes (relaxation, density of nearby protons, etc.). For example, transferring magnetization from an amide proton surrounded by other protons to a more isolated methyl group may not result in an (HN, HMe) cross peak, since the amide proton magnetization is subject to dipolar interactions with an entire network of protons, whereas a transfer from the same isolated methyl to the same amide proton would result in an (HMe, HN) cross-peak. Thus, the complete set of all four experiments must be recorded, with each correlation map requiring many days of spectrometer time. Fortunately, a time-shared method analogous to that described for 3D-NOESY-HSQC spectra can be used to provide all four spectra within a single measurement [155]. The experiment is a straightforward modification of that used for 3D spectra, with a time shared HSQC (or HMQC or HMQC/HSQC [148]) appended to the beginning of the pulse sequence. It provides the following four magnetization transfer elements:
HN(i)→N(i)(t1)→HN(i)(t2)→HN(j)→N(j)(t3)→HN(t4).
HN(i)→N(i)(t1)→HN(i)(t2)→HMe(j)→C(j)(t3)→HMe(t4).
HMe(i)→C(i)(t1)→HMe(i)(t2)→HN(j)→N(j)(t3)→HN(t4).
HMe(i)→C(i)(t1)→HMe(i)(t2)→HMe(j)→C(j)(t3)→HMe(t4).
The information provided by each 4D experiment is best understood by representing the data as a two-dimensional array of two-dimensional planes (Fig. 14a). The analysis of the 4D spectra consists in displaying HSQC-like planes of two out of four dimen-sionsatthe coordinatesofthe remaining two dimensions. For example, the 4D HC-HSQC-NOESY-HN-trosy-HSQC spectrum provides H/ N planes for each point in the HC-HSQC, and vice versa. Each H/N plane displays all (H,N)correlations that are mediated by nOe transfers originating from the {HMe, CMe} spin system matching the (H, C) coordinates in the remaining two dimensions. Fig. 14 shows examples obtained with a 37 kDa protein and depicts the advantages of using 4D experiments for the three pathways HN → HN(I), HN → HMe(II), and HMe → HMe(IV). Panel b shows a strip comparison from 3D spectra used in an attempt to assign an HN → HN cross-peak for residue G172. Three residues, E198, Q199, and V200, display a poten-tial cross-peak with G172. G172 itself displays a cross peak that may belong to K15, Q199, or V314. Thus, Q199 is the most likely candidate for assigning the cross-peakin G172 since Q199 also displays a cross-peak with G172. However, the cross-peak in Q199 had been assigned already to a correlation with residue A201, largely because such an assignment was apparently supported by further correlations involving near-sequence-neighbors of A201 (those between A201 and V200 and between E198 and A201). However, the 4D HN-HSQC-NOESY-HN-trosy-HSQC shown in panel c reveals unambiguously that G172 is in fact correlated to Q199. It thus turns out that the cross-peak provides a long-range distance constraint; such a long-range constraint will be critical during the structure calculation. A similar situation occurs for the HN → HMe nOe cross-peaks (pathway II) shown in panels d and e. The strip comparison in Fig. 14d shows that the amide of A23 is potentially correlated to the γ1 methyl of V77 (the labels γ1 and γ2 are arbitrary in what follows; stereospecific assignment had not been performed at this stage of analysis). However, the corresponding cross-peak has been assigned to the adjacent residue I22. The 4D HC-HSQC-NOESY-HN-trosy-HSQC demonstrates that, while A23 and I22 are indeed correlated, the majority of the signal in the 3D cross-peak originates from a correlation with V77, again providing a long-range distance constraint. Because of the small spectral width of methyl protons, ambiguous correlations abound in the 3D-NOESY-HC-HSQC for (HMe, HMe) correlations. Indeed, the strip-comparison shown in Fig. 14f for the γ1 methyl of V71 shows two candidates that provide cross-peak patterns that could indicate correlations not only with γ1 but also with the γ2 methyl. This pair of methyl proton chemical shifts, Hγ1, Hγ2, is unique in the protein; only one {CMe, HMe, HMe} spin-system has these two proton frequencies. Thus, the methyls of either L54 or L193, or both, could be near those of V71. The 4D-HC-HSQC-NOESY-HC-HSQC spectrum indicates unambiguously that the cross peaks belong to L54 and reveals an additional correlation with I81, that suffered from a partial overlap with the inter-methyl correlation (Hγ1, Hγ2) in the 3D. The same 4D analysis of L193 reveals that the apparent correlations to both methyls of V71 is in fact due to the combination of a correlation with L162 and another with L166. All these observations are representative examples of the limits of 3D NOESY experiments used with selectively methyl protonated samples: while the spectrum is greatly simplified by the labeling scheme, the combination of poor dispersion in the methyl proton dimension and the reduction of the number of nOe cross-peaks, which may otherwise help resolve ambiguities, makes 4D experiments a necessity for structural studies of larger proteins.
Fig. 14.
nOe assignment with time-shared 4D NOESY spectra. For clarity, the labeling of the cross-peaks departs from the convention used in the remainder of the article. Superscripts refer to chemical groups and subscripts refer to residue numbers. (a) Cartoon representing the 4D spectrum as a 2D array of 2D planes. The axis labels are used to describe panels c, e, and g. In (c), ω2/ω3 planes are displayed (i.e. like the shaded planes in panel (a) and in (e and g) ω1/ω4 planes are displayed (i.e. like the unshaded plane in panel (a). (b) HN → HN: Strip comparison of the 3D NOESY-HN-trosy-HSQC obtained with the time-shared technique on a 37 kDa protein, with G172 as a reference strip. Only the amide region is investigated. Based on this information, all candidates could be correlated to either G172 or A201 or both. See text for a detailed description. The circle indicates the (HN, N) correlation of G172 that is used to provide the coordinates in the 4D experiment. (c) HN → HN: 4D HN-HSQC-NOESY-HN-trosy-HSQC. Shown is the HN/N plane along ω2 and ω3 at the coordinates ω1 = ωN(G172) and ω4 = ωH(G172), which unambiguously shows that it is Q199 that has the nOe with G172. (d) HMe → HN: Strip comparison of the 3D NOESY-HN-trosy-HSQC showing both the amide and methyl regions. The circle indicates the (HN, N) correlation that provides the coordinates in (e). Based on this information, it is unclear whether the nOe from A23 NH involves the δ1 methyl of I22, the γ1 methyl of V77, or both (e) HMe → HN: HMe/C plane of the 4D-HN-HSQC-NOESY-HC-HSQC at the coordinates (ω1, ω3) = [ωH(A23), ωN(A23)]. The vertical dashed line indicates the frequency corresponding to the horizontal dashed line in d, and the cross-peaks in this plane show that both candidate methyls identified by analysing the 3D spectrum have an nOe interaction with A23 HN, with the larger contribution coming from Val77 γ1. (f) HMe → HMe: Strip comparison of the 3D NOESY-HC-HSQC showing the methyl region. The circle indicates the (HMe, C) correlation that provides the coordinates in (g). The frequency of the cross-peak inspected in g is denoted by the horizontal dashed line. Two residues, L54 and L193, have proton frequencies matching that of the cross-peak seen in the strip of V71 and feature two cross-peaks at frequencies corresponding to both methyl protons of V71. (g) HMe → HMe: HMe/C plane of the 4D-HC-HSQC-NOESY-HC-HSQC at the coordinates (ω2, ω3) = [ωHγ1(V71), ωCγ1(V71)]. The vertical dashed line indicates the frequency corresponding to the horizontal dashed line in (e) and reveals that the g1 methyl of V71 is near in space to L54 and not L193. See text for details of interpretation. Adapted from “D.P. Frueh, D.A. Vosburg, C.T. Walsh, G. Wagner, Determination of all nOes in 1H–13C–Me–ILV–U–2H–15N proteins with two time-shared experiments, J. Biomol. NMR 34 (2006) 31–40” with kind permission from Springer Science and Business Media [155].
In summary, assignments of distance constraints are possible even in larger proteins when using methyl groups as selective spectroscopic probes. To be accurate, however, such assignments require the combination of all 3D and 4D spectra, which can be obtained in two acquisitions with time-shared techniques. 4D spectra provide unambiguous assignments but suffer from reduced sensitivity, whereas 3D spectra are harder to analyze but may feature additional constraints. The quality of the spectra can be further improved with non-uniform sampling, as recently demonstrated for 4D time-shared experiments on a 35 kDa protein [69]. Thus, optimal correlation maps can be obtained and used in combination to facilitate unambiguous assignment of distance constraints.
8. Other larger biological systems: macromolecular complexes and large unfolded proteins
The methods discussed in this article focused on large monomeric folded proteins, i.e. proteins with a large number of residues in a single polypeptidic chain and forming a defined tertiary structure. Different strategies must be employed when studying very large assemblies (multi-domain proteins or protein complexes of several hundreds of kDa or nearing a weight of 1 MDa) or unfolded proteins. Each topic deserves a dedicated review and, here, we will merely relate the concepts introduced in the previous sections to these two classes of large proteins.
Very large molecular assemblies are typically studied by NMR using a “divide and conquer” approach. The individual components of an assembly are isolated, and their respective NMR spectra are assigned individually as described in Sections 5–7. This assignment must then be transposed to the resonances observed in the complex. This transfer of assignment is best performed by focusing on one component of the complex at a time, i.e. by labeling it selectively so that only the corresponding subset of signals appears in the spectrum. For multi-domain proteins, segmental labeling can often be used to label each domain individually [159–162]. In most cases, only a small subset of signals will have different backbone chemical shifts for the protein isolated and in complex, the remaining signals having identical frequencies, and only the subset of signals showing changes must be re-assigned. The sequential fragments assigned for the isolated protein can be used as a starting point to assign the signals in the complex. In most instances, this fragment will feature both signals of residues that have the same shifts, and others that have different shifts in isolated and bound proteins. Frequently, there is a “transition” fragment of a few residues in which the difference in chemical shifts between the protein in complexed and free forms gradually increases. This transition can be followed residue by residue, with the sequential correlations of an experiment such as trosy-HNCA visualized via strip comparisons (Section 5.2) and H/N plane inspections (Section 5.4). Even in the least favorable case of a residue (i) with identical chemical shifts in both environments [ωHN(i), ωN(i), ωCα(i)] preceded by a residue with a drastic change in chemical shifts, the sequential correlation [ωHN(i), ωN(i), ωCα(i – 1)] will provide the new shifts in the carbon dimension. The H/N plane at this chemical shift can be inspected as shown in Fig. 7a and reveal the new [ωHN(i – 1), ωn(i – 1)] coordinates of residue i – 1; this time, however, only one (H, N) correlation is unassigned and a single spectrum may be enough to complete the assignment. In general, when only few resonances need to be re-assigned, this simplified assignment strategy can be performed with a limited number of experiments. The corresponding savings in spectrometer time can be used to increase the acquisition time of the experiments) and compensate for sensitivity losses, at least in part. The sensitive 3D experiments trosy-HNCA and NOESY-HN-trosy-HSQC are a good starting point for assigning the majority of the signals, for instance by combining the procedures described in Section 5.4, Fig. 7a and Section 7, Fig. 12. We note that for studies of so-called supramolecular assemblies, such as the 72 kDa GroES homohept-amer in complex with the 800 kDa GRoEL homotetradecamer, the assignment of {H, N} systems is limited to residues that can be identified by comparing 2D correlation maps [163]. Backbone resonances of multi-protein complexes, homo-oligomers, and multi-domain proteins can be assigned with procedures built upon the strategy described in this paragraph.
The assignments of methyl resonances and distance constraints can be obtained on isolated components, and transposed to larger assemblies, with procedures virtually identical to those described in Sections 6 and 7, respectively [122]. However, for optimal acquisition of correlation maps in the larger assemblies, it becomes necessary to use selective 13C labeling and protonation of a single methyl in Leu and Val to optimize sensitivity [19]. Such strategies have enabled spectacular studies, such as investigations of the 670 kDa α7β7β7α7 core particle of the proteasome [122,164].
Many of the concepts introduced in this article can be adapted to studies of large intrinsically disordered proteins (IDPs). However, the limit in size for successful assignment cannot be compared to those mentioned in the text. IDPs, which are subject to large amplitude fast (sub-ns) internal motions, typically have slower relaxation rates than folded proteins with the same number of residues (excluding exchange phenomena). On the other hand, the lack of structured environments leads to extensive overlap and numerous degeneracies, even for proteins of modest length. In particular, alpha and beta carbons, which are extensively used during assignment of folded protein signals, are of little use for IDPs because their chemical shifts report primarily on dihedral backbone torsion angles, Φ and ψ [50], which are averaged in such environments. In contrast, carbonyl carbons and amide nitrogens maintain relatively good dispersion in IDPs. Thus, the experiments (H)N(CA)NH, described in Section 5.5 and Fig. 13, as well as (H)N(COCA)NH and the related H(NCA)NH and H(NCOCA)NH, have found numerous applications for assigning unfolded proteins [91]. (H)NCA(N)H, which provides the straightforward stairway backbone walk assignment procedure described in Section 5.5 and Fig. 13, has been modified by Hosur and coworkers to an (H)NCO(CAN)H, thereby replacing the 13Cα dimension with a well dispersed 13CO dimension [165]. A more dramatic change in strategy consists in replacing proton detection by carbon detection in experiments designed for sequential correlations. This strategy was initially developed to combat extreme relaxation, e.g. in proteins with paramagnetic centers, by working with so-called protonless samples, i.e. perdeuterated samples in D2O [137,166–169]. For studies of IDPs, the major improvement is obtained when using carbonyl carbons during detection, to provide better dispersion than other carbons or protons [170]. This nucleus is also less sensitive to line-broadening induced by exchange than amide protons. In contrast to 13C detected experiments used to overcome relaxation, 13C detection of IDP signals can also be used to provide correlations with protons, thus providing much needed additional sets of correlations [171].
In summary, for the two classes of large proteins discussed in this section, the strategies used for resonance assignment depart from those introduced in this article, either because only a limited set of experiments is available (supramolecular assemblies) or because the experiments need to be replaced by others (IDPs). However, the central tenet of this article remains true: for larger proteins, all the data must be combined during analysis to exploit various sets of correlations and overcome the complexity of spectra.
9. Conclusions
Following the development of TROSY techniques, many existing NMR experiments for protein resonance assignment were adapted to provide correlation maps for larger proteins. This enabled sensitivity limitations due to relaxation to be substantially overcome, thus extending the strategies developed in the early 1990s to more challenging systems. However, these methods rapidly proved insufficient for combating the increased complexity of the NMR spectra, and many applications were limited to homo-ligomeric proteins. At the outset, the problem appeared unresolvable: more correlations are needed to resolve ambiguities but fewer experiments are applicable. Fortunately, the last decade has seen the emergence of a number of methods to overcome this complexity, involving new developments in sample preparation, spectroscopic manipulations, or a combination of both. All have the same objective: to resolve ambiguities and make assignment a tractable task. In terms of NMR experiments, the objective was to increase the number of correlation maps available for studies of larger proteins. For backbone assignment, this was achieved either by providing correlations in additional dimensions, as discussed in Section 5.6, or by increasing the number of correlations per dimension, Section 5.5. In effect, the additional correlations were not strictly speaking obtained with completely novel experiments, but rather by redesigning experiments to maximize their information content. Thus, the 3D-trosy-HN(CO)CA and 3D-trosy-HN(CA)CO experiments were modified into the 4D-trosy-HNCOCA, 4D-trosy-HNCACO, and 4D-trosy-HNCOi–1CA experiments, and the 3D-trosy-(H)NCA(N)H experiment was designed by optimizing magnetization pathways in the 3D-trosy-(H)N(CA)NH experiment to provide sequential correlation in three dimensions instead of one. For methyl assignment, the solution turned out to be to replace direct correlations with HN-anchors by correlations with Cα and Cβ, each correlated to HN-anchors. For assignment of distance constraints, multiple spectra are measured in parallel and the saving in time is used to optimize spectral quality. Indeed, for assignments of all types of resonances (backbone, methyl, and distance constraints), a major breakthrough was the ability to exploit the benefits of high-field measurements without compromise in resolution. Thus, throughout the examples discussed in this review, non-uniform sampling has been used either to reach optimal resolution or to maintain good resolution while optimizing sensitivity. Overall, a large number of NMR spectra can be recorded with high quality to enable assignment of NMR signals.
Novel assignment procedures emerged with the availability of an increased number of multidimensional spectra. Thus, the 3D-trosy-(H)NCA(N)H spectrum allows for a sequential backbone stairway assignment procedure and multiple 4D spectra are used to correlate two sets of 2-dimensional correlation maps. As a general rule, the complete set of NMR data accumulated is greater than the sum of its parts, and resonances are often assigned by using several spectra. Thus, experiments used to assign methyl resonances or to provide distance constraints are also used during sequential assignment of backbone resonances. Similarly, 15N and 13C edited 3D NOESY experiments can be combined to assign distance constraints, or four 4D experiments may be used together for assigning backbone resonances. In addition, it becomes important to analyze spectra with alternate strategies. For example, backbone triple resonance experiments are analyzed not only with combined strip comparisons but also by synchronous inspection of H/N planes. Likewise, trosy-(H)N(CA)NH and NOESY-HN-trosy-HSQC can provide assignments by displaying pairs of orthogonal planes.
In summary, the accumulation of novel correlation maps, together with the emergence of alternate assignment strategies, has helped overcome many of the challenges associated with spectral complexity in NMR studies of larger proteins. Strategies such as those presented in this review may help popularize these challenging investigations, and, in the end, allow for harnessing the many powerful advantages of NMR in studies of biological systems.
Acknowledgements
The author is indebted to Drs. Sven Hyberts and Valdislav Orekhov for in-depth discussions regarding the minimal number of sampled points and the quality of reconstructed spectra. I would also like to thank Andrew Goodrich and Dr. Ananya Majumdar for careful reading of the manuscript and insightful comments. Data used for Figs. 3, 5, 6 and 9 were collected while the author was working with Prof. Gerhard Wagner and funded by NIH Grant PO1GM47467. Fig. 10 is adapted with permission from V. Tugarinov, R. Muhandiram, A. Ayed, L.E. Kay, “Four-dimensional NMR spectroscopy of a 723-residue protein: chemical shift assignments and secondary structure of malate synthase g”, J. Am. Chem. Soc. 124 (2002) 10025–10035, Copyright 2002 American Chemical Society. Fig. 14 is adapted from D.P. Frueh, D.A. Vosburg, C.T. Walsh, G. Wagner, “Determination of all nOes in 1H–13C–Me–ILV–U–2H–15N proteins with two time-shared experiments”, J. Biomol. NMR 34 (2006) 31–40 with kind permission from Springer Science and Business Media. All other figures were produced with data supported by Grant NIH R01GM104257.
Glossary
- BMRB
Biological Magnetic Resonance Bank
- CARA
Computer aided resonance assignment
- CCPN
Collaborative computing project for NMR
- COSY
Correlation spectroscopy
- CSA
Chemical shift anisotropy
- DD
Dipole–dipole
- HMQC
Heteronuclear multiple-quantum correlation spectroscopy
- HSQC
Heteronuclear single-quantum correlation spectroscopy
- INEPT
Insensitive nuclei enhanced by polarization transfer
- IST
Iterative soft threshold
- MaxEnt
Maximum Entropy reconstruction (Hoch and Stern)
- MDD
Multi-dimensional decomposition
- MWD
Multi-way decomposition
- nOe
nuclear Overhauser effect
- NUS
Nonuniform sampling
- NOESY
nOe spectroscopy
- ST2-PT
Single transition-to-single, transition polarization transfer
- SW
spectral width
- TRACT
TROSY for rotational correlation times
- TROSY
Transverse relaxation optimized spectroscopy
- trosy-
Prefix indicating use of the TROSY principle within a pulse sequence or pulse sequence element
- TS
Time-shared
- WATERGATE
WATER suppression by GrAdient Tailored Excitation
References
- 1.Keller RLJ. The Computer Aided Resonance Assignment Tutorial, Cantina Verlag, Goldau. 2004 [Google Scholar]
- 2.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins: Struct. Funct. Bioinform. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 3.Goddard T, Kneller D. SPARKY 3. San Fransisco: Univeristy of California; [Google Scholar]
- 4.Johnson BA, Blevins RA. NMR view: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 5.Peti W, Page R. Strategies to maximize heterologous protein expression in Escherichia coli with minimal cost. Protein Exp. Purif. 2007;51:1–10. doi: 10.1016/j.pep.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 6.Ducat T, Declerck N, Gostan T, Kochoyan M, Demene H. Rapid determination of protein solubility and stability conditions for NMR studies using incomplete factorial design. J. Biomol. NMR. 2006;34:137–151. doi: 10.1007/s10858-006-0003-0. [DOI] [PubMed] [Google Scholar]
- 7.Bodenhausen G, Ruben DJ. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 1980;69:185–189. [Google Scholar]
- 8.Pervushin K, Riek R, Wider G, Wüthrich K. Attenuated T 2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piotto M, Saudek V, Sklenárˇ V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 10.Krishnan VV, Thornton KH, Cosman M. An improved experimental scheme to measure self-diffusion coefficients of biomolecules with an advantageous use of radiation damping. Chem. Phys. Lett. 1999;302:317–323. [Google Scholar]
- 11.Ferrage F, Zoonens M, Warschawski DE, Popot JL, Bodenhausen G. Slow diffusion of macromolecular assemblies by a new pulsed field gradient NMR method. J. Am. Chem. Soc. 2003;125:2541–2545. doi: 10.1021/ja0211407. [DOI] [PubMed] [Google Scholar]
- 12.Chou JJ, Baber JL, Bax A. Characterization of phospholipid mixed micelles by translational diffusion. J. Biomol. NMR. 2004;29:299–308. doi: 10.1023/B:JNMR.0000032560.43738.6a. [DOI] [PubMed] [Google Scholar]
- 13.Lee D, Hilty C, Wider G, Wüthrich K. Effective rotational correlation times of proteins from NMR relaxation interference. J. Magn. Reson. 2006;178:72–76. doi: 10.1016/j.jmr.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Riek R, Fiaux J, Bertelsen EB, Horwich AL, Wüthrich K. Solution NMR techniques for large molecular and supramolecular structures. J. Am. Chem. Soc. 2002;124:12144–12153. doi: 10.1021/ja026763z. [DOI] [PubMed] [Google Scholar]
- 15.Gardner KH, Kay LE. The use of 2H, 13C, 15N multidimensional NMR to study the structure and dynamics of proteins. Annu. Rev. Biophys. Biomol. Struct. 1998;27:357–406. doi: 10.1146/annurev.biophys.27.1.357. [DOI] [PubMed] [Google Scholar]
- 16.Nietlispach D, Clowes RT, Broadhurst RW, Ito Y, Keeler J, Kelly M, Ashurst J, Oschkinat H, Domaille PJ, Laue ED. An approach to the structure determination of larger proteins using triple resonance NMR experiments in conjunction with random fractional deuteration. J. Am. Chem. Soc. 1996;118:407–415. [Google Scholar]
- 17.Gardner KH, Kay LE. Production and incorporation of 15N, 13C, 2H (1H–1 methyl) isoleucine into proteins for multidimensional NMR studies. J. Am. Chem. Soc. 1997;119:7599–7600. [Google Scholar]
- 18.Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE. A robust and cost-effective method for the production of Val, Leu, Ile (delta 1) methyl-protonated 15N-, 13C-, 2H-labeled proteins. J. Biomol. NMR. 1999;13:369–374. doi: 10.1023/a:1008393201236. [DOI] [PubMed] [Google Scholar]
- 19.Tugarinov V, Kay LE, Leu Ile. and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J. Am. Chem. Soc. 2003;125:13868–13878. doi: 10.1021/ja030345s. [DOI] [PubMed] [Google Scholar]
- 20.Gans P, Hamelin O, Sounier R, Ayala I, Dura MA, Amero CD, Noirclerc-Savoye M, Franzetti B, Plevin MJ, Boisbouvier J. Stereospecific isotopic labeling of methyl groups for NMR spectroscopic studies of high-molecular-weight proteins. Angew. Chem. 2010;49:1958–1962. doi: 10.1002/anie.200905660. [DOI] [PubMed] [Google Scholar]
- 21.Fischer M, Kloiber K, Häusler J, Ledolter K, Konrat R, Schmid W. Synthesis of a 13C-methyl-group-labeled methionine precursor as a useful tool for simplifying protein structural analysis by NMR spectroscopy. ChemBioChem. 2007;8:610–612. doi: 10.1002/cbic.200600551. [DOI] [PubMed] [Google Scholar]
- 22.Stoffregen Mira C, Schwer Matthias M, Renschler Fabian A, Wiesner S. Methionine scanning as an NMR tool for detecting and analyzing biomolecular interaction surfaces. Structure. 2012;20:573–581. doi: 10.1016/j.str.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 23.Gelis I, Bonvin AMJJ, Keramisanou D, Koukaki M, Gouridis G, Karamanou S, Economou A, Kalodimos CG. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell. 2007;131:756–769. doi: 10.1016/j.cell.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weininger U, Liu Z, McIntyre DD, Vogel HJ, Akke M. Specific 12CβD212CγD2S13CεHD2 isotopomer labeling of methionine to characterize protein Dynamics by 1H and 13C NMR relaxation dispersion. J. Am. Chem. Soc. 2012;134:18562–18565. doi: 10.1021/ja309294u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isaacson RL, Simpson PJ, Liu M, Cota E, Zhang X, Freemont P, Matthews S. A new labeling method for methyl transverse relaxation-optimized spectroscopy NMR spectra of alanine residues. J. Am. Chem. Soc. 2007;129:15428–15429. doi: 10.1021/ja0761784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayala I, Sounier R, Usé N, Gans P, Boisbouvier J. An efficient protocol for the complete incorporation of methyl-protonated alanine in perdeuterated protein. J. Biomol. NMR. 2009;43:111–119. doi: 10.1007/s10858-008-9294-7. [DOI] [PubMed] [Google Scholar]
- 27.Ruschak AM, Velyvis A, Kay LE. A simple strategy for (1)(3)C, (1)H labeling at the Ile-gamma2 methyl position in highly deuterated proteins. J. Biomol. NMR. 2010;48:129–135. doi: 10.1007/s10858-010-9449-1. [DOI] [PubMed] [Google Scholar]
- 28.Takeda M, Jee J, Ono AM, Terauchi T, Kainosho M. Hydrogen exchange study on the hydroxyl groups of serine and threonine residues in proteins and structure refinement using NOE restraints with polar side-chain groups. J. Am. Chem. Soc. 2011;133:17420–17427. doi: 10.1021/ja206799v. [DOI] [PubMed] [Google Scholar]
- 29.Sinha K, Jen-Jacobson L, Rule GS. Specific labeling of threonine methyl groups for NMR studies of protein-nucleic acid complexes. Biochemistry. 2011;50:10189–10191. doi: 10.1021/bi201496d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Velyvis A, Ruschak AM, Kay LE. An economical method for production of 2H,13CH3 - threonine for solution NMR studies of large protein complexes: application to the 670 kDa proteasome. PLoS ONE. 2012;7:e43725. doi: 10.1371/journal.pone.0043725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pervushin KV, Wider G, Wüthrich K. Single transition-to-single transition polarization transfer (ST2-PT) in [15N,1H]-TROSY. JBNMR. 1998 doi: 10.1023/A:1008268930690. [DOI] [PubMed] [Google Scholar]
- 32.Pervushin K, Riek R, Wider G, Wüthrich K. Transverse relaxation-optimized spectroscopy (TROSY) for NMR studies of aromatic spin systems in 13C-labeled proteins. J. Am. Chem. Soc. 1998;120:6394–6400. [Google Scholar]
- 33.Miclet E, Williams DC, Jr, Clore GM, Bryce DL, Boisbouvier J, Bax A. Relaxation-optimized NMR spectroscopy of methylene groups in proteins and nucleic acids. J. Am. Chem. Soc. 2004;126:10560–10570. doi: 10.1021/ja047904v. [DOI] [PubMed] [Google Scholar]
- 34.Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE. Cross-correlated relaxation enhanced 1H[bond]13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J. Am. Chem. Soc. 2003;125:10420–10428. doi: 10.1021/ja030153x. [DOI] [PubMed] [Google Scholar]
- 35.Schanda P, Kupče E¯, Brutscher B. SOFAST-HMQC experiments for recording two-dimensional deteronuclear correlation spectra of proteins within a few seconds. J. Biomol. NMR. 2005;33:199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- 36.Lescop E, Schanda P, Brutscher B. A set of BEST triple-resonance experiments for time-optimized protein resonance assignment. J. Magn. Reson. 2007;187:163–169. doi: 10.1016/j.jmr.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Cai S, Seu C, Kovacs Z, Sherry AD, Chen Y. Sensitivity enhancement of multidimensional NMR experiments by paramagnetic relaxation effects. J. Am. Chem. Soc. 2006;128:13474–13478. doi: 10.1021/ja0634526. [DOI] [PubMed] [Google Scholar]
- 38.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. NMR Spectrosc. 1999;34:93–158. [Google Scholar]
- 39.Loria JP, Rance M, Palmer 3rd AG. Transverse-relaxation-optimized (TROSY) gradient-enhanced triple-resonance NMR spectroscopy. J. Magn. Reson. 1999;141:180–184. doi: 10.1006/jmre.1999.1891. [DOI] [PubMed] [Google Scholar]
- 40.Salzmann M, Wider G, Pervushin K, Wüthrich K. Improved sensitivity and coherence selection for [15N,1H]-TROSY elements in triple resonance experiments. J. Biomol. NMR. 1999;15:181–184. doi: 10.1023/a:1008358030477. [DOI] [PubMed] [Google Scholar]
- 41.Nietlispach D. Suppression of anti-TROSY lines in a sensitivity enhanced gradient selection TROSY scheme. J. Biomol. NMR. 2005;31:161–166. doi: 10.1007/s10858-004-8195-7. [DOI] [PubMed] [Google Scholar]
- 42.Yang D, Kay LE. Improved 1HN-detected triple resonance TROSY-based experiments. J. Biomol. NMR. 1999;13:3–10. doi: 10.1023/A:1008329230975. [DOI] [PubMed] [Google Scholar]
- 43.Brutscher B. Intraresidue HNCA and COHNCA experiments for protein backbone resonance assignment. J. Magn. Reson. 2002;156:155–159. doi: 10.1006/jmre.2002.2546. [DOI] [PubMed] [Google Scholar]
- 44.Nietlispach D, Ito Y, Laue ED. A novel approach for the sequential backbone assignment of larger proteins: selective intra-HNCA and DQ-HNCA. J. Am. Chem. Soc. 2002;124:11199–11207. doi: 10.1021/ja025865m. [DOI] [PubMed] [Google Scholar]
- 45.Meissner A, Sørensen OW. A sequential HNCA NMR pulse sequence for protein backbone assignment. J. Magn. Reson. 2001;150:100–104. doi: 10.1006/jmre.2001.2317. [DOI] [PubMed] [Google Scholar]
- 46.Permi P, Annila A. A new approach for obtaining sequential assignment of large proteins. J. Biomol. NMR. 2001;20:127–133. doi: 10.1023/a:1011208803036. [DOI] [PubMed] [Google Scholar]
- 47.Ohki S-Y, Kainosho M. Stable isotope labeling methods for protein NMR spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 2008;53:208–226. [Google Scholar]
- 48.Muchmore DC, McIntosh LP, Russell CB, Anderson DE, Dahlquist FW. Expression and nitrogen-15 labeling of proteins for proton and nitrogen-15 nuclear magnetic resonance. In: Norman JO, Thomas LJ, editors. Methods Enzymol. Academic Press; 1989. pp. 44–73. [DOI] [PubMed] [Google Scholar]
- 49.Wishart DS, Sykes BD. The 13C chemical shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. JBNMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 50.Havlin RH, Le H, Laws DD, deDios AC, Oldfield E. An ab initio quantum chemical investigation of carbon-13 NMR shielding tensors in glycine, alanine, valine, isoleucine, serine, and threonine: comparisons between helical and sheet tensors, and the effects of χ1 on shielding. J. Am. Chem. Soc. 1997;119:11951–11958. [Google Scholar]
- 51.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 52.Barna JCJ, Laue ED, Mayger MR, Skilling J, Worrall SJP. Exponential sampling an alternative method for sampling, in two-dimensional NMR experiments. J. Magn. Reson. 1987;73:69–77. [Google Scholar]
- 53.Laue ED, Mayger MR, Skilling J, Staunton J. Reconstruction of phase-sensitive two-dimensional NMR spectra by maximum entropy. J. Magn. Reson. 1986;68:14–29. 1969. [Google Scholar]
- 54.Laue ED, Skilling J, Staunton J. Maximum entropy reconstruction of spectra containing antiphase peaks. J. Magn. Reson. 1985;63:418–424. 1969. [Google Scholar]
- 55.Schmieder P, Stern AS, Wagner G, Hoch JC. Improved resolution in triple-resonance spectra by nonlinear sampling in the constant-time domain. J. Biomol. NMR. 1994;4:483–490. doi: 10.1007/BF00156615. [DOI] [PubMed] [Google Scholar]
- 56.Hoch JC, Stern AS. NMR Data Processing. New York: Wiley-Liss; 1996. [Google Scholar]
- 57.Orekhov VY, Ibraghimov I, Billeter M. Optimizing resolution in multidimensional NMR by three-way decomposition. J. Biomol. NMR. 2003;27:165–173. doi: 10.1023/a:1024944720653. [DOI] [PubMed] [Google Scholar]
- 58.Luan T, Jaravine V, Yee A, Arrowsmith CH, Orekhov VY. Optimization of resolution and sensitivity of 4D NOESY using multi-dimensional decomposition. J. Biomol. NMR. 2005;33:1–14. doi: 10.1007/s10858-005-1363-6. [DOI] [PubMed] [Google Scholar]
- 59.Holland DJ, Bostock MJ, Gladden LF, Nietlispach D. Fast multidimensional NMR spectroscopy using compressed sensing. Angew. Chem. 2011;50:6548–6551. doi: 10.1002/anie.201100440. [DOI] [PubMed] [Google Scholar]
- 60.Kazimierczuk K, Orekhov VY. Accelerated NMR spectroscopy by using compressed sensing. Angew. Chem. 2011;50:5556–5559. doi: 10.1002/anie.201100370. [DOI] [PubMed] [Google Scholar]
- 61.Stern AS, Donoho DL, Hoch JC. NMR data processing using iterative thresholding and minimum l(1)-norm reconstruction. J. Magn. Reson. 2007;188:295–300. doi: 10.1016/j.jmr.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hyberts SG, Milbradt AG, Wagner AB, Arthanari H, Wagner G. Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson Gap scheduling. J. Biomol. NMR. 2012;52:315–327. doi: 10.1007/s10858-012-9611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hyberts S, Arthanari H, Wagner G. Applications of non-uniform sampling and processing. In: Billeter M, Orekhov V, editors. Novel Sampling Approaches in Higher Dimensional NMR. Berlin, Heidelberg: Springer; 2012. pp. 125–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maciejewski M, Mobli M, Schuyler A, Stern A, Hoch J. Data sampling in multidimensional NMR: fundamentals and strategies. In: Billeter M, Orekhov V, editors. Novel Sampling Approaches in Higher Dimensional NMR. Berlin, Heidelberg: Springer; 2012. pp. 49–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Orekhov VY, Jaravine VA. Analysis of non-uniformly sampled spectra with multi-dimensional decomposition. Prog. Nucl. Magn. Reson. Spectrosc. 2011;59:271–292. doi: 10.1016/j.pnmrs.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 66.Coggins BE, Venters RA, Zhou P. Radial sampling for fast NMR: concepts and practices over three decades. Prog. Nucl. Magn. Reson. Spectrosc. 2010;57:381–419. doi: 10.1016/j.pnmrs.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Werner-Allen JW, Coggins BE, Zhou P. Fast acquisition of high resolution 4-D amide-amide NOESY with diagonal suppression, sparse sampling and FFT-CLEAN. J. Magn. Reson. 2010;204:173–178. doi: 10.1016/j.jmr.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coggins BE, Zhou P. Sampling of the NMR time domain along concentric rings. J. Magn. Reson. 2007;184:207–221. doi: 10.1016/j.jmr.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coggins BE, Werner-Allen JW, Yan A, Zhou P. Rapid protein global fold determination using ultrasparse sampling, high-dynamic range artifact suppression and time-shared NOESY. J. Am. Chem. Soc. 2012;134:18619–18630. doi: 10.1021/ja307445y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kazimierczuk K, Misiak M, Stanek J, Zawadzka-Kazimierczuk A, Koz´min´ ski W. Top. Curr. Chem. Berlin, Heidelberg: Springer; 2011. Generalized Fourier transform for non-uniform sampled data; pp. 1–46. [DOI] [PubMed] [Google Scholar]
- 71.Kazimierczuk K, Zawadzka-Kazimierczuk A, Kozminski W. Non-uniform frequency domain for optimal exploitation of non-uniform sampling. J. Magn. Reson. 2010;205:286–292. doi: 10.1016/j.jmr.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 72.Marion D. Processing of ND NMR spectra sampled in polar coordinates: a simple Fourier transform instead of a reconstruction. J. Biomol. NMR. 2006;36:45–54. doi: 10.1007/s10858-006-9066-1. [DOI] [PubMed] [Google Scholar]
- 73.Marion D. Fast acquisition of NMR spectra using Fourier transform of non-equispaced data. J. Biomol. NMR. 2005;32:141–150. doi: 10.1007/s10858-005-5977-5. [DOI] [PubMed] [Google Scholar]
- 74.Hoch JC, Maciejewski MW, Filipovic B. Randomization improves sparse sampling in multidimensional NMR. J. Magn. Reson. 2008;193:317–320. doi: 10.1016/j.jmr.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hyberts SG, Takeuchi K, Wagner G. Poisson-gap sampling and forward maximum entropy reconstruction for enhancing the resolution and sensitivity of protein NMR data. J. Am. Chem. Soc. 2010;132:2145–2147. doi: 10.1021/ja908004w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kazimierczuk K, Zawadzka A, Kozminski W. Optimization of random time domain sampling in multidimensional NMR. J. Magn. Reson. 2008;192:123–130. doi: 10.1016/j.jmr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 77.Rovnyak D, Frueh DP, Sastry M, Sun ZY, Stern AS, Hoch JC, Wagner G. Accelerated acquisition of high resolution triple-resonance spectra using non-uniform sampling and maximum entropy reconstruction. J. Magn. Reson. 2004;170:15–21. doi: 10.1016/j.jmr.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 78.Jaravine V, Ibraghimov I, Orekhov VY. Removal of a time barrier for high-resolution multidimensional NMR spectroscopy. Nat. Methods. 2006;3:605–607. doi: 10.1038/nmeth900. [DOI] [PubMed] [Google Scholar]
- 79.Maciejewski MW, Hoch JC. Sample Scheduler. 2007. [Google Scholar]
- 80.Kazimierczuk K, Jaravine V, Mayzel M, Orekhov VY. MddNMR. 2011. [Google Scholar]
- 81.Rovnyak D, Hoch JC, Stern AS, Wagner G. Resolution and sensitivity of high field nuclear magnetic resonance spectroscopy. J. Biomol. NMR. 2004;30:1–10. doi: 10.1023/B:JNMR.0000042946.04002.19. [DOI] [PubMed] [Google Scholar]
- 82.Grzesiek S, Anglister J, Bax A. Correlation of backbone amide and aliphatic side-chain resonances in 13C/15N-enriched proteins by isotropic mixing of 13C magnetization. J. Magn. Reson. B. 1993;101:114–119. [Google Scholar]
- 83.Logan TM, Olejniczak ET, Xu RX, Fesik SW. A general method for assigning NMR spectra of denatured proteins using 3D HC(CO)NH-TOCSY triple resonance experiments. J. Biomol. NMR. 1993;3:225–231. doi: 10.1007/BF00178264. [DOI] [PubMed] [Google Scholar]
- 84.Weisemann R, Rüterjans H, Bermel W. 3D triple-resonance NMR techniques for the sequential assignment of NH and 15N resonances in 15N- and 13C-labelled proteins. J. Biomol. NMR. 1993;3:113–120. doi: 10.1007/BF00242479. [DOI] [PubMed] [Google Scholar]
- 85.Grzesiek S, Anglister J, Ren H, Bax A. Carbon-13 line narrowing by deuterium decoupling in deuterium/carbon-13/nitrogen-15 enriched proteins. Application to triple resonance 4D J connectivity of sequential amides. J. Am. Chem. Soc. 1993;115:4369–4370. [Google Scholar]
- 86.Matsuo H, Kupče Ē, Li H, Wagner G. Use of selective C alpha pulses for improvement of HN(CA)CO-D and HN(COCA)NH-D experiments. J. Magn. Reson. B. 1996;111:194–198. doi: 10.1006/jmrb.1996.0082. [DOI] [PubMed] [Google Scholar]
- 87.Bracken C, Palmer 3rd AG, Cavanagh J. (H)N(COCA)NH and HN(COCA)NH experiments for 1H-15N backbone assignments in 13C/15N-labeled proteins. J. Biomol. NMR. 1997;9:94–100. doi: 10.1023/a:1018679819693. [DOI] [PubMed] [Google Scholar]
- 88.Ikegami T, Sato S, Walchli M, Kyogoku Y, Shirakawa M. An efficient HN(CA)NH pluse scheme for triple-resonance 4D correlation of sequential amide protons and nitrogens-15 in deuterated proteins. J. Magn. Reson. 1997;124:214–217. doi: 10.1006/jmre.1996.7497. [DOI] [PubMed] [Google Scholar]
- 89.Liu A, Riek R, Wider G, von Schroetter C, Zahn R, Wüthrich K. NMR experiments for resonance assignments of 13C, 15N doubly-labeled flexible polypeptides: application to the human prion protein hPrP(23-230) J. Biomol. NMR. 2000;16:127–138. doi: 10.1023/a:1008305022907. [DOI] [PubMed] [Google Scholar]
- 90.Lohr F, Pfeiffer S, Lin YJ, Hartleib J, Klimmek O, Rüterjans H. HNCAN pulse sequences for sequential backbone resonance assignment across proline residues in perdeuterated proteins. J. Biomol. NMR. 2000;18:337–346. doi: 10.1023/a:1026737732576. [DOI] [PubMed] [Google Scholar]
- 91.Panchal SC, Bhavesh NS, Hosur RV. Improved 3D triple resonance experiments, HNN and HN(C)N, for HN and 15N sequential correlations in (13C, 15N) labeled proteins: application to unfolded proteins. J. Biomol. NMR. 2001;20:135–147. doi: 10.1023/a:1011239023422. [DOI] [PubMed] [Google Scholar]
- 92.Frueh DP, Sun ZY, Vosburg DA, Walsh CT, Hoch JC, Wagner G. Non-uniformly sampled double-TROSY hNcaNH experiments for NMR sequential assignments of large proteins. J. Am. Chem. Soc. 2006;128:5757–5763. doi: 10.1021/ja0584222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hyberts SG, Robson SA, Wagner G. Exploring signal-to-noise ratio and sensitivity in non-uniformly sampled multi-dimensional NMR spectra. J. Biomol. NMR. 2013;55:167–178. doi: 10.1007/s10858-012-9698-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Paramasivam S, Suiter CL, Hou G, Sun S, Palmer M, Hoch JC, Rovnyak D, Polenova T. Enhanced sensitivity by nonuniform sampling enables multidimensional MAS NMR spectroscopy of protein assemblies. J. Phys. Chem. 2012;B116:7416–7427. doi: 10.1021/jp3032786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rovnyak D, Sarcone M, Jiang Z. Sensitivity enhancement for maximally resolved two-dimensional NMR by nonuniform sampling. Magn. Reson. Chem. 2011 doi: 10.1002/mrc.2775. [DOI] [PubMed] [Google Scholar]
- 96.Yang D, Kay LE. TROSY triple-resonance four-dimensional NMR spectroscopy of a 46 ns tumbling protein. J. Am. Chem. Soc. 1999;121:2571–2575. [Google Scholar]
- 97.Konrat R, Yang D, Kay LE. A 4D TROSY-based pulse scheme for correlating 1HNi,15Ni,13Calphai, 13C’i – 1 chemical shifts in high molecular weight 15N,13C,2H labeled proteins. J. Biomol. NMR. 1999;15:309–313. doi: 10.1023/A:1008310617047. [DOI] [PubMed] [Google Scholar]
- 98.Tugarinov V, Muhandiram R, Ayed A, Kay LE. Four-dimensional NMR spectroscopy of a 723-residue protein: chemical shift assignments and secondary structure of malate synthase g. J. Am. Chem. Soc. 2002;124:10025–10035. doi: 10.1021/ja0205636. [DOI] [PubMed] [Google Scholar]
- 99.Grzesiek S, Wingfield P, Stahl S, Kaufman JD, Bax A. 4-Dimensional N-15-separated NOESY of slowly tumbling perdeuterated N-15-enriched proteins -application to Hiv-1 Nef. J. Am. Chem. Soc. 1995;117:9594–9595. [Google Scholar]
- 100.Venters RA, Metzler WJ, Spicer LD, Mueller L, Farmer BT. Use of H-1(N)-H-1(N) NOEs to determine protein global folds in perdeuterated proteins. J. Am. Chem. Soc. 1995;117:9592–9593. [Google Scholar]
- 101.Xia Y, Sze K, Zhu G. Transverse relaxation optimized 3D and 4D 15N/15N separated NOESY experiments of 15N labeled proteins. J. Biomol. NMR. 2000;18:261–268. doi: 10.1023/a:1026590201757. [DOI] [PubMed] [Google Scholar]
- 102.Tugarinov V, Kay LE, Ibraghimov I, Orekhov VY. High-resolution four-dimensional 1H-13C NOE spectroscopy using methyl-TROSY, sparse data acquisition, and multidimensional decomposition. J. Am. Chem. Soc. 2005;127:2767–2775. doi: 10.1021/ja044032o. [DOI] [PubMed] [Google Scholar]
- 103.Hyberts SG, Frueh DP, Arthanari H, Wagner G. FM reconstruction of non-uniformly sampled protein NMR data at higher dimensions and optimization by distillation. J. Biomol. NMR. 2009;45:283–294. doi: 10.1007/s10858-009-9368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hyberts S, Arthanari H, Wagner G. Applications of Non-Uniform Sampling and Processing. Berlin, Heidelberg: Springer; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Szyperski T, Wider G, Bushweller JH, Wüthrich K. Reduced dimensionality in triple-resonance NMR experiments. J. Am. Chem. Soc. 1993;115:9307–9308. [Google Scholar]
- 106.Kim S, Szyperski T, GFT NMR. a new approach to rapidly obtain precise high-dimensional NMR spectral information. J. Am. Chem. Soc. 2003;125:1385–1393. doi: 10.1021/ja028197d. [DOI] [PubMed] [Google Scholar]
- 107.Kupče Ē, Freeman R. Projection-reconstruction of three-dimensional NMR spectra. J. Am. Chem. Soc. 2003;125:13958–13959. doi: 10.1021/ja038297z. [DOI] [PubMed] [Google Scholar]
- 108.Mobli M, Stern AS, Hoch JC. Spectral reconstruction methods in fast NMR: reduced dimensionality, random sampling and maximum entropy. J. Magn. Reson. 2006;182:96–105. doi: 10.1016/j.jmr.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 109.Hiller S, Wider G, Wüthrich K. APSY-NMR with proteins: practical aspects and backbone assignment. J. Biomol. NMR. 2008;42:179–195. doi: 10.1007/s10858-008-9266-y. [DOI] [PubMed] [Google Scholar]
- 110.Hiller S, Fiorito F, Wüthrich K, Wider G. Automated projection spectroscopy (APSY) Proc. Natl. Acad. Sci. USA. 2005;102:10876–10881. doi: 10.1073/pnas.0504818102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Braunschweiler L, Ernst RR. Coherence transfer by isotropic mixing - application to proton correlation spectroscopy. J. Magn. Reson. 1983;53:521–528. [Google Scholar]
- 112.Bax A, Clore M, Gronenborn AM. 1H-1H correlation via isotropic mixing of 13C magnetization a new three-dimensional approach for assigning 1H and 13C, spectra of 13C-enriched proteins. J. Magn. Reson. 1990;88:425–431. [Google Scholar]
- 113.Kay LE, Ikura M, Bax A. Proton proton correlation via carbon carbon couplings - a 3-dimensional NMR Approach for the assignment of aliphatic resonances in proteins labeled with C-13. J. Am. Chem. Soc. 1990;112:888–889. [Google Scholar]
- 114.Otten R, Chu B, Krewulak KD, Vogel HJ, Mulder FA. Comprehensive and cost-effective NMR spectroscopy of methyl groups in large proteins. J. Am. Chem. Soc. 2010;132:2952–2960. doi: 10.1021/ja907706a. [DOI] [PubMed] [Google Scholar]
- 115.Palmer III AG, Cavanagh J, Wright PE, Rance M. Sensitivity improvement in proton-detected two-dimensional heteronuclear correlation NMR spectroscopy. J. Magn. Reson. 1991;93:151–170. [Google Scholar]
- 116.Kay LE, Keifer P, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 1992;114:10663–10665. [Google Scholar]
- 117.Wurtz P, Hellman M, Tossavainen H, Permi P. Towards unambiguous assignment of methyl-containing residues by double and triple sensitivity-enhanced HCCmHm-TOCSY experiments. J. Biomol. NMR. 2006;36:13–26. doi: 10.1007/s10858-006-9056-3. [DOI] [PubMed] [Google Scholar]
- 118.Tugarinov V, Kay LE. Side chain assignments of Ile Î1 methyl groups in high molecular weight proteins: an application to a 46 ns tumbling molecule. J. Am. Chem. Soc. 2003;125:5701–5706. doi: 10.1021/ja021452+. [DOI] [PubMed] [Google Scholar]
- 119.Guo C, Tugarinov V. Selective 1H-13C NMR spectroscopy of methyl groups in residually protonated samples of large proteins. J. Biomol. NMR. 2010;46:127–133. doi: 10.1007/s10858-009-9393-0. [DOI] [PubMed] [Google Scholar]
- 120.Sheppard D, Guo C, Tugarinov V. 4D 1H-13C NMR spectroscopy for assignments of alanine methyls in large and complex protein structures. J. Am. Chem. Soc. 2009;131:1364–1365. doi: 10.1021/ja808202q. [DOI] [PubMed] [Google Scholar]
- 121.Sheppard D, Guo C, Tugarinov V. Methyl-detected ‘out-and-back’ NMR experiments for simultaneous assignments of Alab and Ilec2 methyl groups in large proteins. J. Biomol. NMR. 2009;43:229–238. doi: 10.1007/s10858-009-9305-3. [DOI] [PubMed] [Google Scholar]
- 122.Sprangers R, Kay LE. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature. 2007;445:618–622. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]
- 123.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Nuclear magnetic dipole interactions in field-oriented proteins: information for structure determination in solution. Proc. Natl. Acad. Sci. USA. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 125.Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 126.Clore GM, Tang C, Iwahara J. Elucidating transient macromolecular interactions using paramagnetic relaxation enhancement. Curr. Opin. Struct. Biol. 2007;17:603–616. doi: 10.1016/j.sbi.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Paterson Y, Englander S, Roder H. An antibody binding site on cytochrome c defined by hydrogen exchange and two-dimensional NMR. Science. 1990;249:755–759. doi: 10.1126/science.1697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mandell J, Baerga-Ortiz A, Falick A, Komives E. Measurement of solvent accessibility at protein–protein interfaces. In: GUlrich Nienhaus G, editor. Protein–Ligand Interactions. Humana Press; 2005. pp. 65–79. [DOI] [PubMed] [Google Scholar]
- 129.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 130.Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A. Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl. Acad. Sci. USA. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shen Y, Bryan PN, He Y, Orban J, Baker D, Bax A. De novo structure generation using chemical shifts for proteins with high-sequence identity but different folds. Protein Sci. 2010;19:349–356. doi: 10.1002/pro.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shen Y, Vernon R, Baker D, Bax A. De novo protein structure generation from incomplete chemical shift assignments. J. Biomol. NMR. 2009;43:63–78. doi: 10.1007/s10858-008-9288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cavalli A, Montalvao RW, Vendruscolo M. Using chemical shifts to determine structural changes in proteins upon complex formation. J. Phys. Chem. B. 2011;115:9491–9494. doi: 10.1021/jp202647q. [DOI] [PubMed] [Google Scholar]
- 135.Cavalli A, Salvatella X, Dobson CM, Vendruscolo M. Protein structure determination from NMR chemical shifts. Proc. Natl. Acad. Sci. USA. 2007;104:9615–9620. doi: 10.1073/pnas.0610313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Frueh DP, Arthanari H, Koglin A, Vosburg DA, Bennett AE, Walsh CT, Wagner G. Dynamic thiolation-thioesterase structure of a non-ribosomal peptide synthetase. Nature. 2008;454:903–906. doi: 10.1038/nature07162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pervushin K, Eletsky A. A new strategy for backbone resonance assignment in large proteins using a MQ-HACACO experiment. J. Biomol. NMR. 2003;25:147–152. doi: 10.1023/a:1022225711122. [DOI] [PubMed] [Google Scholar]
- 138.Rosen MK, Gardner KH, Willis RC, Parris WE, Pawson T, Kay LE. Selective methyl group protonation of perdeuterated proteins. J. Mol. Biol. 1996;263:627–636. doi: 10.1006/jmbi.1996.0603. [DOI] [PubMed] [Google Scholar]
- 139.Gerstein M. A structural census of genomes: comparing bacterial, eukaryotic, and archaeal genomes in terms of protein structure. J. Mol. Biol. 1997;274:562–576. doi: 10.1006/jmbi.1997.1412. [DOI] [PubMed] [Google Scholar]
- 140.Siemion IZ. Compositional frequencies of amino acids in the proteins and the genetic code. BioSystems. 1994;32:163–170. doi: 10.1016/0303-2647(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 141.McCaldon P, Argos P. Oligopeptide biases in protein sequences and their use in predicting protein coding regions in nucleotide sequences. Proteins: Struct. Funct. Bioinform. 1988;4:99–122. doi: 10.1002/prot.340040204. [DOI] [PubMed] [Google Scholar]
- 142.Conte LL, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 143.Miller S, Janin J, Lesk AM, Chothia C. Interior and surface of monomeric proteins. J. Mol. Biol. 1987;196:641–656. doi: 10.1016/0022-2836(87)90038-6. [DOI] [PubMed] [Google Scholar]
- 144.Farmer B., II Simultaneous [13C,15N]-HMQC a pseudo-triple resonance experiment. J. Magn. Reson. 1991;93:635–641. [Google Scholar]
- 145.Sattler M, Maurer M, Schleucher J, Griesinger C. A simultaneous 15N,1H and 13C,1H-HSQC with sensitivity enhancement and a heteronuclear gradient echo. J. Biomol. NMR. 1995;5:97–102. doi: 10.1007/BF00227475. [DOI] [PubMed] [Google Scholar]
- 146.Kupče Ē, Muhandiram DR, Kay LE. A combined HNCA/HNCO experiment for 15N labeled proteins with 13C at natural abundance. J. Biomol. NMR. 2003;27:175–179. doi: 10.1023/a:1024985219764. [DOI] [PubMed] [Google Scholar]
- 147.Frueh DP, Arthanari H, Wagner G. Unambiguous assignment of NMR protein backbone signals with a time-shared triple-resonance experiment. J. Biomol. NMR. 2005;33:187–196. doi: 10.1007/s10858-005-3204-z. [DOI] [PubMed] [Google Scholar]
- 148.Boelens R, Burgering M, Fogh RH, Kaptein R. Time-saving methods for heteronuclear multidimensional NMR of (13C, 15N) doubly labeled proteins. J. Biomol. NMR. 1994;4:201–213. doi: 10.1007/BF00175248. [DOI] [PubMed] [Google Scholar]
- 149.Pascal S, Muhandiram D, Yamazaki T, Forman-Kay J, Kay L. Simultaneous acquisition of 15N and 13C-edited NOE spectra of proteins dissolved in H2O. J. Magn. Reson. B. 1994;103:197–201. [Google Scholar]
- 150.Jerala R, Rule G. A 3D 1H,15N and 13C NOESY correlating experiment. J. Magn. Reson. 1995;108:294–298. [Google Scholar]
- 151.Uhrín D, Bramham J, Winder SJ, Barlow PN. Simultaneous CT-13C and VT-15N chemical shift labelling: application to 3D NOESY-CH3NH and 3D 13C,15N HSQC-NOESY-CH3NH. J. Biomol. NMR. 2000;18:253–259. doi: 10.1023/a:1026550629595. [DOI] [PubMed] [Google Scholar]
- 152.Xia Y, Zhu G. 3D Haro-NOESY-CH3NH and Caro-NOESY-CH3NH experiments for double labeled proteins. J. Biomol. NMR. 2001;19:355–360. doi: 10.1023/a:1011288324900. [DOI] [PubMed] [Google Scholar]
- 153.Grzesiek S, Bax A. The importance of not saturating H2O in protein NMR. Application to sensitivity enhancement and NOE measurements. J. Am. Chem. Soc. 1993;115:12593–12594. [Google Scholar]
- 154.Pervushin K, Braun D, Fernandez C, Wüthrich K. [15N,1H]/[13C,1H]-TROSY for simultaneous detection of backbone 15N–1H, aromatic 13C–1H and side-chain 15N–1H2 correlations in large proteins. J. Biomol. NMR. 2000;17:195–202. doi: 10.1023/a:1008399320576. [DOI] [PubMed] [Google Scholar]
- 155.Frueh DP, Vosburg DA, Walsh CT, Wagner G. Determination of all nOes in 1H-13C-Me-ILV-U-2H-15N proteins with two time-shared experiments. J. Biomol. NMR. 2006;34:31–40. doi: 10.1007/s10858-005-5338-4. [DOI] [PubMed] [Google Scholar]
- 156.Wurtz P, Aitio O, Hellman M, Permi P. Simultaneous detection of amide and methyl correlations using a time shared NMR experiment: application to binding epitope mapping. J. Biomol. NMR. 2007;39:97–105. doi: 10.1007/s10858-007-9178-2. [DOI] [PubMed] [Google Scholar]
- 157.Guo C, Tugarinov V. Identification of HN-methyl NOEs in large proteins using simultaneous amide-methyl TROSY-based detection. J. Biomol. NMR. 2009;43:21–30. doi: 10.1007/s10858-008-9285-8. [DOI] [PubMed] [Google Scholar]
- 158.Frueh DP, Leed A, Arthanari H, Koglin A, Walsh CT, Wagner G. Time-shared HSQC-NOESY for accurate distance constraints measured at high-field in (15)N-(13)C-ILV methyl labeled proteins. J. Biomol. NMR. 2009;45:311–318. doi: 10.1007/s10858-009-9372-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Sortagging: a versatile method for protein labeling. Nat. Chem. Biol. 2007;3:707–708. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- 160.Muralidharan V, Muir TW. Protein ligation: an enabling technology for the biophysical analysis of proteins. Nat. Methods. 2006;3:429–438. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- 161.Refaei M, Combs A, Kojetin D, Cavanagh J, Caperelli C, Rance M, Sapitro J, Tsang P. Observing selected domains in multi-domain proteins via sortase-mediated ligation and NMR spectroscopy. J. Biomol. NMR. 2011;49:3–7. doi: 10.1007/s10858-010-9464-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Michel E, Skrisovska L, Wüthrich K, Allain FHT. Amino acid-selective segmental isotope labeling of multidomain proteins for structural biology. ChemBioChem. 2013;14:457–466. doi: 10.1002/cbic.201200732. [DOI] [PubMed] [Google Scholar]
- 163.Fiaux J, Bertelsen EB, Horwich AL, Wüthrich K. NMR analysis of a 900K GroEL GroES complex. Nature. 2002;418:207–211. doi: 10.1038/nature00860. [DOI] [PubMed] [Google Scholar]
- 164.Ruschak AM, Religa TL, Breuer S, Witt S, Kay LE. The proteasome antechamber maintains substrates in an unfolded state. Nature. 2010;467:868–871. doi: 10.1038/nature09444. [DOI] [PubMed] [Google Scholar]
- 165.Kumar D, Hosur RV. hNCOcanH pulse sequence and a robust protocol for rapid and unambiguous assignment of backbone ((1) H(N), (15) N and (13) C′) resonances in (15) N/(13) C-labeled proteins. Magn. Reson. Chem. 2011 doi: 10.1002/mrc.2787. [DOI] [PubMed] [Google Scholar]
- 166.Serber Z, Richter C, Dötsch V. Carbon-detected NMR experiments to investigate structure and dynamics of biological macromolecules. ChemBioChem. 2001;2:247–251. doi: 10.1002/1439-7633(20010401)2:4<247::AID-CBIC247>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 167.Eletsky A, Moreira O, Kovacs H, Pervushin K. A novel strategy for the assignment of side-chain resonances in completely deuterated large proteins using 13C spectroscopy. J. Biomol. NMR. 2003;26:167–179. doi: 10.1023/a:1023572320699. [DOI] [PubMed] [Google Scholar]
- 168.Bermel W, Bertini I, Duma L, Felli IC, Emsley L, Pierattelli R, Vasos PR. Complete assignment of heteronuclear protein resonances by protonless NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2005;44:3089–3092. doi: 10.1002/anie.200461794. [DOI] [PubMed] [Google Scholar]
- 169.Bermel W, Bertini I, Felli IC, Piccioli M, Pierattelli R. 13C–detected protonless NMR spectroscopy of proteins in solution. Prog. Nucl. Magn. Reson. Spectrosc. 2006;48:25–45. [Google Scholar]
- 170.Bermel W, Bertini I, Felli IC, Lee YM, Luchinat C, Pierattelli R. Protonless NMR experiments for sequence-specific assignment of backbone nuclei in unfolded proteins. J. Am. Chem. Soc. 2006;128:3918–3919. doi: 10.1021/ja0582206. [DOI] [PubMed] [Google Scholar]
- 171.O’Hare B, Benesi AJ, Showalter SA. Incorporating 1H chemical shift determination into 13C-direct detected spectroscopy of intrinsically disordered proteins in solution. J. Magn. Reson. 2009;200:354–358. doi: 10.1016/j.jmr.2009.07.014. [DOI] [PubMed] [Google Scholar]