Abstract
Phosphorylated sphingolipids [ceramide-1-phosphate (C1P) and sphingosine-1-phosphate (S1P)] have emerged as key regulators of cell growth, survival, migration, and inflammation1–5. C1P (Fig. 1a) produced by ceramide kinase is an activator of group IVA cytosolic phospholipase A2α (cPLA2α), the rate-limiting releaser of arachidonic acid used for pro-inflammatory eicosanoid production3,6–9, which contributes to disease pathogenesis in asthma/airway hyper-responsiveness, cancer, atherosclerosis, and thrombosis. To modulate eicosanoid action and avoid the damaging effects of chronic inflammation, cells require efficient targeting, trafficking, and presentation of C1P to specific cellular sites. Vesicular trafficking is likely10 but nonvesicular mechanisms for C1P sensing, transfer, and presentation remain unexplored11,12. Moreover, the molecular basis for selective recognition and binding among signaling lipids with phosphate headgroups, namely C1P, phosphatidic acid (PA) or their lyso-derivatives, remains unclear. Herein, an ubiquitously-expressed lipid transfer protein (CPTP) is shown to specifically transfer C1P between membranes. Crystal structures establish C1P binding via a novel surface-localized, phosphate headgroup recognition center connected to an interior hydrophobic pocket that adaptively expands to ensheath differing-length lipid chains using a cleft-like gating mechanism. The two-layer, α-helically-dominated ‘sandwich’ topology identifies CPTP as the prototype for a new GLTP-fold13 subfamily. CPTP resides in the cell cytosol but associates with the trans-Golgi/TGN, nucleus, and plasma membrane. RNAi-induced CPTP depletion elevates C1P steady-state levels and alters Golgi cisternae stack morphology. The resulting C1P decrease in plasma membranes and increase in the Golgi complex stimulates cPLA2α release of arachidonic acid, triggering pro-inflammatory eicosanoid generation.
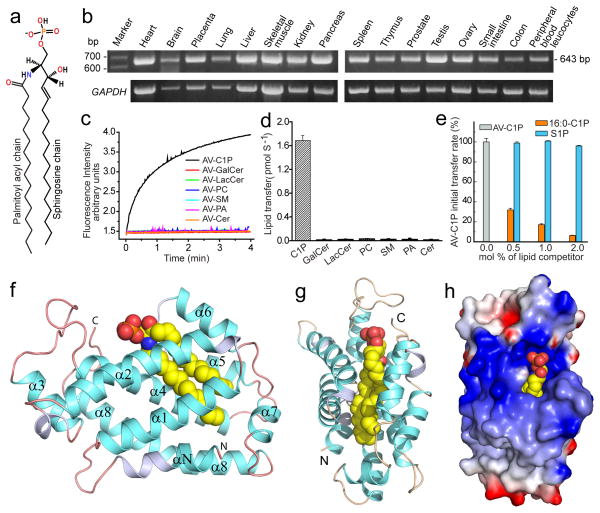
During screening of the NCBI human genome database, we noted an in silico predicted transcript (GenBank NP_001025056.1; glycolipid transfer protein-containing-domain-1; GLTPD1) encoding a protein sharing sequence identity (17%) with glycolipid transfer protein (GLTP). Although annotation indicated glycolipid binding and transport activity, Lys and Arg substitutions occurred at key positions (N52, W96) essential for sugar headgroup recognition by GLTP (Supplementary Fig. S1a; yellow highlights)13–14. We validated GLTPD1 mRNA transcript expression in human tissues, finding wide-spread occurrence and relatively elevated transcript levels in placenta, kidney, pancreas and testis (Fig. 1b). Cloning and heterologous expression revealed that purified GLTPD1 (GenBank JN542538) transfers anthrylvinyl-C1P (AV-C1P) between 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayer vesicles in protein concentration-dependent fashion (Fig. 1c,d,; Supplementary Fig. S1b,d) requiring acceptor membranes (Supplementary Fig. S1c). Testing of lipid specificity revealed no transfer of galactosylceramide (GalCer), lactosylceramide (LacCer), sphingomyelin (SM), phosphatidyl-choline (PC), phosphatidic acid (PA), or ceramide (Cer) by GLTPD1 (Fig. 1c,d). Slow-down of AV-C1P transfer by potential lipid ligands (nonfluorescent) showed no ‘competition effect’ by S1P (Fig. 1e; Supplementary Fig. S1e,f) and a transfer rate of ~4 C1P molecules/min/protein at 37°C. GLTPD1 was designated ceramide-1-phosphate transfer protein (CPTP).
Figure 1. CPTP lipid transfer activity and architecture.
a, 16:0-C1P chemical formula. b, CPTP mRNA transcript levels in various human tissues. c, Lipid transfer in vitro by Förster resonance energy transfer. d, Initial lipid transfer rates for panel c. e, Competition against CPTP-mediated AV-C1P transfer by nonfluorescent lipids. Kinetic traces appear as Supplementary Fig. S1e–g. Data in d and e represent the mean ± s.d. of three independent experiments. f, g, Two views of CPTP structure (ribbon) with bound 16:0-C1P (space-filling). α-helices (cyan), 310-helices (light blue), loops (orange) and bound 16:0-C1P (yellow, red, blue for carbon, oxygen, nitrogen, respectively). α-helices (αN and α1–α8) are numbered from N- to C-termini. h, Surface electrostatics of CPTP with bound 16:0-C1P showing positive- (blue) and negative- (red) charged residues.
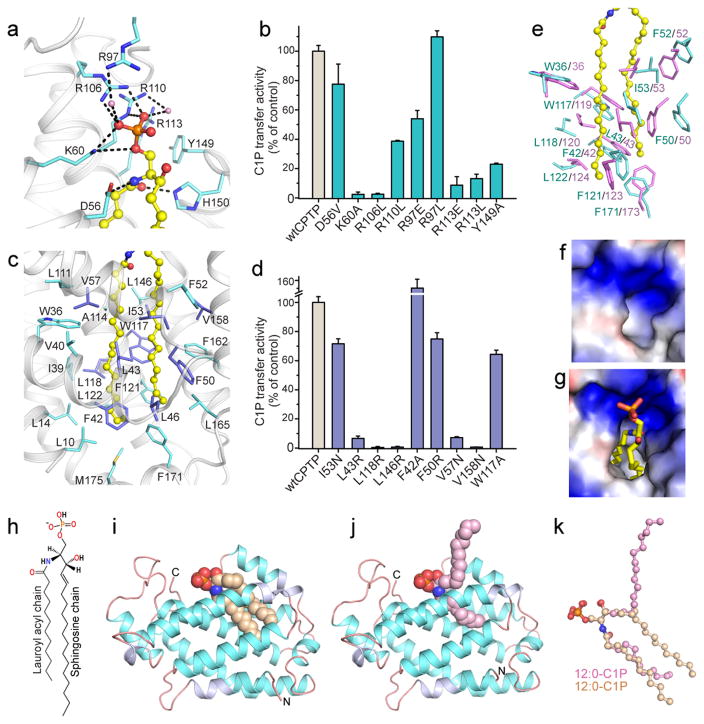
The crystal structure of human CPTP/16:0-C1P complex (1.9 Å, Table S1) revealed a two-layered, all α-helical topology (Fig. 1f,g) homologous with GLTP-fold13. αN, α1 and α2 form one layer, α4, α5 and α8 form another layer, and α3, α6 and α7 localize along the periphery of the two-layer core. A positively-charged surface cavity for anchoring the lipid headgroup (Fig. 1h) extends through a gateway portal, transforming into a deep interior hydrophobic cavity that accommodates the sphingosine and acyl chains. αN-helix (Leu10 and Leu14) impart pocket-like features by sealing the bottom. A triad of cationic residues (K60, R106, R110) in the surface cavity recognizes and binds the C1P phosphate headgroup (Fig. 2a). The anchoring H-bond network is complex, involving bifurcated hydrogen bonding by K60 (α2-helix) with the O1 and O2 atoms, bidentate hydrogen bonding by R106 (α4-helix) with the O2 and O3 atoms, and bidentate hydrogen bonding by R110 (α4-helix) directly and via water bridging to O3. Point mutation supports key roles for K60 and R106 in C1P headgroup recognition with K60A and R106L showing almost no C1P transfer, while R110 mutation (R110L) shows ~40% transfer (Fig. 2b). The positive-charge of this site also is enhanced by R97 (α3–4 loop) which hydrogen bonds to the O2 atom of phosphate via water-bridging. R97 is almost fully active when mutated to L (R97L), but mutation to acidic E (R97E) reduces C1P transfer to ~55%, supporting its role for attracting the lipid phosphate headgroup. Cation-pi interaction between R113 (α4-helix) and Y149 (α5–6 loop) provides stabilizing underpinning for the site (Supplementary Fig. S2c), as R113 mutation (R113L) strongly diminishes activity. Mutants Y149A, R113E, and R113L show poor C1P transfer, as expected by conformational destabilization (Fig. 2b). All key residues of the phosphate recognition site appear to be conserved in eukaryotes (Supplementary Fig. S2a).
Figure 2. CPTP conformation and functional recognition of C1P.
a, CPTP lipid headgroup recognition center residue interaction with phosphate and amide groups of bound 16:0-C1P (ball-and-stick). Hydrogen-bonds = dashed lines. CPTP Cα backbone is light gray; side chains, cyan; and oxygen and nitrogen, red and blue, respectively. Water molecules are pink spheres. b, C1P transfer by CPTP point mutants of phosphate headgroup recognition cavity (cyan). wtCPTP (gray). c, Non-polar residues forming hydrophobic pocket that accommodates 16:0-C1P sphingosine and acyl chains. d, C1P transfer by CPTP point mutants (violet) of the hydrophobic pocket. wtCPTP (gray); Side-chains shown in panel c. Data in b and d represent the mean ± s.d. of three independent experiments. e, Conformational changes in hydrophobic pocket upon 16:0-C1P binding. Side-chains (lavender; stick) of apo-CPTP and human CPTP with bound 16:0-C1P (yellow; ball-and-stick). f, g, Surface electrostatics of hydrophobic pocket opening at lipid headgroup recognition sites in apo-CPTP (f) and CPTP/16:0-C1P complex (g). h, 12:0-C1P chemical formula. i, j Crystal structures of CPTP (ribbon) with bound 12:0-C1P in sphingosine-in i; beige) and sphingosine-out (j; pink) conformations. k, Superposition of bound 12:0-C1P in sphingosine-in and -out conformations.
The hydrophobic pocket is lined by ~25 nonpolar residues, mainly Phe, Leu, Val, and Ile (Fig. 2c; Supplementary Fig. S2b) that prevent water entry while ensheathing the ceramide aliphatic chains. Mutation of L43, L118, or L146 to positively-charged R or V57 or V158 to high polarity N compromises hydrophobic pocket functionality and strongly diminishes C1P transfer (Fig. 2d). More conservative mutation (e.g. W117A) only moderately reduces C1P transfer, while F42A near the pocket bottom stimulates C1P transfer. Mutation near the entry portal (I53N) or in the flexible α1–2 loop (F50R) is well tolerated (75–80% active) (Fig. 2d). Ceramide entry is oriented by hydrogen bonding of the lipid amide oxygen and nitrogen with H150 and D56, respectively. Hydrogen bond disruption between lipid amide nitrogen and D56 (D56V) moderately slows C1P transfer, but H150 mutation (H150L) abolishes activity. Superpositioning of apo- and 16:0-C1P/CPTP structures (rmsd 1.4 Å) shows K60, R106 and R110 nearly identically positioned in the positively-charged surface cavities. Yet, large conformational differences exist for I53, W36, W119 and F52 (Fig. 2e) due to closer packing of certain α-helices in apo-CPTP (Supplementary Fig. S2d,e). Many Leu and Phe are repositioned, reducing the solvent accessible (SA) volume (40 Å3) (Table S4) and effectively collapsing the hydrophobic pocket (Fig. 2f,g) compared to 16:0-C1P/CPTP complex (364 Å3).
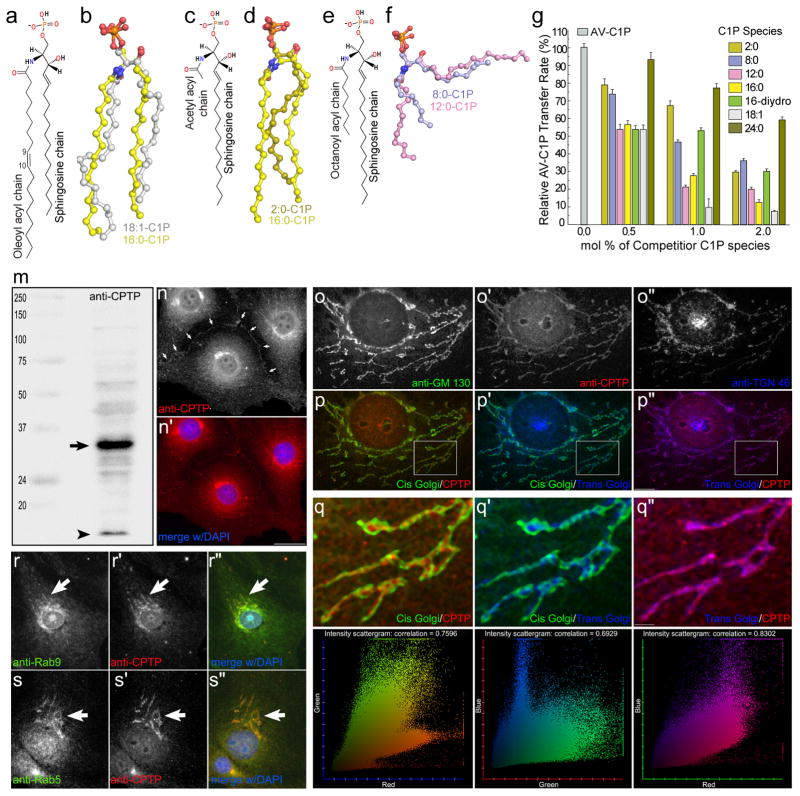
CPTP adaptability for different C1P species is reflected in structures of CPTP complexed with C1P containing differing-length acyl chains (Fig. 2h–k; 3a–f; Supplementary Fig. S4c–f). Two lipid-binding conformational modes are apparent. In ‘sphingosine-in’ mode, both ceramide chains occupy the hydrophobic pocket; whereas, only the acyl chain occupies the pocket in the ‘sphingosine-out’ mode. For 12:0-C1P/CPTP complex (Fig. 2h–j), both binding modes occurred in the same asymmetric unit (Fig. 2i, j; Supplementary Fig. S3c,d; S4a,b) enabling comparison (Fig. 2k) under closest possible conditions. The lipid phosphate headgroups and amide groups bind exactly as in 16:0-C1P/CPTP complex (Supplementary Fig. S2b; S3a,b). In sphingosine-out mode (Fig. 2j; Supplementary Fig. S3d), a bend in sphingosine at C6 is stabilized by hydrophobic interactions with V153, V154 and the D56 Cβ atom, enabling outward projection. Sphingosine cross-bridging interactions with F50, I149, A157 and V153 of neighboring, symmetry-related CPTP stabilize further (Supplementary Fig. S4b). SA pocket volumes reflect the altered sphingosine location, i.e. 261 Å3 for sphingosine-out versus 329 Å3 for sphingosine-in (Table S4). In 18:1-C1P/CPTP complex (Fig. 3a, b), the cis-double bond kink in the acyl chain increases separation from the sphingosine chain, maximally expanding the pocket (stereo view; Supplementary Fig. S4c,d) while leaving the overall chain length in the pocket similar to 16:0-C1P. Accordingly, the SA volume of the hydrophobic pocket of 18:1-C1P/CPTP is larger (387 Å3) (Table S4) than in 16:0-C1P/CPTP where slightly closer packing by the saturated acyl chain decreases the SA volume (364 Å3). Shortening the acyl chain length reduces SA pocket volumes to 104 Å3 for 8:0 and 263 Å3 for 2:0 (Table S4). Structures for 2:0-C1P/CPTP and 8:0-C1P/CPTP are detailed in Supplementary Fig. S3a,b, S4e,f and Discussion.
Figure 3. CPTP accommodation and adaptation for C1P species, functional assessment, and intracellular localization.
a, 18:1-C1P chemical formula. b, Superposition of bound 18:1-C1P (silver) and 16:0-C1P (yellow) lipid chains in their sphingosine-in CPTP complexes. c, 2:0-C1P chemical formula. d, Superposition of bound 2:0-C1P (olive) and 16:0-C1P (yellow) lipid chains in their sphingosine-in CPTP complexes. e, 8:0-C1P chemical formula. f, Superposition of bound 8:0-C1P (light blue) and 12:0-C1P (pink) lipid chains in their sphingosine-out CPTP complexes. g, Competition effects by C1P species containing different acyl chains on the CPTP-mediated initial transfer rate of fluorescent C1P. Data represent the mean ± s.d. of three independent experiments. h–n, CPTP localization to trans-Golgi and endosomes in vivo. h, SDS-PAGE/Immunoblot of BSC1 whole cell lysate, (anti-CPTP label), reveals a single immuno-reactive band (arrow). Dye front position = arrowhead. i–i′, anti-CPTP decorates peri-nuclear membrane stacks, nuclear membrane, and plasma membrane (arrows). j–l, Peri-nuclear membrane stacks detected by anti-CPTP are trans-Golgi. j–j″, BSC1 cell Golgi stacks labeled with anti-GM130 (cis-Golgi marker), anti-TGN 46 (trans-Golgi marker) and anti-CPTP. k–k″, pseudo-color overlay of frames from j–j″. l–l″, high magnification insets from k-k″ show CPTP/TGN-46 co-localization, and intensity scattergram analyses with measured correlation coefficients32. m–n, Anti-CPTP co-localization with late (m–m″, anti-Rab9 co-labeling) and early (n–n″, anti-Rab5 co-labeling) endosomes. Wide-field fluorescence microscopy, bar = 10 μm (i′), 2.5 μm (k″).
The functional consequences of CPTP hydrophobic pocket structural adaptability become clear upon transfer analyses. Pocket expansion accommodates ceramide aliphatic chains in ‘molecular ruler’-like fashion with CPTP adaptability limits optimized for 16:0- or 18:1-C1P species which are particularly effective competitors at slowing the AV-C1P transfer rate (Fig. 3g), consistent with maximal pocket expansion and optimal fit (Table S4). It is noteworthy that C1P containing long lignoceryl (24:0) acyl chains are not very effective competitors, suggesting poor accommodation in the hydrophobic pocket because of adaptation limitations. Also 16:0-C1P with dihydrosphingosine base competes less effectively than 16:0-C1P with naturally-prevalent sphingosine base.
Structure determination of di12:0-PA/CPTP complex elucidated the molecular basis of PA non-transfer (Supplementary Fig. S5a–h; Table S1). PA occupies the same binding site and its phosphate group interacts with the same positively-charged residues as C1P (Supplementary Fig. S5b–d). Yet, K60 hydrogen bonding is single rather than bifurcated, and the lack of the acyl-amide moiety results in no hydrogen bonding with D56, distorting the position of the phosphate headgroup and both lipid chains and loosening PA binding. The distorted interaction mitigates PA transfer by CPTP (Supplementary Fig S5e–h and Discussion).
CPTP architecture not only represents a novel motif for specific binding of phosphosphingo-lipids, but is previously unknown for any phosphate-modified biomolecule15–17. In CPTP, the fixed cationic residues of the phosphate recognition site undergo minimal conformational change upon C1P binding. B-factor distribution analyses show the regions between α1-α2 and α5-α6 are most flexible (Supplementary Fig. S6a,b), consistent with a cleft-like gating mechanism facilitating ceramide chain entry/exit. The conserved lipid orientation in the pocket, with the nonpolar acyl chain always inside regardless of sphingosine being in or out, supports a concerted mechanism of action in which the acyl chain enters first and leaves last during membrane interaction (Supplementary Fig. S6c,d and Discussion).
The conformational adaptability of the inherently flexible, single-cavity, hydrophobic pocket of CPTP contrasts with lipid cavities in fatty acid binding proteins which use β-barrels/β-cups to generate a large, solvent-filled binding site that remains conformationally fixed whether or not occupied by fatty acid18. A single, fixed, lipid binding cavity also is characteristic of START lipid binding domains in PC transfer protein19 and CERT20, which uses an α/β fold built around an incomplete U-shaped β-barrel to bind ceramide21 (Supplementary Fig. S7 and Discussion).
In the human genome, the differing origins of CPTP and GLTP are clear. CPTP (214 aa) is encoded by a 3-exon transcript originating from GLTPD1 on chromosome 1 (locus 1p36.33). GLTP (209 aa) is encoded by a 5-exon transcript originating from GLTP on chromosome 12 (locus 12q24.11)22. The shared folding topology encoded by GLTPD1 and GLTP, despite only limited sequence homology (Supplementary Fig. S8a–e) and different lipid specificity, provides a striking example of evolutionary convergence and emphasizes the structural premium placed by eukaryotes on conservation of this fold23–25. The related architectures of CPTP and GLTP, but with naturally-evolved and remarkably different lipid headgroup specificity (Supplementary Discussion), suggest that ‘Sphingolipid Transfer Protein (SLTP) Superfamily” might better reflect the existence of the two major subfamilies: CPTP, with selectivity for ceramide-linked phosphates; and GLTP, with selectivity for ceramide-linked sugars.
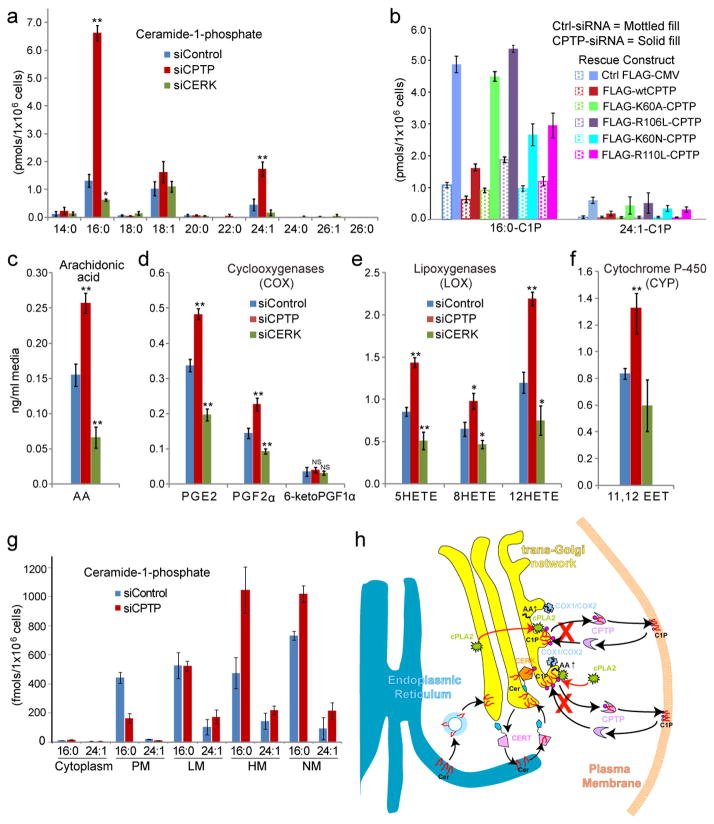
In cells, CPTP tracked by monospecific antibody or fluorescent epitope tag (EGFP) is cytosol-localized but also associates with peri-nuclear membranes (e.g. Golgi/TGN/endosomes), nuclei, and plasma membranes (Fig. 3n–s; Supplementary Fig. S9). No localization to mitochondria, lysosomes, or ER is detected. CPTP co-localization with TGN-46 verified interaction with trans-Golgi/TGN, a site where ceramide kinase (CERK) generates C1P3,26–28 and led us to hypothesize a C1P regulatory/sensing role for CPTP during CERK-mediated metabolic/signaling events. siRNA-induced CPTP down-regulation (~90%; Supplementary Fig. S10a) elevated both 16:0-C1P and 24:1-C1P (~4-fold) (Fig. 4a) and fragmented the Golgi cisternal stacks (Supplementary Fig. S11). RNAi-induced C1P changes were partially rescued with moderately-active R110L and K60N, but not with inactive K60A and R106L mutants (Fig. 4b). CPTP overexpression in the absence of RNAi decreased 16:0-C1P and 24:1-C1P. K60A or R106L overexpression had the opposite effect (Supplementary Fig. S12) and fragmented the Golgi cisternal stacks (not shown) consistent with a dominant-negative effect. CPTP depletion measurably decreased sphingosine and S1P (Supplementary Fig. S10b,c), 14:0-, 22:0-, 24:1-, and 24:0-SM, 16:0-monohexosylceramide, and 24:0-Cer, but modestly increased 24:1-Cer (Supplementary Fig. S10d–f and Discussion). Subcellular fractionation showed increased 16:0-C1P levels in ‘heavy’ membranes (trans-Golgi/endosome-enriched) and nuclear fractions, and decreased levels in plasma membranes without affecting levels in ‘light’ membranes (cis-Golgi/ER-enriched). These CPTP-depletion induced effects were rescued by ectopic wtCPTP expression (Fig. 4g; Supplementary Fig. S13c and Discussion).
Figure 4. CPTP siRNA depletion/rescue effects on cellular C1P levels, arachidonic acid generation, and eicosanoid release and model of CPTP cell biological function.
a, Intracellular C1P levels of siRNA-treated and control A549 cells. X-axis shows acyl composition of C1P species with sphingosine base chains (d18:1). b, Rescue effect by partially-active K60N- and R110L-CPTP mutants, but not inactive K60A- and R106L-CPTP mutants with siRNA targeted to endogenous CPTP 3′-UTR. c, d, e, f, Arachidonic acid (AA) and eicosanoids (COX, LOX, CYP pathways) secreted into media by siRNA-treated and control A549 cells. Data for b and d-g represent averages of 6 experiments (2 procedures) by Student’s t-test (* p<0.05, ** p<0.01, NS not significant). 6keto-PGF1α, the primary prostacyclin metabolite, is nearly unaffected by siCPTP or siCERK suggesting existence of an AA pool derived independently of C1P-activated cPLA2α. g, Changes in C1P levels at various subcellular locations by CPTP-siRNA. X-axis represents C1P species containing sphingosine base chains (d18:1) and either 16:0 or 24:1 acyl chains. PM, plasma membranes; LM, light membranes = ER-/cis-Golgi-enriched; HM, heavy membranes = trans-Golgi-/endosome-/mitochondria-enriched, NM, nuclear membranes. Data represent the mean ± s.d. of three independent experiments. h, Model for CPTP regulation of eicosanoid production. C1P is synthesized by ceramide kinase (CERK) which concentrates in the trans-Golgi network vicinity via its pleckstrin homology (PH) domain during stimulation. To produce C1P, CERK uses ceramide transported from the ER by ceramide transfer protein (CERT) which also contains a targeting PH domain. After synthesis, C1P is transported to subcellular destinations by CPTP and possibly by vesicular trafficking. RNAi knockdown of CPTP (red X) leads to accumulation and elevation of C1P at the Golgi complex, a condition that activates soluble cytosolic phospholipase A2α (cPLA2α), releasing AA needed for generation of downstream, pro-inflammatory eicosanoids. CPTP overexpression has the opposite effect on lipid levels. cPLA2α activation occurs by translocation from the cytoplasm and/or cis-Golgi (red arrows) via enhanced anchoring to C1P generated by CERK in TGN. COX-1 and inducible COX-2 which use AA to produce pro-inflammatory prostaglandins, also concentrate in the trans-Golgi/TGN vicinity during stimulation. For clarity, other eicosanoid generator pathways, i.e. LOX (e.g., cytoplasmic 5-lipoxygenase) and CYP (ER-associated cytochrome P-450), are not depicted. Also not depicted is Golgi cisternal stack fragmentation induced by 24 h of CPTP RNAi.
Because CERK-generated C1P induces Group IVA cPLA2α activity which releases arachidonic acid (AA) used to produce pro-inflammatory eicosanoids26,27, CPTP involvement was assessed. In siRNA-induced CPTP-depleted cells, arachidonic acid increased (Fig. 4c) consistent with C1P accumulation at the Golgi/TGN that activates cPLA2α26. Also elevated were major AA metabolites generated by cyclooxygenase (COX), lipoxygenase (LOX), and cytochrome p450 (CYP) pathways, i.e. PGE2, PGF2α, 6-keto-PGF1α (COX, Fig. 4d); 5HETE, 8HETE, 12HETE (LOX, Fig. 4e); 11,12 EET (CYP, Fig. 4f). By contrast, AA and eicosanoid levels decreased upon overexpression of wtCPTP but not K60A or R106L mutant (Supplementary Fig. S14). Parallel siRNA-induced down-regulation of CERK (Supplementary Fig. S10), the only established producer of C1P in mammals, decreased 16:0-C1P (Fig. 4a), AA (Fig. 4c), and eicosanoids (Fig. 4d–f) elevated by CPTP depletion.
Figure 4h depicts a model showing how CPTP could regulate pro-inflammatory eicosanoid generation. In mammals, the only established pathway for C1P production is via phosphorylation of ceramide by CERK at the cytoplasmic surface of the trans-Golgi/TGN3,28. CERK also contains nuclear localization/export signals and traffics to the plasma membrane via microtubule-driven vesicles in response to hyperosmotic shock28. To produce C1P, CERK uses ceramide delivered from its ER synthetic site to the Golgi by either CERT27 or possibly by vesicular trafficking10. C1P elevation by CERK is known to activate soluble cPLA2α by enhancing translocation to the trans-Golgi/TGN3,26 where cPLA2α action releases AA needed by eicosanoid producers such as COX-1 or COX-2. siRNA-induced CPTP depletion elevates C1P in the Golgi complex and nucleus, but lowers C1P plasma membrane levels. We propose that CPTP prevents excess C1P accumulation after production by CERK, thereby regulating cPLA2α action, diminishing AA release and downstream generation of pro-inflammatory eicosanoids. One destination for CPTP-cargo is the plasma membrane. Models involving CPTP in catabolic C1P generation by sphingomyelinase D are less plausible because mammalian cells lack this enzyme3 (Supplementary Discussion).
Previously, the only identified mechanism for regulating CERK-mediated production of C1P was by control of ceramide availability via ceramide transfer protein27. It is noteworthy that siRNA-induced CPTP depletion yields the highest increase in endogenous C1P reported to date, mostly as 16:0-C1P, and dramatically alters Golgi cisternal stack morphology suggesting CPTP-mediated transport is essential for maintaining proper Golgi organization by safeguarding localized C1P levels. The ensuing stimulation in eicosanoid production triggered by elevated C1P in the Golgi complex potentially implicates CPTP in as of yet unidentified disease states associated with inflammation.
FULL METHODS (for online version)
Protein Expression and Purification
GLTPD1 ORFs encoding human and mouse (GenBank JN542538 & NP_077792.2) CPTP were cloned in pET-SUMO vector (Invitrogen) and expressed in BL21 (DE3) Star cells (Invitrogen). Soluble CPTP tagged N-terminally with His6-SUMO was affinity-purified by Ni-NTA chromatography followed by ubiquitin-like protein 1 (Ulp1) SUMO protease digestion overnight at 4°C to release His6-SUMO tag. Affinity repurification by Ni-NTA chromatography was followed by FPLC gel filtration chromatography. L-selenomethionine (Se-Met)-labeled protein for ab initio phasing was produced by feedback inhibition of the methionine synthesis pathway. Mutants were constructed by QuikChange Site-Directed Mutagenesis (Stratagene) and verified by sequencing.
CPTP Lipid Transfer Activity Involving Membrane Vesicles
Intermembrane lipid transfer by CPTP was measured in real time by Förster resonance energy transfer (FRET) between donor POPC vesicles, containing 1 mole% AV-lipid [acyl chain omega-labeled with anthrylvinyl fluorophore, i.e. (11E)-12-(9-anthryl)-11-dodecenoyl], and 1.5 mole% 1-acyl-2-[9-(3-perylenoyl)-nonanoyl]-3-sn-glycero-3-phosphocholine [Per-PC] and POPC acceptor vesicles at 10-fold excess. In competition assays, donor vesicles also contained competitor lipids at 0.5, 1.0 and 2.0 mole%29. Briefly, CPTP addition produces an exponential increase in AV emission intensity (425 nm) as the protein transports AV-C1P away from the donor vesicles (creating separation from the ‘nontransferable’ Per-PC) and delivers to the POPC acceptor vesicles present in 10-fold excess. The time-dependent increase in 425 nm emission relative to signal in the absence of CPTP reflects lipid transfer kinetics. In the absence of acceptor vesicles, no transfer is observed. The initial lipid transfer rate, νo, is obtained by nonlinear regression analyses (see Supplementary Methods).
Crystallization and Structure Determination
Crystallization hits from initial screens were optimized by the hanging drop vapor diffusion method and systematically varying pH and individual component concentrations (Table S3). For data collection, crystals were flash frozen (100 K) in reservoir solutions containing 20% (v/v) ethylene glycol. Diffraction data sets were collected on 24-ID-C and 24-ID-E beamlines at the Advanced Photon Source (APS) and X29 beamline at Brookhaven National Laboratory. All crystals belonged to different crystal forms. For phasing, single-wavelength anomalous dispersion (SAD) data were collected for Se-Met-labeled, CPTP/8:0-C1P crystal complex at Se peak wavelength (Table S2; see Supplementary Methods). Use of Se-CPTP structure enabled other structures to be solved by molecular replacement (Supplementary Methods). Statistics for data collection, refinement and SAD phasing are provided in Tables S1 and S2.
Epifluorescence Microscopy Analyses
BSC-1 cells on coverslips were fixed in −20 °C methanol and labeled with anti-CPTP (Santa Cruz Biotechnology sc247014), and Golgi markers anti-TGN46, anti-GM 130, and anti-p230, or endosome markers anti-Rab5 and anti-Rab9 (Cell Signaling) followed by secondary antibodies coupled to Alexa-488, Alexa-594, or Alexa-660. Cells were counter-stained with DAPI, mounted in 10% PBS, 90% glycerol, imaged using a Leica DM RXA2 microscope with a 63x 1.4 NA APO C objective, a Hamamatsu ORCA ER CCD camera, and Simple PCI software and analyzed as intensity scattergrams with measured correlation coefficients30. Time-lapse images of living cells expressing EGFP-CPTP were captured with a Leica DM RXA2 microscope stand equipped with a Yokagawa CSU-10 spinning disk confocal head and using illumination from a Coherent 488 nm 200mW “Sapphire” continuous wave optically pumped solid-state laser and 3I Slidebook software31 (see Supplementary Methods for more details).
Immunoblot Analysis
BSC1 cells were grown to semi-confluence, collected by manual scraping, pelleted, and boiled in SDS-PAGE buffer. Proteins were separated on a 10% discontinuous SDS-PAGE gel, transferred to PDVF membrane, and immuno-labeled32. The immunoreactive band was detected by chemiluminescence (Image Quant system, GE Healthcare).
siRNA-mediated CPTP Down-regulation and Rescue in Cells Ectopically Expressing Wildtype and Mutant CPTP Constructs
Low passage A549 cells (5×105) were grown (10-cm plates) in appropriate medium under standard incubator conditions (SIC) overnight. Cells were treated with siRNA (Dharmacon) against CPTP (GLTPD1) or CERK as well as non-targeting siRNA sequence for control per manufacturer’s protocol and incubated for 48 h under SIC. For rescue experiments, cells were transfected with either the empty pFLAG-CMV4 (Neor) plasmid or this vector containing wtCPTP, K60A-CPTP, K60N-CPTP, R106L-CPTP, or R110L-CPTP. Batch cultures of cells stably expressing the transfected constructs were obtained by selection for two weeks in regular medium containing G418 (genticin; 500μg/ml) under SIC. Following selection, cells (5×105) were transferred to 10 cm tissue culture plates and cultured overnight in regular media without G418 under SIC. Cells then were treated with either control siRNA or a mixture of 4 siRNA constructs (Dharmacon) designed against the 3′ UTR of endogenous CPTP (GLTPD1) mRNA following standard manufacturer’s protocol (Supplementary Methods). The 3′UTR was not included in the ectopically-expressed constructs to ensure siRNA targeting only to endogenous CPTP. Cells were incubated 48 h in regular media without G418 under SIC. Full serum media was replaced with media containing 2% serum 15 h prior to harvest.
RNA Isolation, Reverse Transcription-PCR, and Quantitative PCR
To evaluate downregulation of CERK and CPTP, quantitative PCR was performed26. Briefly, total RNA was isolated using RNeasy kits (Qiagen). Total RNA (1 μg) was reverse-transcribed using Superscript III reverse transcriptase (Invitrogen). The level of CERK transcript was monitored using quantitative PCR and TaqMan technology (Applied Biosystems) specific to CERK and CPTP with 18 S as control. cDNA was amplified using an ABI 7900HT with premixed primer-probe sets and TaqMan Universal PCR master mix (Applied Biosystems).
Intracellular Sphingolipid Analyses
Cell lipids were harvested using an improved Bligh-Dyer protocol32 (Supplementary Methods). Sphingolipids were separated by HPLC (Prominence HPLC system, Shimadzu) using a Kinetix-C18 column (50 × 2.1 mm, 2.6μ; Phenomenex) and eluted using a linear gradient (solvent A, methanol:water:formic acid (58:41:1) in 5 mM ammonium formate; solvent B, methanol:formic acid (99:1) in 5 mM ammonium formate, 20–100% B (3.5 min) and at 100% B (4.5 min); flow rate of 0.4 ml/min, 60°C). ESI-tandem-MS (API 4000 QTRAP instrument; Applied Biosystems, MDS Sciex) was used to detect C1P32, ceramide, sphingosine, S1P, sphingomyelin, and monohexosyl ceramide under positive ionization (see Supplementary Methods).
Eicosanoid Analysis
Eicosanoids were analyzed as detailed by the Lipid Maps Consortium33,34. Culture media (4 ml) from siRNA was combined with 10% methanol (400 μl) and glacial acetic acid (20 μl) before spiking with internal standard (100 μl) containing the following deuterated eicosanoids (100 pg/μl, 10 ng total): (d4) 6keto-PGF1α, (d4) PGF2α, (d4) PGE2, (d4) PGD2, (d8) 5-hydroxyeicosa-tetranoic acid (5HETE), (d8) 15-hydroxyeicosatetranoic acid (15HETE), (d8) 14,15 epoxyeicosa-trienoic acid and (d8) arachidonic acid. Samples and vial rinses (5% MeOH; 2 ml) were applied to Strata-X SPE columns (Phenomenex), previously washed with methanol (2 ml) and then dH2O (2 ml). Eicosanoids eluted with isopropanol (2 ml), were dried in vacuuo and reconstituted in EtOH:dH2O (50:50;100 μl) prior to HPLC ESI-MS/MS analysis (see Supplementary Methods).
Subcellular Fractionation
Subcellular fractionation was performed by multi-step centrifugation as detailed and characterized previously27 with minor modifications (see Supplementary Methods). Fraction enrichment was validated by SDS-PAGE/Western blotting (Supplementary Fig. S13b) using organelle markers for: nuclei (anti-lamin AC, trans-Golgi (anti-TGN46), ER (anti-protein disulfide isomerase (PDI)), and plasma membrane (anti-caveolin-1).
Supplementary Material
Acknowledgments
This research was supported by NIH/NCI CA121493 (DJP & REB), NIH/NIGMS GM45928 (REB), NIH/NIGMS GM072754 (EHH), NIH/CA154314 (CEC), VA Merit Award (CEC), VA Research Career Scientist Award (CEC), VA Career Devel. Award (DSW), NRS-T32/NIGMS 008695 (DSW), Spanish Ministerio de Ciencia e Innovacion BFU2010-17711 (LM), Russian Fnd. for Basic Research #12-04-00168 (JGM), Hormel Fnd. (REB), Abby Rockefeller Mauze Trust (DJP), and Maloris Fnd. (DJP). We thank Helen Pike (UMN-Hormel Institute) for expressing and purifying protein used for transfer activity analyses, Kul Karanjeet (UMN-Hormel Institute) for preparing cells for confocal and epifluorescence microscopy, and the staff of X-29 beamline at the National Synchrotron Light Source and ID-24-C/E beamlines at the Advanced Photon Source for help.
Footnotes
Author Contributions
DKS: all structural analyses and provided definitive evidence for C1P binding by CPTP; generated all CPTP point mutants; wrote text. RKK: transfer analyses of wtCPTP and CPTP point mutants; wrote text. DSW: siRNA CPTP knockdown, rescue, and all lipid analyses; wrote text. XZ: cloned wtCPTP; PCR analyses of CPTP transcript distribution in human tissues. XZ: wtCPTP transfer rate analyses. SKM: prepared CPTP RNAi constructs for microscopy and CPTP overexpression constructs for lipidomics analyses. JGM: synthesized all fluorescent lipids. LM: contributed to structural data interpretation. EHH: fluorescence microscopy of CPTP localization in fixed and living cells; finalized the write-up. CEC: directed siRNA knockdown, rescue and related lipidomics analyses; finalized the write-up. DJP: directed CPTP structural analyses; finalized the write-up. REB: directed functional analyses after initial CPTP discovery in his lab; finalized the write-up; coordinated and integrated all section write-ups.
Author Information
Atomic coordinates and structure factors for human CPTP crystal complexes with various lipids and mouse apoCPTP have been deposited in the Protein Data Bank. Accession codes are: 2:0-C1P/CPTP (4K80), 8:0-C1P/CPTP (4KF6), 12:0-C1P/CPTP (4K85), 16:0-C1P/CPTP (4K84), 18:1-C1P/CPTP (4K8N), di12:0-PA/CPTP (4KBS) and mouse apoCPTP (4KBR).
References
- 1.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Bio. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 2.Chalfant CE, Spiegel S. Sphingosine 1-phosphate and ceramide 1-phosphate: expanding roles in cell signaling. J Cell Sci. 2005;118:4605–4612. doi: 10.1242/jcs.02637. [DOI] [PubMed] [Google Scholar]
- 3.Lamour NF, Chalfant CE. Ceramide kinase and the ceramide-1-phosphate/cPLA2α interaction as a therapeutic target. Curr Drug Targets. 2008;9:674–682. doi: 10.2174/138945008785132349. [DOI] [PubMed] [Google Scholar]
- 4.Gangoiti P, et al. Control of metabolism and signaling of simple bioactive sphingolipids: Implications in disease. Prog Lipid Res. 2010;49:316–334. doi: 10.1016/j.plipres.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Maceyka M, Harikumar KB, Milstien S, Spiegel S. Sphingosine-1-phosphate signaling and its role in disease. Trends Cell Biol. 2012;22:50–60. doi: 10.1016/j.tcb.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Bio. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 7.Leslie CC, Gangelhoff TA, Gelb MH. Localization and function of cytosolic phospholipase A2α at the Golgi. Biochimie. 2010;92:620–626. doi: 10.1016/j.biochi.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bechler ME, de Figueiredo P, Brown WJ. A PLA1–2 punch regulates the Golgi complex. Trends Cell Biol. 2012;22:116–124. doi: 10.1016/j.tcb.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harizi H, Corcuff JB, Gualde N. Arachidonic-acid-derived eicosanoids: roles in biology and immunopathology. Trends Mol Med. 2008;14:461–469. doi: 10.1016/j.molmed.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Boath A, et al. Regulation and traffic of ceramide 1-phosphate produced by ceramide kinase: comparative analysis to glucosylceramide and sphingomyelin. J Biol Chem. 2008;283:8517–8526. doi: 10.1074/jbc.M707107200. [DOI] [PubMed] [Google Scholar]
- 11.Lev S. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat Rev Mol Cell Biol. 2010;11:739–750. doi: 10.1038/nrm2971. [DOI] [PubMed] [Google Scholar]
- 12.Prinz WA. Lipid trafficking sans vesicles: where, why, how? Cell. 2010;143:870–874. doi: 10.1016/j.cell.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malinina L, Malakhova ML, Teplov A, Brown RE, Patel DJ. Structural basis for glycosphingolipid transfer specificity. Nature. 2004;430:1048–1053. doi: 10.1038/nature02856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malakhova ML, et al. Point mutational analysis of the liganding site in human glycolipid transfer protein. Functionality of the complex. J Biol Chem. 2005;280:26312–26320. doi: 10.1074/jbc.M500481200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirsch AK, Fischer FR, Diederich F. Phosphate recognition in structural biology. Angewandte Chemie. 2007;46:338–352. doi: 10.1002/anie.200603420. [DOI] [PubMed] [Google Scholar]
- 16.Bourquin F, Riezman H, Capitani G, Grutter MG. Structure and function of sphingosine-1-phosphate lyase, a key enzyme of sphingolipid metabolism. Structure. 2010;18:1054–1065. doi: 10.1016/j.str.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Berna A, et al. For whom the bell tolls? DING proteins in health and disease. Cellular Mol Life Sci: CMLS. 2009;66:2205–2218. doi: 10.1007/s00018-009-0006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furuhashi M, Hotamisligil GS. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nature Rev Drug Discovery. 2008;7:489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roderick SL, et al. Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nature Struct Biol. 2002;9:507–511. doi: 10.1038/nsb812. [DOI] [PubMed] [Google Scholar]
- 20.Hanada K, et al. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- 21.Kudo N, et al. Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide. Proc Natl Acad Sci U S A. 2008;105:488–493. doi: 10.1073/pnas.0709191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou X, et al. Human glycolipid transfer protein (GLTP) genes: organization, transcriptional status and evolution. BMC Genomics. 2008;9:72. doi: 10.1186/1471-2164-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orengo CA, Thornton JM. Protein families and their evolution - a structural perspective. Annu Rev Biochem. 2005;74:867–900. doi: 10.1146/annurev.biochem.74.082803.133029. [DOI] [PubMed] [Google Scholar]
- 24.Galperin MY, Koonin EV. Divergence and convergence in enzyme evolution. J Biol Chem. 2012;287:21–28. doi: 10.1074/jbc.R111.241976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liberles DA, et al. The interface of protein structure, protein biophysics, and molecular evolution. Protein Sci. 2012;21:769–785. doi: 10.1002/pro.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamour NF, et al. Ceramide 1-phosphate is required for the translocation of group IVA cytosolic phospholipase A2 and prostaglandin synthesis. J Biol Chem. 2009;284:26897–26907. doi: 10.1074/jbc.M109.001677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamour NF, et al. Ceramide kinase uses ceramide provided by ceramide transport protein: localization to organelles of eicosanoid synthesis. J Lipid Res. 2007;48:1293–1304. doi: 10.1194/jlr.M700083-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Bornancin F. Ceramide kinase: the first decade. Cell Signal. 2011;23:999–1008. doi: 10.1016/j.cellsig.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 29.Samygina VR, et al. Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein. Structure. 2011;19:1644–1654. doi: 10.1016/j.str.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manders EMM, Stap J, Brakenhoff GJ, van Driel R, Aten JA. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J Cell Sci. 1992;103:857–862. doi: 10.1242/jcs.103.3.857. [DOI] [PubMed] [Google Scholar]
- 31.Hornick JE, et al. Amphiastral mitotic spindle assembly in vertebrate cells lacking centrosomes. Current biology : CB. 2011;21:598–605. doi: 10.1016/j.cub.2011.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durcan T, et al. Tektin 2 is required for central spindle microtubule organization and the completion of cytokinesis. J Cell Biol. 2008;181:595–603. doi: 10.1083/jcb.200711160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wijesinghe DS, et al. Use of high performance liquid chromatography-electrospray ionization-tandem mass spectrometry for the analysis of ceramide-1-phosphate levels. J Lipid Res. 2010;51:641–651. doi: 10.1194/jlr.D000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blaho VA, Buczynski MW, Brown CR, Dennis EA. Lipidomic analysis of dynamic eicosanoid responses during the induction and resolution of Lyme arthritis. J Biol Chem. 2009;284:21599–21612. doi: 10.1074/jbc.M109.003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.