Summary
The integration of synthetic and cell-free biology has made tremendous strides towards creating artificial cellular nanosystems using concepts from solution-based chemistry: only the concentrations of reacting species modulate gene expression rates. However, it is known that macromolecular crowding, a key feature of natural cells, can dramatically influence biochemical kinetics by volume exclusion effects that reduce diffusion rates and enhance binding rates of macromolecules. Here, we demonstrate that macromolecular crowding can increase the robustness of gene expression through integrating synthetic cellular components of biological circuits and artificial cellular nanosystems. In addition, we reveal how ubiquitous cellular modules, including genetic components, a negative feedback loop, and the size of crowding molecules, can fine tune gene circuit response to molecular crowding. By bridging a key gap between artificial and living cells, our work has implications for efficient and robust control of both synthetic and natural cellular circuits.
Keywords: molecular crowding, synthetic biology, gene regulation, artificial cells, robustness
An overarching desire to harness natural biological principles for biotechnological applications has driven synthetic biology towards the creation of cells with completely synthetic genomes 1 or genetic polymers 2, synthetic organisms with hybrid cellular and polymeric structures 3, synthetic in vitro systems 4, and artificial cells with synthetic components 5–12. These synthetic and hybrid systems have tremendous potential for novel applications in the discovery of fundamental biological principles 13, therapeutic treatment 14, and bioenergy production 15 as they enable more precise control and predictive modeling of the systems by minimizing cellular components. However, while reducing the complexity of the systems allows more precise control of the reaction environment, it also has the potential to eliminate features of the cellular micro-environment that are important for the robust functioning of genetic circuits. One key distinguishing feature between living cells and prevailing artificial cellular systems is the density of molecules around cellular components, which is an under-appreciated, yet an important factor in the regulation of cellular dynamics 16, 17.
Molecular crowding is a natural state of cells in which their intracellular environments are densely packed with macromolecules 18, 19 (Fig. 1a). This crowding is absent in solution-based chemistry approaches that are typically used in synthetic genetic systems. Molecular crowding can cause volume exclusion effects that reduce diffusion rates and enhance binding rates of macromolecules 20, which lead to fundamental impact on cell functions such as the optimum number of transcription factors 21, the dynamical order of metabolic pathways 22, and nuclear architecture 23. However, the impacts of molecular crowding on dynamics of gene circuits and the consequences for their activity in heterologous environments have not been established. It remains elusive whether molecular crowding can increase the robustness of gene expression towards perturbations of genetic micro-environments. Furthermore, there have been tremendous controversies regarding whether molecular crowing could indeed impact cellular activities 24–26. Answers to these fundamental questions would enable more precise control of synthetic gene circuits in artificial systems, provide a bridge between in vitro and in vivo systems 24, and facilitate predictive approaches for both synthetic and natural biological systems.
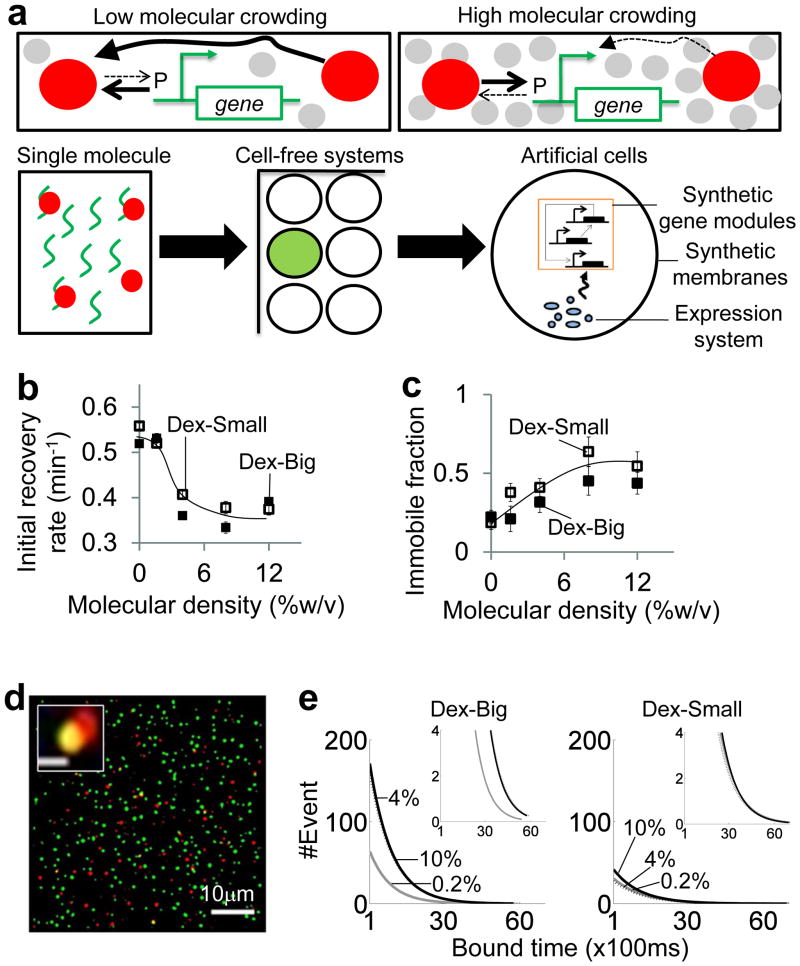
Figure 1. Shaping gene expression in artificial cellular systems by molecular crowding.
a. Macromolecular crowding, a key feature of natural cells, can dramatically influence biochemical kinetics (top panels). Molecular crowding (grey circles) could enhance binding between two molecules (red circles and the promoter P), but decrease diffusion of molecules (top right panel). We studied the impact of molecular crowding on gene expression by bridging between microscopic single molecule dynamics and macroscopic dynamics in both cell-free systems in 96-well plates and artificial cells.
b. Fluorescence recovery after photobleaching was conducted to study diffusion dynamics of RFP-T7RNAP. Both big and small dextran molecules (Dex-Small=6x103g/mol, Dex-Big =2x106g/mol) significantly reduced initial recovery rates of mobile T7 RNAP. Each error bar indicates one standard error of the mean (SEM) of at least nine replicates. See Supplementary Fig. 3a & SI for detailed experimental setup.
c. Both Dex-Small (open squares) and Dex-Big (filled squares) significantly increased immobile fractions of RFP-T7RNAP. Each error bar indicates one SEM of at least nine replicates. We note that the differences in recovery dynamics were not due to altered photo-stability of RFP because crowding molecules did not impact bleaching dynamics of RFP-T7RNAP during the FRAP experiments (results not shown).
d. RFP-T7RNAP (red circles) co-localized with Cy3-PT7 (green circles) molecules. The top panel shows a typical frame of view with approximately 200 Cy3-PT7 molecules. The inset shows a single-molecule view of co-localized RFP-T7 RNAP and Cy3-PT7 molecules (Scale bar = 533nm). See Supplementary Fig. 4a & SI for detailed experimental setup.
e. Dex-Big significantly increased the number of binding events and mean bound time as compared to Dex-Small. Each inset shows a zoomed-in view on the tail of the same distribution. Black lines represent 10% molecular density. Black dotted lines represent 4% molecular density. Grey lines represent 0.2% molecular density.
Molecular crowding impacts the diffusion of T7 RNA polymerase
To investigate the impact of molecular crowding on gene expression, we used genetic components from phage T7 due to their well-characterized kinetics and functions in synthetic systems (Supplementary Fig. 1). We quantified interactions between T7 RNA polymerase (RNAP) and DNA in the presence of crowding agents because molecular crowding can increase excluded volumes, which would affect large molecules more significantly than small molecules 19. To visualize the response, we constructed a red fluorescent protein (RFP)-T7RNAP fusion RNA polymerase that transcribed a cyan fluorescent protein (CFP) from a PT7 promoter (Supplementary Fig. 2a–b and Supplementary Information (SI)).
Since transcriptional activities are sensitive to the diffusion of macromolecules 21, we first focused on diffusion dynamics of T7 RNAP in different crowding environments outside of the artificial cell environment. We modulated crowding molecular size because a previous work has suggested that natural cells could use crowding molecules as molecular sieves to filter molecules and modulate system dynamics 27. Specifically, we performed fluorescence recovery after photobleaching (FRAP, Supplementary Fig. 3, Supplementary Movie 1) using either small (6x103g/mol, Dex-Small) or big (2x106g/mol, Dex-Big) dextran polymers (Fig. 1b&c). These molecules are commonly used as inert molecules that mimic crowded intracellular environments, but do not react with the systems of interest 28. We found that initial recovery rates of mobile RFP-T7RNAP were significantly reduced by increasing densities of both Dex-Small and Dex-Big (Fig. 1b and Supplementary Fig. 3b). This result suggests that diffusion of the mobile fraction of RFP-T7RNAP were affected by the crowding molecules. Both Dex-Small and Dex-Big also resulted in higher immobile fractions of RFP-T7RNAP with increasing crowding densities (Fig. 1c). Based on existing diffusion models 29, the increased immobile fractions are likely due to increased sub-diffusion fractions of RFP-T7RNAP in crowded environments, in which the polymerase explores a smaller space than predicted by classical Brownian dynamics. Therefore, our findings suggest that both Dex-Small and Dex-Big reduced the diffusion of RFP-T7RNAP.
Molecular crowding impacts the binding of the T7 RNAP to a T7 promoter
In addition to diffusion of macromolecules, gene transcription is sensitive to the binding between transcription factors and promoters 30, 31, which was requisite to understand the impact of molecular crowding on gene expression. To measure the binding between RFP-T7RNAP and a PT7 promoter, we built on our FRAP experiments and conjugated biotinylated-Cy3-PT7 DNA molecules onto PEG-surfaces through avidin-biotin linkages (Supplementary Fig. 4). When Cy3-PT7 was immobilized on the surface, RFP-T7RNAP would freely diffuse in the chamber and bind to PT7 on the surface. We used this approach 30 to measure co-localization of RFP-T7RNAP with Cy3-PT7, which would suggest that there was binding between them (Fig. 1d, Supplementary Fig. 5a, Supplementary Movie 2). We expected that a decreased dissociation rate constant would shift the mean of bound time distribution to a higher bound time and an increased association rate constant would increase detected binding events at low bound time. With Cy3-PT7, 10% Dex-Big significantly increased the number of binding events by ~100% compared to the control (0.2% Dex-Big) (Fig. 1e), suggesting an increased association rate constant. 10 %Dex-Big also increased the mean of the bound time distribution by ~100ms compared to the control (0.2% Dex-Big), suggesting a decreased dissociation rate constant. In contrast, 10% Dex-Small only increased the number of binding events by ~25% compared to the control (0.2% Dex-Small). These observations suggest that Dex-Big decreased dissociation rate constants and increased association rate constants between T7 RNAP and PT7 more significantly than Dex-Small, which is consistent with a general theory of molecular crowding that suggests a larger enhancement of molecular binding by large crowding molecules than small crowding molecules 20. Using a control DNA without the PT7 promoter, co-localization of RFP-T7RNAP with Cy3 was negligible (Supplementary Fig. 5b), suggesting that the observation was not due to non-specific interactions between T7 RNAP and either the PEG surface or DNA.
Molecular crowding impacts gene expression dynamics
To gain insight into connections between microscopic dynamics observed using single molecule imaging and gene expression dynamics, we created a parsimonious model based on previous models of molecular crowding 17 and qualitative results obtained from the FRAP and single molecule experiments (SI, Fig. 2a & Supplementary Fig. 6). Based on the model, we first predicted expression dynamics of a genetic module with a native PT7 promoter that regulates the expression of a cyan fluorescent protein (CFP) as a reporter of gene expression (Fig. 2b). Our model predicts that a small crowding molecule would result in a biphasic response, where gene expression rates would first increase with increasing crowding densities, but then would decrease after a specific crowding density (Fig. 2c, black line). The biphasic response arises because Dex-Small only increased the number of binding events by ~25% (Fig. 1e), but significantly reduced the diffusion of T7RNAP (Fig. 1b&c). The predicted biphasic dynamic response was indeed observed with our cell-free expression system, which consisted of an S12 bacterial extract, a S12 supplement, and Dex-Small (Fig. 2c, black squares & SI). The cell-free expression system is a synthetic system that couples both transcription and translation and has been used to create artificial cells 13. In contrast, both our model and experiments show that Dex-Big caused a monotonic increase of gene expression rates with increasing crowding densities (Fig. 2d and Supplementary Fig. 7) because Dex-Big significantly increased the association rate constant and reduced the dissociation rate constant between T7RNAP and PT7 promoter (Fig. 1e). Additional tests using alternative crowding agents, including Ficoll 400 (400x103g/mol), PEG-100k (100x103g/mol), and PEG8000 (8x103g/mol), suggested that the size-dependent impact of molecule crowding on gene expression rates is likely a generic phenomenon (Supplementary Fig. 8a). Specifically, the size of Ficoll-400 is approximately the same as the size of Dex-Big and resulted in monotonically increasing expression rates; PEG-8000, a small crowding molecule, caused biphasic expression rates; and PEG-100k, a crowding molecule with an intermediate size, significantly increased gene expression rates as compared to PEG8000. Similar to the observed impact of molecular crowding on dynamics of nuclear proteins in heterochromatin 32, our results suggest that molecular composition of genetic microenvironments can distinctly modulate gene expression rates.
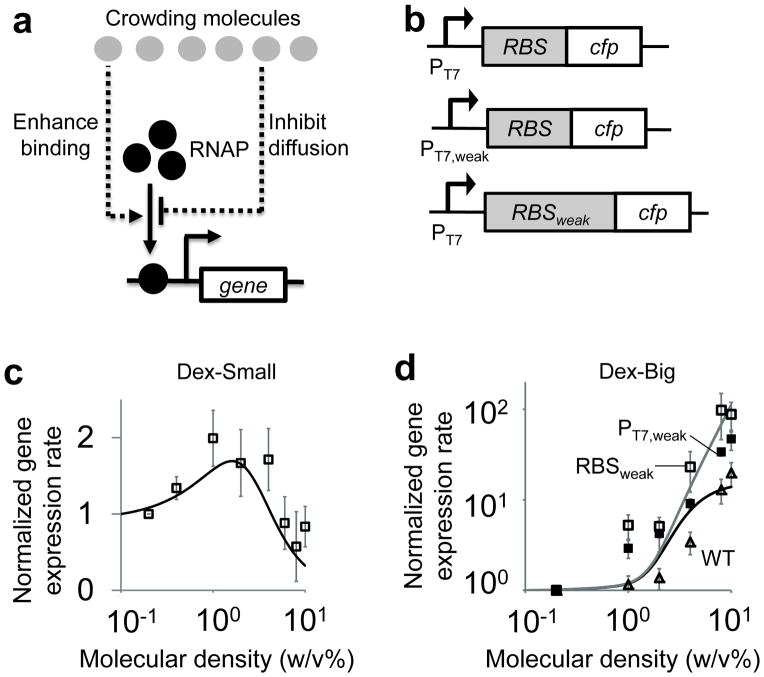
Figure 2. Molecular crowding modulated dynamics of gene expression.
a. A parsimonious model of gene expression in crowded environments. See Eq. S1–10 for detailed mathematical models.
b. Three genetic modules were constructed to study the impact of genetic elements on crowding effects. The first module is a base module that has the wildtype genetic components. The second and third modules have either a weaker T7 promoter or a weaker ribosomal binding site (RBS).
c. Gene expression rates in environments containing small crowding molecules. Both modeling (black line) and experimental (open black squares) results show that, gene expression rates exhibited a biphasic response with increasing densities of Dex-Small. A paired t-test shows that gene expression rates at 1% density are significantly higher than gene expression rates at both 0.2% density (p-value=0.02) and 10% density (p-value=0.0008). Each error bar indicates one SEM of at least four replicates. Each gene expression rate was normalized by the basal rate with 0.2% crowding molecules.
d. Gene expression rates in environments containing big crowding molecules. Our model predicts that a big crowding molecule would increase gene expression rates (black line) and that a weaker genetic component (Fig. 2b) would result in a higher fold increase of gene expression rates (grey line). Consistent with the predictions, gene expression rates increased with increasing densities of Dex-Big (open triangles). With either a weak T7 promoter (pT7weak, filled black squares) or a weak RBS (RBSweak, open black squares), gene expression rates increased more significantly than with the wild-type module (WT, open triangles). A paired t-test shows that for each molecular densities (≥1%), normalized gene expression rates of both pT7weak and RBSweak modules are significantly higher than normalized gene expression rates of the wild-type module (p-value<0.04). Each error bar indicates one SEM of at least five replicates.
To respond to changing environments, natural cells could couple crowding densities with both promoters and ribosomal binding sites (RBS) to modulate gene expression rates 33. To investigate this link between genetic components and molecular crowding, we constructed two additional genetic modules, with either a weak PT7 promoter (PT7,weak) or a weak RBS (RBSweak) (Fig. 2b), which resulted in lower gene expression rates (Supplementary Fig. 8b). Previous studies have also demonstrated that these genetic components exhibit higher dissociation rate constants 34, 35 between T7 RNAP and PT7 and ribosome and RBS as compared to the components used in the original module (Fig. 2b). Based on the kinetic information, mathematical modeling predicts that these modules would result in higher fold changes in gene expression rates with increasing crowding densities as compared to the base module (Fig. 2d), which arises due to faster decrease in dissociation constants of macromolecules and their respective binding sites with increasing crowding densities. Consistent with our model predictions, gene expression rates of both PT7,weak and RBSweak increased more than those of the base module with increasing crowding densities (Fig. 2d).
Molecular crowding increases the robustness of gene expression
Molecular crowding is a cellular feature that has been maintained throughout evolution 36, suggesting that molecular crowding could be essential in maintaining robustness of system dynamics, which is a hallmark of biological systems. To allow for a more systematic assessment of this hypothesis than would be possible experimentally, we first created two parameter sets using the same mathematical model, each representing either a low or a high crowding environment. Next, we used computer simulations to emulate perturbations of these base models by randomizing their kinetic constants within 10-fold of their base values (SI). The changes in gene expression rates of perturbed models as compared to the base models are quantified as “fold perturbations” to estimate robustness of the systems (Fig. 3a): a high absolute value indicates a significant perturbation to a system. The fold perturbations are approximately zero in highly crowded conditions. In contrast, in a low crowding environment, fold perturbations are non-zero, showing that the system is more sensitive to the parameter perturbations. These simulation results suggest that molecular crowding may support the robustness of gene expression in natural cells. Furthermore, these results are consistent with simulation results of a stochastic model that includes intrinsic noise (SI & Supplementary Fig. S9a).
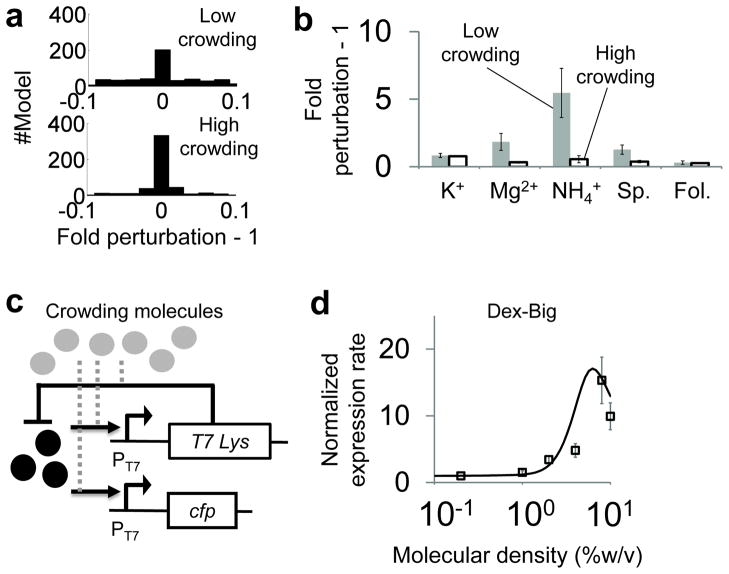
Figure 3. Molecular crowding increased robustness of gene expression.
a. Modeling prediction of gene expression robustness. The highly crowded environment resulted in a narrow distribution of fold gene-expression perturbation (bottom panel), suggesting that molecular crowding decreases fluctuation of gene expression rates due to the perturbation of gene environmental factors. In contrast, the low crowding environment resulted in significant perturbation of the system (top panel). See SI and Eq. S11 for detailed model description.
b. Fold perturbation of gene expression rates using five chemicals. The system was perturbed by changing concentrations of potassium glutamate, magnesium acetate, ammonium acetate, spermidine, and folinic acids. Consistent with our model predictions, gene expression rates were less perturbed in a highly crowded environment than in a low crowded environment. Grey bars represent low crowding environments without Dex-Big. Black open bars represent high crowding environments with 10% Dex-Big. Each error bar indicates one SEM of four replicates.
c. Schematic of interactions between molecular crowding and a negative feedback loop. A negative feedback loop was constructed by using a T7 lysozyme that binds to T7 RNAP and inhibits transcription from the PT7 promoter. Molecular crowding could impact both the binding of T7 RNAP to the promoter and the binding between T7 RNAP and T7 Lys (grey dotted lines).
d. Gene expression rates of a negative feedback loop with increasing crowding densities. Both modeling (black line) and experimental (open squares) results show that the negative feedback circuit generated a biphasic response of gene expression rates that peaked at 8% Dex-Big (open squares). A t-test shows that gene expression rates at 8% density are significantly higher than gene expression rates at both 0.2% density (p-value=0.003) and 10% density (p-value=0.001). Each error bar represents the SEM of seven replicates. Each gene expression rate was normalized by the basal rate with 0.2% crowding molecules.
We tested the impact of molecular crowding on system robustness experimentally by changing concentrations of potassium, magnesium, ammonium, spermidine, and folinic acid (Fig. 3b) in cell-free expression systems. These chemicals modulate both stability and binding of macromolecules 37. Our experimental results show that a low crowding environment (0.2% Dex-Big) resulted in higher perturbations of gene expression relative to a highly crowded environment (10% Dex-Big) for magnesium, ammonium, and spermidine. Furthermore, we tested if the robustness of gene expression could be fine-tuned using intermediate crowding densities. We observed that ammonium indeed caused a monotonic decrease in fold perturbations with increasing crowding densities (Supplementary Fig. 9b).
A negative feedback loop modulates the impact of molecular crowding on gene expression
In addition to genetic elements, gene circuit dynamics are known to be modulated by negative feedback loops, which are the most ubiquitous motif in natural gene circuits 38, likely due to their noise reduction and auto-regulatory roles. Here, we tested if molecular crowding can modulate gene expression of a negative feedback loop. To gain insight into interactions between feedback loops and molecular crowding, we simulated expression dynamics of a negative feedback loop that consisted of a T7 lysozyme that binds to T7 RNA polymerase and inhibits T7 RNAP transcription activities (Fig. 3c). We assume that binding between T7 RNAP and PT7 would be enhanced at a smaller crowding density relative to the binding between T7 RNAP and T7 Lysozyme, which is based on existing theories of molecular crowding 20. As a result, our model predicts that the negative feedback loop would cause biphasic expression rates in environments with large crowding molecules (Fig. 3d, black line). Our experimental results support the model prediction as expression rates were maximized at 8% crowding density (Fig. 3d, black squares). We note that this was not due to increased metabolic burden in the expression systems (Supplementary Fig. 10a). To further corroborate our model, we investigated the impact of a reduced T7 RNAP concentration on gene expression rates. Based on our model, we expected similar biphasic gene expression rates with a negligible shift in the crowding density that maximizes gene expression rates (Supplementary Fig. 10b, black line). To test the prediction, we modulated the amount of T7RNAP by controlling the ratio of cell free expression systems that either contains T7 RNAP or does not contain T7 RNAP, which resulted in a 75% reduction of gene expression rates. Based on this system, we observed the same biphasic gene expression rates with a maximum rate at 8% crowding density (Supplementary Fig. 10b, black squares).
Reaction volumes modulate the impact of molecular crowding on gene expression
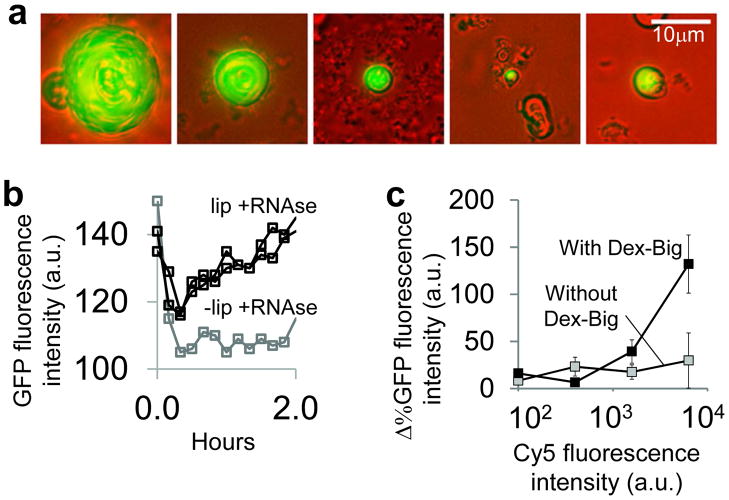
In natural cells, compartmentalization is the hallmark for creating unique functional units such as in Golgi, nucleus, and mitochondria. Based on our results with cell free systems, we hypothesized that crowding could similarly affect gene expression in physiologically-relevant volumes. To test this hypothesis, we constructed artificial cells that consist of phospholipid membranes, synthetic expression systems, and a genetic construct (Fig. 4a & Supplementary Fig. 11). We created artificial cells of different radii ranging from 100nm-10μm that expressed green fluorescent proteins (GFP) using a genetic construct. To ensure that GFP was expressed inside artificial cells, we added RNAse to inhibit RNA synthesis outside artificial cells (Fig. 4b & S12a).Without liposomes, RNAse inhibited GFP expression (Fig. 4b); with liposomes that protect the enclosed volumes, RNAse did not inhibit GFP expression. Next, we investigated the influence of cell volume on the impact of molecular crowding. Specifically, we used the integrated intensity of a Cy5 fluorescent dye inside artificial cells as a surrogate of encapsulation volume (Supplementary Fig. 12b–d)39. Based on our results using cell-free expression systems that essentially resulted in semi-infinite reaction volumes, we expect that molecular crowding would exert a larger impact on gene expression in large volumes as compared to small volumes. Indeed, we observed that Dex-Big increased GFP expression in artificial cells of large volumes as compared to artificial cells without Dex-Big (Fig. 4c, fluorescence intensity of Cy5>2000a.u. & Supplementary Fig. 12e). This is likely due to reduced reaction rates in large volumes without molecular crowding. As the size of artificial cells decreased, GFP levels in artificial cells with Dex-Big approached GFP levels of artificial cells without Dex-Big. These findings indicate that in our synthetic cellular systems, compartmentalization and crowding can play essential roles in controlling gene expression, factors that need to be considered in both mimicking genetic systems for applications in synthetic biology and accurately modeling cellular biochemistry for systems biology.
Figure 4. Volume-dependent impact of molecular crowding in artificial cells.
a. Fluorescence images of artificial cells. The artificial cells encapsulated a synthetic genetic construct that expressed green fluorescent proteins (GFP) within lipid bilayers. See SI for experimental protocol. The scale bar in the last panel applies to all panels.
b. Temporal dynamics of GFP expression. With liposomes (black squares), the fluorescence intensity of GFP increased after the 30th minute. In contrast, without liposomes (grey squares), the fluorescence intensity of GFP remained constant. RNAse A inhibited gene expression outside artificial cells by degrading RNAs. RNAse A was added in the media to ensure that the observed GFP expression inside the artificial cells was not due to either transport of GFP from the outside of artificial cells or attachment of GFP on the surfaces of artificial cells.
c. GFP expression inside artificial cells with and without Dex-Big crowding molecules. Molecular crowding enhanced gene expression in large artificial cells (fluorescence intensity of Cy5>2000a.u.). Each Δ%GFP intensity was calculated by using the percentage differences between mean GFP fluorescence intensity of artificial cells with and without pT7-GFP plasmids (SI). Each error bar represents one SEM of five replicates.
Discussion
In this study, we demonstrated that molecular crowding increases robustness of gene expression and that molecular crowding can be harnessed for the control of a basic genetic construct in artificial cells. We have also shown the impact of molecular crowding on the dynamics of T7 RNAP and how the influence of molecular crowding on gene expression rates can be modulated using genetic components, a negative feedback loop, and the size of crowding molecules. Our results suggest that the construction of both in vitro circuits 4 and programmable artificial cells 13 would have to take into account molecular crowding effects, which would be important in modulating system dynamics. Our findings underscore how scientists could harness molecular crowding for developing advantages in engineered cell functions such as gene expression, metabolic pathways, and cellular computing. For natural systems 36, our conclusions with respect to the impact of molecular crowding sizes, strength of genetic components, and network architectures could potentially explain how gene circuit dynamics are modulated by changing environments. Our studies would also impact in vitro studies of gene expression and their extension to in vivo environments that are crowded with molecules. As we move towards a bottom up and a priori approach of constructing either artificial cells or cells with completely designed genomes, it is critical to understand the impact of crowded environments on system robustness and then exploit these design principles to our advantage just as natural cells have already accomplished. Furthermore, systems biology has begun to take advantage of synthetic cellular systems as high throughput methods to characterize interactions between cellular components 40. In this context, it is critical to understand how molecular crowding can fundamentally impact interactions between cellular components and generate emergent dynamics that cannot be predicted or controlled without considering molecular crowding.
Supplementary Material
Acknowledgments
We thank members of the LeDuc and Schwartz labs for discussions and comments. We are grateful to Dr. Robert Murphy, Dr. Aaron Mitchell, Dr. Bruce Armitage, Dr. Tina Lee, Dr. Frederick Lanni, and Molecular Biosensor and Imaging Center for providing access to equipment. This work was partially supported by Lane Postdoctoral Fellowship (C.T.), Society in Science - Branco Weiss Fellowship (C.T.), NIH 1R01GM086327 (M.B.), NIH 8U54GM103529 (M.B), NIH 1R01AI076318 (R.S), NIH 1R01CA140214 (R.S), NSF CMMI-1100430 (P.L.), and NSF CPS- 1135850 (P.L.).
Footnotes
AUTHOR CONTRIBUTIONS
CT, SS, RS, and PL conceived research and designed experiments. CT and SS performed experiments. CT performed modeling analysis. CT, SS, MB, and PL provided materials and reagents. CT, RS, and PL interpreted results and wrote the manuscript with critical inputs from SS and MB. All authors approved the manuscript.
COMPETING FINANCIAL INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Gibson DG, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 2.Pinheiro VB, et al. Synthetic genetic polymers capable of heredity and evolution. Science. 2012;336:341–344. doi: 10.1126/science.1217622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nawroth JC, et al. A tissue-engineered jellyfish with biomimetic propulsion. Nat Biotechnol. 2012;30:792–797. doi: 10.1038/nbt.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J, Winfree E. Synthetic in vitro transcriptional oscillators. Mol Syst Biol. 2011;7:465. doi: 10.1038/msb.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandes R, Roy V, Wu HC, Bentley WE. Engineered biological nanofactories trigger quorum sensing response in targeted bacteria. Nat Nanotechnol. 2010;5:213–217. doi: 10.1038/nnano.2009.457. [DOI] [PubMed] [Google Scholar]
- 6.Murtas G, Kuruma Y, Bianchini P, Diaspro A, Luisi PL. Protein synthesis in liposomes with a minimal set of enzymes. Biochemical and biophysical research communications. 2007;363:12–17. doi: 10.1016/j.bbrc.2007.07.201. [DOI] [PubMed] [Google Scholar]
- 7.Schroeder A, et al. Remotely activated protein-producing nanoparticles. Nano Lett. 2012;12:2685–2689. doi: 10.1021/nl2036047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martino C, et al. Protein Expression, Aggregation, and Triggered Release from Polymersomes as Artificial Cell-like Structures. Angew Chem Int Edit. 2012;51:6416–6420. doi: 10.1002/anie.201201443. [DOI] [PubMed] [Google Scholar]
- 9.Ishikawa K, Sato K, Shima Y, Urabe I, Yomo T. Expression of a cascading genetic network within liposomes. FEBS letters. 2004;576:387–390. doi: 10.1016/j.febslet.2004.09.046. [DOI] [PubMed] [Google Scholar]
- 10.Leduc PR, et al. Towards an in vivo biologically inspired nanofactory. Nat Nanotechnol. 2007;2:3–7. doi: 10.1038/nnano.2006.180. [DOI] [PubMed] [Google Scholar]
- 11.Gardner PM, Winzer K, Davis BG. Sugar synthesis in a protocellular model leads to a cell signalling response in bacteria. Nature Chemistry. 2009;1:377–383. doi: 10.1038/nchem.296. [DOI] [PubMed] [Google Scholar]
- 12.Mansy SS, et al. Template-directed synthesis of a genetic polymer in a model protocell. Nature. 2008;454:122–125. doi: 10.1038/nature07018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noireaux V, Maeda YT, Libchaber A. Development of an artificial cell, from self-organization to computation and self-reproduction. Proc Natl Acad Sci U S A. 2011;108:3473–3480. doi: 10.1073/pnas.1017075108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang TM. Therapeutic applications of polymeric artificial cells. Nat Rev Drug Discov. 2005;4:221–235. doi: 10.1038/nrd1659. [DOI] [PubMed] [Google Scholar]
- 15.Jewett MC, Calhoun KA, Voloshin A, Wuu JJ, Swartz JR. An integrated cell-free metabolic platform for protein production and synthetic biology. Mol Syst Biol. 2008;4:220. doi: 10.1038/msb.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 17.Morelli MJ, Allen RJ, Wolde PR. Effects of macromolecular crowding on genetic networks. Biophys J. 2011;101:2882–2891. doi: 10.1016/j.bpj.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmerman SB, Trach SO. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J Mol Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 19.Minton AP. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J Biol Chem. 2001;276:10577–10580. doi: 10.1074/jbc.R100005200. [DOI] [PubMed] [Google Scholar]
- 20.Minton AP. The Effect of Volume Occupancy Upon the Thermodynamic Activity of Proteins - Some Biochemical Consequences. Mol Cell Biochem. 1983;55:119–140. doi: 10.1007/BF00673707. [DOI] [PubMed] [Google Scholar]
- 21.Li GW, Berg OG, Elf J. Effects of macromolecular crowding and DNA looping on gene regulation kinetics. Nature Physics. 2009;5:294–297. [Google Scholar]
- 22.Beg QK, et al. Intracellular crowding defines the mode and sequence of substrate uptake by Escherichia coli and constrains its metabolic activity. P Natl Acad Sci USA. 2007;104:12663–12668. doi: 10.1073/pnas.0609845104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richter K, Nessling M, Lichter P. Experimental evidence for the influence of molecular crowding on nuclear architecture. J Cell Sci. 2007;120:1673–1680. doi: 10.1242/jcs.03440. [DOI] [PubMed] [Google Scholar]
- 24.Elcock AH. Models of macromolecular crowding effects and the need for quantitative comparisons with experiment. Curr Opin Struct Biol. 2010;20:196–206. doi: 10.1016/j.sbi.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoen I, Krammer H, Braun D. Hybridization kinetics is different inside cells. P Natl Acad Sci USA. 2009;106:21649–21654. doi: 10.1073/pnas.0901313106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillip Y, Sherman E, Haran G, Schreiber G. Common Crowding Agents Have Only a Small Effect on Protein-Protein Interactions. Biophysical Journal. 2009;97:875–885. doi: 10.1016/j.bpj.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mika JT, Poolman B. Macromolecule diffusion and confinement in prokaryotic cells. Curr Opin Biotechnol. 2011;22:117–126. doi: 10.1016/j.copbio.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 28.Laurent TC. The Interaction between Polysaccharides and Other Macromolecules. 5. The Solubility of Proteins in the Presence of Dextran. Biochem J. 1963;89:253–257. doi: 10.1042/bj0890253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feder TJ, Brust-Mascher I, Slattery JP, Baird B, Webb WW. Constrained diffusion or immobile fraction on cell surfaces: a new interpretation. Biophys J. 1996;70:2767–2773. doi: 10.1016/S0006-3495(96)79846-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedman LJ, Gelles J. Mechanism of transcription initiation at an activator-dependent promoter defined by single-molecule observation. Cell. 2012;148:679–689. doi: 10.1016/j.cell.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Guo L, Golding I, Cox EC, Ong NP. Quantitative transcription factor binding kinetics at the single-molecule level. Biophys J. 2009;96:609–620. doi: 10.1016/j.bpj.2008.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bancaud A, et al. Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. EMBO J. 2009;28:3785–3798. doi: 10.1038/emboj.2009.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burg MB, Kwon ED, Kultz D. Osmotic regulation of gene expression. FASEB J. 1996;10:1598–1606. doi: 10.1096/fasebj.10.14.9002551. [DOI] [PubMed] [Google Scholar]
- 34.Martin CT, Coleman JE. Kinetic analysis of T7 RNA polymerase-promoter interactions with small synthetic promoters. Biochemistry. 1987;26:2690–2696. doi: 10.1021/bi00384a006. [DOI] [PubMed] [Google Scholar]
- 35.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 36.Spitzer J, Poolman B. The role of biomacromolecular crowding, ionic strength, and physicochemical gradients in the complexities of life’s emergence. Microbiology and molecular biology reviews : MMBR. 2009;73:371–388. doi: 10.1128/MMBR.00010-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen HZ, Zubay G. Prokaryotic coupled transcription-translation. Methods Enzymol. 1983;101:674–690. doi: 10.1016/0076-6879(83)01047-2. [DOI] [PubMed] [Google Scholar]
- 38.Milo R, et al. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 39.Sunami T, et al. Femtoliter compartment in liposomes for in vitro selection of proteins. Anal Biochem. 2006;357:128–136. doi: 10.1016/j.ab.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 40.Harris DC, Jewett MC. Cell-free biology: exploiting the interface between synthetic biology and synthetic chemistry. Curr Opin Biotechnol. 2012;23:672–678. doi: 10.1016/j.copbio.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.