Abstract
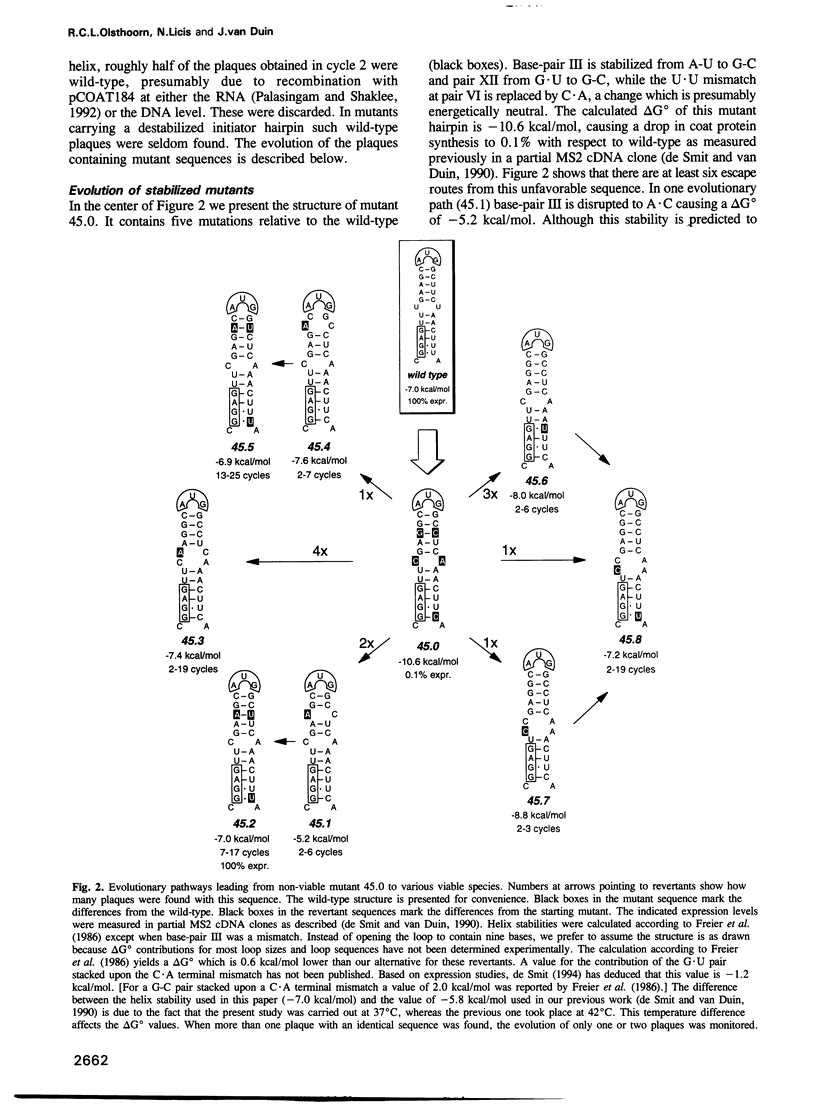
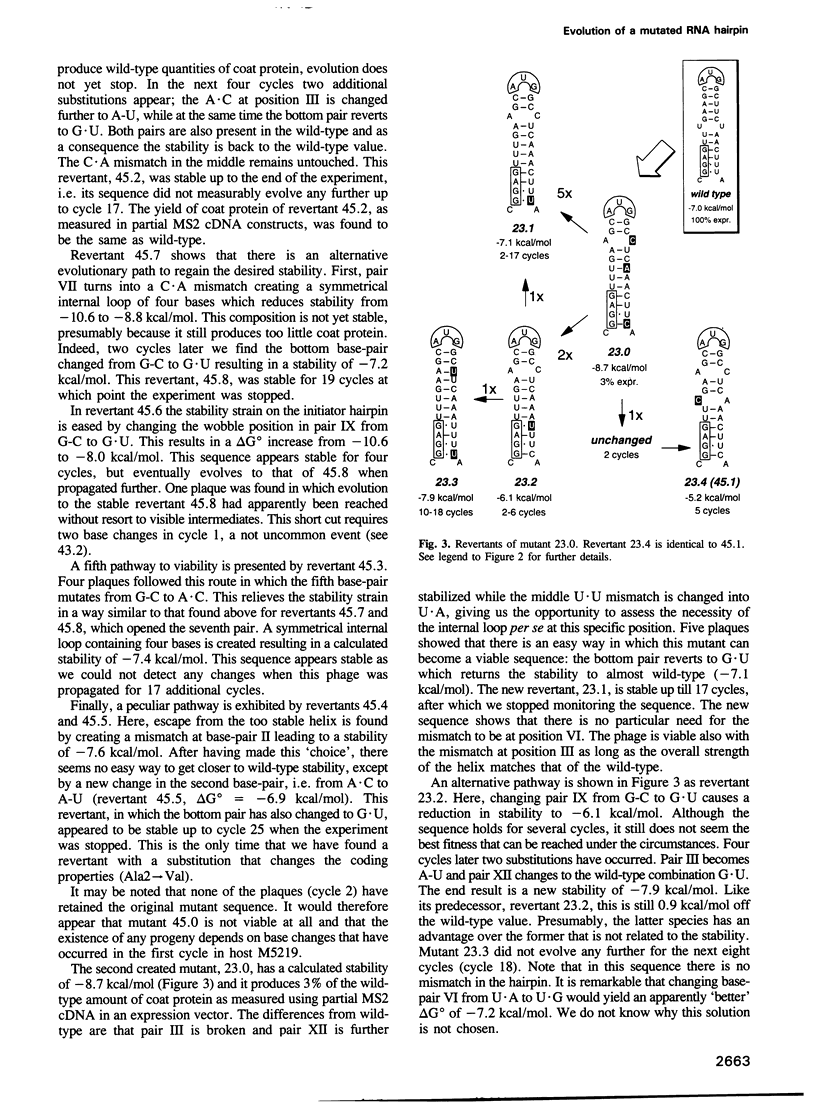
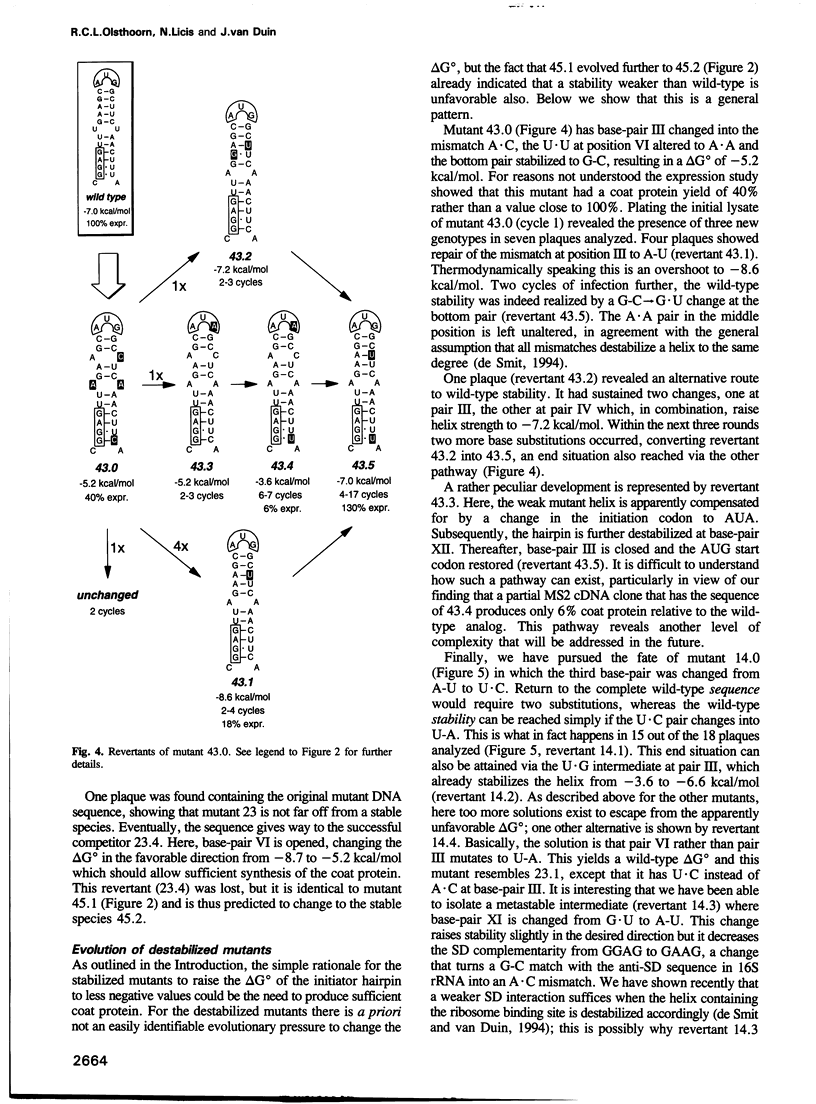
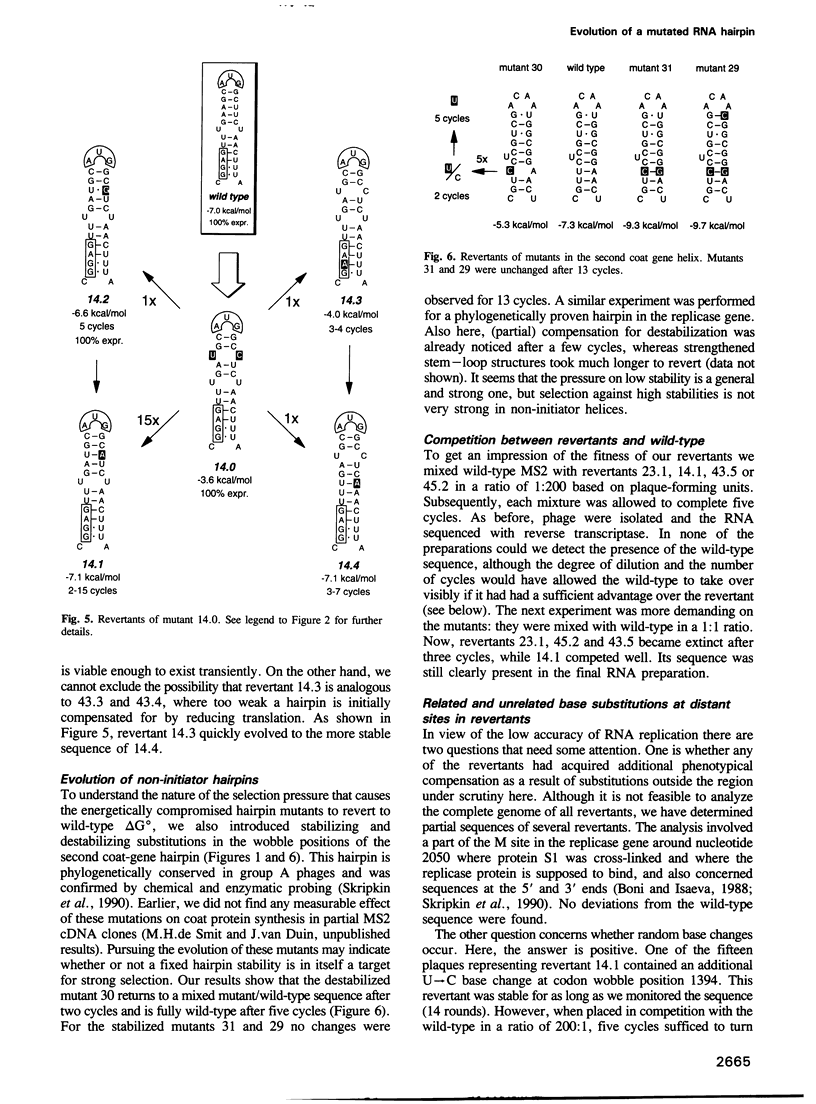
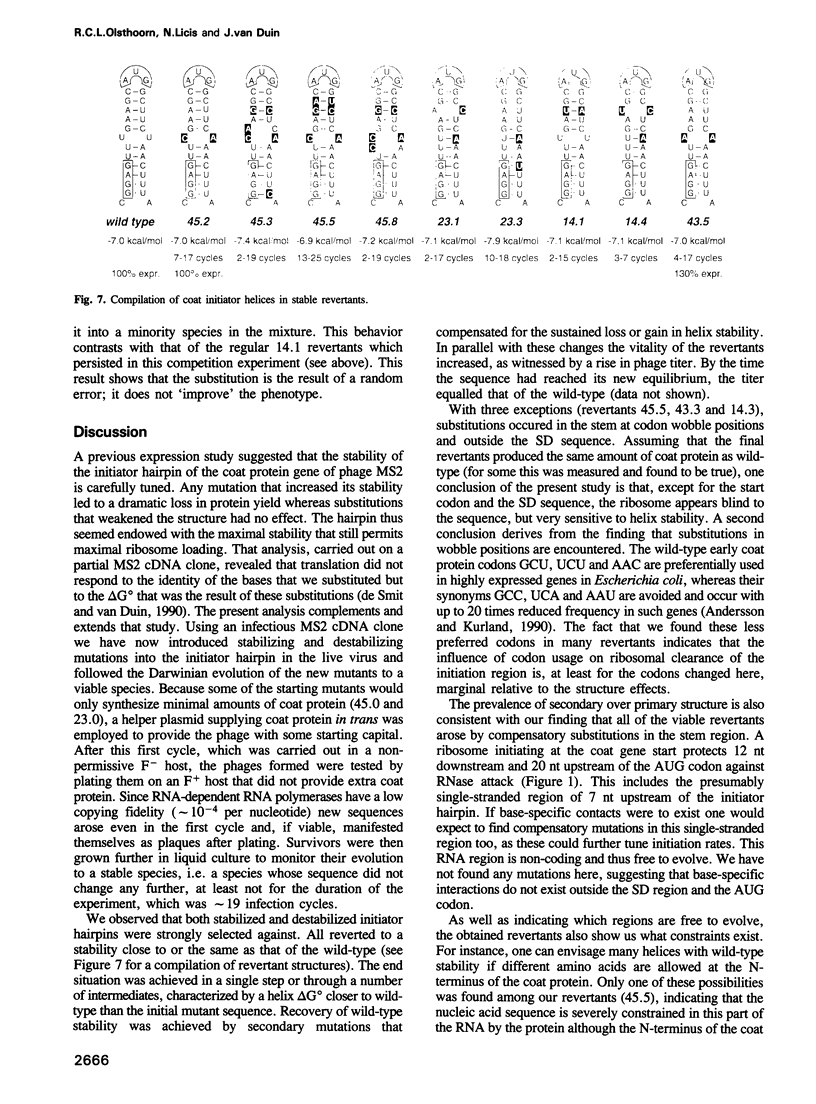
The start of the coat protein gene of RNA phage MS2 adopts a well-defined hairpin structure of 12 bp (including one mismatch) in which the start codon occupies the loop position. An earlier expression study using partial MS2 cDNA clones had indicated that the stability of this hairpin is important for gene expression. For every -1.4 kcal/mol increase in stability a 10-fold reduction in coat protein was obtained. Destabilizations beyond the wild-type value did not affect expression. These results suggested that the hairpin was tuned in the sense that it has the highest stability still compatible with maximal ribosome loading. Employing an infectious MS2 cDNA clone, we have now tested the prediction that the delta G 0 of the coat protein initiator helix is set at a precise value. We have introduced stabilizing and destabilizing mutations into this hairpin in the intact phage and monitored their evolution to viable species. By compensatory mutations, both types of mutants quickly revert along various pathways to wild-type stability, but not to wild-type sequence. As a rule the second-site mutations do not change the encoded amino acids or the Shine-Dalgarno sequence. The return of too strong hairpins to wild-type stability can be understood from the need to produce adequate supplies of coat protein. The return of unstable hairpins to wild-type stability is not self-evident and is presently not understood. The revertants provide an evolutionary landscape of slightly suboptimal phages, that were stable at least for the duration of the experiment (approximately 20 infection cycles).(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersson S. G., Kurland C. G. Codon preferences in free-living microorganisms. Microbiol Rev. 1990 Jun;54(2):198–210. doi: 10.1128/mr.54.2.198-210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod V. D., Brown E., Priano C., Mills D. R. Coliphage Q beta RNA replication: RNA catalytic for single-strand release. Virology. 1991 Oct;184(2):595–608. doi: 10.1016/0042-6822(91)90430-j. [DOI] [PubMed] [Google Scholar]
- Barrera I., Schuppli D., Sogo J. M., Weber H. Different mechanisms of recognition of bacteriophage Q beta plus and minus strand RNAs by Q beta replicase. J Mol Biol. 1993 Jul 20;232(2):512–521. doi: 10.1006/jmbi.1993.1407. [DOI] [PubMed] [Google Scholar]
- Biebricher C. K., Luce R. In vitro recombination and terminal elongation of RNA by Q beta replicase. EMBO J. 1992 Dec;11(13):5129–5135. doi: 10.1002/j.1460-2075.1992.tb05620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni I. V., Isaeva D. M. Lokalizatsiia uchastkov RNK bakteriofagov Qbeta, fr i MS2, vovlechennykh vo vzaimodeistvie s ribosomnym belkom S1 pri obrazovanii kompleksov s 30S ribosomnoi subchastitsei. Dokl Akad Nauk SSSR. 1988;298(4):1015–1018. [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisez Y., Robbens J., Remaut E., Fiers W. Folding of the MS2 coat protein in Escherichia coli is modulated by translational pauses resulting from mRNA secondary structure and codon usage: a hypothesis. J Theor Biol. 1993 May 21;162(2):243–252. doi: 10.1006/jtbi.1993.1085. [DOI] [PubMed] [Google Scholar]
- James B. D., Olsen G. J., Pace N. R. Phylogenetic comparative analysis of RNA secondary structure. Methods Enzymol. 1989;180:227–239. doi: 10.1016/0076-6879(89)80104-1. [DOI] [PubMed] [Google Scholar]
- Macadam A. J., Ferguson G., Burlison J., Stone D., Skuce R., Almond J. W., Minor P. D. Correlation of RNA secondary structure and attenuation of Sabin vaccine strains of poliovirus in tissue culture. Virology. 1992 Aug;189(2):415–422. doi: 10.1016/0042-6822(92)90565-7. [DOI] [PubMed] [Google Scholar]
- Meyer F., Weber H., Weissmann C. Interactions of Q beta replicase with Q beta RNA. J Mol Biol. 1981 Dec 15;153(3):631–660. doi: 10.1016/0022-2836(81)90411-3. [DOI] [PubMed] [Google Scholar]
- Mills D. R., Dobkin C., Kramer F. R. Template-determined, variable rate of RNA chain elongation. Cell. 1978 Oct;15(2):541–550. doi: 10.1016/0092-8674(78)90022-3. [DOI] [PubMed] [Google Scholar]
- Palasingam K., Shaklee P. N. Reversion of Q beta RNA phage mutants by homologous RNA recombination. J Virol. 1992 Apr;66(4):2435–2442. doi: 10.1128/jvi.66.4.2435-2442.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody D. S. The RNA binding site of bacteriophage MS2 coat protein. EMBO J. 1993 Feb;12(2):595–600. doi: 10.1002/j.1460-2075.1993.tb05691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priano C., Kramer F. R., Mills D. R. Evolution of the RNA coliphages: the role of secondary structures during RNA replication. Cold Spring Harb Symp Quant Biol. 1987;52:321–330. doi: 10.1101/sqb.1987.052.01.037. [DOI] [PubMed] [Google Scholar]
- Remaut E., Stanssens P., Fiers W. Plasmid vectors for high-efficiency expression controlled by the PL promoter of coliphage lambda. Gene. 1981 Oct;15(1):81–93. doi: 10.1016/0378-1119(81)90106-2. [DOI] [PubMed] [Google Scholar]
- Remaut E., Waele P. D., Marmenout A., Stanssens P., Fiers W. Functional expression of individual plasmid-coded RNA bacteriophage MS2 genes. EMBO J. 1982;1(2):205–209. doi: 10.1002/j.1460-2075.1982.tb01148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaklee P. N., Miglietta J. J., Palmenberg A. C., Kaesberg P. Infectious positive- and negative-strand transcript RNAs from bacteriophage Q beta cDNA clones. Virology. 1988 Mar;163(1):209–213. doi: 10.1016/0042-6822(88)90250-4. [DOI] [PubMed] [Google Scholar]
- Shaklee P. N. Negative-strand RNA replication by Q beta and MS2 positive-strand RNA phage replicases. Virology. 1990 Sep;178(1):340–343. doi: 10.1016/0042-6822(90)90417-p. [DOI] [PubMed] [Google Scholar]
- Skripkin E. A., Adhin M. R., de Smit M. H., van Duin J. Secondary structure of the central region of bacteriophage MS2 RNA. Conservation and biological significance. J Mol Biol. 1990 Jan 20;211(2):447–463. doi: 10.1016/0022-2836(90)90364-R. [DOI] [PubMed] [Google Scholar]
- Skripkin E. A., Jacobson A. B. A two-dimensional model at the nucleotide level for the central hairpin of coliphage Q beta RNA. J Mol Biol. 1993 Sep 20;233(2):245–260. doi: 10.1006/jmbi.1993.1503. [DOI] [PubMed] [Google Scholar]
- Steitz J. A. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature. 1969 Dec 6;224(5223):957–964. doi: 10.1038/224957a0. [DOI] [PubMed] [Google Scholar]
- Taniguchi T., Palmieri M., Weissmann C. QB DNA-containing hybrid plasmids giving rise to QB phage formation in the bacterial host. Nature. 1978 Jul 20;274(5668):223–228. doi: 10.1038/274223a0. [DOI] [PubMed] [Google Scholar]
- Tsai C. H., Dreher T. W. Second-site suppressor mutations assist in studying the function of the 3' noncoding region of turnip yellow mosaic virus RNA. J Virol. 1992 Sep;66(9):5190–5199. doi: 10.1128/jvi.66.9.5190-5199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valegård K., Liljas L., Fridborg K., Unge T. The three-dimensional structure of the bacterial virus MS2. Nature. 1990 May 3;345(6270):36–41. doi: 10.1038/345036a0. [DOI] [PubMed] [Google Scholar]
- Varenne S., Buc J., Lloubes R., Lazdunski C. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J Mol Biol. 1984 Dec 15;180(3):549–576. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- Witherell G. W., Gott J. M., Uhlenbeck O. C. Specific interaction between RNA phage coat proteins and RNA. Prog Nucleic Acid Res Mol Biol. 1991;40:185–220. doi: 10.1016/s0079-6603(08)60842-9. [DOI] [PubMed] [Google Scholar]
- de Smit M. H., van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smit M. H., van Duin J. Translational initiation at the coat-protein gene of phage MS2: native upstream RNA relieves inhibition by local secondary structure. Mol Microbiol. 1993 Sep;9(5):1079–1088. doi: 10.1111/j.1365-2958.1993.tb01237.x. [DOI] [PubMed] [Google Scholar]
- de Smit M. H., van Duin J. Translational initiation on structured messengers. Another role for the Shine-Dalgarno interaction. J Mol Biol. 1994 Jan 7;235(1):173–184. doi: 10.1016/s0022-2836(05)80024-5. [DOI] [PubMed] [Google Scholar]
- van Himbergen J., van Geffen B., van Duin J. Translational control by a long range RNA-RNA interaction; a basepair substitution analysis. Nucleic Acids Res. 1993 Apr 25;21(8):1713–1717. doi: 10.1093/nar/21.8.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]