Abstract
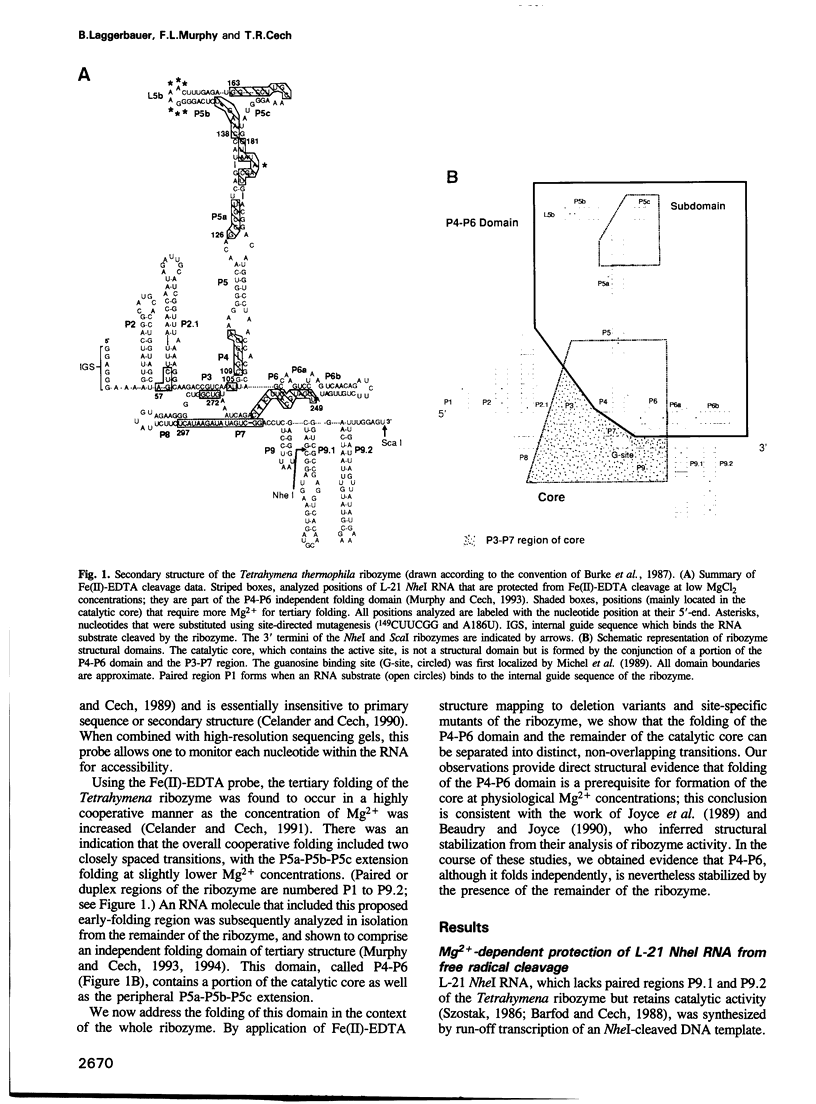
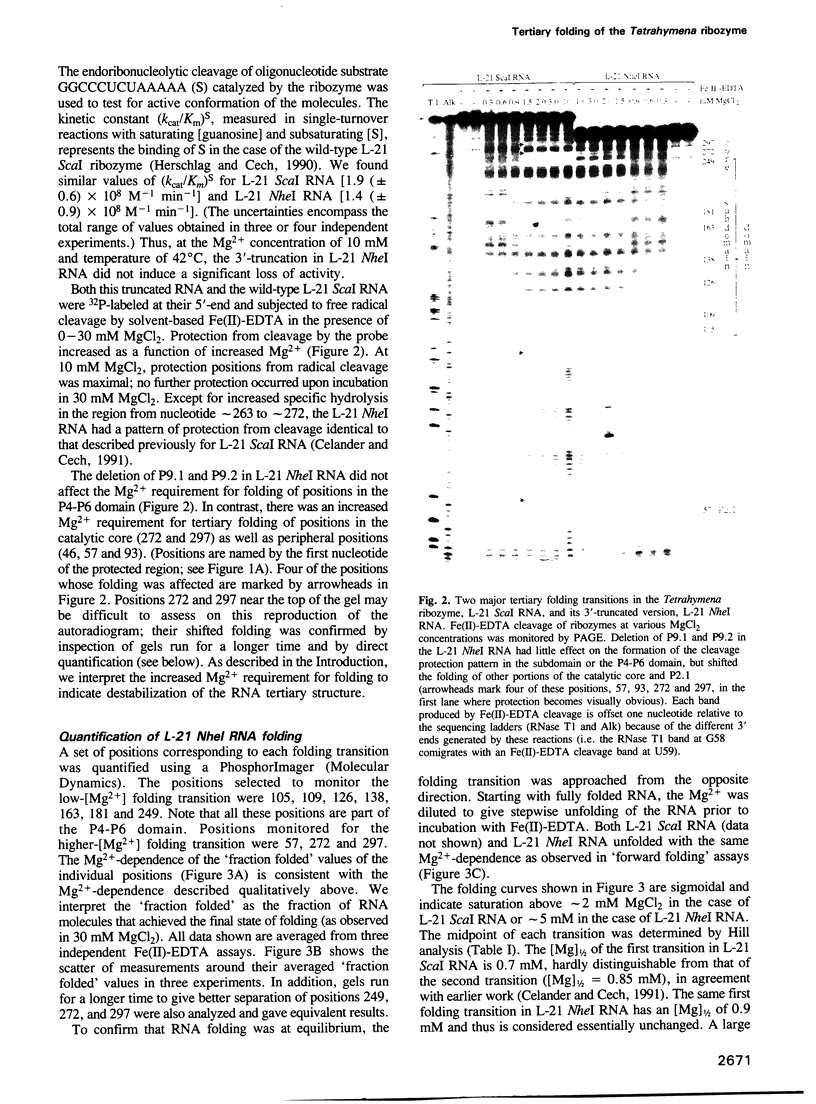
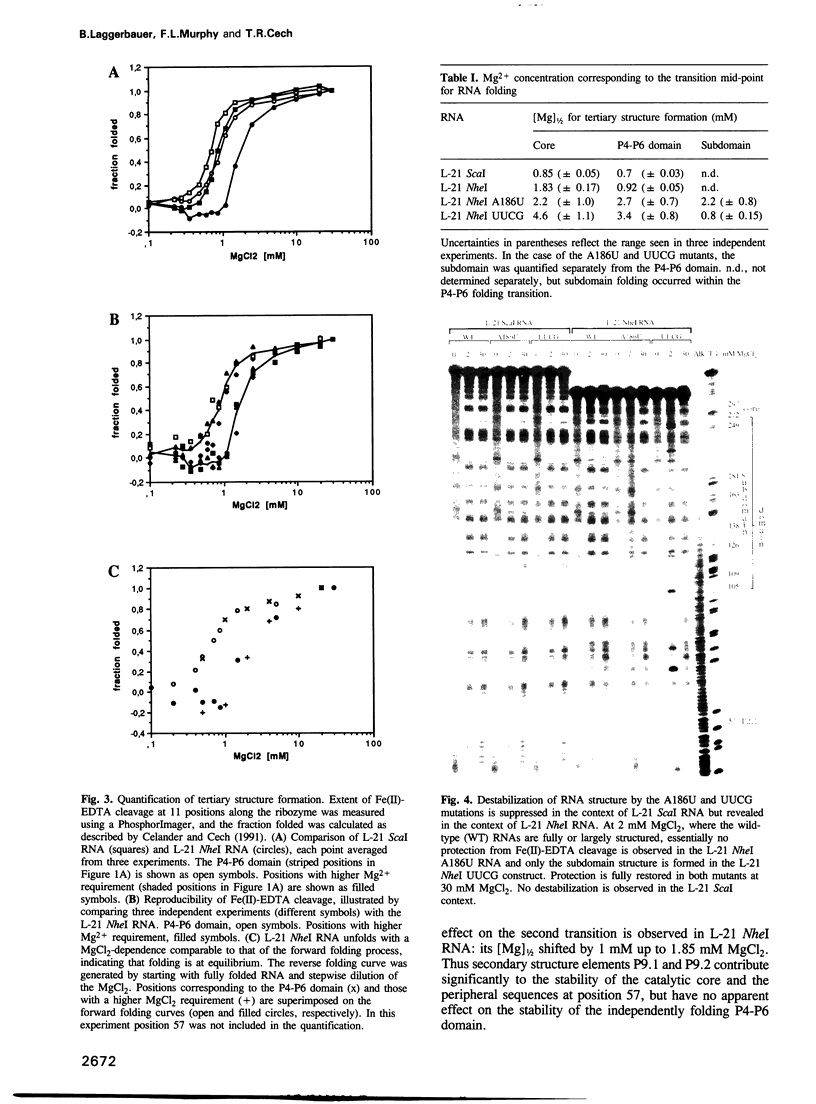
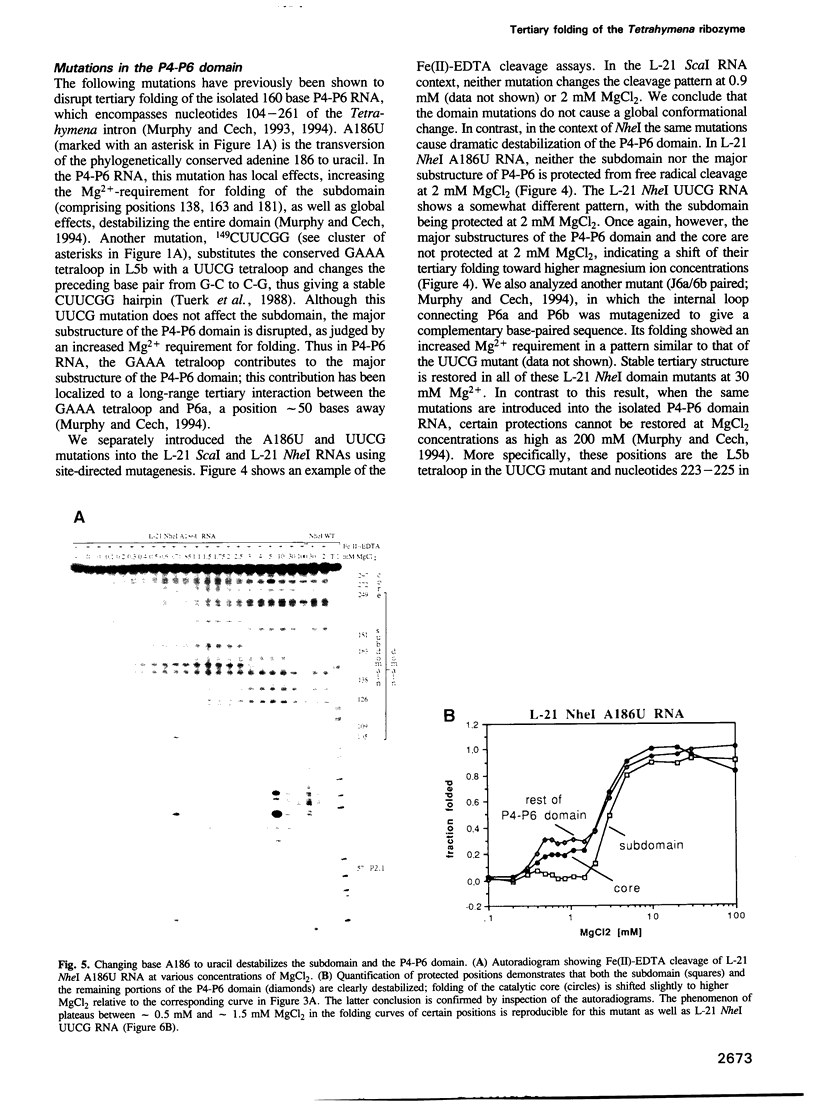
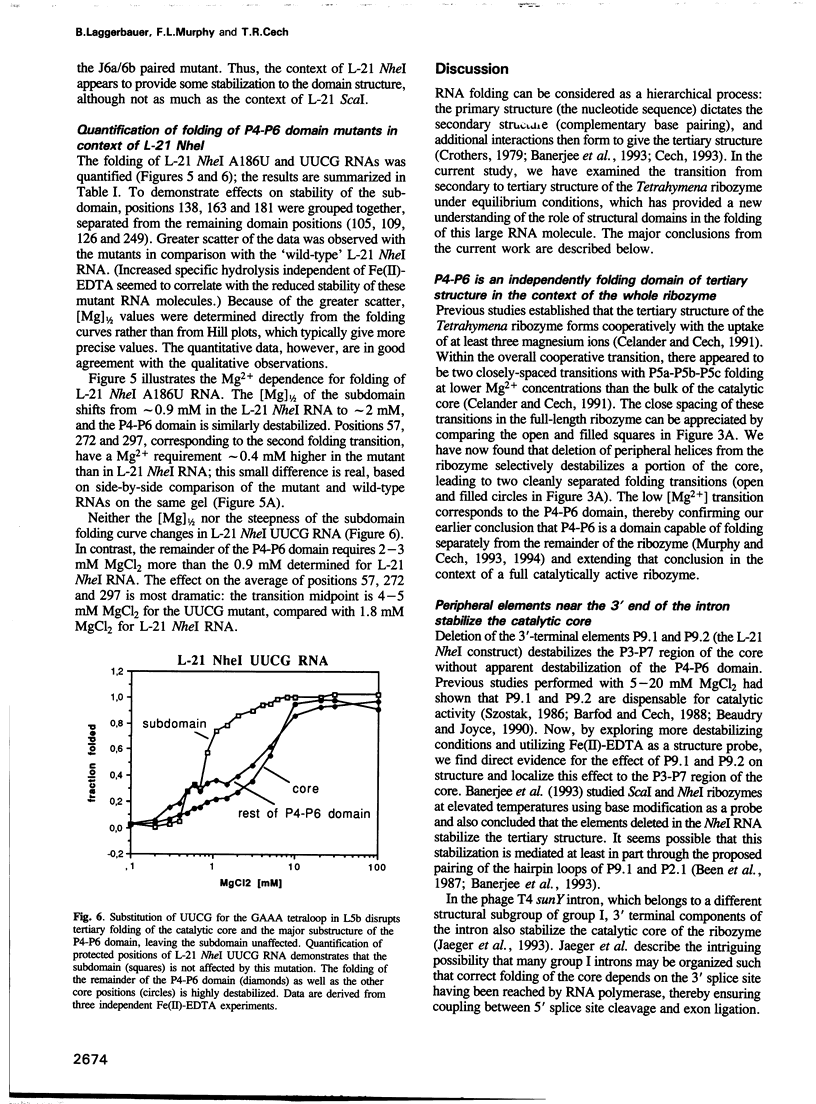
The L-21 Tetrahymena ribozyme, an RNA molecule with sequence-specific endoribonuclease activity derived from a self-splicing group I intron, provides a model system for studying the RNA folding problem. A 160 nucleotide, independently folding domain of tertiary structure (the P4-P6 domain) comprises about half of the ribozyme. We now apply Fe(II)-EDTA cleavage to mutants of the ribozyme to explore the role of individual structural elements in tertiary folding of the RNA at equilibrium. Deletion of peripheral elements near the 3' end of the ribozyme destabilizes a region of the catalytic core (P3-P7) without altering the folding of the P4-P6 domain. Three different mutations within the P4-P6 domain that destabilize its folding also shift the folding of the P3-P7 region of the catalytic core to higher MgCl2 concentrations. We conclude that the role of the extended P4-P6 domain and of the 3'-terminal peripheral elements is at least in part to stabilize the catalytic core. The organization of RNA into independently folding domains of tertiary structure may be common in large RNAs, including ribosomal RNAs. Furthermore, the observation of domain-domain interactions in a catalytic RNA supports the feasibility of a primitive spliceosome without any proteins.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerjee A. R., Jaeger J. A., Turner D. H. Thermal unfolding of a group I ribozyme: the low-temperature transition is primarily disruption of tertiary structure. Biochemistry. 1993 Jan 12;32(1):153–163. doi: 10.1021/bi00052a021. [DOI] [PubMed] [Google Scholar]
- Barfod E. T., Cech T. R. Deletion of nonconserved helices near the 3' end of the rRNA intron of Tetrahymena thermophila alters self-splicing but not core catalytic activity. Genes Dev. 1988 Jun;2(6):652–663. doi: 10.1101/gad.2.6.652. [DOI] [PubMed] [Google Scholar]
- Beaudry A. A., Joyce G. F. Minimum secondary structure requirements for catalytic activity of a self-splicing group I intron. Biochemistry. 1990 Jul 10;29(27):6534–6539. doi: 10.1021/bi00479a027. [DOI] [PubMed] [Google Scholar]
- Been M. D., Barfod E. T., Burke J. M., Price J. V., Tanner N. K., Zaug A. J., Cech T. R. Structures involved in Tetrahymena rRNA self-splicing and RNA enzyme activity. Cold Spring Harb Symp Quant Biol. 1987;52:147–157. doi: 10.1101/sqb.1987.052.01.019. [DOI] [PubMed] [Google Scholar]
- Been M. D., Cech T. R. Selection of circularization sites in a group I IVS RNA requires multiple alignments of an internal template-like sequence. Cell. 1987 Sep 11;50(6):951–961. doi: 10.1016/0092-8674(87)90522-8. [DOI] [PubMed] [Google Scholar]
- Belfort M. An expanding universe of introns. Science. 1993 Nov 12;262(5136):1009–1010. doi: 10.1126/science.7694364. [DOI] [PubMed] [Google Scholar]
- Burke J. M., Belfort M., Cech T. R., Davies R. W., Schweyen R. J., Shub D. A., Szostak J. W., Tabak H. F. Structural conventions for group I introns. Nucleic Acids Res. 1987 Sep 25;15(18):7217–7221. doi: 10.1093/nar/15.18.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J. M., Irvine K. D., Kaneko K. J., Kerker B. J., Oettgen A. B., Tierney W. M., Williamson C. L., Zaug A. J., Cech T. R. Role of conserved sequence elements 9L and 2 in self-splicing of the Tetrahymena ribosomal RNA precursor. Cell. 1986 Apr 25;45(2):167–176. doi: 10.1016/0092-8674(86)90380-6. [DOI] [PubMed] [Google Scholar]
- Cech T. R. Conserved sequences and structures of group I introns: building an active site for RNA catalysis--a review. Gene. 1988 Dec 20;73(2):259–271. doi: 10.1016/0378-1119(88)90492-1. [DOI] [PubMed] [Google Scholar]
- Cech T. R. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- Celander D. W., Cech T. R. Iron(II)-ethylenediaminetetraacetic acid catalyzed cleavage of RNA and DNA oligonucleotides: similar reactivity toward single- and double-stranded forms. Biochemistry. 1990 Feb 13;29(6):1355–1361. doi: 10.1021/bi00458a001. [DOI] [PubMed] [Google Scholar]
- Celander D. W., Cech T. R. Visualizing the higher order folding of a catalytic RNA molecule. Science. 1991 Jan 25;251(4992):401–407. doi: 10.1126/science.1989074. [DOI] [PubMed] [Google Scholar]
- Christian E. L., Yarus M. Metal coordination sites that contribute to structure and catalysis in the group I intron from Tetrahymena. Biochemistry. 1993 May 4;32(17):4475–4480. doi: 10.1021/bi00068a001. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor P. J., Flanegan J. B., Cech T. R. A conserved base pair within helix P4 of the Tetrahymena ribozyme helps to form the tertiary structure required for self-splicing. EMBO J. 1989 Nov;8(11):3391–3399. doi: 10.1002/j.1460-2075.1989.tb08503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans C. A., Cech T. R. A hammerhead ribozyme allows synthesis of a new form of the Tetrahymena ribozyme homogeneous in length with a 3' end blocked for transesterification. Nucleic Acids Res. 1991 Jul 25;19(14):3875–3880. doi: 10.1093/nar/19.14.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans C. A., Cech T. R. Metal ion requirements for sequence-specific endoribonuclease activity of the Tetrahymena ribozyme. Biochemistry. 1989 Aug 22;28(17):6888–6894. doi: 10.1021/bi00443a017. [DOI] [PubMed] [Google Scholar]
- Guo Q., Lambowitz A. M. A tyrosyl-tRNA synthetase binds specifically to the group I intron catalytic core. Genes Dev. 1992 Aug;6(8):1357–1372. doi: 10.1101/gad.6.8.1357. [DOI] [PubMed] [Google Scholar]
- Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991 Jul 12;253(5016):157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- Herschlag D., Cech T. R. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990 Nov 6;29(44):10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- Hertzberg R. P., Dervan P. B. Cleavage of DNA with methidiumpropyl-EDTA-iron(II): reaction conditions and product analyses. Biochemistry. 1984 Aug 14;23(17):3934–3945. doi: 10.1021/bi00312a022. [DOI] [PubMed] [Google Scholar]
- Holbrook S. R., Sussman J. L., Warrant R. W., Church G. M., Kim S. H. RNA-ligant interactions. (I) Magnesium binding sites in yeast tRNAPhe. Nucleic Acids Res. 1977 Aug;4(8):2811–2820. doi: 10.1093/nar/4.8.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack A., Ladner J. E., Rhodes D., Brown R. S., Klug A. A crystallographic study of metal-binding to yeast phenylalanine transfer RNA. J Mol Biol. 1977 Apr 15;111(3):315–328. doi: 10.1016/s0022-2836(77)80054-5. [DOI] [PubMed] [Google Scholar]
- Jaeger L., Westhof E., Michel F. Monitoring of the cooperative unfolding of the sunY group I intron of bacteriophage T4. The active form of the sunY ribozyme is stabilized by multiple interactions with 3' terminal intron components. J Mol Biol. 1993 Nov 20;234(2):331–346. doi: 10.1006/jmbi.1993.1590. [DOI] [PubMed] [Google Scholar]
- Joyce G. F., van der Horst G., Inoue T. Catalytic activity is retained in the Tetrahymena group I intron despite removal of the large extension of element P5. Nucleic Acids Res. 1989 Oct 11;17(19):7879–7889. doi: 10.1093/nar/17.19.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Latham J. A., Cech T. R. Defining the inside and outside of a catalytic RNA molecule. Science. 1989 Jul 21;245(4915):276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- Latham J. A., Zaug A. J., Cech T. R. Self-splicing and enzymatic cleavage of RNA by a group I intervening sequence. Methods Enzymol. 1990;181:558–569. doi: 10.1016/0076-6879(90)81151-j. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W., Tollervey D., Séraphin B. Small nuclear RNAs in messenger RNA and ribosomal RNA processing. FASEB J. 1993 Jan;7(1):47–53. doi: 10.1096/fasebj.7.1.8422974. [DOI] [PubMed] [Google Scholar]
- Michel F., Hanna M., Green R., Bartel D. P., Szostak J. W. The guanosine binding site of the Tetrahymena ribozyme. Nature. 1989 Nov 23;342(6248):391–395. doi: 10.1038/342391a0. [DOI] [PubMed] [Google Scholar]
- Michel F., Jaeger L., Westhof E., Kuras R., Tihy F., Xu M. Q., Shub D. A. Activation of the catalytic core of a group I intron by a remote 3' splice junction. Genes Dev. 1992 Aug;6(8):1373–1385. doi: 10.1101/gad.6.8.1373. [DOI] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990 Dec 5;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Mohr G., Zhang A., Gianelos J. A., Belfort M., Lambowitz A. M. The neurospora CYT-18 protein suppresses defects in the phage T4 td intron by stabilizing the catalytically active structure of the intron core. Cell. 1992 May 1;69(3):483–494. doi: 10.1016/0092-8674(92)90449-m. [DOI] [PubMed] [Google Scholar]
- Murphy F. L., Cech T. R. An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry. 1993 May 25;32(20):5291–5300. doi: 10.1021/bi00071a003. [DOI] [PubMed] [Google Scholar]
- Murphy F. L., Cech T. R. GAAA tetraloop and conserved bulge stabilize tertiary structure of a group I intron domain. J Mol Biol. 1994 Feb 11;236(1):49–63. doi: 10.1006/jmbi.1994.1117. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Hoffarth V., Zimniak L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992 Jun 5;256(5062):1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Piccirilli J. A., Vyle J. S., Caruthers M. H., Cech T. R. Metal ion catalysis in the Tetrahymena ribozyme reaction. Nature. 1993 Jan 7;361(6407):85–88. doi: 10.1038/361085a0. [DOI] [PubMed] [Google Scholar]
- Price J. V., Kieft G. L., Kent J. R., Sievers E. L., Cech T. R. Sequence requirements for self-splicing of the Tetrahymena thermophila pre-ribosomal RNA. Nucleic Acids Res. 1985 Mar 25;13(6):1871–1889. doi: 10.1093/nar/13.6.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich C., Olsen G. J., Pace B., Pace N. R. Role of the protein moiety of ribonuclease P, a ribonucleoprotein enzyme. Science. 1988 Jan 8;239(4836):178–181. doi: 10.1126/science.3122322. [DOI] [PubMed] [Google Scholar]
- Smith D., Burgin A. B., Haas E. S., Pace N. R. Influence of metal ions on the ribonuclease P reaction. Distinguishing substrate binding from catalysis. J Biol Chem. 1992 Feb 5;267(4):2429–2436. [PubMed] [Google Scholar]
- Szostak J. W. Enzymatic activity of the conserved core of a group I self-splicing intron. Nature. 1986 Jul 3;322(6074):83–86. doi: 10.1038/322083a0. [DOI] [PubMed] [Google Scholar]
- Tuerk C., Gauss P., Thermes C., Groebe D. R., Gayle M., Guild N., Stormo G., d'Aubenton-Carafa Y., Uhlenbeck O. C., Tinoco I., Jr CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Iron(II) EDTA used to measure the helical twist along any DNA molecule. Science. 1985 Nov 8;230(4726):679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- Wise J. A. Guides to the heart of the spliceosome. Science. 1993 Dec 24;262(5142):1978–1979. doi: 10.1126/science.8266091. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Been M. D., Cech T. R. The Tetrahymena ribozyme acts like an RNA restriction endonuclease. Nature. 1986 Dec 4;324(6096):429–433. doi: 10.1038/324429a0. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Grosshans C. A., Cech T. R. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry. 1988 Dec 13;27(25):8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]
- van der Horst G., Christian A., Inoue T. Reconstitution of a group I intron self-splicing reaction with an activator RNA. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):184–188. doi: 10.1073/pnas.88.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]