Summary
Deregulated JAK2 signaling plays an important role in the pathogenesis of myeloproliferative neoplasms (MPN). We and others have shown constitutive activation of JAK2 and STAT3 in diffuse large B cell lymphomas (DLBCL). We sought to determine the mechanism of JAK2 signaling in DLBCL tumors with a genetic approach. The most common JAK2 activating mutation present in most MPNs is V617F (exon 14 within the pseudokinase-domain); however, this mutation is absent in lymphoid malignancies. We bi-directionally sequenced all the domains of the JAK2 gene and performed mutational analysis. No novel non-synonymous mutations were detected in the 40 DLBCL tumors tested. However synonymous and non-synonymous single nucleotide polymorphism (SNPs) were detected within the exons 6, 9 and 19 in the majority of patients. Taken together, these data suggest that other mechanisms for altered JAK2 signaling aside from activating JAK2 mutations are present in DLBCL. Targeting JAK2 activation could be an important therapeutic target for DLBCL. Indeed, the JAK2 inhibitor ruxolitinib is now approved for the treatment of MPN. Our study indicates that JAK2 targeted clinical trials in lymphoma should not be confined to only JAK2 mutation cases but rather based on pathway activation.
Emerging evidence indicates that JAK2-STAT3 signaling is frequent in DLBCL cell lines and half of the activated B cell –like (ABC) subtype of DLBCL tumors exhibits a STAT3 gene signature [1, 2]. We have recently demonstrated that 52% of DLBCL tumors exhibit pSTAT3 activation by use of immunohistochemical staining (Gupta. M, AACR 2012 Abstract). Our previous work demonstrated that JAK2 is constitutively activated in DLBCL tumors by cytokines [3]. While high serum IL-10 was associated with JAK2 activation, not all pJAK2+ cases showed high serum IL-10. We hypothesize that there are other mechanisms such as genetic mutation that are responsible for dysregulated JAK2-STAT3 signaling in these cases. For example in melanoma somatic mutations of B-RAF result in constitutive activation of Ras-ERK signaling [4] [5] In this report, we determined the underlying genetic mechanism for constitutive JAK2-STAT3 pathway signaling in DLBCL by evaluating activating JAK2 mutations in DLBCL tumors.
To date, most of the reported mutations in the JAK2 gene have been the 1849G>T in exon 14. This mutation results in an amino acid substitution of valine to phenylalanine (V617F) within the JH2 pseudokinase domain [6]. It occurs in approximately 95% of patients with polycythemia vera (PV), 55% of essential thrombocythemia (ET), and 65% in primary myelofibrosis (PMF) [7]. Studies of the JAK2 (V617F) mutation in other hematological diseases such as de novo acute myeloid leukemia, acute/chronic lymphocytic leukemia, classical Hodgkin lymphoma (cHL), primary mediastinal B cell lymphoma, follicular and mantle cell lymphoma have been negative [8] [9] [10] [11]. Despite the absence of JAK2 activating mutations in lymphoma cells, constitutive activation of the JAK/STAT signaling pathway have been reported in DLBCL cell lines [12] [1]. We hypothesized that lymphomas might harbor other JAK2 mutations in other important domains of the JAK2 gene such as FERM (part of the JH4-JH7), SH2 (JH3-part of the JH4), pseudokinase (JH2) and/or the kinase domains (JH1) [13]. In fact, mutations in the FERM domain (responsible for binding the cytoplasmic tails of cytokine receptor) have been described in 11% of patients with adult T-cell leukemia/lymphoma leading to a gain of function in JAK3 gene [14].
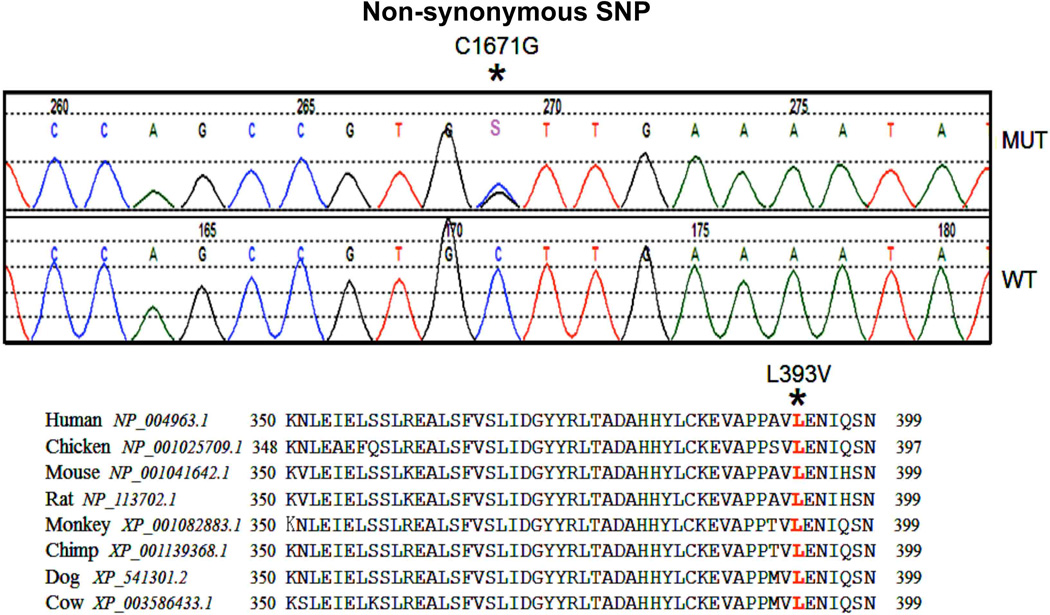
To explore this hypothesis, we PCR amplified and bidirectionally sequenced exons 1–25 (encompassing all seven JAK2 JH domains) of the JAK2 gene from 40 DLBCL tumors from the Iowa/Mayo Lymphoma SPORE Biobank. There were no novel non-synonymous mutations (missense mutations) found in these 40 DLBCL tumors; however, polymorphisms in the JAK2 gene and a series of SNPs (single nucleotide sequence) were found (Table 1). Fourteen DLBCL tumors in our cohort had point mutations within the FERM domain at exon 6; eight of these were heterozygous and six homozygous (C>T; Histidine163Histidine). This was a known SNP (rs10429491) with reported genotype frequency C/C-0.408; CT-0.452 and T/T-0.140. Another 12 patients had a heterozygous point mutation (G>A; Leucine830Leucine) in the pseudokinase domain at exon 19. This point mutation was also reported as a SNP (rs2274649) with A/A-0.483; AT-0.467; T/T-0.050 frequency. Both of these SNPs were synonymous (silent) mutations in that they do not predict an amino acid substitution (Table 1). Interestingly, a heterozygous cytosine to guanine (C>G) transition was identified at position 1671 in two patient specimens. This C>G transition predicts a missense substitution of leucine to valine in codon 393 (L393V) of the FERM domain in exon 9 (Table 1; Figure 1). This JAK2 (L393V) genetic variant was previously identified (rs2230723 NHLB1 SNP database) in data derived from the NHLBI Exome Sequencing Project (ESP-Cohort; Northwest Genomic Center (NWGC)/Broad Institute Genome Sequencing Center). The reported genotype frequency was C/C-0.971; C/G - 0.029; G/G-0.000 with an allele frequency of C-0.985; G-0.015 (Table 1). Our overall genomic frequency in DLBCL tumor DNA for this JAK2 (L393V) SNP was higher at 5%. An additional 1 patient had a rare variant in the 3’UTR (3’UTRhet_insCAT) that did not predict amino acid changes (Table 1). No evidence of the JAK2 (V617F) mutation was found in this cohort of DLBCL samples, suggesting that JAK2 V617 mutations are absent in DLBCL tumors.
Table 1.
Data summarizing single nucleotide polymorphisms in JAK2 associated with primary diffuse large B-cell lymphoma (n=40). Data was generated by bidirectional sequencing of all 25 JAK2 exons covering all 7 JAK2 JH domains.
| Point mutations |
Amino Acid |
Effect | RS# | Published Frequency | DLBCL (n) |
Location | |
|---|---|---|---|---|---|---|---|
| Genotype Detail | Alleles | ||||||
| C>T | 163 | His>His | rs10429491 | 0.408 / 0.452 / 0.140 (CC/CT/TT) 1 | 0.634 / 0.366 (C/T) 1 | 8 het; 6 homo | Exon 6 |
| C>T | n/a | n/a | rs41303235 | No data | No data | 3 het; 0 homo | UTR-5 |
| T>C | n/a | n/a | rs2274472 | 0.345/0.522/0.133 (AA/AG/GG) 2 | 0.606/0.394 (A/G) 2 | 16 het; 4 homo | UTR-5 |
| *C>G | 393 | Leu>Val | rs2230723 | 0.971 / 0.029 / 0.000 (CC/CG/GG) 1 | 0.985/ 0.015 (C/G) 1 | 2 het; 0 homo | Exon 9 |
| G>A | 830 | Leu>Leu | rs2230724 | 0.458/0.383/0.158 (AA/AG/GG) 1 | 0.650/ 0.350 (A/G) 1 | 12 het; 0 homo | Exon19 |
| A>T | n/a | n/a | rs2274649 | 0.483/0.467/0.050 (AA/AT/TT) 2 | 0.717/0.283 (A/T) 2 | 8 het; 4 homo | intron |
| 3’UTRhet_insCAT | n/a | n/a | No data | No data | No data | 1 het; 0 homo | 3’UTR |
ESP_Cohort_Populations
HapMap-CEU
non-synonymous SNP
Figure 1. Identification of JAK2 non-synonymous single nucleotide polymorphism (Leu393Val) in DLBCL.
JAK2 was bidirectionally sequenced and a heterozygous cytosine-to-guanine (C>G) SNP was identified which lead to a missense SNP at position 1671 (C1671G) (upper panel). Alignment of mammalian JAK2 amino acid sequence in the FERM (JH4) domain among various species shows conserved sequences (Lower panel).
This report describes the JAK2 mutational analysis of all 25 exons of the JAK2 gene by bidirectional sequencing. Although JAK2 and STAT3 are constitutively activated in DLBCL tumors and this pathway is an important therapeutic target in DLBCL, we did not find any novel JAK2 activating mutation in our data set. Our data did demonstrate known polymorphism (dbSNP) in JAK2. While this manuscript was in preparation, Lohr JG et al published the whole exome sequencing results in DLBCL and identified several non-synonymous somatic mutations in several genes (MYD88, EGH2, CARD11 and CREBBP but not JAK2) [15]. Our report in a different set of DLBCL tumors confirms their findings regarding the lack of JAK2 mutations in DLBCL and suggests that JAK2 activation in DLBCL tumors is independent of genetic regulation. The role of these SNPs in JAK2 regarding tumor signal pathway activation will need further evaluation in larger cohorts, preferably from patients participating in trials of JAK/STAT inhibitors. In addition, other JAK2-STAT3 pathway activating mechanisms in DLBCL such as epigenetic silencing of negative regulators of the JAK/STAT pathway such as the suppressors of cytokine signaling (SOCS) and the SH2-containing phosphatases (SHP) need to be studied. Several JAK2 inhibitors such as ruxolitinib and TG101348 (SARS302503) have demonstrated clinical activity in MPNs with minimal toxicity [16] [17]. We have recently demonstrated that the JAK2 inhibitor TG101348 inhibits the survival of phospho-JAK2 positive DLBCL cells with minimal effect on phospho-JAK2 negative DLBCL cells [3]. Our data suggest that clinical trials of these novel JAK2 inhibitors need not be restricted to tumors with JAK2 mutations but rather should focus on tumors with demonstrated aberrant JAK2-STAT3 signaling.
Material and Methods
Patient specimens
Frozen tumor cells from DLBCL patients (n=40) were acquired through Iowa/Mayo Lymphoma SPORE. This study was reviewed and approved by the Institutional Review Board of the Mayo Clinic. DNA from frozen tumor cells was extracted by using Puragene kit (Qiagen).
Bidirectional sequencing of JAK2 and mutation analysis
Purified DNA was amplified by PCR using primer pairs that span the JAK2 each exons (from 1–25). Sequencing was performed at the Mayo Clinic DNA Sequencing Core Facility and analyzed using Mutation Surveyor software.
Acknowledgments
Funding: This work was supported in part by a Career Development Award to MG from the University of Iowa/Mayo Clinic Lymphoma Specialized Program of Research Excellence (P50 CA097274); a Goodwin Foundation Pilot award (MG).
Footnotes
Conflict of interest: The authors declare no conflict of interest.
Authorship: Contribution: MG designed the research, analyzed and interpreted data and prepared the manuscript. The sequencing and mutational analysis was performed by the Mayo Clinic Sequencing Core. TPT provided assistance in analyzing data. MJS extracted DNA from DLBCL tumor cells. TEW provided clinical samples and prepared the manuscript.
References
- 1.Ding BB, Yu JJ, Yu RY, et al. Constitutively activated STAT3 promotes cell proliferation and survival in the activated B-cell subtype of diffuse large B-cell lymphomas. Blood. 2008;111:1515–1523. doi: 10.1182/blood-2007-04-087734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lam LT, Wright G, Davis RE, et al. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-{kappa}B pathways in subtypes of diffuse large B-cell lymphoma. Blood. 2008;111:3701–3713. doi: 10.1182/blood-2007-09-111948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta M, Han JJ, Stenson M, et al. Elevated serum IL-10 levels in diffuse large B-cell lymphoma: a mechanism of aberrant JAK2 activation. Blood. 2012;119:2844–2853. doi: 10.1182/blood-2011-10-388538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 5.Libra M, Malaponte G, Navolanic PM, et al. Analysis of BRAF mutation in primary and metastatic melanoma. Cell Cycle. 2005;4:1382–1384. doi: 10.4161/cc.4.10.2026. [DOI] [PubMed] [Google Scholar]
- 6.James C, Ugo V, Le Couedic JP, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 7.Levine RL, Wadleigh M, Cools J, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 8.Steensma DP, McClure RF, Karp JE, et al. JAK2 V617F is a rare finding in de novo acute myeloid leukemia, but STAT3 activation is common and remains unexplained. Leukemia. 2006;20:971–978. doi: 10.1038/sj.leu.2404206. [DOI] [PubMed] [Google Scholar]
- 9.Levine RL, Loriaux M, Huntly BJ, et al. The JAK2V617F activating mutation occurs in chronic myelomonocytic leukemia and acute myeloid leukemia, but not in acute lymphoblastic leukemia or chronic lymphocytic leukemia. Blood. 2005;106:3377–3379. doi: 10.1182/blood-2005-05-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melzner I, Weniger MA, Menz CK, et al. Absence of the JAK2 V617F activating mutation in classical Hodgkin lymphoma and primary mediastinal B-cell lymphoma. Leukemia. 2006;20:157–158. doi: 10.1038/sj.leu.2404036. [DOI] [PubMed] [Google Scholar]
- 11.Chim CS, Wong KY, Loong F, et al. SOCS1 and SHP1 hypermethylation in mantle cell lymphoma and follicular lymphoma: implications for epigenetic activation of the Jak/STAT pathway. Leukemia. 2004;18:356–358. doi: 10.1038/sj.leu.2403216. [DOI] [PubMed] [Google Scholar]
- 12.Kube D, Holtick U, Vockerodt M, et al. STAT3 is constitutively activated in Hodgkin cell lines. Blood. 2001;98:762–770. doi: 10.1182/blood.v98.3.762. [DOI] [PubMed] [Google Scholar]
- 13.Ghoreschi K, Laurence A, O'Shea JJ. Janus kinases in immune cell signaling. Immunol Rev. 2009;228:273–287. doi: 10.1111/j.1600-065X.2008.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott NE, Cleveland SM, Grann V, et al. FERM domain mutations induce gain of function in JAK3 in adult T-cell leukemia/lymphoma. Blood. 2011;118:3911–3921. doi: 10.1182/blood-2010-12-319467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lohr JG, Stojanov P, Lawrence MS, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verstovsek S, Kantarjian H, Mesa RA, et al. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N Engl J Med. 2010;363:1117–1127. doi: 10.1056/NEJMoa1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pardanani A, Gotlib JR, Jamieson C, et al. Safety and Efficacy of TG101348, a Selective JAK2 Inhibitor, in Myelofibrosis. J Clin Oncol. 2011;29:789–796. doi: 10.1200/JCO.2010.32.8021. [DOI] [PMC free article] [PubMed] [Google Scholar]