Abstract
The gene encoding dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A) is located within the Down syndrome (DS) critical region of chromosome 21. DYRK1A interacts with a plethora of substrates in the cytosol, cytoskeleton, and nucleus. Its overexpression is a contributing factor to the developmental alterations and age-associated pathology observed in DS. We hypothesized that the intracellular distribution of DYRK1A and cell-compartment–specific functions are associated with DYRK1A posttranslational modifications. Fractionation showed that in both human and mouse brain, almost 80% of DYRK1A was associated with the cytoskeleton, and the remaining DYRK1A was present in the cytosolic and nuclear fractions. Co-immunoprecipitation revealed that DYRK1A in the brain cytoskeleton fraction forms complexes with filamentous actin, neurofilaments, and tubulin. Two-dimensional gel analysis of the fractions revealed DYRK1A with distinct isoelectric points: 5.5–6.5 in the nucleus, 7.2–8.2 in the cytoskeleton, and 8.7 in the cytosol. Phosphate-affinity gel electrophoresis demonstrated several bands of DYRK1A with different mobility shifts for nuclear, cytoskeletal, and cytosolic DYRK1A, indicating modification by phosphorylation. Mass spectrometry analysis disclosed one phosphorylated site in the cytosolic DYRK1A, and multiple phosphorylated residues in the cytoskeletal DYRK1A, including two not previously described. This study supports the hypothesis that intracellular distribution and compartment-specific functions of DYRK1A may depend on its phosphorylation pattern.
Keywords: DYRK1A, brain, phosphorylation, cytoskeleton, nucleus
INTRODUCTION
The dual-specificity tyrosine phosphorylation-regulated kinase 1A gene (Entrez Gene: human DYRK1A gene ID 1859; mouse Dyrk1A gene ID 13548) is encoded within the Down syndrome (DS) critical region of chromosome 21. Overexpression of its product is detected in DS (Guimera et al., 1999; Dowjat et al., 2007), and an excessive amount of DYRK1A protein is a contributing factor to the developmental (Hämmerle et al., 2003; Canzonetta et al., 2008; Tejedor and Hämmerle, 2011) and age-associated (Shi et al., 2008; Liu et al., 2008; Wegiel et al., 2008, 2011a, 2011b) pathology observed in DS. In spite of its nuclear targeting sequence, in human and mouse brain, DYRK1A is also present within the cell cytoplasm (Martí et al., 2003; Wegiel et. al., 2004). Both nuclear and cytoplasmic substrates for this kinase have been identified. It has been shown that DYRK1A regulates splicing factors such as cyclin L2, SF3b/SAP155, and ASF (de Graaf et al., 2004, 2006; Shi et al., 2008) and interacts with the transcription factors FKHR, CREB, Gli-1, and NFAT (Woods et al., 2001; Yang et al., 2001; Mao et al., 2002; Arron et al., 2006; Gwack et al., 2006). In the cytoplasm, DYRK1A phosphorylates synaptic proteins— dynamin (Chen-Hwang et al., 2002), synaptojanin 1 (Adayev et al., 2006), and amphiphysin I (Murakami et al., 2006)—and cytoskeletal targets: tau (Woods et al., 2001), MAP1B (Scales et al., 2009), and α-synuclein (Kim et al., 2006). The diversity of DYRK1A substrates and their localization in different cell compartments suggests that mechanisms regulating intracellular trafficking of this kinase play an important role in its function.
Phosphorylation is a known mechanism regulating DYRK1A activity but function of only two phosphorylated residues in vivo has been studied in detail. Autophosphorylation of tyrosine residue in the activation loop initiates DYRK1A enzymatic activity (Becker and Joost, 1999; Himpel et al., 2001). Autophosphorylation of serine residue (S520) contributes to the binding of scaffolding protein 14-3-3β, which in turn stimulates the catalytic activity by 100% (Alvarez et al., 2007).
The aim of this study was to characterize the proportions and properties of DYRK1A in cytosolic, cytoskeletal, and nuclear fractions obtained from human and mouse brain homogenates, as well as the interactions of DYRK1A with cytoskeletal proteins. The study revealed binding of the majority of DYRK1A to cytoskeletal proteins, including β-actin and neurofilaments; the distinctive isoelectric points (pIs) of cytoskeletal, cytosolic, and nuclear DYRK1A; and differences in phosphorylation in cytosolic and cytoskeletal DYRK1A. Our findings suggest that phosphorylation affects intracellular DYRK1A distribution and function.
MATERIALS AND METHODS
Human and Mouse Tissue
The samples of frontal cortex of four control subjects from 31–65 years of age were examined. Mouse brain tissue was collected from B6 x C3H/HeJ F1 mice (Jackson Laboratory, Bar Harbor, ME), 2–6 months old (n = 6) and 10–16 months old (n = 16). Mice were anesthetized and transcardially perfused with phosphate-buffered saline (PBS). Removed brains were collected on wet ice.
Human brain tissue was obtained from the New York State Institute for Basic Research in Developmental Disabilities (IBR) Brain Bank and the University of Maryland Brain Bank. All experimental procedures involving human tissues were performed in accordance with the Declaration of Helsinki. Experimental protocols were approved by IBR’s Institutional Review Board. Experiments on the animals were performed in accordance with the National Research Council’s Guide for the Care and Use of Laboratory Animals. IBR’s Animal Care and Use Committee approved the protocols.
Antibodies
Monoclonal antibody (mAb) 8D9 was produced by IBR’s Monoclonal Antibody Facility, as described for another anti-DYRK1A mAb (Wegiel et al., 2004). 8D9 characteristics were reported by Kida et al. (2011). At the concentration of 0.1μg/ml, this antibody recognized DYRK1A of human and mouse origin in Western blots (Table 1). The rabbit polyclonal antibody R420 against DYRK1A, produced at IBR, was generated against the synthetic peptide QQPLTNQRRMPQTC conjugated to KLH carrier protein. This sequence overlaps the alternative splicing junction of exons 5 and 6b that is present in the most prevalent isoform 2 of DYRK1A (754 amino acids). Antibody was affinity-purified using a column prepared by coupling this peptide to SulfoLink gel (Thermo Fisher Scientific, Rockford, IL). Specificity of the antibody for the isoform 2 of DYRK1A was tested using fusion proteins GST-Iso1 (for detecting 763 amino acids isoform 1) and GST-Iso2 (for detecting isoform 2), each containing a short fragment of the respective DYRK1A isoform. To create the GST-Iso1, oligonucleotide adapters—5′-gatccactaaccaggtgatgcctgatattgtcatgttacagaggcggatgtg and 5′-aattcacatccgcctctgtaacatgacaatatcaggcatcacctggttagtg—were annealed and ligated into BamHI-EcoRI restriction sites of pGEX-2T vector. Correspondingly, GST-Iso2 was constructed from the adapters 5′-gatcccagcaacctctaactaaccagaggcggatgccccaaacctg and 5′-aattcaggtttggggcatccgcctctggttagttagaggttgctgg annealed and ligated into the same restriction sites of pGEX-2T vector. Recombinant fusion proteins were extracted by lysis with the aid of B-PER reagent (Pierce) and purified by affinity adsorption to Glutathione-Sepharose resin (GE Healthcare, Piscataway, NJ). R420 exclusively detected the 28kDa fusion protein GST-Iso2 with a fragment specific to the 754-aa DYRK1A isoform 2 (Fig. 1A).
Table 1.
Primary antibodies used for Western blotting (WB)
| Antibody target | Vendor | Concentration |
|---|---|---|
| DYRK1A, 8D9 | IBR, Staten Island, NY | 0.1 μg/ml |
| DYRK1A, R420 | IBR, Staten Island, NY | 1 μg/ml |
| β-actin, AC-15 | Sigma, St. Louis, MO | 1:20,000 |
| α-actinin, BM-75.2 | Sigma, St. Louis, MO | 1:5,000 |
| α-tubulin, B511 | Sigma, St. Louis, MO | 1:20,000 |
| Vinculin, VIN-11-5 | Sigma, St. Louis, MO | 1:10,000 |
| Drebrin, ab60932 | Abcam, Cambridge, MA | 1:2,000 |
| Neurofilament 200, E14 | Sigma, St. Louis, MO | 1:20,000 |
| Lamin A | IBR, Staten Island, NY | 1:2,000 |
| Integrin- α2 | BD Biosciences, Palo Alto, CA | 0.5 μg/ml |
| Tau, Tau-5 | Chemicon, Temecula, CA | 1:1000 |
| Synapsin I | BD Biosciences, Palo Alto, CA | 1:10,000 |
| Synaptotagmin I | BD Biosciences, Palo Alto, CA | 1:10,000 |
| Syntaxin | BD Biosciences, Palo Alto, CA | 1:10,000 |
| SNAP25 | BD Biosciences, Palo Alto, CA | 1:10,000 |
Fig. 1.
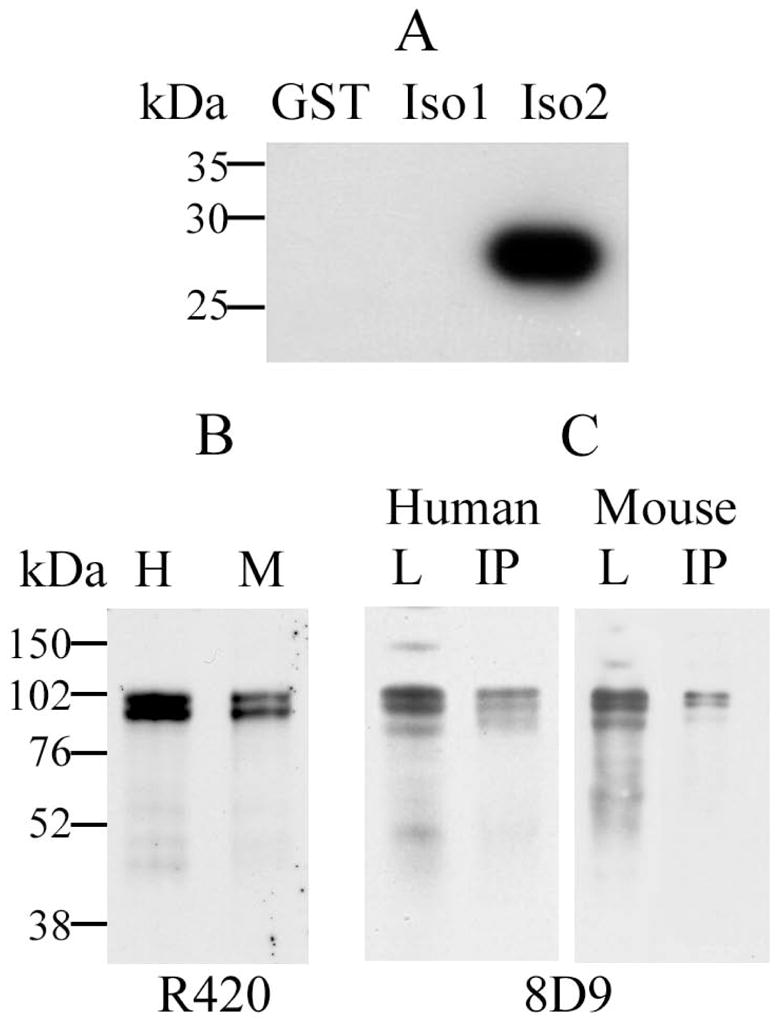
Evaluation of the specificity of affinity-purified pAb R420 by immunoblotting and immunoprecipitation. A: Immunodetection of a recombinant GST (lane GST), and GST fusion proteins containing short fragments of DYRK1A 763-aa isoform 1 (lane Iso1) and 754-aa isoform 2 (lane Iso2) using R420. Twenty ng of antigen was loaded in each lane of 10% SDS-polyacrylamide gel. Specificity of R420 for the DYRK1A isoform 2 was demonstrated by absence of cross-reactivity with the DYRK1A isoform 1. B: DYRK1A protein was detected by R420 as 94–97 kDa bands in lysates from human (H) and mouse (M) brains. C: Specificity of DYRK1A recognition by R420 was confirmed by immunoprecipitation from human brain lysates (Human), and Dyrk1A from mouse brain lysates (Mouse) with R420 (IP); the lysate loads for IP are shown in lanes L. DYRK1A in lanes IP and L was detected with mAb 8D9.
Isolation of Nuclear, Cytosolic and Cytoskeletal Fractions
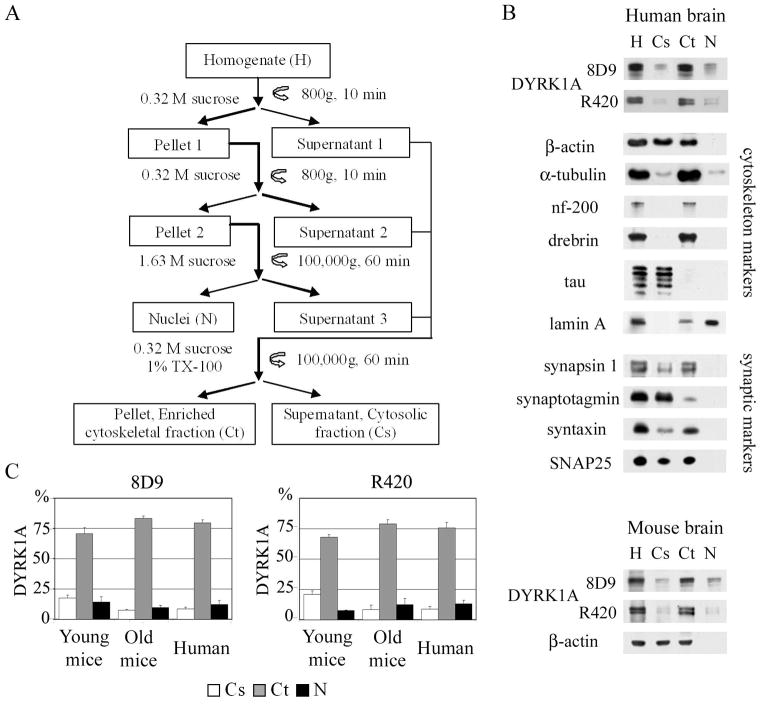
The scheme of fractionation is presented in Fig. 2A. All of the following steps were done on ice or at 4° C. Brain tissue was homogenized with a glass homogenizer (Kontes Glass, Vineland, NJ) in 10 vol of 0.32 M sucrose in TKM buffer (50 mM Tris-Cl, pH 7.4, 25 mM KCl, 5 mM MgCl2, cØmplete EDTA-free) (Roche, Indianapolis, IN). Homogenate (H) was filtered through 30-μm nylon mesh and centrifuged at 800 × g for 10 min. The supernatant 1 was saved, and the pellet 1 was re-homogenized in 5 vol of 0.32 M sucrose in TKM buffer and centrifuged in the same conditions. The supernatant 2 was saved and for each volume of pellet 2, containing crude nuclei, 2 vol of 2.3 M sucrose in TKM were added, and this suspension was overlaid on a 1.8M sucrose cushion in TKM buffer. After 60 min centrifugation at 100,000 × g, the floating ring of cytoplasmic contaminations was collected (supernatant 3). Nuclei–rich bottom portion of sample was collected, slowly resuspended in 10 ml 0.32 M sucrose in TKM, and centrifuged for 10 min at 800 × g. The purity of nuclear fraction (Nu) was monitored by light and electron microscopy and β-actin level. Combined postnuclear fractions (supernatants 1–3) were homogenized in 0.32 M sucrose in TKM supplemented with 1% Triton X-100 (TX-100) and centrifuged for 90 min at 100,000 × g. The supernatant (Cs) yielded a soluble cytosolic fraction whereas detergent-resistant pellet (Ct) was enriched in cytoskeletal proteins.
Fig. 2.
A: Brain homogenate fractionation scheme. B: Characterization of DYRK1A distribution in human brain homogenate (H), and the cytosolic (Cs), cytoskeletal (Ct), and nuclear (N) fractions, by Western blotting with mAb 8D9 and R420. The majority of DYRK1A was revealed in cytoskeletal fraction. Absence of cytoskeletal markers from the nuclear fraction and their presence in the cytosolic and cytoskeletal fractions confirmed the purity of the nuclear fraction. The lower panel shows Dyrk1A in fractions obtained from mouse brain from 10- to 16-month-old animals. C: Densitometric measurements of DYRK1A distribution in lysates of human brains and Dyrk1A in young and old mouse brains. The majority of DYRK1A was detected with both mAb 8D9 and pAb R420 in the cytoskeletal fraction. The graphs show the mean values and standard deviations from three independent experiments. The differences between young and old mice distributions of DYRK1A in Cs, Ct, and N fractions were not significant.
Immunoprecipitation
A 2-mg sample of protein of human brain cytoskeletal fraction was homogenized in 1 ml of RIPA buffer (10 mM sodium phosphate, pH 7.2, 1% NP40, 1% sodium deoxycholate, 0.15% SDS, 150 mM NaCl, cØmplete), centrifuged at 14,000 x g for 10 min, and pre-cleared by 30 min rotation with Protein A Dynabeads (Invitrogen, Carlsbad, CA). The sample was divided into two equal parts for incubation with polyclonal antibody (pAb) R420 or with control non-immune rabbit IgG at 10 μg/ml for 30 min with rotation. Protein A Dynabeads were added, 50 μl to each tube for 15 min incubation, and the beads were washed three times with RIPA. Equal fractions from DYRK1A immunoprecipitation (IP) were analyzed by Western blotting, along with control IgG IP and 10% of input sample.
Density Gradient Subcellular Fractionation
To detect and characterize cytoplasmic DYRK1A, the combined postnuclear fraction was treated with TKM buffer supplemented with 1% of Triton X–100 (TX–100), and 1-ml aliquots were subfractionated by centrifugation on 12 ml of 0.4–2.0 M linear sucrose gradient in TKM buffer with 1% of TX–100 at 200,000 × g for 90 min. One-ml aliquots were collected from the top to the bottom of the gradient. Equal volumes of each fraction were analyzed by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting.
Gel Filtration Chromatography
A 0.2-ml aliquot of cytosolic fraction was separated on Superose 6 10/30 (GE Healthcare, Piscataway, NJ) equilibrated with 50 mM Tris–HCl, pH 7.4, and 300 mM NaCl at 0.4 ml/min. The cytoskeletal fraction was solubilized in 50 mM Tris–HCl, pH 7.4, and 8 M urea and separated on Superose 6 10/30 column pre-equilibrated with the same buffer. The progress of chromatography was monitored by absorption at 280 nm. One-half–ml aliquots were collected, and proteins were precipitated with 2.5 volume of cold acetone and reconstituted in Tricine sample buffer (Bio-Rad Laboratories, Hercules, CA). The molecular weight of DYRK1A and its complexes were determined on the basis of Molecular Weight Gel Filtration Calibration Kits (GE Healthcare) containing proteins in the range of 25–669 kDa and Blue Dextran 2000.
DYRK1A Purification on Immobilized Ni2+-nitrilotriacetate (Ni-NTA) Affinity Chromatography
All protein preparations were performed at 0–4° C except when indicated otherwise. Twenty-five ml of soluble cytosolic fraction from brain homogenate was adjusted to 50 mM Tris–HCl, pH 8.0; 300 mM NaCl; 50 mM imidazole; and cØmplete EDTA-free and filtered through a 0.2-μm membrane filter. This solution was rotated for 3 hr with 1 ml of 50 % Ni-NTA Superflow resin slurry (Qiagen, Alameda, CA), previously equilibrated with the same buffer. Ni-NTA unbound proteins were washed with 20 ml of the same buffer and eluted with 10 ml of buffer containing 50 mM Tris–HCl, pH 7.0; 300 mM NaCl; 350 mM imidazole; and cØmplete EDTA-free. The cytoskeletal fraction was solubilized in buffer containing 50 mM Tris-HCl, pH 8.0; 8M guanidine chloride; and 20 mM imidazole. Twenty-five ml of filtered solution was rotated with Ni-NTA resin in a fashion similar to above, except that the temperature was kept at 20° C. The resin was washed with 10 ml of solubilization buffer, followed by 10 ml of 50 mM Tris-HCl, pH 8.0. DYRK1A was eluted from resin by boiling in tricine sample buffer supplemented with 100 mM EDTA. Eluted DYRK1A, enriched ~1,000-fold from cytosolic and ~3-fold from cytoskeletal fractions, was used for mass spectrometry (MS) analysis.
Gel Electrophoresis and Immunoblotting
Proteins were resolved on 7% or 10% SDS–PAGE with Tris-Tricine running buffer, and electroblotted onto 0.2-μm nitrocellulose membranes (Bio-Rad Laboratories). Membranes were blocked with 5% fat-free milk in PBS with 0.05% Tween-20, pH 7.4, followed by incubation with the primary antibody overnight at 4° C. Incubation with polyclonal peroxidase-conjugated species-specific antibodies was performed for 1 hr at room temperature. Anti-mouse antibody linked to horseradish peroxidase (GE Healthcare) was used at 0.2 μg/ml. Membranes were developed with HyGlo chemiluminescent detection reagents (Denville Scientific, Metuchen, NJ). Molecular weights of proteins in gels and Western blots were determined by comparing their relative mobility to EZ-Run Rec Protein Ladder standards (ThermoFisher BioReagents, Fair Lawn, NJ). Quantitative analysis of chemiluminescent data was done in the linear range of detection by using 1DScan EX software (BD Biosciences, Rockville, MD). An equal percentage of each fraction vol was analyzed by Western blotting, and the distribution of DYRK1A in each fraction was calculated as the percentage of DYRK1A in the whole brain homogenate. The data were obtained from three independent experiments using 8D9 and R420 antibodies. Distribution of DYRK1A among fractions was presented as mean value +/− standard deviation.
Dephosphorylation
An aliquot of enriched soluble DYRK1A protein was subjected to dephosphorylation by lambda protein phosphatase (λPPase) (New England BioLabs, Ipswich, MA). The reaction was conducted for 2 hr at 30° C with 100 U of enzyme in 50 μl of buffer containing 50 mM HEPES, pH 7.5; 0.1 mM EDTA; 5 mM DTT; and 0.01% Brij-35. DYRK1A from the cytoskeletal and nuclear fractions was enriched by separation on semi-preparative denaturing gel and subsequent electroelution of proteins from gel pieces. Proteins were precipitated with 9 vol of ethyl alcohol, dried and reconstituted prior to reaction in dephosphorylation buffer (50 mM HEPES, pH 7.5; 0.1 mM EDTA; 5 mM DTT; and 0.01% Brij-35).
Two-dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE)
Five hundred μg of protein was precipitated with 10% trichloroacetic acid at 0° C for 1 hr and washed with 90% ethanol. For the first dimension, 250-μg protein samples, dissolved in a buffer containing 7 M urea, 2 M thiourea,65 mM DTT, 0.125% (v/v) Biolytes 3–10, 2% CHAPS, and 0.1% bromophenol blue, were applied to a dehydrated, immobilized pH gradient strip (7 cm, pH 3–10) (Bio-Rad Laboratories), and isoelectric focusing was conducted at 20° C for 10,000 volt-hours. Before the proteins were separated by SDS-PAGE, the isoelectric-focusing gel strips were equilibrated for 15 min in a buffer consisting of 37.5 mM Tris-HCl at pH 8.8, 6 M urea, 2% (w/v) SDS, 30% (w/v) glycerol, 0.5% DTT, and 0.1% bromophenol blue. Next, to achieve alkylation, the gel strips were re-equilibrated for 15 min in the same buffer containing 2% iodoacetamide in place of DTT. The second-dimension separation was carried out by placing the strips on 10% polyacrylamide gels, after which gels were electroblotted onto nitrocellulose membranes or stained directly with Coomassie Brilliant Blue R-250 (ThermoFisher BioReagents).
Phosphate-affinity Gel Electrophoresis (Phos-tag SDS–PAGE)
Phosphoprotein mobility shift was done in neutral pH SDS-PAGE as described (Kinoshita and Kinoshita-Kikuta, 2011), with the exception of reduced Zn2+–Phos-tag concentration at 10 μM. After electrophoresis, the divalent cations were removed from gel by incubation for 15 min with transfer buffer containing 2 mM EDTA and were transferred to nitrocellulose.
Mass Spectrometry Analysis
Samples were processed either at the Keck Laboratory’s Mass Spectrometry Resources at Yale University on LTQ Orbitrap (ThermoFisher Scientific) or at the Molecular Structure and Function Laboratory’s Mass Spectrometry Facility at IBR on Q-TOF micro (Waters, Milford, MA). Protein spots, stained with Coomassie Blue R250, were excised, cut into 1-mm pieces, and processed for trypsin digestion with the Trypsin Profile IGD Kit (Sigma, St. Louis, MO). Tryptic peptides analyzed on LTQ Orbitrap were subjected to titanium dioxide column, producing phosphopeptide-enriched and -depleted fractions that were analyzed separately. All MS/MS spectra were searched using the automated Mascot algorithm against the IPI mouse database for possible methionine oxidation; acrylamide modification of cysteine; and serine, threonine, and tyrosine phosphorylation. Peptides extracted from in-gel tryptic digests were dissolved in 25 μl LC-MS grade water containing 0.1% formic acid and separated on a Waters BEH130 C18 column (75 μm × 250 mm) using a complex gradient of 0% to 90% acetonitrile in aqueous 0.1% formic acid (solvent B) at a flow rate of 250 nL/min over 76 min, with a total run time of 124 min essentially as described previously (Adayev et al., 2007; Murakami et al., 2009). Continuum MS and MS/MS spectra were collected, stored, and analyzed offline using the ProteinLynx software package (Waters). Proteins identified in the database search were subsequently analyzed with the Automod search function, using the same tolerances but allowing for nonspecific cleavage and one modification or substitution per peptide, with the substitution likelihood value set to 11. All identified sequences contained a series of at least three consecutive y″ ions in the MS/MS spectrum, as specified in the validation rules set by the program.
Prediction of Putative DYRK1A Phosphorylation Sites
NetPhos 2.0 program (Blom et al., 1999; http://www.cbs.dtu.dk/services/NetPhos) was used to predict putative DYRK1A phosphorylation sites in human and mouse DYRK1A orthologues. To quantify the number of phosphate groups needed to change the DYRK1A pI, the Protein Modification Screening Tool (ProMoST; Halligan et al., 2004) was applied.
RESULTS
Relative Quantity of DYRK1A in Subcellular Fractions
Immunodetection of DYRK1A with R420, specific for the isoform 2 (Fig. 1A), revealed two major bands in the range of 97 and 94 kDa (Fig. 1B) and efficiently immunoprecipitated DYRK1A (Fig. 1C – lanes IP) from human and mouse brain homogenates. The mAb 8D9 detected the two major bands and a much weaker 87-kDa band (Fig 1C, lanes L). The cytoskeletal fraction obtained in the protocol shown in Fig. 2A contained the cytoskeletal proteins drebrin, nf-200, α-tubulin, and filamentous actin, but not tau protein (Fig. 2B). This fraction also contained synaptic proteins interacting with the cytoskeleton, particularly synapsin 1 and syntaxin. SNAP25 was recovered in both the cytosolic and cytoskeletal fractions, whereas synaptotagmin I was mainly recovered in the cytosol. The nuclear matrix protein lamin A was detected almost exclusively in the nuclear fraction (Fig. 2B).
Two antibodies—mAb8D9 and pAbR420—were used to determine the relative amounts of DYRK1A in three fractions. The relative amount of DYRK1A in each fraction was determined as the percentage of DYRK1A in whole-brain homogenate. In human brain, DYRK1A was detected with mAb 8D9 mainly in the cytoskeletal fraction—79% of the total amount of DYRK1A—and in the nuclear and cytosolic fractions, 12% and 8%, respectively. The distribution of Dyrk1A in mouse brain from young and old animals was similar—Dyrk1A was predominantly detected in the cytoskeletal fraction (Fig. 2B, C). The pattern of subcellular DYRK1A distribution detected with mAb 8D9 was comparable to those detected with pAb R420 (Fig. 2C). The differences between Dyrk1A distribution in the mice aged 2–6 months and 10–16 months were not significant. Because DS is associated with accelerated aging, further studies of distribution and phosphorylation of murine Dyrk1A were done in the material from the older mouse group.
Detection of DYRK1A Complexes
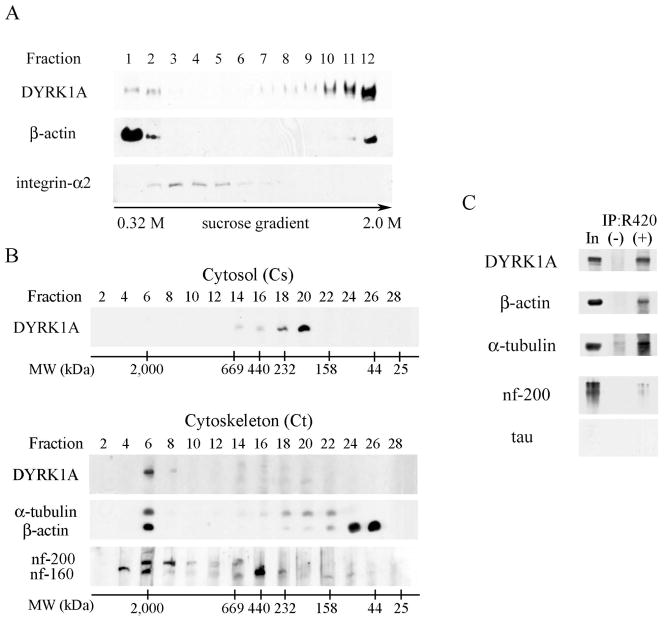
Postnuclear fraction treated with TX-100 was subjected to sucrose density gradient sedimentation (Fig. 3A). In the presence of non-ionic detergent, DYRK1A was found mainly in high sucrose densities (fraction 12) known to be highly enriched in the cytoskeleton (Funchal et al., 2003). This detergent-resistant fraction was also enriched in β-actin. A small amount of DYRK1A was detected in low sucrose densities (fractions 1 and 2) considered to be rich in cytosolic proteins (Nebl et al., 2002) and positive for soluble β-actin. Fractions 3–5 enriched in detergent-resistant membranes—membrane lipid rafts—that were revealed by the presence of integrin-α2 did not contain DYRK1A.
Fig. 3.
A: Sucrose density fractionation of postnuclear supernatant in the presence of non-ionic detergent (1% TX-100) revealed weak DYRK1A immunoreactivity in detergent-soluble fractions 1 and 2 (containing cytosol proteins) and a strong immunoreactivity in β-actin–rich and detergent-resistant fractions 11 and 12 (cytoskeletal proteins). Fractions 3–5 enriched in membrane lipid rafts (shown by the presence of integrin-α2) did not contain DYRK1A. Equal vols of 20 μL of each fraction were loaded per lane. B. To estimate the size of DYRK1A, 100,000 × g supernatant (cytosol) and 100,000 × g pellet (cytoskeleton fraction) were subjected to gel filtration on Superose 6. Cytosolic DYRK1A that peaked in fractions 18–20 comprised proteins in the range of 232–158 kDa (upper panel). DYRK1A from the detergent-resistant cytoskeletal fraction (lower panel), solubilized in 8 M urea and subjected to gel filtration on Superose 6, peaked in fraction 6 and comprised protein complexes larger than 2,000 kDa and cytoskeletal proteins: neurofilament heavy (nf-200) and medium (nf-160), α-tubulin, and β-actin. Images of mAb 8D9–detected Western blots are shown with the position of molecular weight markers of 2,000, 669, 440, 232, 158, 44, and 25 kDa. C: Western blots of DYRK1A immunoprecipitates revealed several cytoskeletal proteins co-precipitated with DYRK1A. The material immunoprecipitated with pAb R420 was divided in five equal parts to detect DYRK1A (mAb 8D9), β-actin, α-tubulin, heavy neurofilament, and tau proteins in WB. Non-immune rabbit IgG—a specificity control of IP—is shown in lane (−), lane In marks 10% of the reaction input.
The supernatant and pellet from 100,000g centrifugation—the cytosol and cytoskeleton fractions—were further used to study DYRK1A distribution. Superose 6–based gel filtration of the cytosol identified the size of the DYRK1A to be in the range of 158–232 kDa (Fig. 3B), indicating that DYRK1A in the soluble fraction forms either dimers or relatively small complexes with other proteins, but does not exist in the monomeric 85–kDa form. In contrast to cytosol, the majority of DYRK1A detected in the cytoskeletal detergent-resistant fraction was part of large protein complexes, and even in the presence of high concentrations of urea co-migrated on Superose 6 column with markers larger then 2,000 kDa. DYRK1A-containing fractions were also enriched in cytoskeletal proteins: β-actin, α-tubulin, and neurofilament 200 and neurofilament 160 proteins (Fig. 3B). Immunoprecipitation of DYRK1A from cytoskeletal fractions with R420 resulted in co-precipitation of β-actin, α-tubulin, neurofilament 200 but not tau protein (Fig. 3C).
Isoelectric Point of DYRK1A in Cellular Subfractions
Two-dimensional analysis of the cytosolic, cytoskeletal, and nuclear fractions revealed fraction-specific distribution of DYRK1A pIs. In the cytosolic fraction, the DYRK1A was focused exclusively around pI 8.7, in close proximity to the predicted spot for the calculated pI value of unmodified DYRK1A protein (Fig. 4). Stable complexes in cytoskeletal and nuclear fractions were dissociated by denaturing in the presence of SDS and reducing agent. In the cytoskeletal fraction, 2D Western blot developed with mAb 8D9 revealed DYRK1A focused in the range of pI 7.2–8.2 in the form of multiple distinguished spots, whereas nuclear DYRK1A was focused in the range of pI 5.5–6.5, as several overlapping spots.
Fig. 4.
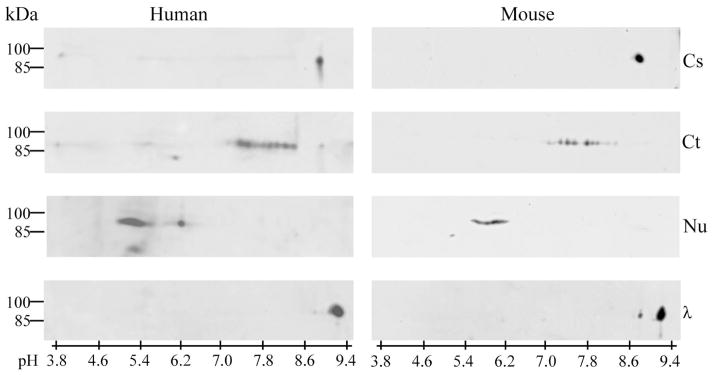
Brain cytosolic (Cs), cytoskeletal (Ct), and nuclear (Nu) fractions of human (left panel) and mouse (right panel) were denatured prior to separation by 2-D electrophoresis, and DYRK1A was detected in Western blots with mAb 8D9. In cytosolic fraction, DYRK1A was observed exclusively at pI 8.7, and after dephosphorylation with λPPase (λ), its pI shifted to 9.0. In the cytoskeletal and nuclear fractions of both human and mouse brains, different patterns were observed: several discrete focused spots of DYRK1A with pI 7.2–8.2 in the cytoskeletal fraction, and several overlapping spots with pI 5.0–6.5 in the nuclear fraction.
DYRK1A Phosphorylation Analysis
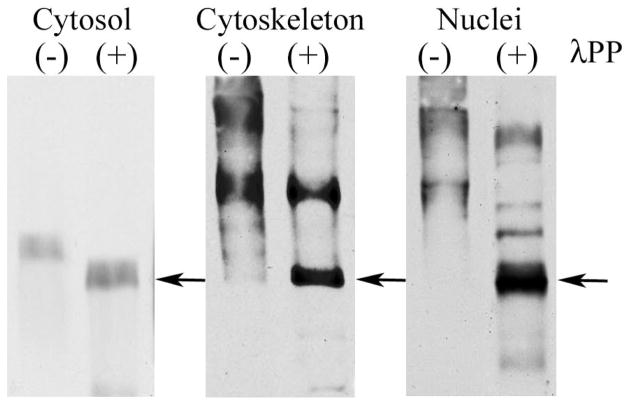
The observed moderate pI shift for cytosolic DYRK1A suggested modification of the kinase by a single phosphate molecule. The cytosolic DYRK1A pI increase by 0.3 unit, from 8.7 to 9.0, in samples treated with λPPase indicated the loss of one phosphate group and was in agreement with the acidic shift of approximately 0.2 pH units for mono-phosphorylated DYRK1A predicted by ProMoST (Fig. 4). Finally, the phosphorylation stoichiometry of cytosolic DYRK1A was examined using phosphate-affinity SDS-PAGE (Phos-tag). This experiment revealed a single band of DYRK1A with electrophoretic mobility moderately retarded in comparison to mobility of the bands observed in λPPase-dephosphorylated cytosolic DYRK1A (Fig. 5). The phosphorylated residue was identified by MS/MS analysis of the DYRK1A isolated from the spot at pI 8.7. Of 37 unique DYRK1A peptides identified from that spot (data not shown)—covering over 64% of the protein sequence—only a single modified site, the conserved autophosphorylated Y321 (isoform 1 numbering) in the activation loop, was found.
Fig. 5.
Phosphate-affinity gel electrophoresis. Mouse brain cytosolic, cytoskeletal, and nuclear fractions were separated in SDS-PAGE in the presence of 10 μM Zn2+–Phos-tag, and DYRK1A was detected with mAb 8D9. The mobility of untreated DYRK1A from the cytosolic fraction, lane (−), was moderately shifted, as compared to the sample dephosphorylated with λPPase (arrow), lane (+). For DYRK1A from the cytoskeletal and nuclear fractions, the shift of the electrophoretic mobility was more pronounced, and numerous distinct bands were detected, lanes (−). Treatment with λPPase, lanes (+), resulted in a partial dephosphorylation of DYRK1A, with the main protein band regaining electrophoretic mobility similar to the dephosphorylated DYRK1A from the cytosol fraction (arrow).
In contrast, in the cytoskeletal and nuclear fractions, the significant acidic shift of DYRK1A pI values from the pI predicted for unmodified protein that was observed could be attributed to phosphorylation at multiple sites. The pI shift by 1.5 units, as observed for the cytoskeletal DYRK1A, was predicted to require the addition of approximately eight phosphate residues. In the cytoskeletal and nuclear fractions, Phos-tag SDS-PAGE revealed several DYRK1A-phosphorylated bands whose mobilities were shifted to a much higher extent, indicating the presence of many heterogeneously phosphorylated forms of DYRK1A (Fig. 5). After dephosphorylation, the electrophoretic mobility of DYRK1A was significantly increased. The fastest band from cytoskeletal and nuclear DYRK1A migrated to similar position as the dephosphorylated band of DYRK1A from the cytosolic fraction. Using MS/MS, we identified the Y321 phosphorylation sites in cytosolic DYRK1A from human and murine samples. Murine cytoskeletal Dyrk1A contained phosphorylated residues S47, S49, and Y321 (Fig. 6). This finding confirmed the presence of multiple sites of phosphorylation in the cytoskeletal DYRK1A and partially explained the multi-spot pattern observed in 2D. The S47 and S49 phosphorylation sites in Dyrk1A protein have not yet been reported.
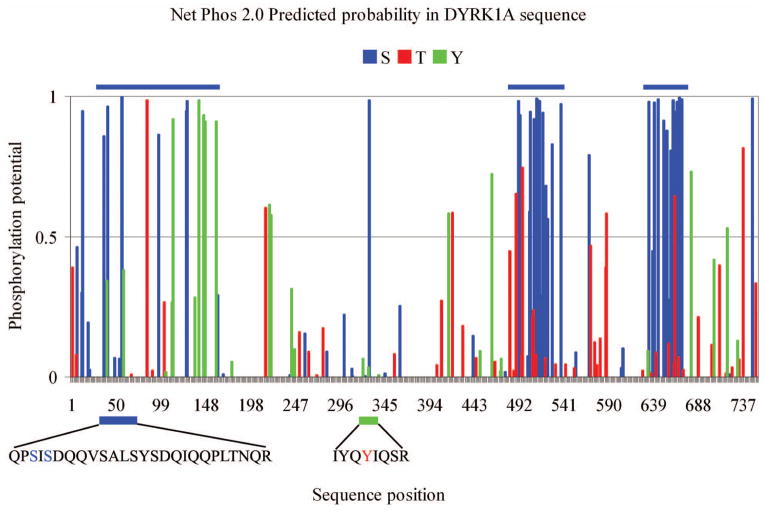
Fig. 6.
Analysis of DYRK1A sequence using NetPhos 2.0 demonstrates three clusters of Ser/Thr with a high probability of phosphorylation localized at 20–180 aa, 480–540 aa, and 630–670 aa (blue bars). MS/MS identified in Dyrk1A isolated from the murine cytoskeletal fraction three phosphorylation sites: S47, S49, and Y321.
DISCUSSION
Detection of DYRK1A in the cytosol, cytoskeleton, synapses, and nuclei indicates involvement of this kinase in regulation of multiple cellular processes. The results of this study indicate that distinct phosphorylation of DYRK1A is associated with its intracellular distribution and interactions. These posttranslational modifications may regulate DYRK1A interactions with nuclear, cytoskeletal, and cytosolic substrates and diverse DYRK1A functions in physiological and pathological conditions.
Intracellular DYRK1A distribution
DYRK1A contains two nuclear localization signals and the protein has been detected in the nucleus (Song et al., 1997; Becker et al., 1998; Mao et al., 2002; Alvarez et al., 2003, 2007; Slepak et al., 2012). Depending on the methods applied, DYRK1A has been reported both in the nucleus and in the cytoplasm (Martí et al., 2003). However, confocal microscopy studies revealed DYRK1A in neurons mainly in the cytoplasm (Hämmerle at al., 2003; Wegiel et al., 2008). Fractionation based study revealed the majority (almost 80%) of cellular DYRK1A in the detergent-resistant insoluble fractions enriched with cytoskeletal proteins in human brains and mouse brains from the younger and older group. This cytoskeletal fraction also contained some synaptic proteins strongly interacting with the cytoskeletal network that were not released by the detergent used in our isolation protocol, e.g., synapsin 1—known to bind actin and tubulin (Petrucci and Morrow, 1991),—syntaxin, and SNAP25 (Chin et al., 2000). Thus, some cytoskeletal Dyrk1A shown in this study may have a synaptic location, consistently with a previous detection of Dyrk1A in crude synaptosomal preparations (Martí et al., 2003; Aranda et al., 2008) and the reported involvement of Dyrk1A in synaptic vesicle–recycling processes (Murakami et al., 2009). It should be noted, however, that crude synaptosomal preparations studied by these authors may contain Dyrk1A loosely bound to vesicles, plasma membranes and cytoskeleton, while the fraction obtained with the use of a detergent (as in this study) provides better purified cytoskeletal proteins and a cytosolic fraction containing proteins released from plasma membranes. Expression and subcellular distribution of DYRK1A is cell-type– and structure-specific (Martí et al., 2003; Wegiel et al., 2004). Hence, the DYRK1A distribution in subcellular fractions of the brain reported here reflects an average of the distribution of DYRK1A in different types of neurons, glia, and cells of vascular wall.
The differences in the reported proportions between nuclear and cytoplasmic DYRK1A may depend on: (i) the type of cells examined (Wegiel et al., 2004); (ii) mainly nuclear location of DYRK1A in over-expression culture systems (Becker et al., 1998; Mao et al., 2002; Slepak et al., 2012); (iii) the antibodies used (Wegiel et al., 2008; Wegiel et al., 2011b; Mazur-Kolecka et al., 2012). Detection of DYRK1A with the antibody 8D9 is dependent on both tyrosines in its epitope (corresponding to Tyr-145/147 in the isoform 1) not being phosphorylated (Kida et al., 2011). The antibody R420 specifically detects the isoform 2 (Fig 1A), which is the most abundant DYRK1A isoform in the brain, as indicated by the studies of human fetal brain cDNA library, which revealed that about 98% of the DYRK1A transcript codes for the 754 aa protein, i.e., for the isoform 2 [Guimera et al., 1999]. Differences in the band patterns detected by mAb 8D9 (three bands) and pAb R420 (two bands) may be caused by the different access of each antibody to its epitope and the existence of various forms of DYRK1A, as has already been reported (Kida et al., 2011). The data presented here show that distinct posttranslational modifications of DYRK1A in cell nucleus, cytoskeleton, and cytosol, may also affect detection of DYRK1A by different antibodies.
The increased expression of DYRK1A is associated with intensive cytoskeleton formation in migrating and differentiating neuronal progenitor cells (Mazur-Kolecka et al., 2012), and with growth and differentiation of dendritic tree (Hämmerle at al., 2003). The association of DYRK1A with cytoskeletal β-actin and neurofilaments in differentiating brain cells, shown by confocal microscopy suggests DYRK1A involvement in cytoskeleton formation during neuronal differentiation and maturation (Mazur-Kolecka et al., 2012). Overexpression of DYRK1A in DS (Guimera et al., 1999; Dowjat et al., 2007) may affect neuronal migration, differentiation and maturation, and may contribute to DS functional deficits. It has been recently shown that in the brain DYRK1A is co-immunoprecipitated with actin and the DYRK1A-actin interaction is dependent on the phosphorylation state of cytoskeletal proteins. DYRK1A overexpression in DS is associated with its reduced complexing with actin (Dowjat et al., 2012).
Recently, it was demonstrated that Dyrk1A may influence neuronal morphogenesis through the regulation of actin dynamics within dendritic spines (Martinez de Lagran et al., 2012). This finding supported the former report of the role of minibrain (mnb/DYRK1A) kinase in the regulation of actin-based protrusions in CNS-derived cell lines (Liu et al., 2009). The growing list of DYRK1A cytoskeletal targets—tau, α-synuclein, SEPT4, and MAP1B (Woods et al., 2001; Kim et al., 2006; Sitz et al., 2008, Scales et al., 2009), and N-WASP (Park et al., 2012)—indicates that DYRK1A plays a significant role in the regulation of cytoskeleton structure and function. Studies of tau suggest that DYRK1A functions as a priming kinase for glycogen synthase kinase 3 (Woods et al., 2001; Liu et al., 2008) and contributes to intracellular accumulation of abnormal cytoskeleton, including neurofibrillary tangles (NFTs) (Liu et al., 2008; Shi et al., 2008; Wegiel et al., 2008, 2011a, 2011b) and Lewy bodies (Kim et al., 2006). The presence of DYRK1A in NFTs in the brains of individuals with sporadic Alzheimer disease (AD) and a several-fold increase in the number of DYRK1A-positive NFTs in the brains of individuals with DS/AD suggest a direct contribution of DYRK1A to the accelerated and enhanced neurofibrillary degeneration and neuronal loss in DS (Wegiel et al., 2008).
DYRK1A Phosphorylation
The phosphorylation prediction tool NetPhos 2.0 revealed a high concentration of potential phosphorylation target amino acids in DYRK1A clustered in three regions grouping serine/threonine and tyrosine residues (Fig. 6). The PhosphoSitePlus database (www.phosphosite.org) lists 17 confirmed phosphorylated residues in human DYRK1A: 10 Tyr and 7 Ser.
This study shows a different pattern of phosphorylation of cytosolic (soluble) and cytoskeleton-bound (insoluble) DYRK1A. The cytosolic kinase, shown by its pI of 8.7 with mAb 8D9, apparently has a single phosphorylation site, at least for the DYRK1A species unphosphorylated on Tyr145/147 (Tyr136/138 in isoform 2) (Kida et al., 2011). Single phosphorylation was confirmed by Phos-tag SDS-PAGE, and MS analysis of a single phosphorylated site—Y321—that is essential for DYRK1A activation (Himpel et al., 2001). In contrast, cytoskeletal DYRK1A is characterized by a more acidic pI (7.2–8.2) than the soluble form and is phosphorylated in multiple sites, as revealed by Phos-tag SDS-PAGE. The MS/MS analysis identified at least three phosphorylated amino acid residues—S47, S49, and Y321—in mouse Dyrk1A.
Nuclear DYRK1A, represented by multiple forms, is characterized by a strong acidic shift of pI to 5.5–6.5. Similar to the cytoskeletal form, nuclear DYRK1A was resistant to detergent extraction and was associated with the insoluble matrix (data not shown, and Becker et al., 1998). Thus, the transition of DYRK1A from the soluble to the insoluble states may be regulated by phosphorylation. To date, the roles of only a few phosphorylated sites have been identified for DYRK1A. The in vivo autophosphorylation of DYRK1A in the activation loop of the catalytic domain, at the second tyrosine residue (Y321) in the Y-X-Y motif (Becker et al., 1999), was demonstrated as essential for its activation (Himpel et al., 2001). Autophosphorylation of DYRK1A on serine residue (S520) initiates binding of the 14-3-3β scaffolding protein (Alvarez et al., 2007). We confirmed the presence of phosphorylation at Y321 in all of the cytoplasmic forms of DYRK1A analyzed. However, the two novel phosphorylation sites reported here—S47 and S49—appear to be characteristic of the cytoskeletal DYRK1A. These data are consistent with evidence of endogenous DYRK1A phosphorylation on serine residues (Kentrup et al., 1996; Adayev et al., 2007).
Both cytoskeletal and nuclear DYRK1A appear to contain subfractions with distinct phosphorylation states, as indicated by the presence of multiple forms with distinct pIs (Fig. 4) and by a partial resistance to dephosphorylation of some DYRK1A subfractions (Fig. 5). The mechanisms of DYRK1A resistance to dephosphorylation are not known, but in other protein kinases, it was linked to certain primary sequences (Jideama et al., 2006), the presence of nucleophilic cysteine close to the phosphorylation site (Humphries et al., 2005), and shielding by conformational changes (Chan et al., 2011).
This study demonstrated that approximately 80% of cellular DYRK1A in the human and mouse brain is detected in the cytoskeletal fraction. DYRK1A in the nuclear, cytosolic, and cytoskeletal fractions exists in fraction-specific pI forms, probably reflecting its phosphorylation. The majority of DYRK1A is complexed with other molecules, as shown by sedimentation and size exclusion studies. Co-immunoprecipitation revealed that DYRK1A forms complexes with cytoskeleton filamentous actin, neurofilaments, and tubulin. The study shows that distinct phosphorylation of DYRK1A is associated with its intracellular distribution and interactions.
Acknowledgments
Contract grant sponsor: the New York State Office for People With Developmental Disabilities. Contract grant sponsor: National Institutes of Health; Contract grant number: R01 HDO43960 (to JW).
The authors thank Ms. Maureen Marlow for editorial corrections, and Mr. James C.M. Chen and Ms. Heni Hong of IBR’s Monoclonal Antibody Core Facility for producing antibody 8D9. For producing antibody R420 authors thank Mr. Marc Barshatsky and Mr. Bruce Patrick. The tissue was provided by the Aging and Dementia Research Center at New York University, the Brain and Tissue Bank at the New York State Institute for Basic Research in Developmental Disabilities, and the Brain and Tissue Bank for Developmental Disorders at the University of Maryland.
References
- Adayev T, Chen-Hwang MC, Murakami N, Wegiel J, Hwang YW. Kinetic properties of a MNB/DYRK1A mutant suitable for the elucidation of biochemical pathways. Biochemistry. 2006;45:12011–12019. doi: 10.1021/bi060632j. [DOI] [PubMed] [Google Scholar]
- Adayev T, Chen-Hwang MC, Murakami N, Lee E, Bolton DC, Hwang YW. Dual-specificity tyrosine phosphorylation-regulated kinase 1A does not require tyrosine phosphorylation for activity in vitro. Biochemistry. 2007;46:7614–7624. doi: 10.1021/bi700251n. [DOI] [PubMed] [Google Scholar]
- Alvarez M, Estivill X, de la Luna S. DYRK1A accumulates in splicing speckles through a novel targeting signal and induces speckles disassembly. J Cell Sci. 2003;116:3099–3107. doi: 10.1242/jcs.00618. [DOI] [PubMed] [Google Scholar]
- Alvarez M, Altafaj X, Aranda S, de la Luna S. Dyrk1A autophosphorylation on serine residue 520 modulates its kinase activity via 14-3-3 binding. Mol Biol Cell. 2007;18:1167–1178. doi: 10.1091/mbc.E06-08-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda S, Alvarez M, Turró S, Laguna A, de la Luna S. Sprouty2-mediated inhibition of fibroblast growth factor signaling is modulated by the protein kinase DYRK1A. Mol Cell Biol. 2008;28:5899–911. doi: 10.1128/MCB.00394-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arron JR, Winslow MM, Polleri A, Chang C-P, Wu H, Gao X, Neilson JR, Chen L, Heit JJ, Kim SK, Yamasaki N, Miyakawa T, Francke U, Graef IA, Crabtree GR. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441:595–600. doi: 10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- Becker W, Weber Y, Wetzel K, Eimbter K, Tejedor FJ, Joost HG. Sequence characteristics, subcellular localization, and substrate specificity of DYRK-related kinases, a novel family of dual specificity protein kinases. J Biol Chem. 1998;273:25893–25902. doi: 10.1074/jbc.273.40.25893. [DOI] [PubMed] [Google Scholar]
- Becker W, Joost HG. Structural and functional characteristics of Dyrk, a novel subfamily of protein kinases with dual specificity. Prog Nucleic Acid Res Mol Biol. 1999;62:1–17. doi: 10.1016/s0079-6603(08)60503-6. [DOI] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Canzonetta C, Mulligan C, Deutsch S, Ruf S, O’Doherty A, Lyle R, Borel C, Lin-Marq N, Delom F, Groet J, Schnappauf F, De vita S, Averill S, Priestley JV, Martin JE, Shipley J, Denyer G, Epstein CJ, Fillat C, Estivill X, Tybulewicz VLJ, Fisher EMC, Antonarakis SE, Nizetic D. DYRK1A-dosage imbalance perturbs NRSF/REST levels, deregulating pluripotency and embryonic stem cell fate in Down syndrome. Am J Hum Genet. 2008;83:388–400. doi: 10.1016/j.ajhg.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TO, Zhang J, Rodeck U, Pascal JM, Armen RS, Spring M, Dumitru CD, Myers V, Li X, Cheung JY, Feldman AM. Resistance of Akt kinases to dephosphorylation through ATP-dependent conformational plasticity. Proc Natl Acad Sci U S A. 2011;108:E1120–1127. doi: 10.1073/pnas.1109879108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Hwang MC, Chen HR, Elzinga M, Hwang YW. Dynamin is a minibrain kinase/dual specificity Yak 1-related kinase 1A substrate. J Biol Chem. 2002;277:17597–17604. doi: 10.1074/jbc.M111101200. [DOI] [PubMed] [Google Scholar]
- Chin LS, Nugent RD, Raynor MC, Vavalle JP, Li L. SNIP, a novel SNAP-25-interacting protein implicated in regulated exocytosis. J Biol Chem. 2000;275:1191–1200. doi: 10.1074/jbc.275.2.1191. [DOI] [PubMed] [Google Scholar]
- de Graaf K, Hekerman P, Spelten O, Herrmann A, Packman LC, Büssow K, Müller-Newen G, Becker W. Characterization of Cyclin L2, a novel cyclin with an arginine/serine-rich domain: phosphorylation by DYRK1A and colocalization with splicing factors. J Biol Chem. 2004;279:4612–4624. doi: 10.1074/jbc.M310794200. [DOI] [PubMed] [Google Scholar]
- de Graaf K, Czajkowska H, Rottmann S, Packman LC, Lilischkis R, Lüscher B, Becker W. The protein kinase DYRK1A phosphorylates the splicing factor SF3b1/SAP155 at Thr434, a novel in vivo phosphorylation site. BMC Biochem. 2006;7:7–19. doi: 10.1186/1471-2091-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowjat WK, Adayev T, Kuchna I, Nowicki K, Palminiello S, Hwang YW, Wegiel J. Trisomy-driven overexpression of DYRK1A kinase in the brain of subjects with Down syndrome. Neurosci Lett. 2007;413:77–81. doi: 10.1016/j.neulet.2006.11.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowjat K, Adayev T, Kaczmarski W, Wegiel J, Hwang YW. Gene dosage-dependent association of DYRK1A with the cytoskeleton in the brain and lymphocytes of Down syndrome patients. J Neuropathol Exp Neurol. 2012;71:1100–12. doi: 10.1097/NEN.0b013e31827733c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funchal C, de Almeida LM, Oliveira Loureiro S, Vivian L, de Lima Pelaez P, Dall Bello Pessutto F, Rosa AM, Wajner M, Pessoa Pureur R. In vitro phosphorylation of cytoskeletal proteins from cerebral cortex of rats. Brain Res Protoc. 2003;11:111–118. doi: 10.1016/s1385-299x(03)00022-9. [DOI] [PubMed] [Google Scholar]
- Guimera J, Casas C, Estivill X, Pritchard M. Human minibrain homologue (MNBH/DYRK1): characterization, alternative splicing, differential tissue expression, and overexpression in Down syndrome. Genomics. 1999;57:407–418. doi: 10.1006/geno.1999.5775. [DOI] [PubMed] [Google Scholar]
- Gwack Y, Sharma S, Nardone J, Tanasa B, Iuga A, Srikanth S, Okamura H, Bolton D, Feske S, Hogan PG, Rao A. A genome-wide Drosophila RNAi screen identifies DYRK-family kinases as regulators of NFAT. Nature. 2006;441:646–650. doi: 10.1038/nature04631. [DOI] [PubMed] [Google Scholar]
- Halligan BD, Ruotti V, Jin W, Laffoon S, Twigger SN, Dratz EA. ProMoST (Protein Modification Screening Tool): a web-based tool for mapping protein modifications on 2 dimensional gels. Nucl Acids Res. 2004;32:W638–644. doi: 10.1093/nar/gkh356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hämmerle B, Carnicero A, Elizalde C, Ceron J, Martínez S, Tejedor FJ. Expression patterns and subcellular localization of the Down syndrome candidate protein MNB/DYRK1A suggest a role in late neuronal differentiation. Eur J Neurosci. 2003;17:2277–2286. doi: 10.1046/j.1460-9568.2003.02665.x. [DOI] [PubMed] [Google Scholar]
- Himpel S, Panzer P, Eirmbter K, Czajkowski H, Sayed M, Packman LC, Blundell T, Kentrup H, Grötzinger J, Joost H-G, Becker W. Identification of the autophosphorylation sites and characterization of their effects in the protein kinase DYRK1A. Biochem J. 2001;359:497–505. doi: 10.1042/0264-6021:3590497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries KM, Deal MS, Taylor SS. Enhanced dephosphorylation of cAMP-dependent protein kinase by oxidation and thiol modification. J Biol Chem. 2005;280:2750–2758. doi: 10.1074/jbc.M410242200. [DOI] [PubMed] [Google Scholar]
- Jideama NM, Crawford BH, Hussain AKMA, Raynor RL. Dephosphorylation specificities of protein phosphatase for cardiac troponin I, troponin T, and sites within troponin T. Int J Biol Sci. 2006;2:1–9. doi: 10.7150/ijbs.2.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentrup H, Becker W, Heukelbach J, Wilmes A, Schurmann A, Huppertz C, Kainulainen H, Joost HG. Dyrk, a dual specificity protein kinase with unique structural features whose activity is dependent on tyrosine residues between subdomains VII and VIII. J Biol Chem. 1996;271:3488–3495. doi: 10.1074/jbc.271.7.3488. [DOI] [PubMed] [Google Scholar]
- Kida E, Walus M, Jarząbek K, Palminiello S, Albertini G, Rabe A, Hwang YW, Golabek AA. Form of dual-specificity tyrosine-(Y)-phosphorylation-regulated kinase 1A nonphosphorylated at tyrosine 145 and 147 is enriched in the nuclei of astroglial cells, adult hippocampal progenitors, and some cholinergic axon terminals. Neuroscience. 2011;195:112–127. doi: 10.1016/j.neuroscience.2011.08.028. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Sung JY, Lee HJ, Rhim H, Hasegawa M, Iwatsubo T, Min do S, Kim J, Paik SR, Chung KC. Dyrk1A phosphorylates alpha-synuclein and enhances intracellular inclusion formation. J Biol Chem. 2006;281:33250–33257. doi: 10.1074/jbc.M606147200. [DOI] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E. Improved Phos-tag SDS-PAGE under neutral pH conditions for advanced protein phosphorylation profiling. Proteomics. 2011;11:319–323. doi: 10.1002/pmic.201000472. [DOI] [PubMed] [Google Scholar]
- Liu F, Liang Z, Wegiel J, Hwang YW, Iqbal K, Grundke-Iqbal I, Ramakrishna N, Gong CX. Over-expression of Mnb/Dyrk1A contributes to neurofibrillary degeneration in Down syndrome. FASEB J. 2008;22:3224–3233. doi: 10.1096/fj.07-104539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Sims D, Baum B. Parallel RNAi screens across different cell lines identify generic and cell type-specific regulators of actin organization and cell morphology. Genome Biol. 2009;10:R26. doi: 10.1186/gb-2009-10-3-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Maye P, Kogerman P, Tejedor FJ, Toftgard R, Xie W, Wu G, Wu D. Regulation of Gli1 transcriptional activity in the nucleus by Dyrk1. J Biol Chem. 2002;277:35156–35161. doi: 10.1074/jbc.M206743200. [DOI] [PubMed] [Google Scholar]
- Martí E, Altafaj X, Dierssen M, de la Luna S, Fotaki V, Alvarez M, Perez-Riba M, Ferrer I, Estivill X. Dyrk1A expression pattern supports specific roles of this kinase in the adult central nervous system. Brain Res. 2003;964:250–263. doi: 10.1016/s0006-8993(02)04069-6. [DOI] [PubMed] [Google Scholar]
- Martinez de Lagran M, Benavides-Piccione R, Ballesteros-Yañez I, Calvo M, Morales M, Fillat C, Defelipe J, Ramakers GJ, Dierssen M. Dyrk1A influences neuronal morphogenesis through regulation of cytoskeletal dynamics in mammalian cortical neurons. Cerebral Cortex. 2012 doi: 10.1093/cercor/bhr362. [DOI] [PubMed] [Google Scholar]
- Mazur-Kolecka B, Golabek A, Kida E, Rabe A, Hwang Y-W, Adayev T, Wegiel J, Flory M, Kaczmarski W, Marchi E, Frackowiak J. The effect of Dyrk1A activity inhibition on development of neuronal progenitors isolated from Ts65Dn mice. J Neurosci Res. 2012;90:999–1010. doi: 10.1002/jnr.23007. [DOI] [PubMed] [Google Scholar]
- Murakami N, Xie W, Lu RC, Chen-Hwang MC, Wieraszko A, Hwang YW. Phosphorylation of amphiphysin I by minibrain kinase/dual-specificity tyrosine phosphorylation-regulated kinase, a kinase implicated in Down syndrome. J Biol Chem. 2006;281:23712–23724. doi: 10.1074/jbc.M513497200. [DOI] [PubMed] [Google Scholar]
- Murakami N, Bolton D, Hwang Y-W. Dyrk1A bind to multiple endocytic proteins required for clathrin coated vesicle formation. Biochemistry. 2009;48:9297–9305. doi: 10.1021/bi9010557. [DOI] [PubMed] [Google Scholar]
- Nebl T, Pestonjamasp KN, Leszyk JD, Crowley JL, Oh SW, Luna EJ. Proteomic analysis of a detergent-resistant membrane skeleton from neutrophil plasma membranes. J Biol Chem. 2002;277:43399–43409. doi: 10.1074/jbc.M205386200. [DOI] [PubMed] [Google Scholar]
- Park J, Sung JY, Park J, Song WJ, Chang S, Chung KC. Dyrk1A negatively regulates the actin cytoskeleton through threonine phosphorylation of N-WASP. J Cell Sci. 2012;125:67–80. doi: 10.1242/jcs.086124. [DOI] [PubMed] [Google Scholar]
- Petrucci TC, Morrow JS. Actin and tubulin binding domains of synapsins Ia and Ib. Biochemistry. 1991;30:413–422. doi: 10.1021/bi00216a016. [DOI] [PubMed] [Google Scholar]
- Scales TME, Lin S, Kraus M, Goold RG, Gordon-Weeks PR. Nonprimed and DYRK1A-primed GSK3β-phosphorylation sites on MAP1B regulate microtubule dynamics in growing axons. J Cell Sci. 2009;122:2424–2435. doi: 10.1242/jcs.040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhang T, Zhou C, Chohan MO, Gu X, Wegiel J, Zhou J, Hwang Y-W, Iqbal K, Grundke-Iqbal I, Gong C-X, Liu F. Increased dosage of Dyrk1A alters ASF-regulated alternative splicing of tau in Down syndrome. J Biol Chem. 2008;283:28660–28669. doi: 10.1074/jbc.M802645200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitz JH, Baumgärtel K, Hämmerle B, Papadopoulos C, Hekerman P, Tejedor FJ, Becker W, Lutz B. The Down syndrome candidate dual-specificity tyrosine phosphorylation-regulated kinase 1A phosphorylates the neurodegeneration-related septin 4. Neuroscience. 2008;157:596–605. doi: 10.1016/j.neuroscience.2008.09.034. [DOI] [PubMed] [Google Scholar]
- Slepak TI, Salay LD, Lemmon VP, Bixby JL. Dyrk kinases regulate phosphorylation of doublecortin, cytoskeletal organization, and neuronal morphology. Cytoskeleton. 2012;69:514–527. doi: 10.1002/cm.21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WJ, Chung SH, Kurnit DM. The murine Dyrk protein maps to chromosome 16, localizes to the nucleus, and can form multimers. Biochem Biophys Res Commun. 1997;231:640–644. doi: 10.1006/bbrc.1997.6154. [DOI] [PubMed] [Google Scholar]
- Tejedor FJ, Hämmerle B. MNB/DYRK1A: a multiple regulator of neuronal development. FEBS J. 2011;278:223–235. doi: 10.1111/j.1742-4658.2010.07954.x. [DOI] [PubMed] [Google Scholar]
- Wegiel J, Kuchna I, Nowicki K, Frackowiak J, Dowjat K, Silverman WP, Reisberg B, DeLeon M, Wisniewski T, Adayev T, Chen-Hwang M-C, Hwang Y-W. Cell type- and brain structure-specific patterns of distribution of minibrain kinase in human brain. Brain Res. 2004;1010:69–80. doi: 10.1016/j.brainres.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Wegiel J, Dowjat K, Kaczmarski W, Kuchna I, Nowicki K, Frackowiak J, Mazur-Kolecka B, Wegiel J, Silverman WP, Reisberg B, DeLeon M, Wisniewski T, Gong C-X, Liu F, Adayev T, Chen-Hwang M-C, Hwang Y-W. The role of overexpressed DYRK1A protein in the early onset of neurofibrillary degeneration in Down syndrome. Acta Neuropathol. 2008;116:391–407. doi: 10.1007/s00401-008-0419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegiel J, Gong CX, Hwang YW. The role of DYRK1A in neurodegenerative diseases. FEBS J. 2011a;278:236–245. doi: 10.1111/j.1742-4658.2010.07955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegiel J, Kaczmarski W, Barua M, Kuchna I, Nowicki K, Wang KC, Wegiel J, Yang S, Frackowiak J, Mazur-Kolecka B, Silverman WP, Reisberg B, Monteiro I, de Leon M, Wisniewski T, Dalton A, Lai F, Hwang YW, Adayev T, Liu F, Iqbal K, Iqbal IG, Gong CX. Link between DYRK1A overexpression and several-fold enhancement of neurofibrillary degeneration with 3-repeat tau protein in Down syndrome. J Neuropathol Exp Neurol. 2011b;70:36–50. doi: 10.1097/NEN.0b013e318202bfa1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods YL, Cohen P, Becker W, Jakes R, Goedert M, Wang X, Proud CG. The kinase DYRK phosphorylates protein-synthesis initiation factor elF2Bepsilon at Ser539 and the microtubule-associated protein tau at Thr212: potential role for DYRK as a glycogen synthase kinase 3-priming kinase. Biochem J. 2001;355:609–615. doi: 10.1042/bj3550609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang EJ, Ahn YS, Chung KC. Protein kinase Dyrk1 activates cAMP response element-binding protein during neuronal differentiation in hippocampal progenitor cells. J Biol Chem. 2001;276:39819–39824. doi: 10.1074/jbc.M104091200. [DOI] [PubMed] [Google Scholar]