Abstract
The mammalian kidney forms from several populations of progenitors that only persist during embryogenesis. The epithelial nephron progenitors reside in the cap mesenchyme (CM), whereas mesangial and endothelial cell progenitors reside in the neighboring stromal mesenchyme (SM). After a ureteric bud (UB) signal induces mesenchymal to epithelial transition of some CM cells, they form a nascent epithelial ball (a renal vesicle or RV) that require signals mediated by Notch receptors to separate proximal from distal fates. Two Notch receptors (Notch1 and Notch2) and two ligands (Jagged1 and Delta1) are expressed in the RV. Notably, instead of providing sufficient redundancy to ensure that losing any one allele will be inconsequential to human health, a reduction in the dose of one ligand (Jagged1) or one receptor (Notch2) is causally associated with a rare developmental syndrome (Alagille syndrome, ALGS) affecting eye, kidney, liver, and cranio-facial development. Here we will discuss our current understanding of the molecular basis for the non-redundant role of Notch2 in this process, and the avenue for new therapeutic strategies that these insights provide.
Keywords: Notch, Alagille, Jagged, Dll1, Kidney, nephron
Introduction
The intermediate mesoderm forms discrete cellular populations that interact to produce the metanephric kidney
The metanephric kidney is derived entirely from the intermediate mesoderm (IM). The dorsal IM will form the nephric duct (ND) or Wolffian duct [1], which extends in a rostral to caudal direction during development until it reaches the pelvis, connecting to the urethra. In contrast, the developmental potential of cells in the rostral/ventral IM becomes progressively restricted; at the level of the hind limb they form a bean-shaped territory called the metanephric mesenchyme (or MM) around E10. At that time the MM contains distinct populations of mesenchymal progenitors for the epithelial [2, 3], stromal [4] and vascular [5] components of the metanephric (adult) kidney. The epithelial progenitors begin to secrete GDNF, FGF10 and other inductive signals that activate receptor tyrosine kinases (RTKs) in the ND, triggering pseudostratification and evagination of a single ureteric bud (UB) from the ND. UB evagination is led by tip stem cells experiencing high levels of Ret activity [6, 7]. Continuous Ret activation triggered by MM signals causes the UB to extend and branch repeatedly, contributing to the future collecting duct system. At the same time, UB signals back to the MM to induce their differentiation into nephrons. Detailed discussion of the molecular mechanisms involved in ND and UB formations are discussed elsewhere [8, 9].
Wnt signals induce epithelia in a reiterative process
Two Wnt ligands, WNT9b and WNT4, are sequentially required to induce differentiation of nephron progenitors into renal epithelia. The UB produces WNT9b, which signals to the CM to perform two functions: first, Wnt9b supports expansion of the progenitors [10, 11]. Then, in cells conditioned by BMP7-mediated SMAD signaling [12], Wnt9b initiates mesenchymal to epithelial transition [13] and activates target genes including Fgf8 and Wnt4. Wnt4 then drives epithelialization [14] and Fgf8 drives proliferation leading to the formation of the renal vesicles (RV) [15, 16].
Genetic and pharmacological manipulations suggested that β-catenin/Lef/TCF signaling pathway is activated by both Wnt9b and Wnt4 signaling during CM differentiation [10, 11, 17, 18]. CM specific removal of β-catenin activity completely blocked the formation of RVs and abolished expression of genes such as Fgf8, Pax8 and Wnt4. In contrast, ectopic expression of stabilized β-catenin in the CM leads to differentiation of CM in vitro and rescued some differentiation defects in Wnt9b and Wnt4 mutants in vivo. Together, these results indicate that canonical Wnt signaling mediates the majority of signaling activity of Wnt9b and Wnt4 during nephrogenesis. However, non-canonical Wnt signals are also contributing [19], perhaps by antagonizing the canonical signal: only transient, low-level stabilization of β-catenin could induce epithelization [11, 20], whereas continuous signal or a higher dosage blocks this process [18]. This highlights the requirement for tightly regulated Wnt activity for proper nephron differentiation.
Nephron segmentation: the separation of proximal fates from mostly distal-fated epithelia
In order to form functional nephrons, newly formed epithelia need to acquire a proximal-distal axis (PDA) in a process called segmentation and elongate giving rise to more than 15 different cells types [21] within the mature nephron (Fig. 1C). Morphologically, the progenitor niche remains at the surface of the developing metanephric kidney, while the nascent RVs always appear under the UB branch until the last day of nephrogenesis [22]. Beginning as an un-polarized cell cluster, RV quickly acquires apical-basal polarity to create a lumen [23]. At this stage, differential gene expression along the PDA in the RV suggests that nephron segmentation has already been initiated. Notch ligands (Jag1, Dll1) and Wnt signaling components (Wnt4, Dkk1, and Ccnd1) are all expressed in the distal RV, while Notch receptor (Notch1, Notch2) and transcription factor Wt1 are highly expressed on the proximal RV. The RV PDA is always aligned with the duct tip at the distal end, facilitating fusion with the duct that occurs before a burst of proliferation that contorts the nascent tube into an S-shaped body [24].
Figure 1.
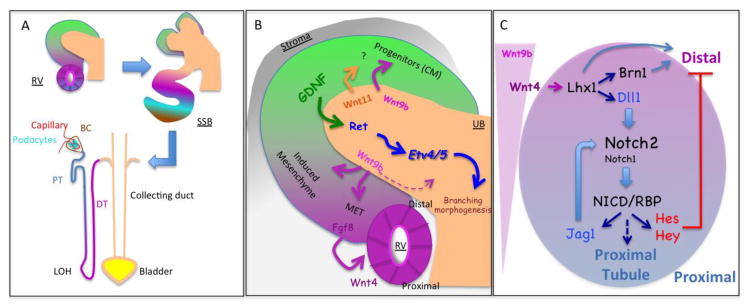
Schematic representation of steps in nephrogenesis. (A) Renal vesicles (RV) form beneath the ureteric bud (UB) tip near the periphery of the developing kidney. These polarized epithelial ball elongate into an S shape body (SSB) that will give rise to the entire nephron. Proximal-distal axis (PDA) organization in the RV and SSB seems to be preserved in the nephron with the most proximal [Bowman capsule (BC) and podocytes] followed by intermediate structures [proximal tubules (PT), loop of Henle (LOH)] and distal one [distal tubule (DT), connecting segment and collecting duct]. (B). GDNF/Ret/Etv4-5 signaling controls UB branching whereas Wnt signaling controls progenitor expansion in the cap mesenchyme (CM) as well as their differentiation via mesenchymal to epithelial transition (MET). Wnt4 is produced by the epithelia. (C) Wnt9b could create a morphogen gradient initiating the formation of PDA but Notch signaling is required to fix the PDA and allow proximal fate selection. The role of the genes listed in C is reviewed elsewhere [8, 26]
The earliest molecular evidence of a PDA include changes in the expression of Pax2 and Wt1 which are seen even in mutants that fail to establish a proximal domain [25]. This PDA alignment is reminiscent of a process involving a morphogen gradient, with Wnt as a candidate morphogen produced by the UB [26]. A morphogen gradient would be sufficient to establish a PD axis in other system, but not here: a signal exchanged between RV cells is required to establish the emerging proximal domain to generate the proximal tubule, podocytes, and partial epithelium lining the bowman capsule. This process requires a signal generated by Notch receptors [27, 28].
The Notch signaling pathway
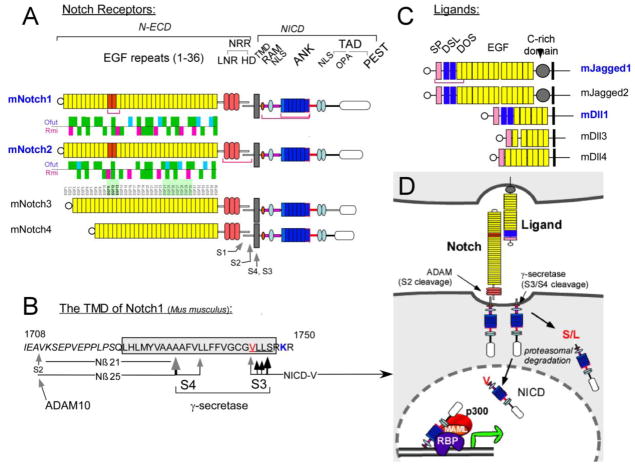
Notch proteins are large, single-pass molecules with an extracellular domain containing 29–36 EGF repeats and a negative control region (NRR) that folds into a structure unique to Notch proteins (Fig. 2; for reviews, see [29, 30]). Activation occurs when ligand binding exerts a force that unfolds the NRR [31], exposing a hidden target (called S2) to the protease ADAM10 [29, 32] which cleaves ligand-bound Notch receptors shedding their extracellular domains. This cleavage generates a truncated Notch substrates for the enzyme -secretase [33] which cleaves the Notch transmembrane domain at multiple peptide bonds, starting near the inner plasma membrane leaflet at S3 (Fig. 2B,D). Notch intracellular domain (NICD) fragments containing a Val or Met residues at their amino terminus are longer-lived and translocate to the nucleus, where they bind the protein RBPjk and recruit the adaptor Mastermind-like (MAML), p300, the mediator complex, and RNA polymerase II [29]. The Notch pathway is used throughout development and in adult tissue to enable cells to choose one of two cellular options in coordination with their neighbors [34].
Figure 2.
Type I proteins belonging to the mammalian Notch receptor and ligand families. A) Notch proteins contain multiple extracellular EGF-like repeats. Repeats 11–12 (orange) mediate interactions with ligands. 4 mammalian Notch receptors differ in the number of repeats (29–36). EGF repeats are followed by three cysteine-rich Lin12-Notch repeats (LNR-A, B and C) and a heterodimerization domain (HD) which together fold into one globular domain, the NRR. Notch proteins are cleaved by furin-like convertases at site 1 (S1). EGF repeats may contains consensus motifs for fucosylation by O-Fut1 followed by glycosylation by Fringe; Rumi, another glycosyltransferase, has its own conserved sites. The distribution of potential fringe (Cyan) and rumi (magenta) sites in the extracellular domain are shown for mNotch1 and mNotch2 (Green- same in by both receptors). Note the ligand binding regions differ in their modification patterns. B) Details of mouse Notch1 proteolysis. After ligand binding, cleavage at S2 is followed by cleavage at the S3 region. Multiple scissile bonds are cleaved but only peptides initiating at Val 1744 evade Notch-end rule degradation. Cleavage then proceeds towards the cell surface until the short Nβ peptides can escape the lipid bilayer; most are 21 amino acids long. Two amino acid substitutions (V1744G and K1749R, colorized) change the choice of scissile bonds. Only antibodies that recognize the underlined sequence detect the active form of Notch1. C) Ligands of Notch receptors can be divided into two groups based on the length and subtype of EGF-like repeats they contain (DSL, DOS and EGF motifs; see [29]. Red brackets mark domains that have been crystallized alone or in combination with binding partners. Abbreviated domain names are: SP- signal peptide. DSL- delta, serrate, lag1. DOS- Delta and OSM-11-like. D). a schematic diagram of the Notch activation mechanism. Explained in the text and in [29].
The two Notch receptors expressed in the RV are not equivalent
In the RV, two of four mammalian Notch receptors (Notch1 and Notch2) are co-expressed. Despite the great degree of conservation between Notch1 and Notch2 and overlap in their expression domain, the two receptors are not functionally equivalent: in mice, loss of both Notch1 alleles is tolerated, but loss of two Notch2 alleles is not; in humans, loss of one Notch2 allele leads to Alagille syndrome [25, 35–38]. In the mouse, Alagille-like symptoms were seen only when the dose of Notch2 and the ligand Jag1 are simultaneously reduced [35]. In contrast, no Notch1 or Dll1 mutations have been linked to Allagile. Notch1 protein does provide some activity during RV segmentation, but this can only be demonstrated in a sensitized background in which Notch2 levels are greatly reduced [39]. Given that all NICD proteins contain highly conserved RBPjk binding domain and Ankyrin repeats, and that all bind to RBPjk with the same affinity [40–42] without changing RBPjk sequence preferences [43], a nagging question remained: why is Notch1 incapable of rescuing loss of one Notch2 allele? And, given that that loss of one Notch2 allele causes Alagille syndrome, what can be done to elevate Notch1 activity in affected organs without causing other systemic problems?
To address the mechanism behind Alagille, we created two new mouse strains harboring genes we call Notch12 and Notch21 (Fig. 3). These alleles were created by swapping the genomic sequences coding for the signaling element (the intracellular domains or ICDs) between the two receptors, leaving the promoter, regulatory elements, the extracellular domain, the transmembrane domain and the UTR of the original locus intact [44]. Thus, expression domains, transcript stability and protein levels, ligand binding and cleavage are all controlled by the locus (the Notch1 locus controlling Notch12 and the Notch2 locus controlling Notch21) but the signaling entity released after γ-secretase cleavage has been swapped. Given that the few amino acids distinguishing N1ICD from N2ICD are all on exposed surfaces of the molecule (Fig. 4), this experiment will ask whether these differences can drive the assembly of different transcription complexes by Notch1 vs Notch2. If the two ICD are equivalent, it may help identify other elements within the locus that establish Notch2 as the main signaling receptor during renal development.
Figure 3.
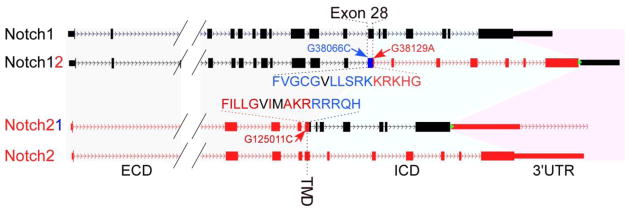
The Notch12 and Notch21 alleles. (A) Schematic illustration of Notch1 (black) and Notch2 (red) loci before and after the ICD swap. The swapped fragments between the Notch1 and Notch2 loci range from after the transmembrane domain in Exon 28 to the stop codon in Exon 34. The Notch1 ICD encompasses 5,926bp on chromosome 2, ranging from nucleotide +38,103 to +44,028 (numbering the A in ATG as +1) and encoding amino acid 1,750 to 2,531; for Notch2, the ICD encompasses 8,699bp on chromosome 3, ranging from nucleotide +125,048 to +133,746 and encoding amino acid 1,705 to 2,473. G38066C, G38129A and G125011C denote silent single nucleotide mutations introduced into the targeting constructs for pyrosequencing-based ES cell screening and mRNA comparisons. Amino acids in black denote the S3 cleavage sites.
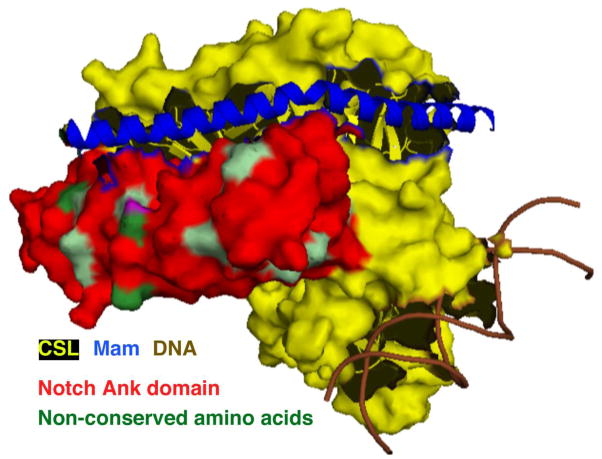
Figure 4.
The superimposed crystal structure of Notch1, Notch2, RBP, Mastermind-like (MAML) and DNA shows that non-conserved amino acids (light and dark green) between Notch1 and Notch2 ICDs exist at the surface of the transcription complex, which might serve to recruit different binding partners and determine specificity of target genes.
Analysis of these mice has just been completed, and the results are unequivocal [44]. We detected no differences in mRNA abundance or in the overall levels of Notch1 and Notch2 protein expression in nascent renal epithelia. The surface distribution of the Notch1 and Notch12 proteins was identical, as was the surface levels of Notch2 and Notch21 indicating the domain swap did not affect cellular expression, stability, or transport. Only Notch2 was expressed in CM cells but we have established previously that the CM does not use Notch signaling to self renew or to execute MET [25, 28, 45]. Importantly, as long as it was released from a Notch21 protein, a single Notch21 allele producing N1ICD was sufficient to support the formation of glomeruli domains in every single nephrons, whereas two alleles of Notch12, releasing N2ICD, could not support the formation of even one glomerulus. In short, the Notch1 and Notch2 ICD are interchangeable. The difference is due to other elements in the Notch1 and Notch2 locus.
Two differences between Notch21 and Notch12 did emerge. First, two different pairs of antibodies to the Notch extracellular domains (ECDs) established that Notch2 was between 2 and 3 fold more abundant than Notch1 on the cell surface of nascent renal epithelia. Second, when we measured how many NICD entered the nucleus in cells containing equivalent surface levels of Notch1 and Notch21, we noticed again a 2 fold advantage to Notch21 proteins activated by either Dll1 or Jag1 ligands. These small differences could synergize to create a significant overall difference in the amounts of NICD present in the nucleus, reaching a threshold necessary for nephron segmentation only when Notch2 molecules are present.
What was the molecular basis of these differences between Notch1 and Notch2? Since overall expression levels were the same, and the swap of ICD did not alter the surface distribution of Notch1 relative to Notch12 or of Notch2 relative to Notch21 (Fig. 3 in [44]), the ECD holds the key. Not only does the Notch 2 ECD contain signals that promote exocytosis but it is also better at unfolding the NRR in response to ligand. Notch exocytosis involves passage through a furin-containing compartment, which cleaves Notch at an unstructured loop protruding from the folded NRR at S1 (see [46–48] for NMR and crystal structures of the Notch1 and Notch2 NRRs). This cleavage generates a form of Notch1 or Notch2 that is longer-lived at the surface than the un-processed, full-length protein [49, 50]. Interestingly, when the S1 containing loop was removed, no structural changes in the NRRs were detected but surface expression of Notch1 lacking the S1 loop was reduced and its response to ligand was reduced as well [46]. In contrast, Notch2 lacking the S1 loop reached the surface and accumulated there to a similar degree with WT Notch2, and had an improved response to ligand. It thus appears safe to speculate that the NRR may regulate differential exocytosis as well as ligand responsiveness, and that the details of transporting Notch molecules to the surface will explain the functional differences between the two proteins.
The two Notch ligands expressed in the RV are also not equivalent
Similar to what has been observed with the receptors, the Notch ligands Jag1 and Dll1 are co-expressed in the developing RVs yet their functions are not redundant. In human, whereas 94% of Alagille patients harbor Jag1 mutations, not one Dll1 mutation has been identified so far [51]. Consistently, deletion of Jag1 in mouse results in dramatic reduction of nephron numbers, while deletion of Dll1 produced a milder phenotype [44]. Clearly, Jag1 ligand and Notch2 receptor produce the key signals during RV segmentation in both mouse and human kidneys.
Why does Jag1 play a more important role than Dll1 in this process? After all, Dll1 is a dominant ligand, activating Notch1 during the maintenance of gut stem cells [52]. It is indispensable for the activation of Notch2 during marginal zone B cell development in the spleen [53, 54]. The answer lies in the modification status of the Notch extracellular domains by Fringe glycosyltransferases. The three related enzymes (lunatic fringe (Lfng), manic fringe (Mfng) and radial fringe (Rfgn)) mediate glycosylation of specific, fucosylated Ser/Thr residues on Notch receptors (Fig. 2A and [55]) and result in qualitatively different interaction between the Notch receptors and their ligands. In general, these modifications enhance the response to Dll ligands but suppress the response to Jag ligands; however, these effects are highly cell-context specific. At least in one system the activation of Notch2 by Jag1 was enhanced, not suppressed, by Lfng modification and subsequent glycosylation [56].
In the developing kidney, Lfng is expressed in the same expression domain of Notch receptors and ligands [44]. More importantly, tests in the human embryonic kidney 293 cell line show that Lfng expression significantly potentiates the response of Notch1 to Dll1, inhibits its response to Jag1, but enhances the responses of Notch2 to both ligands. These data suggest that Lfng modification underlie the unequal contributions of different receptor-ligand pairs during nephrogenesis.
Implications for the clinic
At present, several Notch antagonists have entered clinical trials, mostly targeting T-cell acute lymphoblastic leukemia and other tumors [57]. No agonists have been developed with the exception of EDTA [58]. Agonists will not present a meaningful intervention in Alagille because of the many untoward effects anticipated with elevating Notch activity in all the developmental processes in which Notch is involved, which may literally be too many to count. However, the observation that differential transport differentiates Notch2 from Notch1 opens a possible avenue for therapeutic intervention. We know that not all cells display the same transport bias, and it may be that the tissues affected in Alagille are all the ones favoring Notch2 transport over Notch1. Investigation into the mechanism preventing Notch1 from efficiently reaching the surface in these cells but not in others may lead to new strategy for augmenting the surface presence of Notch1 without a negative impact on other tissues.
References
- 1.Saxen L. Organogenesis of the kidney. Cambridge University Press; Cambridge: 1987. [Google Scholar]
- 2.Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, McMahon AP. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3:169–181. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle S, Misfeldt A, Chandler KJ, Deal KK, Southard-Smith EM, Mortlock DP, Baldwin HS, de Caestecker M. Fate mapping using Cited1-CreERT2 mice demonstrates that the cap mesenchyme contains self-renewing progenitor cells and gives rise exclusively to nephronic epithelia. Dev Biol. 2008;313:234–245. doi: 10.1016/j.ydbio.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Humphreys BD, Lin SL, Kobayashi A, Hudson TE, Nowlin BT, Bonventre JV, Valerius MT, McMahon AP, Duffield JS. Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am J Pathol. 2010;176:85–97. doi: 10.2353/ajpath.2010.090517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sequeira Lopez ML, Gomez RA. Development of the renal arterioles. J Am Soc Nephrol. 2011;22:2156–2165. doi: 10.1681/ASN.2011080818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chi X, Michos O, Shakya R, Riccio P, Enomoto H, Licht JD, Asai N, Takahashi M, Ohgami N, Kato M, Mendelsohn C, Costantini F. Ret-dependent cell rearrangements in the Wolffian duct epithelium initiate ureteric bud morphogenesis. Dev Cell. 2009;17:199–209. doi: 10.1016/j.devcel.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shakya R, Watanabe T, Costantini F. The role of GDNF/Ret signaling in ureteric bud cell fate and branching morphogenesis. Dev Cell. 2005;8:65–74. doi: 10.1016/j.devcel.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Costantini F, Kopan R. Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev Cell. 2010;18:698–712. doi: 10.1016/j.devcel.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costantini F. Genetic controls and cellular behaviors in branching morphogenesis of the renal collecting system. Wiley Interdiscip Rev Dev Biol. 2012;1:693–713. doi: 10.1002/wdev.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karner CM, Das A, Ma Z, Self M, Chen C, Lum L, Oliver G, Carroll TJ. Canonical Wnt9b signaling balances progenitor cell expansion and differentiation during kidney development. Development. 2011;138:1247–1257. doi: 10.1242/dev.057646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park J-S, Ma W, O’Brien LL, Chung E, Guo J-J, Cheng J-G, Valerius MT, McMahon JA, Wong W-H, McMahon AP. Six2 and Wnt Regulate Self-Renewal and Commitment of Nephron Progenitors through Shared Gene Regulatory Networks. Dev Cell. 2012;23:637–651. doi: 10.1016/j.devcel.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown AC, Muthukrishnan SD, Guay JA, Adams DC, Schafer DA, Fetting JL, Oxburgh L. Role for compartmentalization in nephron progenitor differentiation. Proc Natl Acad Sci U S A. 2013;110:4640–4645. doi: 10.1073/pnas.1213971110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll TJ, Park JS, Hayashi S, Majumdar A, McMahon AP. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell. 2005;9:283–292. doi: 10.1016/j.devcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 14.Stark K, Vainio S, Vassileva G, McMahon AP. Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature. 1994;372:679–683. doi: 10.1038/372679a0. [DOI] [PubMed] [Google Scholar]
- 15.Grieshammer U, Cebrian C, Ilagan R, Meyers E, Herzlinger D, Martin GR. FGF8 is required for cell survival at distinct stages of nephrogenesis and for regulation of gene expression in nascent nephrons. Development. 2005;132:3847–3857. doi: 10.1242/dev.01944. [DOI] [PubMed] [Google Scholar]
- 16.Perantoni AO, Timofeeva O, Naillat F, Richman C, Pajni-Underwood S, Wilson C, Vainio S, Dove LF, Lewandoski M. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development. 2005;132:3859–3871. doi: 10.1242/dev.01945. [DOI] [PubMed] [Google Scholar]
- 17.Karner CM, Merkel CE, Dodge M, Ma Z, Lu J, Chen C, Lum L, Carroll TJ. Tankyrase is necessary for canonical Wnt signaling during kidney development. Dev Dyn. 2010;239:2014–2023. doi: 10.1002/dvdy.22340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park JS, Valerius MT, McMahon AP. Wnt/{beta}-catenin signaling regulates nephron induction during mouse kidney development. Development. 2007;134:2533–2539. doi: 10.1242/dev.006155. [DOI] [PubMed] [Google Scholar]
- 19.Tanigawa S, Wang H, Yang Y, Sharma N, Tarasova N, Ajima R, Yamaguchi TP, Rodriguez LG, Perantoni AO. Wnt4 induces nephronic tubules in metanephric mesenchyme by a non-canonical mechanism. Dev Biol. 2011;352:58–69. doi: 10.1016/j.ydbio.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuure S, Popsueva A, Jakobson M, Sainio K, Saino H. Glycogen Synthase Kinase-3 Inactivation and Stabilization of β-Catenin Induce Nephron Differentiation in Isolated Mouse and Rat Kidney Mesenchymes. J Am Soc Nephrol. 2007;18:1130–1139. doi: 10.1681/ASN.2006111206. [DOI] [PubMed] [Google Scholar]
- 21.Herzlinger D, Koseki C, Mikawa T, al-Awqati Q. Metanephric mesenchyme contains multipotent stem cells whose fate is restricted after induction. Development. 1992;114:565–572. doi: 10.1242/dev.114.3.565. [DOI] [PubMed] [Google Scholar]
- 22.Rumballe BA, Georgas KM, Combes AN, Ju AL, Gilbert T, Little MH. Nephron formation adopts a novel spatial topology at cessation of nephrogenesis. Dev Biol. 2011;360:110–122. doi: 10.1016/j.ydbio.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Z, Zimmerman S, Brakeman PR, Beaudoin GM, 3rd, Reichardt LF, Marciano DK. De novo lumen formation and elongation in the developing nephron: a central role for afadin in apical polarity. Development. 2013;140:1774–1784. doi: 10.1242/dev.087957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georgas K, Rumballe B, Valerius MT, Chiu HS, Thiagarajan RD, Lesieur E, Aronow BJ, Brunskill EW, Combes AN, Tang D, Taylor D, Grimmond SM, Potter SS, McMahon AP, Little MH. Analysis of early nephron patterning reveals a role for distal RV proliferation in fusion to the ureteric tip via a cap mesenchyme-derived connecting segment. Dev Biol. 2009;332:273–286. doi: 10.1016/j.ydbio.2009.05.578. [DOI] [PubMed] [Google Scholar]
- 25.Cheng HT, Kim M, Valerius MT, Surendran K, Schuster-Gossler K, Gossler A, McMahon AP, Kopan R. Notch2, but not Notch1, is required for proximal fate acquisition in the mammalian nephron. Development. 2007;134:801–811. doi: 10.1242/dev.02773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kopan R, Cheng HT, Surendran K. Molecular Insights into Segmentation along the Proximal-Distal Axis of the Nephron. J Am Soc Nephrol. 2007;18:2014–2020. doi: 10.1681/ASN.2007040453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang P, Pereira FA, Beasley D, Zheng H. Presenilins are required for the formation of comma- and S-shaped bodies during nephrogenesis. Development. 2003;130:5019–5029. doi: 10.1242/dev.00682. [DOI] [PubMed] [Google Scholar]
- 28.Cheng H, Miner J, Lin M, Tansey MG, Roth KA, Kopan R. g-Secretase Activity is Dispensable for the Mesenchyme-to-Epithelium Transition but Required for Proximal Tubule Formation in Developing Mouse Kidney. Development. 2003;130:5031–5041. doi: 10.1242/dev.00697. [DOI] [PubMed] [Google Scholar]
- 29.Kopan R, Ilagan MX. The Canonical Notch Signaling Pathway: Unfolding the Activation Mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon WR, Arnett KL, Blacklow SC. The molecular logic of Notch signaling--a structural and biochemical perspective. J Cell Sci. 2008;121:3109–3119. doi: 10.1242/jcs.035683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Musse AA, Meloty-Kapella L, Weinmaster G. Notch ligand endocytosis: Mechanistic basis of signaling activity. Semin Cell Dev Biol. 2012;23:429–436. doi: 10.1016/j.semcdb.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groot AJ, Cobzaru C, Weber S, Saftig P, Blobel CP, Kopan R, Vooijs M, Franzke CW. Epidermal ADAM17 Is Dispensable for Notch Activation. J Invest Dermatol. 2013;133:2286–2288. doi: 10.1038/jid.2013.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jorissen E, De Strooper B. Gamma-secretase and the intramembrane proteolysis of Notch. Curr Top Dev Biol. 2010;92:201–230. doi: 10.1016/S0070-2153(10)92006-1. [DOI] [PubMed] [Google Scholar]
- 34.Artavanis-Tsakonas S, Muskavitch MA. Notch: the past, the present, and the future. Curr Top Dev Biol. 2010;92:1–29. doi: 10.1016/S0070-2153(10)92001-2. [DOI] [PubMed] [Google Scholar]
- 35.McCright B, Lozier J, Gridley T. A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development. 2002;129:1075–1082. doi: 10.1242/dev.129.4.1075. [DOI] [PubMed] [Google Scholar]
- 36.Kamath BM, Bauer RC, Loomes KM, Chao G, Gerfen J, Hutchinson A, Hardikar W, Hirschfield G, Jara P, Krantz ID, Lapunzina P, Leonard L, Ling S, Ng VL, Hoang PL, Piccoli DA, Spinner NB. NOTCH2 mutations in Alagille syndrome. J Med Genet. 2012;49:138–144. doi: 10.1136/jmedgenet-2011-100544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, Spinner NB. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet. 2006;79:169–173. doi: 10.1086/505332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrissette JD, Colliton RP, Spinner NB. Defective intracellular transport and processing of JAG1 missense mutations in Alagille syndrome. Hum Mol Genet. 2001;10:405–413. doi: 10.1093/hmg/10.4.405. [DOI] [PubMed] [Google Scholar]
- 39.Surendran K, Boyle S, Barak H, Kim M, Stromberski C, McCright B, Kopan R. The contribution of Notch1 to nephron segmentation in the developing kidney is revealed in a sensitized Notch2 background and can be augmented by reducing Mint dosage. Dev Biol. 2010;337:386–395. doi: 10.1016/j.ydbio.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lubman OY, Ilagan MX, Kopan R, Barrick D. Quantitative Dissection of the Notch:CSL Interaction: Insights into the Notch-mediated Transcriptional Switch. J Mol Biol. 2007;365:577–589. doi: 10.1016/j.jmb.2006.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friedmann DR, Wilson JJ, Kovall RA. RAM-induced allostery facilitates assembly of a notch pathway active transcription complex. J Biol Chem. 2008;283:14781–14791. doi: 10.1074/jbc.M709501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Del Bianco C, Aster JC, Blacklow SC. Mutational and energetic studies of Notch 1 transcription complexes. J Mol Biol. 2008;376:131–140. doi: 10.1016/j.jmb.2007.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Del Bianco C, Vedenko A, Choi SH, Berger MF, Shokri L, Bulyk ML, Blacklow SC. Notch and MAML-1 complexation do not detectably alter the DNA binding specificity of the transcription factor CSL. PLoS One. 2010;5:e15034. doi: 10.1371/journal.pone.0015034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Z, Chen S, Boyle S, Ilagan MX, Zhu Y, Zhang A, Kopan R. The Extracellular Domain of Notch2 Increases its Cell surface Abundance and Ligand Responsiveness During Kidney. Dev Cell. 2013;25:585–598. doi: 10.1016/j.devcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyle SC, Kim M, Valerius MT, McMahon AP, Kopan R. Notch pathway activation can replace the requirement for Wnt4 and Wnt9b in mesenchymal-to-epithelial transition of nephron stem cells. Development. 2011;138:4245–4254. doi: 10.1242/dev.070433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gordon WR, Vardar-Ulu D, L’Heureux S, Ashworth T, Malecki MJ, Sanchez-Irizarry C, McArthur DG, Histen G, Mitchell JL, Aster JC, Blacklow SC. Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PLoS One. 2009;4:e6613. doi: 10.1371/journal.pone.0006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gordon WR, Roy M, Vardar-Ulu D, Garfinkel M, Mansour MR, Aster JC, Blacklow SC. Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL. Blood. 2009;113:4381–4390. doi: 10.1182/blood-2008-08-174748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- 49.Blaumueller CM, Qi HL, Zagouras P, Artavanistsakonas S. Intracellular Cleavage Of Notch Leads to a Heterodimeric Receptor On the Plasma Membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 50.Logeat F, Bessia C, Brou C, Lebail O, Jarriault S, Seidah NG, Israel A. The Notch1 Receptor Is Cleaved Constitutively By a Furin-Like Convertase. Proc Natl Acad Sci U S A. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kamath BM, Spinner NB, Rosenblum ND. Renal involvement and the role of Notch signalling in Alagille syndrome. Nat Rev Nephrol. 2013;9:409–418. doi: 10.1038/nrneph.2013.102. [DOI] [PubMed] [Google Scholar]
- 52.Pellegrinet L, Rodilla V, Liu Z, Chen S, Koch U, Espinosa L, Kaestner KH, Kopan R, Lewis J, Radtke F. Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology. 2011;140:1230–1240. e1231–1237. doi: 10.1053/j.gastro.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hozumi K, Negishi N, Suzuki D, Abe N, Sotomaru Y, Tamaoki N, Mailhos C, Ish-Horowicz D, Habu S, Owen MJ. Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat Immunol. 2004;5:638–644. doi: 10.1038/ni1075. [DOI] [PubMed] [Google Scholar]
- 54.Saito T, Chiba S, Ichikawa M, Kunisato A, Asai T, Shimizu K, Yamaguchi T, Yamamoto G, Seo S, Kumano K, Nakagami-Yamaguchi E, Hamada Y, Aizawa S, Hirai H. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 2003;18:675–685. doi: 10.1016/s1074-7613(03)00111-0. [DOI] [PubMed] [Google Scholar]
- 55.Stanley P, Okajima T. Roles of glycosylation in Notch signaling. Curr Top Dev Biol. 2010;92:131–164. doi: 10.1016/S0070-2153(10)92004-8. [DOI] [PubMed] [Google Scholar]
- 56.Hicks C, Johnston SH, diSibio G, Collazo A, Vogt TF, Weinmaster G. Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nat Cell Biol. 2000;2:515–520. doi: 10.1038/35019553. [DOI] [PubMed] [Google Scholar]
- 57.Aster JC, Blacklow SC. Targeting the Notch pathway: twists and turns on the road to rational therapeutics. J Clin Oncol. 2012;30:2418–2420. doi: 10.1200/JCO.2012.42.0992. [DOI] [PubMed] [Google Scholar]
- 58.Rand DM, Grimm MLM, Artavanis-Tsakonas S, Patriub V, Blacklow CS, Sklar CJ, Aster CJ. Calcium depletion dissociates and activates heterodimeric Notch receptors. Mol Cell Biol. 2000;20:1825–1835. doi: 10.1128/mcb.20.5.1825-1835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]