Abstract
Sequencing of pediatric gliomas has identified missense mutations Lys27Met (K27M) and Gly34Arg/Val (G34R/V) in genes encoding histone H3.3 (H3F3A) and H3.1 (HIST3H1B). We report that human diffuse intrinsic pontine gliomas (DIPGs) containing the K27M mutation display significantly lower overall amounts of H3 with trimethylated lysine 27 (H3K27me3) and that histone H3K27M transgenes are sufficient to reduce the amounts of H3K27me3 in vitro and in vivo. We find that H3K27M inhibits the enzymatic activity of the Polycomb repressive complex 2 through interaction with the EZH2 subunit. In addition, transgenes containing lysine-to-methionine substitutions at other known methylated lysines (H3K9 and H3K36) are sufficient to cause specific reduction in methylation through inhibition of SET-domain enzymes. We propose that K-to-M substitutions may represent a mechanism to alter epigenetic states in a variety of pathologies.
Somatic mutations in genes encoding proteins that modify chromatin dynamics frequently contribute to tumorigenesis (1). Mutations in subunits of the Polycomb repressive complex 2 (PRC2) are often associated with tumor progression (2). PRC2 normally helps maintain epigenetic gene silencing and X chromosome inactivation through enzymatic di- and trimethylation of K27 on histone H3 (3). In addition to enzymatic machinery, histone H3 missense mutations in pediatric gliomas represent direct evidence that alterations of the histones themselves can promote cancer. In two pediatric brain cancers, diffuse intrinsic pontine gliomas (DIPGs) and supratentorial glioblastoma multiforme (GBMs), 60% of patients studied exhibited one of two mutually exclusive mutations in either H3F3A, one of two genes encoding the histone H3 variant H3.3, or HIST1H3B, one of several genes encoding H3.1 (4–6). The K27M mutation occurring in either H3F3A or HIST1H3B was observed in nearly 80% of DIPGs, and 22% of non–brain stem gliomas (6).
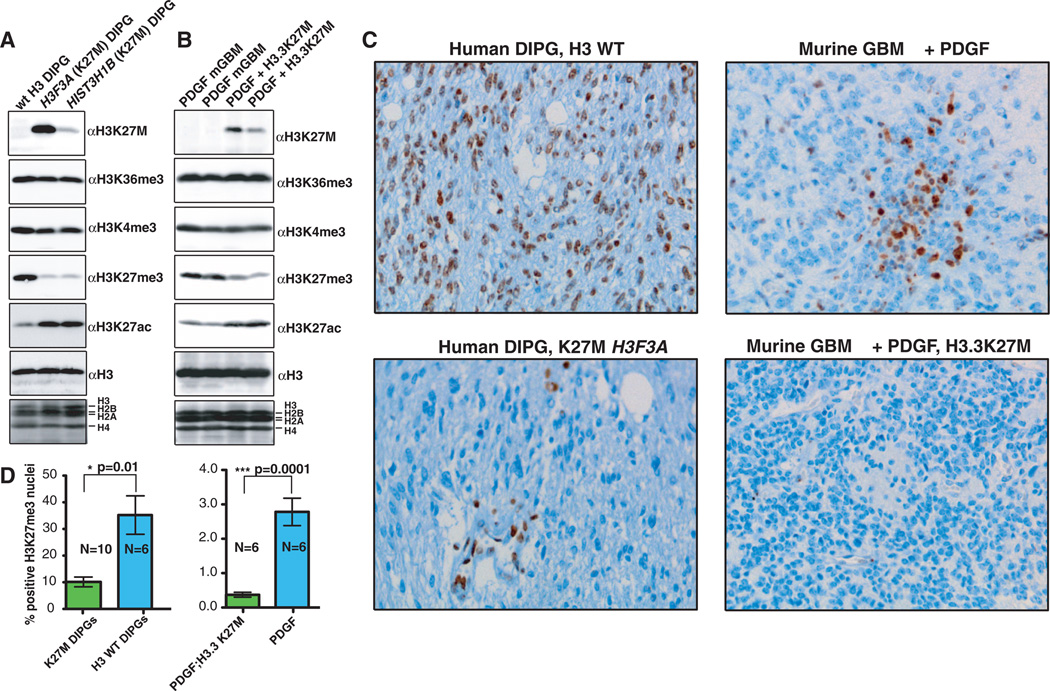
We sought to determine whether DIPG samples that contain the K27M mutation exhibit global changes in key regulatory histone modifications. Immunoblots with antisera raised against the K27M substitution (fig. S1A) indicated the presence of H3 K27M protein in DIPG samples containing H3F3A (H3.3) or HIST3H1B (H3.1) alleles (Fig. 1A). DIPG tumors containing H3K27M mutations exhibited both a decrease in H3 with trimethylated lysine 27 (H3K27me3) and a modest increase in amounts of H3 with acetylated lysine 27 (H3K27ac) by immunoblot with modification-specific antisera (Fig. 1, A and C). Quantification of H3K27me3 by immunohistochemistry demonstrated that K27M mutant DIPGs contained significantly less H3K27me3 than non-K27M mutant DIPGs (Fig. 1D). The H3.1/3 K27M protein is 3.63% (±0.33) to 17.61% (±1.11) of total H3 in human DIPG samples, as measured by quantitative mass spectrometry (fig. S1B). Two histone modifications related to transcription and/or activation (H3K4me3 and H3K36me3) were similar in DIPG samples regardless of tumor genotype, which could be used to argue that global changes in posttranslational modifications were specific to H3K27. Using a platelet-derived growth factor (PDGF)–induced brain stem glioma model to characterize this mutation in vivo (7), we found that transgenic H3.3K27M was sufficient to increase H3K27ac and to significantly reduce H3K27me3 (Fig. 1, B to D). Expression of H3.3K27M with p53 loss in nestin progenitors of the neonatal mouse brain stem was not sufficient to generate gliomas but did induce proliferating ectopic cell clusters in 72% (21 out of 29) of the mice (fig. S1C), whereas expression of the wild-type H3.3 with p53 loss (n = 8) in nestin progenitors of the neonatal mouse brain stem did not result in the induction of ectopic proliferating cell clusters (fig. S1D).
Fig. 1. Histones extracted from human DIPG containing H3F3A K27M (H3.3) or HIST3H1B K27M (H3.1) mutations exhibit decreased H3K27me3.
(A) Immunoblots of acid-extracted histones from DIPGs of different indicated H3 genotypes. (B) Immunoblots of histones from PDGF-induced gliomas with and without the H3.3K27M transgene. (C) Immunohistochemistry of H3K27me3 in human or murine gliomas containing wild-type H3.3 or H3.3K27M. (D) Immunohistochemistry quantification of nuclei staining positive for H3K27me3 (P=0.01 for human DIPG, and P = 0.0001 for murine gliomas with the unpaired t test).
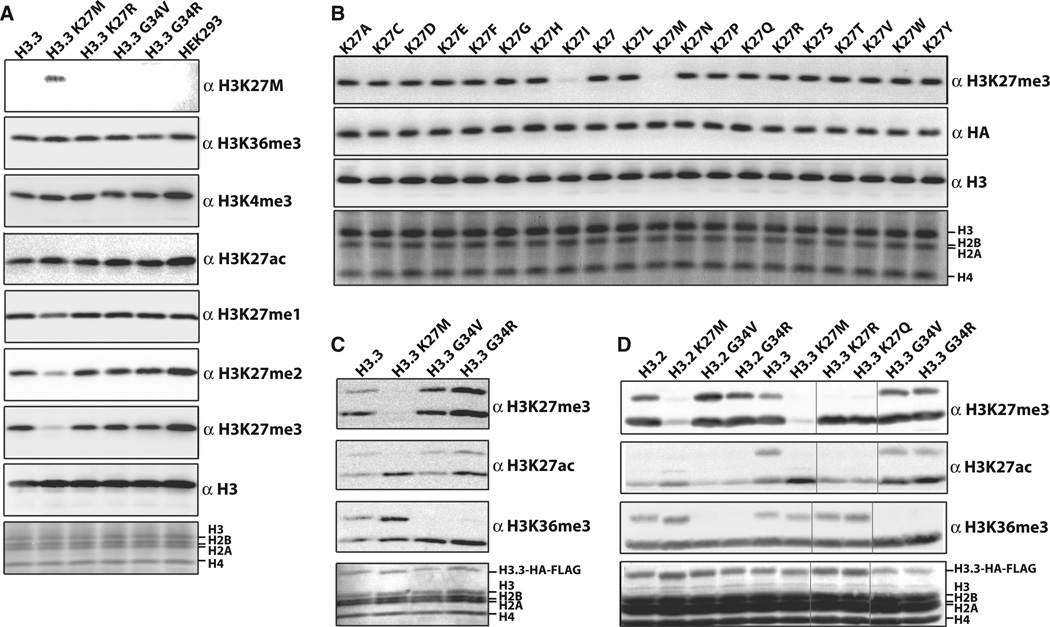
To investigate the mechanism by which H3K27M decreased overall H3K27me3, we generated stable human embryonic kidney–293T (HEK293T) cell lines that express FLAG- and hemagglutinin (HA) epitope–tagged wild-type histone H3.3 or K27M and G34R/V mutants, which contributed to about 1% of total cellular histone H3, as measured by immunoblot. Cells expressing the H3K27M mutant histone exhibited a profound reduction in H3K27me2/3, with little changes in the amounts ofH3K4me3 or H3K36me3 (Fig. 2A). This reduction was specific to the H3K27M mutation, as amounts of H3K27me2/3 remained unchanged in H3.3G34R/V transgenic cell lines. We did not observe change in the amounts of PRC2 complex components in H3.3K27M, PDGF-induced glioblastoma cell lines (fig. S2A).
Fig. 2. H3K27M transgenes cause a global decrease of H3K27me3.
(A) Immunoblots of whole-cell extract from lentivirus-transduced 293T cells expressing the indicated H3.3 transgenes. (B) Immunoblots of whole extract from H3.3 K27-to-X transduced 293T cells. The antibody against HA (anti-HA) blot shows the relative H3.3 transgene amounts. (C) Immunoblots of anti-HA–immunoprecipitated heterotypic H3.3 K27M or G34R/V mononucleosomes from the indicated H3.3 transgenic 293T cell lines. (D) Immunoblots of anti-HA–immunoprecipitated oligonucleosomes (>95%of four to five nucleosomes in length, with a median of two to three nucleosomes) from the indicated H3.3 transgenic 293T cell lines (vertical black lines indicate where control lanes were removed for clarity).
The heterozygous and invariant nature of the lysine-to-methionine mutation at residue 27 in nearly 80% of pediatric DIPGs strongly suggests that this specific amino acid substitution imparts a unique gain-of-function to the mutant histone. To further test the specificity of this substitution, we performed a survey of all amino acid substitutions at H3K27. Nearly all substitutions had little effect, if any, on the amounts of K27me3, with the exception of methionine, and to a lesser extent isoleucine (Fig. 2B).
The global reduction in H3K27me2/3 suggested that the H3K27M transgene reduced methylation on endogenous wild-type H3 histones. Indeed, purified heterotypic mononucleosomes (1:1 ratio of H3.3-FLAG-HA: endogenous H3.3), oligonucleosomes containing H3K27M, or wild-type oligonucleosomes from K27M-expressing cells displayed a marked decrease in H3K27me3 on the endogenous H3 protein (fig. S2B). Concomitantly, these nucleosomes exhibited a modest increase in the acetylation of H3K27 (H3K27ac) on the untagged H3 in the heterotypic K27M nucleosomes (Fig. 2C), and, similar to the modification pattern observed on mononucleosomes, oligonucleosome arrays showed elevated H3K27ac when they contained H3.3K27M (Fig. 2D).
We investigated amounts of H3K36me3 in cell lines carrying H3.3G34R/V mutations to determine whether these mutations behave similarly to H3K27M. Although overall amounts of H3K36me3 remained unchanged in H3.3G34R/V cell lines, a marked loss of H3K36m3 was exclusive to the epitope-tagged H3.2/3 of purified mono- or oligonucleosomes (Fig. 2, B and C). The epitope-tagged H3 exhibited a decrease in K36me2 (~2.7-fold) and K36me3 (~18.5-fold) on the G34R/V mutants, compared with wild type, as measured by mass spectrometry (fig. S2C).Heterotypic H3.3 G34R or G34V mononucleosomes were methylated to a lesser extent by recombinant SETD2 in vitro, which indicated that nonglycine residues at position 34 on the substrate peptide decreases SETD2 methylation at K36 (fig. S2, D and E).
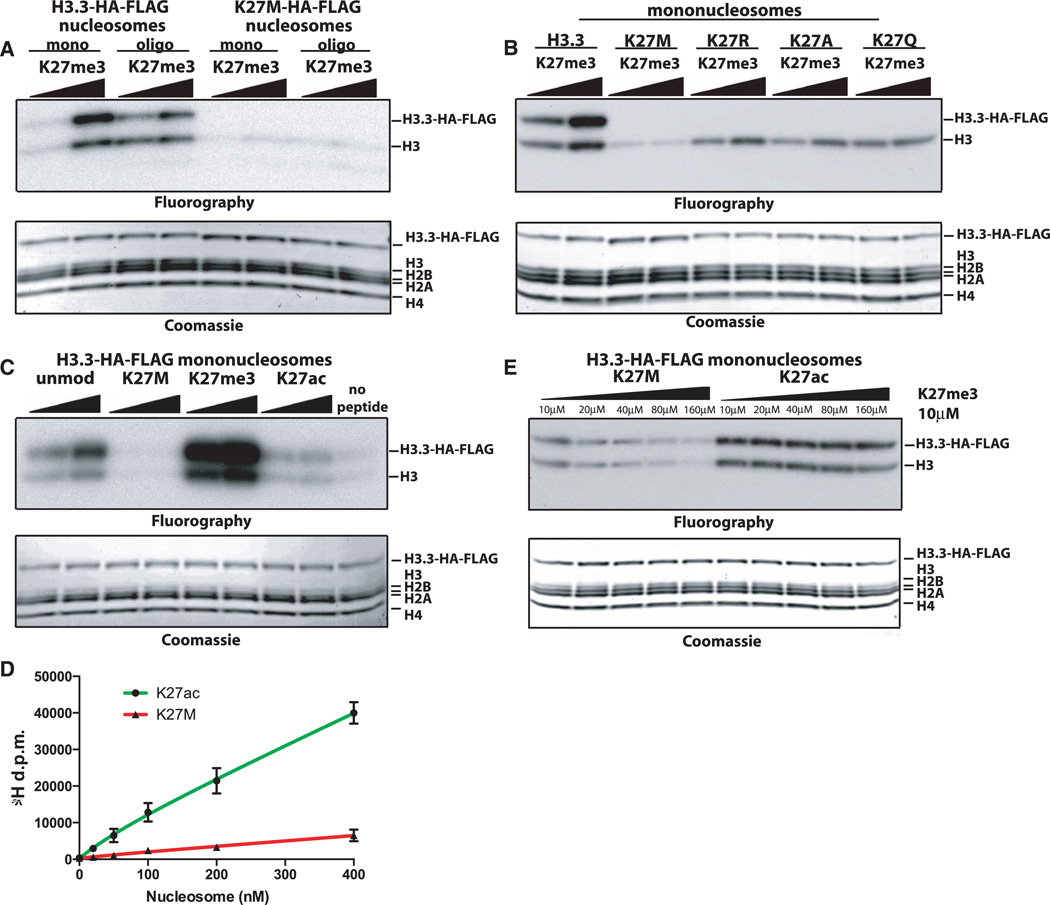
H3K27me3 peptides allosterically stimulate PRC2 methyltransferase activity on nucleosome substrates (8, 9), and we found that incubation of 10 or 100 µM of H3K27me3 peptide strongly stimulated PRC2 activity toward mononucleosome or oligonucleosome templates (Fig. 3A). Heterotypic H3K27M mono- or oligonucleosomes containing H3K27M (fig. S3, A and B) were methylated to a lesser extent than wild-type nucleosome substrates when purified human PRC2 was used (Fig. 3A and fig. S3C). This reduction in PRC2-dependent signal was unique to chromatin containing H3K27M, as heterotypic K27A, K27R, or K27Q mononucleosomes showed no decrease in methylation on the endogenous histone (Fig. 3B).
Fig. 3. H3K27M inhibits PRC2 methyltransferase activity in cis and trans.
(A) Fluorography of PRC2-mediated methylation of wild-type H3.3 or heterotypic H3.3K27M nucleosome substrates. (B) Fluorography of PRC2-mediated methylation of heterotypic mononucleosomes containing the indicated amino acid substitution. (C) Fluorography of PRC2-mediated methylation of wild-type mononucleosomes in the presence of 10 or 100 µM of the indicated peptide. (D) Quantification of PRC2 methyltransferase activity with varying concentrations of wild-type oligonucleosomes in the presence of 45 µM of H3K27ac or H3K27M peptides. Error bars represent the standard deviation for three repeats of the experiment. (E) Fluorography of PRC2-mediated nucleosome methylation with titration of K27M or K27ac peptides.
The reduced methylation on nucleosome templates suggested that the K27M peptide might interfere with PRC2 activity normally stimulated by H3K27me3. Incubation of K27M peptide in trans decreased PRC2 activity on wild-type nucleosomes to below the no-peptide signal (Fig. 3C), whereas the unmodified or K27ac peptide exhibited little stimulation relative to no-peptide control (Fig. 3C). The H3K27M peptide reduced PRC2-dependent methylation by 6.2-fold at 45 µM as measured by scintillation counting (Fig. 3D). Peptide titration showed that µM concentrations of H3K27M, but not the K27ac peptide, could inhibit PRC2-dependent methyltransferase activity in a dose-dependent manner (Fig. 3E and fig. S3, D and E).
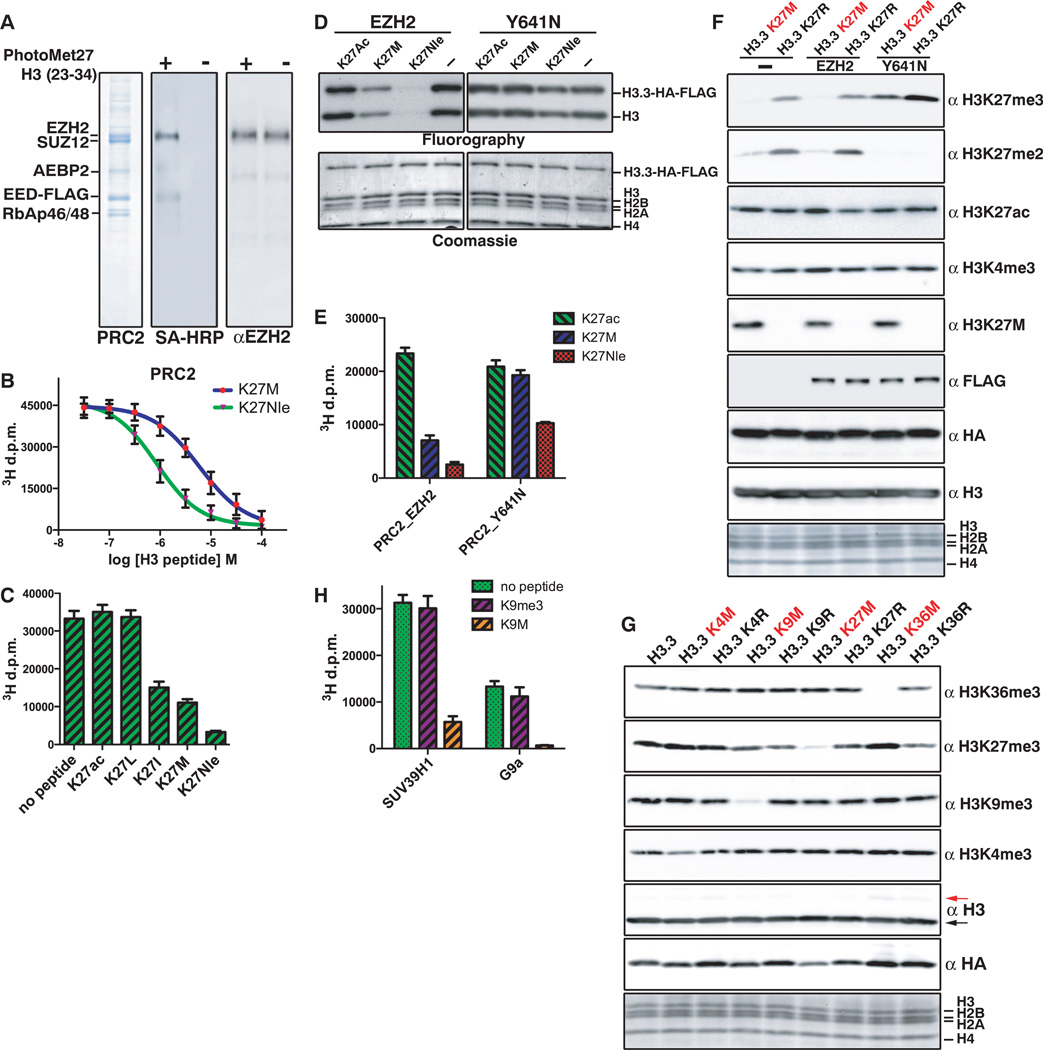
We synthesized biotinylated H3 peptides carrying either the diazirine-containing methionine isostere l-photo-Met at position 27 (peptide 1) or a benzophenylalanine residue at position 31 that retains the K27M mutation (peptide 2) (fig. S4, F and G).Upon irradiation, both peptides cross-linked effectively to the EZH2 subunit and less so to the AEBP2 and EED subunits (Fig. 4A and fig. S4E), which suggested that K27M peptides inhibit PRC2 through interaction with the EZH2 active site. Although the EED subunit is needed for enzymatic activity and K27me3 allosteric activation (8), we did not observe interaction between EED and H3K27M peptides by surface plasmon resonance or peptide pull-down assays (fig. S4, A to D). We additionally found the K27M peptide could inhibit a PRC2 complex that contained a mutant EED (Y365A) that cannot interact with K27me3 (fig. S5A).
Fig. 4. H3K27M-mediated inhibition of PRC2 occurs through interaction with EZH2.
(A) Coomassie gel of purified PRC2 cross-linked to peptide 1 (fig. S4F) and immunoblots with streptavidin–horseradish peroxidase (SA-HRP) or antibody against EZH2. (B) IC50 measurement for methionine or norleucine substitution at K27. Titration reactions of H3.3K27M or K3.3K27Nle peptides with 70 ng PRC2 and 0.8 µg wild-type oligonucleosomes. Error bars represent the standard deviation of five repeats. (C) Quantification of PRC2 methyltransferase activity in the presence of 50 µM of H3 peptides (18 to 37) containing K27acetyl, K27Leu, K27Ile, K27Met, or K27Nle. Error bars represent standard deviation of three repeats. (D) Fluorography of methyltransferase reactions with wild-type or EZH2-Y641N PRC2 on wild-type H3.3 nucleosomes in the presence of 50 µM H3 peptides (18 to 37) with K27acetyl, K27Met, or K27Nle. Error bars represent the standard deviation of three repeats. (E) Quantification of methyltransferase activity of reactions in (D). Error bars represent the standard deviation of five repeats. (F) Immunoblots of 293T cells expressing the indicated EZH2 and H3.3 transgenes. The anti-HA blot shows relative H3.3 transgene amounts, and the anti-FLAG shows the relative EZH2 or Y641N amounts. (G) Immunoblots of whole-cell extract from transduced 293T cells expressing the indicated H3.3 transgenes. The arrows to the right of the total H3 blot point to the transgenic histone (red) or endogenous wild-type histone (black). (H) Quantification of histone methyltransferase activity on H3 peptides (1 to 20) by SUV39H1 or G9a in the presence of 10 µM H3K9me3 or H3K9M or no peptide. Error bars represent the standard deviation for five repeats.
Titration of K27M peptide to in vitro methylation reactions revealed a median inhibitory concentration (IC50) of 5.9 µM [95% confidence interval (CI) of 1.10 to 6.42]. To evaluate whether the thioether moiety of methionine was required for inhibition of PRC2, a norleucine derivative (K27Nle) was prepared. The K27Nle variant proved to be an even better inhibitor of PRC2 (Fig. 4B) (IC50 for K27Nle = 0.85 µM) (95% CI: 0.57 to 1.27) (fig. S6A). Peptides containing Lys27 replaced by Ile inhibited PRC2 to a lesser extent than K27M (IC50 for K27I = 8.9 µM (95% CI: 4.12 to 11.2), whereas Lys27 replaced by Leu had no inhibitory effect on the amounts of H3K27me3 in vivo or PRC2 in vitro (Fig. 4C and fig. S6, B to D). Thus, a long, hydrophobic residue suffices for EZH2 binding, and methionine—and to a slightly lesser extent isoleucine—represents the ideal biochemically accessible choices.
SET domain histone methyltransferases contain several highly conserved aromatic residues that serve in substrate binding and catalysis. The Tyr641 residue in EZH2 is hypothesized to restrict higher-order lysine methylation by acting as a steric gatekeeper (10, 11). Recombinant Y641N EZH2-containing PRC2 was less sensitive to inhibition by the H3K27M peptide compared with wild-typePRC2 (Fig. 4, D to F, and fig. S6,E and F), which furthers the argument that hydrophobic interactions between the methionine side chain and aromatic residues in the EZH2 active site are important for K27M inhibition of PRC2.
We tested whether the highly conserved active sites of other SET-domain proteins may be similarly sensitive tomethionine substitution at their cognate peptidyl-lysine substrate by constructing H3.3 transgenes containing methionine at K4, K9, and K36 (12). H3K9M and H3K36M transgenes decrease overall amounts of H3K9me2/3 and H3K36me2/3, respectively (Fig. 4G and fig. S7A). Although H3K4M transgenes did not diminish overall amounts of H3K4me2/3, the lack of reduction in methylation may be a consequence of reported inhibition of LSD1/2 histone H3K4me3 demethylases by H3K4M peptides (13). Histone H3K9M (1–20) peptides effectively inhibit recombinant histone H3K9 methyltransferases SUV39H1 and G9a (Fig. 4H) [IC50 for K9M = 2.2 µM for SUV39H1 (95% CI: 1.7 to 2.6); IC50 for K9M = 3.6 µM for G9a (95% CI: 2.1 to 5.9)] (fig. S7, B and C). These data argue that the H3K9-specific SET-domain inhibition likely causes the reduction ofH3K9me2/3 in H3.3K9M-expressing cells.
To determine whether the chromatin context of K-to-M mutants is important for in vivo inhibition of trimethylation, we placed the H3 tail (amino acids 1 to 42) onto the core domain (amino acids 23 to 102) of histone H4-HA-FLAG.H3K9me3 was reduced in cells expressing the K9M H3–H4 hybrid (fig. S8A). However, K27M and K36M failed to reduce H3K27me3 and K3K36me3 when expressed in the context of H3-H4 hybrid transgenes (fig. S8, B, C, and D). These results argue that SET-domain protein interactions with non–N-terminal nucleosomal surfaces are important for the full inhibitory activity of some K-to-M, but not all, mutants in vivo.
In summary, the data shown here provide compelling evidence that a missense histone mutation can dramatically alter nuclear biochemical processes through a gain-of-function mechanism. Moreover, our data indicate that K-to-M mutations target the active sites of diverse SET domain–containing methyltransferases and, thereby, effectively compete with substrate binding and turnover. Notably, in their proper chromosomal context, K-to-M mutant histones that make up only a few percent of total histones suffice to prevent global methylation at their cognate residues. This reduction in histone methylation is expected to disrupt positive-feedback loops that contribute to the regulation of PRC2 and thus to enhance the inhibitory effect of mutant histones. We propose a model whereby aberrant epigenetic silencing through H3K27M-mediated inhibition of PRC2 activity promotes gliomagenesis. The broadly adaptable, yet highly specific, inhibition of SET-domain proteins through K-to-M mutation offers the intriguing possible existence of other etiological missense mutations in histones. Additionally, our work has uncovered a potentially useful mechanism to exclusively inhibit individual SET-domain methyltransferases, and conceivably other chromatin-modifying enzymes, implicated in a variety of malignancies.
Supplementary Material
Acknowledgments
B.A.G. acknowledges funding from NIH Innovator grant (DP2OD007447), Office of the Director, NIH. O.J.B. is supported by the U.S. Department of Defense; The Pediatric Brain Tumor Foundation; and the Damon Runyon, Rory David Deutsch, and Cristian Rivera Foundations. C.D.A. acknowledges funding from NIH (GM040922) and Starr Cancer Consortium (grant SCC I6-A614). We thank D. Reinberg for reagents, and EMD Millipore for its role in H3K27M antibody generation.
Footnotes
Supplementary Materials
www.sciencemag.org/cgi/content/full/science.1232245/DC1
Materials and Methods
Supplementary Text
Figs. S1 to S8
Reference (14)
References and Notes
- 1.Elsässer SJ, Allis CD, Lewis PW. Science. 2011;331:1145. doi: 10.1126/science.1203280. [DOI] [PubMed] [Google Scholar]
- 2.Simon JA, Lange CA. Mutat. Res. 2008;647:21. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 3.Margueron R, Reinberg D. Nature. 2011;469:343. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khuong-Quang DA, et al. Acta Neuropathol. 2012;124:439. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartzentruber J, et al. Nature. 2012;482:226. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 6.Wu G, et al. Nat. Genet. 2012;44:251. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becher OJ, et al. Cancer Res. 2010;70:2548. doi: 10.1158/0008-5472.CAN-09-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Margueron R, et al. Nature. 2009;461:762. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu C, et al. Proc. Natl. Acad. Sci. U.S.A. 2010;107:19266. [Google Scholar]
- 10.Yap DB, et al. Blood. 2011;117:2451. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sneeringer CJ, et al. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20980. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. Nat. Struct. Mol. Biol. 2007;14:1025. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karytinos A. J. Biol. Chem. 2009;284:17775. doi: 10.1074/jbc.M109.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.