Abstract
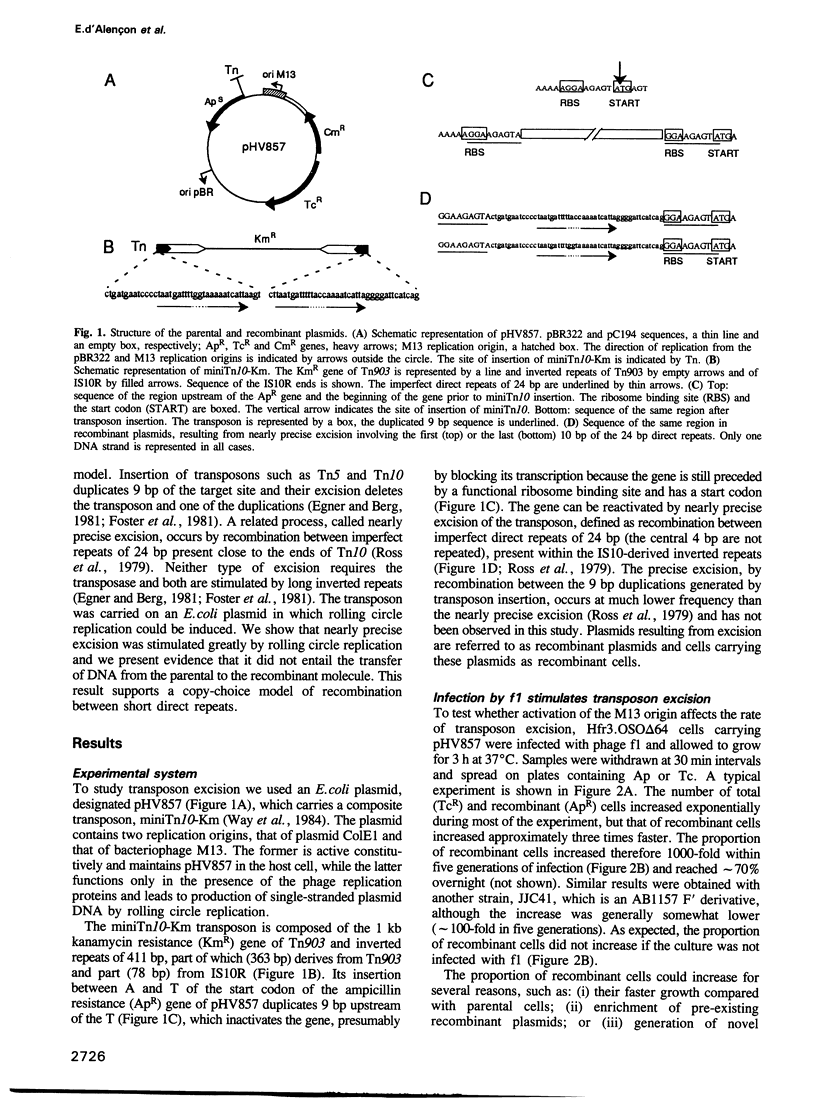
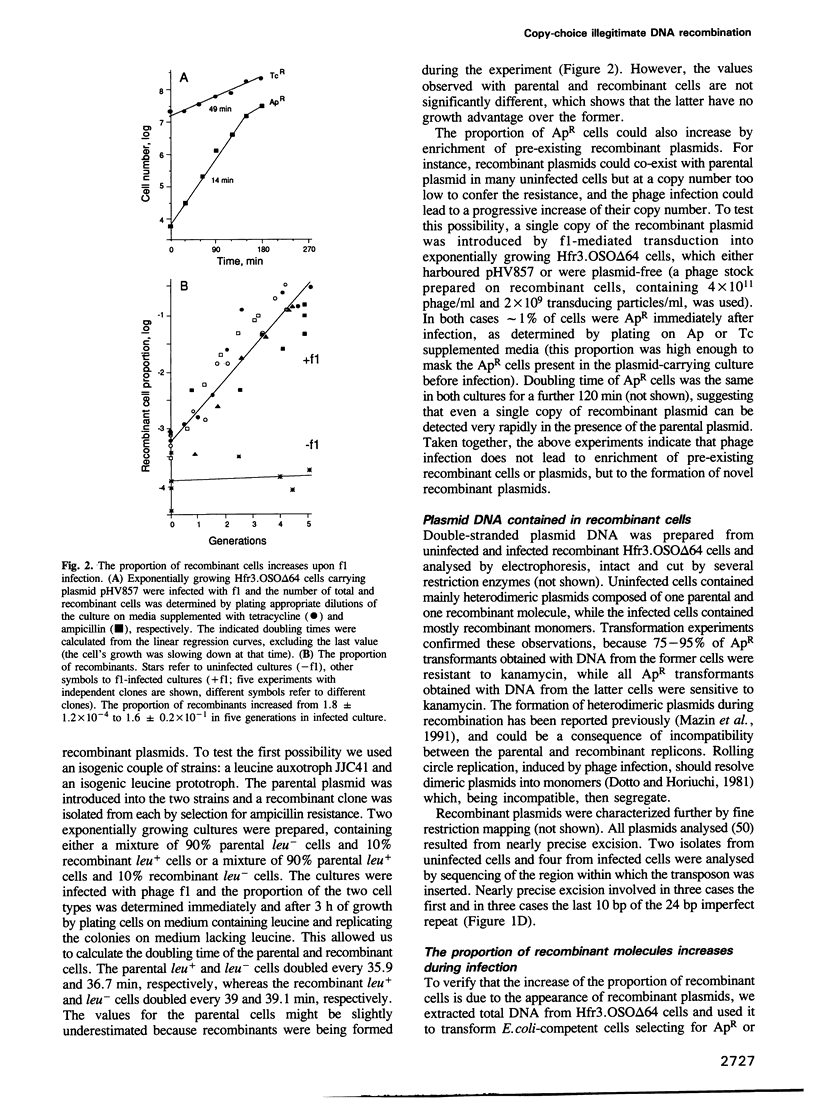
Nearly precise excision of a transposon related to Tn10 from an Escherichia coli plasmid was used as a model to study illegitimate DNA recombination between short direct repeats. The excision was stimulated 100-1000 times by induction of plasmid single-stranded DNA synthesis and did not involve transfer of DNA from the parental to the progeny molecule. We conclude that it occurred by copy-choice DNA recombination, and propose that other events of recombination between short direct repeats might be a result of the same process.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Alikhanian S. I., Iljina T. S., Kaliaeva E. S., Kameneva S. V., Sukhodolec V. V. A genetical study of thymineless mutants of E. coli K12. Genet Res. 1966 Aug;8(1):83–100. doi: 10.1017/s0016672300009939. [DOI] [PubMed] [Google Scholar]
- Anderson P. Anecdotal, historical and critical commentaries on genetics twenty years of illegitimate recombination. Genetics. 1987 Apr;115(4):581–583. doi: 10.1093/genetics/115.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R. P., Roth J. R. Tandem genetic duplications in phage and bacteria. Annu Rev Microbiol. 1977;31:473–505. doi: 10.1146/annurev.mi.31.100177.002353. [DOI] [PubMed] [Google Scholar]
- Bashkirov V. I., Khasanov F. K., Prozorov A. A. Illegitimate recombination in Bacillus subtilis: nucleotide sequences at recombinant DNA junctions. Mol Gen Genet. 1987 Dec;210(3):578–580. doi: 10.1007/BF00327215. [DOI] [PubMed] [Google Scholar]
- Bazaral M., Helinski D. R. Replication of a bacterial plasmid and an episome in Escherichia coli. Biochemistry. 1970 Jan 20;9(2):399–406. doi: 10.1021/bi00804a029. [DOI] [PubMed] [Google Scholar]
- Beck E., Zink B. Nucleotide sequence and genome organisation of filamentous bacteriophages fl and fd. Gene. 1981 Dec;16(1-3):35–58. doi: 10.1016/0378-1119(81)90059-7. [DOI] [PubMed] [Google Scholar]
- Berg D. E., Egner C., Lowe J. B. Mechanism of F factor-enhanced excision of transposon Tn5. Gene. 1983 Apr;22(1):1–7. doi: 10.1016/0378-1119(83)90058-6. [DOI] [PubMed] [Google Scholar]
- Bierne H., Ehrlich S. D., Michel B. The replication termination signal terB of the Escherichia coli chromosome is a deletion hot spot. EMBO J. 1991 Sep;10(9):2699–2705. doi: 10.1002/j.1460-2075.1991.tb07814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunier D., Michel B., Ehrlich S. D. Copy choice illegitimate DNA recombination. Cell. 1988 Mar 25;52(6):883–892. doi: 10.1016/0092-8674(88)90430-8. [DOI] [PubMed] [Google Scholar]
- Calvin N. M., Hanawalt P. C. High-efficiency transformation of bacterial cells by electroporation. J Bacteriol. 1988 Jun;170(6):2796–2801. doi: 10.1128/jb.170.6.2796-2801.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley E. C., Saunders V. A., Jackson V., Saunders J. R. Mechanism of intramolecular recyclization and deletion formation following transformation of Escherichia coli with linearized plasmid DNA. Nucleic Acids Res. 1986 Nov 25;14(22):8919–8932. doi: 10.1093/nar/14.22.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad B., Bashkirov V. I., Hofemeister J. Imprecise excision of plasmid pE194 from the chromosomes of Bacillus subtilis pE194 insertion strains. J Bacteriol. 1992 Nov;174(21):6997–7002. doi: 10.1128/jb.174.21.6997-7002.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D. N., Schmidtke J. Diagnosis of genetic disease using recombinant DNA. Second edition. Hum Genet. 1989 Nov;83(4):307–334. doi: 10.1007/BF00291376. [DOI] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- DasGupta U., Weston-Hafer K., Berg D. E. Local DNA sequence control of deletion formation in Escherichia coli plasmid pBR322. Genetics. 1987 Jan;115(1):41–49. doi: 10.1093/genetics/115.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey L. A., Dubnau D. A. Identification of plasmid and Bacillus subtilis chromosomal recombination sites used for pE194 integration. J Bacteriol. 1989 May;171(5):2856–2865. doi: 10.1128/jb.171.5.2856-2865.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto G. P., Horiuchi K. Replication of a plasmid containing two origins of bacteriophage. J Mol Biol. 1981 Nov 25;153(1):169–176. doi: 10.1016/0022-2836(81)90532-5. [DOI] [PubMed] [Google Scholar]
- Edlund T., Normark S. Recombination between short DNA homologies causes tandem duplication. Nature. 1981 Jul 16;292(5820):269–271. doi: 10.1038/292269a0. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Egner C., Berg D. E. Excision of transposon Tn5 is dependent on the inverted repeats but not on the transposase function of Tn5. Proc Natl Acad Sci U S A. 1981 Jan;78(1):459–463. doi: 10.1073/pnas.78.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich S. D., Michel B. Retraction: copy choice illegitimate DNA recombination. Cell. 1990 Aug 10;62(3):409–409. doi: 10.1016/0092-8674(90)90004-x. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Foster T. J., Lundblad V., Hanley-Way S., Halling S. M., Kleckner N. Three Tn10-associated excision events: relationship to transposition and role of direct and inverted repeats. Cell. 1981 Jan;23(1):215–227. doi: 10.1016/0092-8674(81)90286-5. [DOI] [PubMed] [Google Scholar]
- Fulford W., Model P. Bacteriophage f1 DNA replication genes. II. The roles of gene V protein and gene II protein in complementary strand synthesis. J Mol Biol. 1988 Sep 5;203(1):39–48. doi: 10.1016/0022-2836(88)90089-7. [DOI] [PubMed] [Google Scholar]
- Fulford W., Model P. Regulation of bacteriophage f1 DNA replication. I. New functions for genes II and X. J Mol Biol. 1988 Sep 5;203(1):49–62. doi: 10.1016/0022-2836(88)90090-3. [DOI] [PubMed] [Google Scholar]
- Gordenin D. A., Malkova A. L., Peterzen A., Kulikov V. N., Pavlov Y. I., Perkins E., Resnick M. A. Transposon Tn5 excision in yeast: influence of DNA polymerases alpha, delta, and epsilon and repair genes. Proc Natl Acad Sci U S A. 1992 May 1;89(9):3785–3789. doi: 10.1073/pnas.89.9.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989 Jun;53(2):231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzesiuk E., Carroll D. Recombination of DNAs in Xenopus oocytes based on short homologous overlaps. Nucleic Acids Res. 1987 Feb 11;15(3):971–985. doi: 10.1093/nar/15.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyina T. V., Koonin E. V. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992 Jul 11;20(13):3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannière L., Bruand C., Ehrlich S. D. Structurally stable Bacillus subtilis cloning vectors. Gene. 1990 Mar 1;87(1):53–61. doi: 10.1016/0378-1119(90)90495-d. [DOI] [PubMed] [Google Scholar]
- LEDERBERG J. Recombination mechanisms in bacteria. J Cell Physiol Suppl. 1955 May;45(Suppl 2):75–107. doi: 10.1002/jcp.1030450506. [DOI] [PubMed] [Google Scholar]
- LaDuca R. J., Fay P. J., Chuang C., McHenry C. S., Bambara R. A. Site-specific pausing of deoxyribonucleic acid synthesis catalyzed by four forms of Escherichia coli DNA polymerase III. Biochemistry. 1983 Oct 25;22(22):5177–5188. doi: 10.1021/bi00291a018. [DOI] [PubMed] [Google Scholar]
- Lopez P., Espinosa M., Greenberg B., Lacks S. A. Generation of deletions in pneumococcal mal genes cloned in Bacillus subtilis. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5189–5193. doi: 10.1073/pnas.81.16.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MESELSON M., WEIGLE J. J. Chromosome brekage accompanying genetic recombination in bacteriophage. Proc Natl Acad Sci U S A. 1961 Jun 15;47:857–868. doi: 10.1073/pnas.47.6.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin A. V., Kuzminov A. V., Dianov G. L., Salganik R. I. Mechanisms of deletion formation in Escherichia coli plasmids. II. Deletions mediated by short direct repeats. Mol Gen Genet. 1991 Aug;228(1-2):209–214. doi: 10.1007/BF00282467. [DOI] [PubMed] [Google Scholar]
- Meselson M., Stahl F. W., Vinograd J. EQUILIBRIUM SEDIMENTATION OF MACROMOLECULES IN DENSITY GRADIENTS. Proc Natl Acad Sci U S A. 1957 Jul 15;43(7):581–588. doi: 10.1073/pnas.43.7.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., D'Alençon E., Ehrlich S. D. Deletion hot spots in chimeric Escherichia coli plasmids. J Bacteriol. 1989 Apr;171(4):1846–1853. doi: 10.1128/jb.171.4.1846-1853.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., Niaudet B., Ehrlich S. D. Intramolecular recombination during plasmid transformation of Bacillus subtilis competent cells. EMBO J. 1982;1(12):1565–1571. doi: 10.1002/j.1460-2075.1982.tb01356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris T., Thacker J. Formation of large deletions by illegitimate recombination in the HPRT gene of primary human fibroblasts. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1392–1396. doi: 10.1073/pnas.90.4.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbantoglu J., Hartley D., Phear G., Tear G., Meuth M. Spontaneous deletion formation at the aprt locus of hamster cells: the presence of short sequence homologies and dyad symmetries at deletion termini. EMBO J. 1986 Jun;5(6):1199–1204. doi: 10.1002/j.1460-2075.1986.tb04347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirot P., Petit M. A., Ehrlich S. D. Plasmid replication stimulates DNA recombination in Bacillus subtilis. J Mol Biol. 1987 Jul 5;196(1):39–48. doi: 10.1016/0022-2836(87)90509-2. [DOI] [PubMed] [Google Scholar]
- Oehler S., Eismann E. R., Krämer H., Müller-Hill B. The three operators of the lac operon cooperate in repression. EMBO J. 1990 Apr;9(4):973–979. doi: 10.1002/j.1460-2075.1990.tb08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters B. P., de Boer J. H., Bron S., Venema G. Structural plasmid instability in Bacillus subtilis: effect of direct and inverted repeats. Mol Gen Genet. 1988 Jun;212(3):450–458. doi: 10.1007/BF00330849. [DOI] [PubMed] [Google Scholar]
- Pierce J. C., Kong D., Masker W. The effect of the length of direct repeats and the presence of palindromes on deletion between directly repeated DNA sequences in bacteriophage T7. Nucleic Acids Res. 1991 Jul 25;19(14):3901–3905. doi: 10.1093/nar/19.14.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross D. G., Swan J., Kleckner N. Nearly precise excision: a new type of DNA alteration associated with the translocatable element Tn10. Cell. 1979 Apr;16(4):733–738. doi: 10.1016/0092-8674(79)90089-8. [DOI] [PubMed] [Google Scholar]
- Scearce L. M., Pierce J. C., McInroy B., Masker W. Deletion mutagenesis independent of recombination in bacteriophage T7. J Bacteriol. 1991 Jan;173(2):869–878. doi: 10.1128/jb.173.2.869-878.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R. H., Dominska M., Petes T. D. Transformation of Saccharomyces cerevisiae with nonhomologous DNA: illegitimate integration of transforming DNA into yeast chromosomes and in vivo ligation of transforming DNA to mitochondrial DNA sequences. Mol Cell Biol. 1993 May;13(5):2697–2705. doi: 10.1128/mcb.13.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyamala V., Schneider E., Ames G. F. Tandem chromosomal duplications: role of REP sequences in the recombination event at the join-point. EMBO J. 1990 Mar;9(3):939–946. doi: 10.1002/j.1460-2075.1990.tb08192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R. R., Zheng G. X., Brankamp R. G., Allen K. N. On the deletion of inverted repeated DNA in Escherichia coli: effects of length, thermal stability, and cruciform formation in vivo. Genetics. 1991 Dec;129(4):991–1005. doi: 10.1093/genetics/129.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B. S., Westlye J. Deletion formation in bacteriophage T4. J Mol Biol. 1988 Jul 20;202(2):233–243. doi: 10.1016/0022-2836(88)90454-8. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Trinh T. Q., Sinden R. R. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature. 1991 Aug 8;352(6335):544–547. doi: 10.1038/352544a0. [DOI] [PubMed] [Google Scholar]
- Uzest M., Ehrlich S. D., Michel B. The Escherichia coli terB sequence affects maintenance of a plasmid with the M13 phage replication origin. J Bacteriol. 1991 Dec;173(23):7695–7697. doi: 10.1128/jb.173.23.7695-7697.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilette D., Uzest M., Ehrlich S. D., Michel B. DNA transcription and repressor binding affect deletion formation in Escherichia coli plasmids. EMBO J. 1992 Oct;11(10):3629–3634. doi: 10.1002/j.1460-2075.1992.tb05447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. C. Diseases of the mitochondrial DNA. Annu Rev Biochem. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- Weston-Hafer K., Berg D. E. Deletions in plasmid pBR322: replication slippage involving leading and lagging strands. Genetics. 1991 Apr;127(4):649–655. doi: 10.1093/genetics/127.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston-Hafer K., Berg D. E. Palindromy and the location of deletion endpoints in Escherichia coli. Genetics. 1989 Apr;121(4):651–658. doi: 10.1093/genetics/121.4.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whoriskey S. K., Nghiem V. H., Leong P. M., Masson J. M., Miller J. H. Genetic rearrangements and gene amplification in Escherichia coli: DNA sequences at the junctures of amplified gene fusions. Genes Dev. 1987 May;1(3):227–237. doi: 10.1101/gad.1.3.227. [DOI] [PubMed] [Google Scholar]
- Willetts N., Wilkins B. Processing of plasmid DNA during bacterial conjugation. Microbiol Rev. 1984 Mar;48(1):24–41. doi: 10.1128/mr.48.1.24-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams W. L., Müller U. R. Effects of palindrome size and sequence on genetic stability in the bacteriophage phi X174 genome. J Mol Biol. 1987 Aug 20;196(4):743–755. doi: 10.1016/0022-2836(87)90401-3. [DOI] [PubMed] [Google Scholar]
- Yen P. H., Allen E., Marsh B., Mohandas T., Wang N., Taggart R. T., Shapiro L. J. Cloning and expression of steroid sulfatase cDNA and the frequent occurrence of deletions in STS deficiency: implications for X-Y interchange. Cell. 1987 May 22;49(4):443–454. doi: 10.1016/0092-8674(87)90447-8. [DOI] [PubMed] [Google Scholar]
- Yunis J. J. The chromosomal basis of human neoplasia. Science. 1983 Jul 15;221(4607):227–236. doi: 10.1126/science.6336310. [DOI] [PubMed] [Google Scholar]
- Zipursky S. L., Marians K. J. Identification of two Escherichia coli factor Y effector sites near the origins of replication of the plasmids (ColE1 and pBR322. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6521–6525. doi: 10.1073/pnas.77.11.6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zamaroczy M., Faugeron-Fonty G., Bernardi G. Excision sequences in the mitochondrial genome of yeast. Gene. 1983 Mar;21(3):193–202. doi: 10.1016/0378-1119(83)90002-1. [DOI] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Are single-stranded circles intermediates in plasmid DNA replication? EMBO J. 1986 Mar;5(3):631–637. doi: 10.1002/j.1460-2075.1986.tb04257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2541–2545. doi: 10.1073/pnas.83.8.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wezenbeek P. M., Hulsebos T. J., Schoenmakers J. G. Nucleotide sequence of the filamentous bacteriophage M13 DNA genome: comparison with phage fd. Gene. 1980 Oct;11(1-2):129–148. doi: 10.1016/0378-1119(80)90093-1. [DOI] [PubMed] [Google Scholar]