Abstract
The development of the vertebrate nervous system requires a switch of ATP-dependent chromatin remodeling mechanisms, which occures by substituting subunits within these complexes near cell cycle exit. This switching involves a triple negative genetic circuitry in which REST represses miR-9 and miR-124, which in turn repress BAF53a, which in turn repress the homologous neuron-specific BAF53b. Recapitulation of this microRNA/chromatin switch in human fibroblasts converts them to neurons. The genes involved in this fate-determining chromatin switch play genetically dominant roles in several human neurologic diseases suggesting that they are rate-limiting for aspects of human neural development. We review how this switch in ATP-dependent chromatin complexes might interface with traditional ideas about neural determination and reprogramming.
Introduction
The discovery that mutation of a single homeotic gene could convert one body part to another in Drosophila Melanogaster opened the possibility that tissues could be interconverted in other species [1–5]. Later the discovery of myoD [6] demonstrated that even late in development, tissues could be interconverted if one gained an understanding of the genes that operate at the pinnacle of a development circuitry. Presently, the direct conversion of many cell types has been accomplished. The route to these accomplishments have come from either systematic screens to find the factors that have the ability to convert one tissue to another [7–9] or through detailed delineation of pathways of development. Often the two approaches are not entirely separate and studies of direct conversion provide an understanding of a biologic process. The focus of this review illustrates how the understanding of a mechanism underlying an aspect of the development of the nervous system lead to a method of converting human fibroblasts to neurons.
A microRNA-Chromatin Switch Underlying Neural Development
Neurons have features not found in any other cell type. Perhaps over a thousand subtypes of neurons are generated, each with common features such as the ability to extend axons, dendrites and form synapses between each other. Another unique feature is their extraordinary morphologic stability, which is the basis of stable, long-term memory. These unique features lead us to look for an epigenetic state that would characterize neurons as opposed to other cell types. Purification of ATP-dependent chromatin remodeling complexes led to the realization that neurons from adult brains do indeed have a chromatin remodeling complex that is not found in any other cell type, which we named nBAF for its neural specific subunits (BAF53b, BAF45b/c and CREST) and the fact that it contains either the Brg or Brm ATPases [10]. These neuronal specific complexes appear near cell cycle exit in the mammalian nervous system and additional purification studies revealed that neural progenitors also have a npBAF complex with a specific subunit composition not found to date in any other cell type. Finally, purification of the complexes from pluripotent ES cells revealed that they also have a unique subunit composition not found to date in any other cell type [11]. The separate complexes apparently reflect a developmental series of epigenetic stages required for pluriopotency, neural multipotency and finally the differentiated state of post mitotic neurons. The transitition between these complexes is illustrated in Figure 1. Six of 15 subunits of these complexes have homologues in the yeast SWI/SNF complex and when we initially purified the complexes we called them mSWI/SNF [12,13] for what we took to be the mammalian form of the yeast SWI/SNF complex. However, upon further purification and cloning we found that 9 of the subunits do not have homologues in yeast SWI/SNF and that substantial evolutionary differences exist that apparently reflect the changing strategies of chromatin regulation over the past 1.5 billion years. These changes include multicellularity and the need to repress the majority of genes, DNA methylation, polycomb mediated repression and also the need to compact the mammalian genome by several hundred-fold more than the yeast genome is compacted. Additionally, the changes in BAF chromatin remodeling complexes appear to reflect the need for a complex nervous system, since the neural specific subunits appear in fish or possibly even later (Table 1).
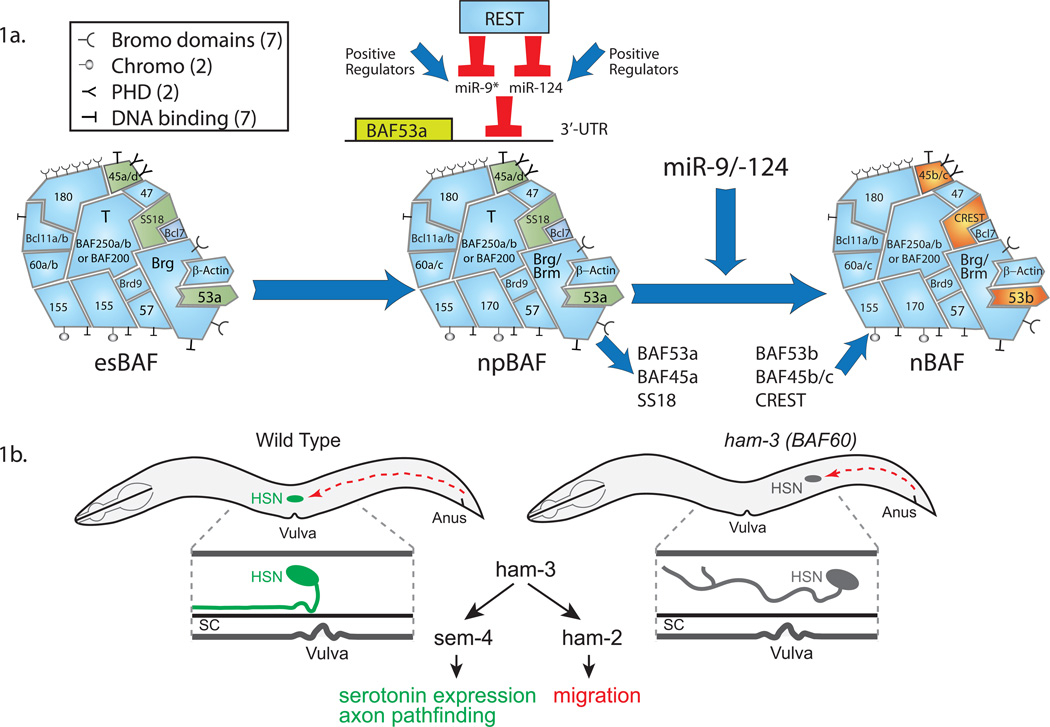
Figure 1. Roles of BAF complexes and their subunit switching during vertebrate neural development.
1a. A triple negative genetic circuitry regulation of microRNA/chromatin switch underlying vertebrate neural development. As cells progress from pluripotent ES cells to neural progenitors to neurons, BAF complexes switch subunits and assume new functions. In neural progenitors, REST represses miR-9/9* and miR-124 to release their inhibitory effect on BAF53a expression. The subunit compostions shown are based on the criteria that each subunit is dedicated to the complex, stable to 2.5 M urea and not exchangable during the course of density centrifugation analysis. Note the large number of domains that bind histone modifications or DNA. Not all domains are shown. 1b. BAF60 (ham-3) and its role in the generation and function of a specific neuronal cell type. A specific serotonergic neuron known as the hermaphrodite specific neuron (HSN), requires BAF60 for migration, all steps in serotonin synthesis and axonal pathfinding.
Table 1.
Evolution and nomenclature of BAF subunits from yeast to human.
| Subunit | Alias | SMARC | esBAF | npBAF | nBAF | S.cerevisiae | D.melanogaster | C.elegans | D.rerio | G.gallus | H.sapiens |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BAF250a | Arid1a | x | x | x | N | Osa/eyelid | Psa-10/let-526 | Arid1aa(b) | Arid1a | BAF250a | |
| BAF250b | Arid1b | x | x | N | Arid1b | Arid1b | BAF250b | ||||
| BAF200 | Arid2 | x | x | x | N | BAP170 | Swsn-7 | Arid2 | Arid2 | BAF200 | |
| BAF190a (brg) | Snf2β | SMARCA4 | x | x | x | Swi2 | Yng/smarca4 | Brg1 | BAF190a (brg) | ||
| BAF190b (brm) | Snf2α | SMARCA2 | x | x | Brahma | Psa-4/Swsn-4 | brm | Brm | BAF190b (brm) | ||
| BAF180 | PBRM1 | x | x | x | N | BAP180 | Pbrm-1 | Pbrm-1 | Pbrm-1 | BAF180 | |
| BAF170 | SMARCC2 | x | x | Swi3 | moira | Psa-1/Swsn-1 | Smarcc2 | Smarcc2 like | BAF170 | ||
| BAF155 | SMARCC1 | x | x | x | Smarcc1 | Smarcc1 | BAF155 | ||||
| BAF100a | Bcl11a | x | x | x | N | CG9650? | Bcl11aa(b) | Bcl11a | BAF100a | ||
| BAF100b | Bcl11b | x | x | x | N | F13H6.1? | Bcl11b | Bcl11b | BAF100b | ||
| BAF60a | SMARCD1 | x | x | x | Swp73 | BAP60 | Ham-3/swsn-2.1 | Smarcd1 | Smarcd1 | BAF60a | |
| BAF60b | SMARCD2 | x | Swsn-2.2 | Smarcd2 | Smarcd2 | BAF60b | |||||
| BAF60c | SMARCD3 | x | Smarcd3a(b) | Smarcd3 | BAF60c | ||||||
| BAF57 | SMARCE1 | x | x | x | N | Dalao/BAP111 | Swsn-3 | Smarce1 | Smarce1 | BAF57 | |
| BAF55a | SS18 | x | x | N | N | N | SS18 | SS18 | BAF55 | ||
| BAF55b | CREST | x | N | N | N | N | SS18l1 | BAF55b | |||
| β-Actin | x | x | x | N | Actin 5C | ACT-2 | ACTB | ACTB | β-Actin | ||
| BAF53a | Actl6a | x | x | Arp7/9? | BAP55 | Psa-13/Swsn-6 | Actl6a | Actl6a | BAF53a | ||
| BAF53b | Actl6b | x | Actl6b | N | BAF53b | ||||||
| BAF47 | (INI1/hSNF5) | SMARCB1 | x | x | x | Snf5 | Snr1 | Snfc-5/Swsn-5 | Smarcb1a(b) | Smarcb1/MMP11 | BAF47 |
| BAF45a | PHF10 | x | x | N | SAYP | Phf-10 | XAP135 | LOC100857350 | BAF45a | ||
| BAF45b | DPF1 | x | N | D4 | Dpff-1 | Dpf1 | Neuro-d4/Dpf1 | BAF45b | |||
| BAF45c | DPF3 | x | N | Dpf3 | Dpf3 | BAF45c | |||||
| BAF45d | DPF2 | x | x | N | Dpf2 | Dpf2/REQ | BAF45d | ||||
| BAF40a | Bcl7a | x | x | x | N | Bcl-7 like? | Bcl-7 like? | Bcl7a | Bcl7a | BAF40a | |
| BAF40b | Bcl7b | x | x | x | N | Bcl7ba(b) | LOC100857890 | BAF40b | |||
| BAF40c | Bcl7c | x | x | x | N | N | N | BAF40c | |||
| Brd7 | Brd7 | ? | ? | ? | N | CG7154 | Swsn-9 | Brd7 | Brd7 | Brd7 | |
| Brd9 | Brd9 | x | ? | ? | N | Brd9 | Brd9 | Brd9 |
Nomenclature, tissue specificity and evolutionary relationships of different BAF complexes subunits. The subunits of BAF complexes are listed in order of size as they appear on gels of the purified complex (Wang W, et al. Genes Dev 1996; Wang W, et al. Embo J 1996). The homologues shown for SWI/SNF complex are from various species including S.cerevisa(yeast), D.melanogaster(fruit fly), C.elegans(nematode), D.rerio(zebrafish), G.gallus(chicken). N-no significant similarity found by Blast based on mice proteins. “Genename”a(b) means there are two family members a and b for that gene. ? means genes with low homology, but still considered as homolog or not confirmed yet.
The developmental transition between the npBAF and nBAF complexes appears to occur by a triple negative genetic circuit, in which REST represses two microRNAs, miR-9* and miR-124, which in turn repress BAF53a (Fig. 1a) [14]. The microRNAs bind to three sequences in the 3'UTR of BAF53a resulting in its repression. Interestingly, the microRNAs are first expressed in the last progenitor divisions along the ventricular surface of the growing neural tube [15–17]. The removal of BAF53a somehow results in the rapid activation of BAF53b, which is essential for dendritic morphogenesis and synapogenesis [18,19]. While we are beginning to know something of the mechanism underlying the switch of BAF53a-to-BAF53b [14], we know much less of the mechanisms underlying the other switches that result in assembly of nBAF complexes. Pointing to an instructive role in neural development, recent genetic screens in flys have shown that BAF53 is capable of directing the dendritic targeting specificity of olfactory neurons [20]. Indeed it was the only gene found in this demanding screen that gave rise to perfect retargeting, a robust phenotype with nearly 100% penetrance.
At nearly the same developmental time that BAF53a switches to BAF53b in the nBAF complex, SS18 (BAF55a) is replaced with CREST (BAF55b) [21](Fig. 1a). CREST is expressed only in neurons [22] and like BAF53b is required for dendritic morphogenesis and perhaps other aspects of post mitotic neural function [22]. Also, like BAF53b, CREST is expressed in all post mitotic neurons thus far examined, consistent with its broad function in promoting fundamental characteristics of post-mitotic neurons rather than a single neural subtype. Interestingly SS18, which occupies the position of CREST in neural progenitor complexes (npBAF) is an oncogene that is translocated to the SSX locus resulting in the formation of a specialized oncogenic complex, ssBAF [23]. The mechanism of oncogenesis gives important clues as to how these complexes might reprogram fibroblasts to neurons and will be described below.
The third switching subunit is encoded by a family of genes, BAF45a, b, c, d. (Fig. 1a). npBAF complexes have either BAF45a or BAF45d at this position. BAF45b/c takes the place of BAF45a as neural progenitor approach their final mitotic divisions [21,24]. This is followed shortly by the replacement of BAF45b in many of the nBAF complexes. However, at this point we do not know if different types of neurons switch BAF45a for BAF45b and other types of cells switch BAF45a for BAF45c reflecting cell type specific regulation. The other possibility is that within the same cell both switches are made perhaps reflecting different functions at different genetic loci.
The Specificity of the Neural Progenitor (npBAF) Complex
BAF complexes found in ES cells are characterized by the presence of BAF155 but not BAF170, Brg but not Brm [11]. esBAF complexes also contain BAF60a and BAF60b, but do not express BAF60c, which appears in neural progenitors [25]. Interestingly, BAF250b, which is commonly mutated in human neurologic diseases [26–28] also appears as ES cells differentiate into neurons [21]. BAF60 was recently shown to be essential for a program of serotonergic neuron development that regulates axon pathfinding, cellular migration and all steps in the enzymatic pathway of serotonin synthesis (Fig. 1b) [29]. These studies point to a programmatic and instructive function in controlling the formation of a specific type of post mitotic neuron in C elegans. The function of ham-3 (cBAF60) is not redundant with the closely related BAF60 subunit (swsn-2.2) in C elegans (Table 1). Thus it appears that the extraordinary specificity of the ham-3 phenotype, much in the same way as nBAF compelx, is an emergent feature of the polymorphic complex most likely based on a function provided by the composite surface of ham-3 with its neighboring subunits. In mammals BAF60c has been shown to be a specific subunit of neural progenitor npBAF complexes [25]. The recent discoveries of specific subunit assemblies and that highly specific programmatic events such as olfactory neuron target selection, serotonerigic neuron generation and axonal targeting are under the control of specific subunits (BAF53 and BAF60, respectively) has contributed to a conceptual change in the chromatin remodeling field. Until recently chromatin remodeling complexes were thought to play non-specific permissive functions such as helping RNA polymerase bind to the promoters of most genes in a fairly indiscriminant way.
Reconstituting the microRNA Chromatin Switch Converts Human Fibroblasts to Neurons
During the course of defining the genetic circuitry underlying the npBAF to nBAF switch [14] we noted that expression of the miR-9/9* and miR-124 in a variety of cell types resulted in their assuming neuronal morphologies [30]. Indeed, mouse and human fibroblasts readily began to take on neuronal characteristics when the two microRNAs were ectopically expressed. The converted cells expressed a wide variety of neuronal genes including those encoding ion channels and synaptic vesicle components and even made synaptic vesicles that could take up and release their components upon depolarization [30]. Addition of NeuroD, which had been found in the frog by Weintraub and colleagues [31] for its ability to expand the field of neural fate determination in frog embryo increased the frequency of conversion but did not drive the neurons to maturity and the development of repetitive action potentials. However, the pioneering studies of Wernig and colleagues were published at this time [32,33] showing that three factors could convert fibroblasts to neurons in the mouse. In agreement with their results, we found that two of these factors (ASCL1 and MYT1L) in conjuction with NeuroD2 were sufficient to help the microRNAs obtain effective conversion to mature neurons producing repetitive action potentials [30].
The combination of miR-9/9*, miR-124 (expressed on a single vector) with additional neural factors, was quite robust in that it could induce the formation of neurons from skin fibroblasts of older human individuals and was somewhat more rapid than the fibroblast-to-neuron conversion resulting from the actions of the 3 transcription factors. Interestingly, conversion of human fibroblasts was substantially more effective with the microRNA mix than conversion of murine fibroblasts. Since the opposite is true for conversion with 3 transcription factors, this observation may reflect fundamental differences in the underlying genetic circuitry involved. Interestingly, we found that conversion of human fibroblasts to neurons was slower than the conversion of murine fibroblasts, an observation that several groups have made.
Another feature that appears to be characteristic of the microRNA-mediated conversion is the range of neuron types obtained. For example, we found that both excitatory and inhibitory neurons appeared in about equal numbers [30]. This contrasted with methods of preparing neurons either from ES cells or using transcription factors, which produced rather a large population of excitatory neurons [32–34]. Further studies have revealed other rarer types of neurons appear in the cultures and their abundance can be increased by adding back specific transcription factors characteristic of a specific lineage [35].
Genetically Dominant Mutations in BAF Subunits Underlying Several Neurologic Diseases
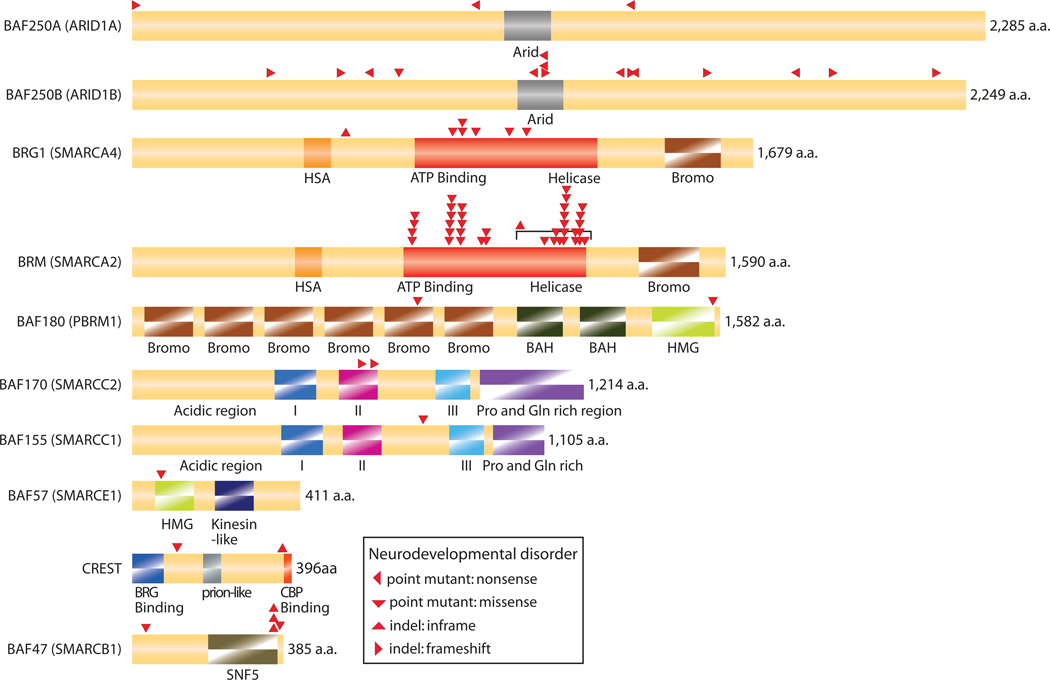
Within the past year many exome sequencing studies have found mutations in BAF subunits that cause or contribute to human neurologic diseases. Many of these mutations appear to produce null alleles by premature stop codons from either nonsense mutations or frame-shifts in the amino-terminus of the gene (Table 2). The neurologic diseases are remarkably diverse and include syndromic and non-syndromic mental retardation [26–28,36–38], including disorders such as Nicolaides-Baraitser and Coffin-Siris syndromes where most of the patients have mutations in one of 7 BAF subunits (Fig 2). However, mutations in the nBAF subunit CREST are found in ALS [39], a disease that is due to motor neuron degeneration. Also, 4 subunits (BAF180, BAF170, BAF155 and Brm) and REST were found to be mutated in autism, and two subunits in schizophrenia [40–43]. Again, many of these mutations are present on only one allele and appear to be genetically dominant (Table 2).
Table 2.
BAF complexes subunit mutations in neurologic diseases
| SMARCA2/Brm (BAF complex) | Coffin-Siris syndrome, Nicolaides-Baraitser syndrome, Schizophrenia | partial deletion, missense, in frame deletion, intronic alteration |
Tsurusaki, Y., N. Okamoto, et al. (2012). Nat Genet. Wolff D ea al. (2012) Mol Syndromol. Van Houdt, J. K., B. A. Nowakowska, et al. (2012). Nat Genet; Koga, M., H. Ishiguro, et al. (2009). Human molecular genetics. Loe-Mie, Y., A. M. Lepagnol-Bestel, et al. (2010). Human molecular genetics |
Chromatin remodeling complex ATPase |
| SMARCA4/Brg (BAF complex) | Coffin-Siris syndrome | Missense, inframe deletion | Tsurusaki, Y., N. Okamoto, et al. (2012). Nat Genet. | Chromatin remodeling complex ATPase |
| SMARCB1/BAF47 (BAF complex) | Coffin-Siris syndrome, Kleefstra syndrome phenotypic spectrum | In-frame deletion, missense, | Tsurusaki, Y., N. Okamoto, et al. (2012). Nat Genet; Kleefstra, T., J. M. Kramer, et al. (2012). Am J Hum Genet | Chromatin remodeling complex subunit |
| SMARCC1/BAF155 (BAF complex) | Autism | Missense | Neale, B. M., Y. Kou, et al. (2012). Nature. | Chromatin remodeling complex subunit |
| SMARCC2/BAF170 (BAF complex) | Autism | Splice site mutation | Neale, B. M., Y. Kou, et al. (2012). Nature. | Chromatin remodeling complex subunit |
| PBRM/BAF180 (BAF complex) | Autism | missense | O'Roak, B. J., L. Vives, et al. (2012). Nature. | Chromatin remodeling complex subunit |
| BAF57 (BAF complex) | Coffin-Siris syndrome | missense | Tsurusaki, Y., N. Okamoto, et al. (2012). Nat Genet; | Chromatin remodeling complex subunit |
| Arid1A/BAF250A (BAF complex) | Coffin-Siris syndrome | Nonsense, frame shift deletion | Tsurusaki, Y., N. Okamoto, et al. (2012). Nat Genet; | Chromatin remodeling complex subunit |
| Arid1B/BAF250B (BAF complex) | Coffin-Siris syndrome, Autism, Intellectual Disability | nonsense, missense, frame shift deletion, microdeletion |
Tsurusaki, Y., N. Okamoto, et al. (2012). Nat Genet; Santen, GWE., et al. (2012). Nat Genet. O'Roak, B. J., L. Vives, et al. (2012). Nature. Hoyer, J. et al. (2012). Am. J. Hum. Genet. |
Chromatin remodeling complex subunit |
Nonsense mutations and frame-shift mutations in one allele of BAF subunits in human neurologic diseases that suggest dosage sensitivity and also define critical residues for human neural development. Other missense and inframe shift mutations imply gain-of-function or dominant-negative effects.
Figure 2. BAF subunit mutations in human neurologic diseases indicate dosage-sensitive roles in human neural development.
Dosage sensitivity is likely to reflect a rate limiting step in neural development and may help explain why recapitulating the microRNA-chromatin switch can reprogram human fibroblasts to neurons. Missense mutations in BAF subunits also imply gain-of-function or dominant-negative effects in neurologic diseases.
Genetic dominance can have several underlying mechanisms, however genetic dominance often points to a rate-limiting role for a step in a pathway. The aspects of neurogenesis where BAF subunits might be dosage-sensitive and perhaps rate-limiting are unknown. However, the presence of microcephally in many of the syndromic intellectual disability diseases caused by dominant BAF subunit mutations suggests a dosage sensitive role in neuronal proliferation or survival. Another process that appears to be affected by BAF subunit mutations in worms, flys and mice has been dendritic morphogenesis and targeting and at least one study has found that CREST mutations found in ALS produce defective dendritic morphogenesis [39]. Interestingly, expression of the alternative npBAF subunit, SS18 in post mitotic neurons produces rather similar dendritic outgrowth defects as the ALS mutations indicating that timing of the inactivation of SS18 might also lead to neurologic defects [21]. Although speculative at this point, the genetic dominance of BAF subunit mutations in human neural development is consistent with an instructive role and might contribute to the ability of recapitulating the microRNA-chromatin switch to produce neurons from fibroblasts.
Does the MicroRNA Chromatin Switch Produce an Epigenetic Neuronal Ground State?
The ability to produce different types of neurons using the microRNA-chromatin switch may be a reflection of the normal developmental roles of these complexes. While the transcription factors that have been used for reprogramming are expressed in isolated and selected populations of post mitotic neurons, we find that BAF53b, BAF45b, BAF45c and CREST are expressed in all types of post mitotic neurons. In addition, these genes have functions in promoting fundamental aspects of neuronal maturation such as dendritogenesis [18], synaptogenesis [19] and neurotransmitter production [29]. Thus we feel that the role of the microRNA-chromatin switch might be to provide a base state that is then acted upon by neuronal transcription factors expressed more narrowly, which then drive the cell to a specific identity and the ability to produce repetitive action potentials and other features of mature neurons.
Additional evidence that the apparent instructive roles of the microRNA-chromatin switch are related to the establishment of a neural specific epigenetic state comes from the role of BAF complexes in development. Early studies in Drosophila demonstrated that the BAP complex subunit, Brm opposed polycomb-mediated repression of homeotic genes [44]. Brm is the ATPase of the fly BAF or BAP complex (also called dSWI/SNF). Genome wide studies in murine ES cells using conditional deletion of Brg (one of two Brm homologues in mammals) lead to rapid accumulation of H3K27Me3 over genes that are targets of pluripotency-signaling pathways such as STAT3 and BMP [45]. These results suggested that nBAF complexes might oppose polycomb, which places the H3K27Me3 mark and these complexes play a fundamental role in maintaining epigenetic states necessary to allow critical signals to engage their targets. Further evidence for this mode of action was obtained when STAT3 was mapped across the ES cell genome after Brg deletion. Remarkably, BAF function was necessary for STAT3 to bind to 2000 of 2400 of its sites after LIF treatment [45]. Additionally, in the absence of BAF function, STAT3 often went to incorrect sites. Thus esBAF complexes are necessary to allow this critical pluripotency signaling pathway to access its correct binding sites. This observation raises the question of whether nBAF complexes direct transcription factors functioning at the terminus of ubiquitious signaling pathways to sites used in neurons? Supporting this role was the observation that BAF complexes are also able to help replace polycomb function and repress nearby genes [45].
Another observation that bears upon the mechanism of neuronal conversion comes from a malignancy where the npBAF subunit, SS18 is translocated to the SSX gene [46]. This translocation produces an inframe fusion that precisely substitutes the C-terminal 9 amino acids of SS18 for 78 amino acids of SSX. The fusion protein enters BAF complexes evicting BAF47, but the remaining complex is intact. Remarkably, this retargets the complex to many neurogenic genes and importantly activates the Sox2 gene, which drives the proliferation of the malignancy [23]. The mechanism by which the ssBAF complex activates Sox 2 is the eviction of polycomb over the Sox2 locus, thereby reversing H3K27Me3-mediated repression.
The studies mentioned above suggest that the essential feature of the switch from esBAF to npBAF to nBAF is the ability of the specific complex to be retargeted to different loci reversing H3K27me3-mediated repression. We propose that this might have two effects. First, it would allow signaling pathways in use in fibroblasts to divert their output to neuronal loci, thereby activating neuronal genes as in the case of the SS18-SSX fusion proteins. Second, it would provide continuity to the differentiated state by virtue of the fact that polycomb-mediated repression spreads both along the chromosome and over cellular generations by binding the mark that it produces [47]. While this senario is consistent with the dominant and possibly rate-limiting role of nBAF complexes in human neural development (Table 2) additional experimental evidence is necessary to support these speculations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Frischer LE, Hagen FS, Garber RL. An inversion that disrupts the Antennapedia gene causes abnormal structure and localization of RNAs. Cell. 1986;47:1017–1023. doi: 10.1016/0092-8674(86)90816-0. [DOI] [PubMed] [Google Scholar]
- 2.Schneuwly S, Klemenz R, Gehring WJ. Redesigning the body plan of Drosophila by ectopic expression of the homoeotic gene Antennapedia. Nature. 1987;325:816–818. doi: 10.1038/325816a0. [DOI] [PubMed] [Google Scholar]
- 3.Schneuwly S, Kuroiwa A, Gehring WJ. Molecular analysis of the dominant homeotic Antennapedia phenotype. EMBO J. 1987;6:201–206. doi: 10.1002/j.1460-2075.1987.tb04739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Postlethwait JH, Schneiderman HA. A clonal analysis of determination in Antennapedia a homoeotic mutant of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1969;64:176–183. doi: 10.1073/pnas.64.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott MP, Weiner AJ, Hazelrigg TI, Polisky BA, Pirrotta V, Scalenghe F, Kaufman TC. The molecular organization of the Antennapedia locus of Drosophila. Cell. 1983;35:763–776. doi: 10.1016/0092-8674(83)90109-5. [DOI] [PubMed] [Google Scholar]
- 6.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 9.Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olave I, Wang W, Xue Y, Kuo A, Crabtree GR. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev. 2002;16:2509–2517. doi: 10.1101/gad.992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. Embo J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 14.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akerblom M, Sachdeva R, Barde I, Verp S, Gentner B, Trono D, Jakobsson J. MicroRNA-124 is a subventricular zone neuronal fate determinant. J Neurosci. 2012;32:8879–8889. doi: 10.1523/JNEUROSCI.0558-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 19. Vogel-Ciernia A, Matheos DP, Barrett RM, Kramar EA, Azzawi S, Chen Y, Magnan CN, Zeller M, Sylvain A, Haettig J, et al. The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nat Neurosci. 2013;16:552–561. doi: 10.1038/nn.3359. * By analyzing BAF53b heterozygous mice and transgenic mice with a dominant negative BAF53b, the authors a requirement for long-term memory and hippocampal synaptic plasticity.
- 20. Tea JS, Luo L. The chromatin remodeling factor Bap55 functions through the TIP60 complex to regulate olfactory projection neuron dendrite targeting. Neural Dev. 2011;6:5. doi: 10.1186/1749-8104-6-5. * The authors found that mutation BAF53 resulted in perfect retargeting of olfactory projection neurons to the incorrect glomulus suggesting an instructive role for these chromatin remodeling complexes.
- 21. Staahl BT, Tang J, Wu W, Sun A, Gitler AD, Yoo AS, Crabtree GR. Kinetic Analysis of npBAF to nBAF Switching Reveals Exchange of SS18 with CREST and Integration with Neural Developmental Pathways. J Neurosci. 2013;33:10348–10361. doi: 10.1523/JNEUROSCI.1258-13.2013. * The authors demonstrate that CREST is a dedicated non-exchangable subuit of nBAF complexes and that CREST takes the place of SS18 in these complexes near cell cycle exit. They also relate this switch to other more well known genes involved in neural development.
- 22.Aizawa H, Hu SC, Bobb K, Balakrishnan K, Ince G, Gurevich I, Cowan M, Ghosh A. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science. 2004;303:197–202. doi: 10.1126/science.1089845. [DOI] [PubMed] [Google Scholar]
- 23.Kadoch C, Crabtree GR. Reversible Disruption of mSWI/SNF (BAF) Complexes by the SS18-SSX Oncogenic Fusion in Synovial Sarcoma. Cell. 2013;153:71–85. doi: 10.1016/j.cell.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamba DA, Hayes S, Karl MO, Reh T. Baf60c is a component of the neural progenitor-specific BAF complex in developing retina. Dev Dyn. 2008;237:3016–3023. doi: 10.1002/dvdy.21697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Santen GW, Aten E, Sun Y, Almomani R, Gilissen C, Nielsen M, Kant SG, Snoeck IN, Peeters EA, Hilhorst-Hofstee Y, et al. Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nat Genet. 2012;44:379–380. doi: 10.1038/ng.2217. * The authors found that most patients with Coffin-Siris syndrome have mutations in BAF250(Arid1b).
- 27. Tsurusaki Y, Okamoto N, Ohashi H, Kosho T, Imai Y, Hibi-Ko Y, Kaname T, Naritomi K, Kawame H, Wakui K, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 2012;44:376–378. doi: 10.1038/ng.2219. ** The authors report de novo mutations in one of 6 subunits of BAF complexes in most patients with this disease. Mutations are often domanent suggesting a dosage sensitive role for BAF complexes in human neural development.
- 28. Hoyer J, Ekici AB, Endele S, Popp B, Zweier C, Wiesener A, Wohlleber E, Dufke A, Rossier E, Petsch C, et al. Haploinsufficiency of ARID1B, a Member of the SWI/SNF-A Chromatin-Remodeling Complex, Is a Frequent Cause of Intellectual Disability. Am J Hum Genet. 2012;90:565–572. doi: 10.1016/j.ajhg.2012.02.007. * The authors estimate that up to 3% of all human intellectual deficiency might be caused by mutations in BAF250b.
- 29. Weinberg P, Flames N, Sawa H, Garriga G, Hobert O. The SWI/SNF Chromatin Remodeling Complex Selectively Affects Multiple Aspects of Serotonergic Neuron Differentiation. Genetics. 2013;194:189–198. doi: 10.1534/genetics.112.148742. * This study showed that ham-3 (BAF60) is essential for serotonin production and migration of specific serotonergic neurons, The remarkable specificity of the phenotype is informative and demonstrates that the older notion that BAF or mSWI/SNF function as a non-specific regulator of PolII binding is not tentable.
- 30. Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. ** This paper shows that recapitulating the micro-RNA chromatin switch can convert fibroblasts to neurons. This is the first paper to show that microRNAs are also important cell fate determinants in humans.
- 31.Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 32.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bibel M, Richter J, Lacroix E, Barde YA. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat Protoc. 2007;2:1034–1043. doi: 10.1038/nprot.2007.147. [DOI] [PubMed] [Google Scholar]
- 35.Vierbuchen T, Wernig M. Molecular roadblocks for cellular reprogramming. Mol Cell. 2012;47:827–838. doi: 10.1016/j.molcel.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Houdt JK, Nowakowska BA, Sousa SB, van Schaik BD, Seuntjens E, Avonce N, Sifrim A, Abdul-Rahman OA, van den Boogaard MJ, Bottani A, et al. Heterozygous missense mutations in SMARCA2 cause Nicolaides-Baraitser syndrome. Nat Genet. 2012;44:445–449. S441. doi: 10.1038/ng.1105. [DOI] [PubMed] [Google Scholar]
- 37.Wolff D, Endele S, Azzarello-Burri S, Hoyer J, Zweier M, Schanze I, Schmitt B, Rauch A, Reis A, Zweier C. In-Frame Deletion and Missense Mutations of the C-Terminal Helicase Domain of SMARCA2 in Three Patients with Nicolaides-Baraitser Syndrome. Mol Syndromol. 2012;2:237–244. doi: 10.1159/000337323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14:347–359. doi: 10.1038/nrg3413. * This review gives an overview of the recent burst of genome-sequencing studies, leading to the realization that mutations in multiple chromatin regulators cause human neurological diseases including microcephaly.
- 39.Chesi A, Staahl BT, Jovicic A, Couthouis J, Fasolino M, Raphael AR, Yamazaki T, Elias L, Polak M, Kelly C, et al. Exome sequencing to identify de novo mutations in sporadic ALS trios. Nat Neurosci. 2013;16:851–855. doi: 10.1038/nn.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koga M, Ishiguro H, Yazaki S, Horiuchi Y, Arai M, Niizato K, Iritani S, Itokawa M, Inada T, Iwata N, et al. Involvement of SMARCA2/BRM in the SWI/SNF chromatin-remodeling complex in schizophrenia. Hum Mol Genet. 2009;18:2483–2494. doi: 10.1093/hmg/ddp166. [DOI] [PubMed] [Google Scholar]
- 41.Loe-Mie Y, Lepagnol-Bestel AM, Maussion G, Doron-Faigenboim A, Imbeaud S, Delacroix H, Aggerbeck L, Pupko T, Gorwood P, Simonneau M, et al. SMARCA2 and other genome-wide supported schizophrenia-associated genes: regulation by REST/NRSF, network organization and primate-specific evolution. Hum Mol Genet. 2010;19:2841–2857. doi: 10.1093/hmg/ddq184. [DOI] [PubMed] [Google Scholar]
- 42.Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kennison JA, Tamkun JW. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 1988;85:8136–8140. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol. 2011;13:903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crew AJ, Clark J, Fisher C, Gill S, Grimer R, Chand A, Shipley J, Gusterson BA, Cooper CS. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. Embo J. 1995;14:2333–2340. doi: 10.1002/j.1460-2075.1995.tb07228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]