Abstract
TopBP1, a multiple-BRCT-containing protein, plays diverse functions in DNA metabolism including DNA replication, DNA damage response and transcriptional regulation. The cytoplasmic localization of TopBP1 has been found associated with breast cancer susceptibility in clinical studies, suggesting the biological significance of TopBP1’s sub-cellular localization. However, it remains elusive how TopBP1 is shuttled into nucleus and recruited to chromatin under normal or stressful conditions. Taking advantage of Xenopus egg extract, we identified Importin β as a new interacting protein of the TopBP1 C-terminus. We verified the TopBP1-Importin β association via GST pulldown and coimmunoprecipitation assays. We then demonstrated that TopBP1’s C-terminal motif (designated as CTM, 23 amino acids) containing a putative NLS (Nuclear Localization Signal) was required for Importin β interaction and that CT100 of Importin β (100 amino acids of extreme C-terminus of Importin β) was required for TopBP1 interaction. Further structure-function analysis reveals that the CTM of TopBP1 is essential for TopBP1’s nuclear import and subsequent chromatin recruitment, thereby playing important roles in DNA replication and mitomycin C (MMC)-induced Chk1 phosphorylation. In addition, Importin β-specific inhibitor importazole inhibits TopBP1’s nuclear import and the MMC-induced Chk1 phosphorylation. With ongoing DNA replication, the Importin β-dependent nuclear import of TopBP1 was indeed required for the MMC-induced Chk1 phosphorylation. Our data also suggest that checkpoint activation requires more TopBP1 than DNA replication does. The requirement of TopBP1’s CTM motif for ATR-Chk1 checkpoint can be bypassed in a nucleus-free AT70 system. Taken together, our findings suggest the CTM motif-mediated TopBP1 shuttling into nucleus via Importin β plays an important role in the ATR-Chk1 checkpoint signaling in Xenopus egg extracts.
Keywords: ATR-Chk1 checkpoint, DNA damage response, DNA replication, Importin β, nuclear import, TopBP1
1. Introduction
The DNA damage response (DDR) is an elaborate surveillance mechanism that senses and responds to DNA damage [1]. Deficiencies in the DDR lead to genomic instability, whose pathological consequences include cancer development, neurological defects, infertility and immunological diseases [2]. While ATM (ataxia telangiectasia mutated)-Chk2 (Checkpoint kinase 2) checkpoint signaling is activated primarily in response to double-strand breaks (DSB), ATR (ATM and Rad3 related)-Chk1 (Checkpoint kinase 1) checkpoint signaling is activated by a variety of DNA damaging agents and replication stress [3,4]. Accumulating evidence suggests that there is also crosstalk and transition between these two pathways [5–8]. Primed single-strand DNA (ssDNA) coupled with RPA (Replication Protein A) is the structural determinant of ATR checkpoint signaling in response to stalled DNA replication forks [9,10]. Activated ATR kinase phosphorylates its downstream substrates including Chk1 [11,12]. Phosphorylated Chk1 is the activated form of Chk1, which then phosphorylates its own downstream substrates such as Cdc25 and regulates cell cycle progression [13]. Targeting ATR and Chk1 kinases has been used as a strategy for cancer therapy [14,15].
In recent years, intense attention has been drawn to mechanistic studies of ATR-Chk1 checkpoint activation [2, 9]. The functional uncoupling of MCM (mini-chromosome maintenance) helicase and DNA polymerase activities is demonstrated as a mechanism for generating a long stretch of ssDNA, thereby activating ATR-Chk1 checkpoint [16]. ssDNA can also be generated after DSB end resection by exonuclease enzymes such as CtIP and Exo1 or after SSB (single-strand breaks) end resection by AP endonuclease 2 (APE2) [17–19]. In addition, ATR-Chk1 checkpoint activation requires several mediator proteins such as ATRIP (ATR Interacting Protein), the 9-1-1 complex (Rad9-Rad1-Hus1 complex), Claspin, and TopBP1 (Topoisomerase IIβ binding protein 1) [20–27]. TopBP1 contains eight BRCT (BRCA1 C-terminus) domains and is implicated in many aspects of DNA metabolism including DNA replication, DNA damage response, transcriptional regulation and DNA repair [24,28–30]. A fragment between the BRCT6 and BRCT7 in TopBP1 is a direct activator for ATR activation [24,25]. The BRCT1 and BRCT2 domains of TopBP1 interact with the 9-1-1 complex, playing an important role in ATR checkpoint [23]. Recently, accumulating evidence demonstrates that the TopBP1 C-terminus (i.e., C-terminal ~330 amino acids) including BRCT7-8 is important for the ATR-Chk1 checkpoint signaling. The TopBP1 C-terminus is essential for ATR-Chk1 checkpoint in response to mitomycin C (MMC)-induced interstrand crosslinks, γ-irradiation, or checkpoint-activating structure AT70 [26,31]. The interaction between TopBP1 C-terminus and the helicase BACH1 is required for the extension of ssDNA and RPA hyperloading following replication stress [32]. The AKT-dependent phosphorylation of TopBP1 induces TopBP1 oligomerization via its C-terminus, suggesting an essential role in cell survival [33]. TopBP1 C-terminus interacts with p53, leading to p53 promoter binding activity inhibition and aggressive tumor behavior [34]. Interestingly, an in vitro reconstitution study has shown that TopBP1 C-terminus is directly required for RPA-ssDNA-mediated ATR activation [35]. All these studies demonstrate a myriad of essential roles for the TopBP1 C-terminus in the DDR via various distinct mechanisms.
Genomic instability is considered as one enabling characteristic of cancer and the DDR has been proposed as a candidate anti-cancer barrier in early human cancer development [36,37]. Therefore, it is pivotal to determine how the DDR is activated in response to DNA damage. Recent immunohistochemical and immunoblotting analyses demonstrated that TopBP1 was expressed and localized in nuclei of normal human breast cells. However, TopBP1 was aberrantly expressed and localized in cytoplasmic compartment of breast cancer cells [38,39]. The percentage of breast cancer patients with cytoplasmic localization of TopBP1 also rose with an increasing histological grade of tumors [38]. These findings suggest that the abnormal localization of TopBP1 to cytoplasm may play a role in the development of breast cancer; however, an understanding of the molecular mechanism involved in this process is lacking. Because TopBP1 plays multiple roles in DDR primarily in the nucleus, we reason that the aberrant cytosolic localization of TopBP1 may have defects in triggering appropriate DNA damage response pathways, leading to possible genomic instability and subsequent cancer development. Therefore, it is vital to determine how TopBP1 is shuttled from cytoplasm into nucleus. Typically, protein import from cytosol into nucleus is mediated by soluble receptors that recognize cargos and carry them through the nuclear pore complex (NPC) [40]. A well-characterized receptor Importin β directly interacts with its cargo for import or indirectly recognizes cargo via a nuclear localization signal (NLS) through adaptor protein Importin α [41,42].
Taking advantage of the cell-free Xenopus egg extract system, we have investigated the roles of TopBP1 in DNA metabolism including DNA replication and DDR through a series of studies [26,31,43–45]. Here, we report the Importin β-dependent nuclear import of TopBP1 in the DDR. We identified a novel interaction between TopBP1 and Importin β with a pulldown assay using TopBP1 C-terminus and confirmed the physical association between TopBP1 and Importin β via coimmunoprecipitation. We demonstrated that the CTM of TopBP1 in its extreme C-terminus, which contains a putative NLS motif, was required for the interaction with Importin β. A CTM-deletion mutant of TopBP1 failed to shuttle into nucleus and onto chromatin. We further revealed that the TopBP1-Importin β interaction is important for DNA replication and DNA damage response via distinct mechanisms. Together, these data suggest that the Importin β-dependent shuttling of TopBP1 into nucleus plays an important role in the ATR-Chk1 checkpoint signaling in Xenopus.
2. Materials and Methods
2.1. Xenopus egg extract preparation and related procedures
The use of Xenopus laevis was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of North Carolina at Charlotte. Xenopus egg extract preparation and sperm chromatin preparation were performed as described previously [19,45]. Immunodepletion of TopBP1 was performed as described [26,43]. To elicit a checkpoint response, extracts were treated with 0.5 mM Mitomycin C (MMC) or 50 μg/ml of annealed oligonucleotides poly(dA)70-poly (dT)70 (designated as AT70) in the presence of 3 μM tautomycin as described previously [26,31]. Importazole was added to egg extracts to a final concentration of 2 mM to specifically inhibit Importin β-mediated nuclear import of its cargo proteins. Chromatin isolation and DNA synthesis analysis were performed as previously described [19,43]. For nuclei isolation, reaction mixture containing egg extracts and sperm chromatin was resuspended in 1 mL Nuclear Isolation Buffer (NIB, 50 mM KCl, 25 mM MgCl2, 25 mM EGTA, 2.5 mM spermidine, 0.75 mM spermine, 50mM HEPES-KOH, pH 7.6) and was then overlayed on the top of 200 μL of 15% sucrose in NIB. After 5-minute centrifugation at 5,000 rpm, the supernatant was carefully removed and the pellet was collected as nuclei fraction for immunoblotting or fluorescence microscopy analysis.
2.2. Recombinant proteins and antibodies
GST-CT333 and WT TopBP1 were described previously [26,43]. The ΔCTM TopBP1 was cloned via PCR on WT TopBP1 with two primers (5′-GGGGGCCATGGCTTCAAGTGAAAAC-3′, and 5′-GGGGGCTCGAGTCATGTAACCCGGACATATG-3′) and ligated into NcoI- and XhoI-digested pCS2+MT vector. Recombinant WT and ΔCTM TopBP1 were expressed in SP6 TnT transcription/translation coupled quick master mix kit (Promega) as described previously [43]. GST-Importin β, the full-length WT Importin β constructed in pGEX6P, was expressed and purified as described previously [46]. GST-ΔCT100 was constructed via PCR on pGEX6P-Importin β with primers (5′-CCCCCGGATCCATGGAGCTCGTCACCATCCTCGAGAAGACC-3′ and 5′-CCCCGCGGCCGCTCAGAGCCCCTGGATTATTCCTGTATAGGC-3′) and inserted into BamHI- and NotI-digested pGEX6P vector. Recombinant GST-ΔCT100 was expressed and purified with a similar approach as GST-Importin β. Antibodies against Xenopus TopBP1 and Orc2 have been described previously [43]. Additionally, antibodies against Chk1 P-S344 (Cell Signaling Technology), Chk1 (Santa Cruz Biotechnology), C-myc (Santa Cruz Biotechnology), GST (Santa Cruz Biotechnology) and Importin β (Abcam) were purchased from respective vendors. Peroxidase-conjugated monoclonal mouse anti-rabbit IgG light chain specific (Jackson ImmunoResearch Laboratories), Peroxidase-conjugated ImmunoPure goat anti-rabbit IgG (H + L; Thermo Fisher Scientific) and peroxidase-linked sheep anti-mouse IgG (GE Healthcare) were used as secondary antibodies in immunoblotting analysis when appropriate.
2.3. GST pulldown assays
For experiments in Fig. 1B and 1D, purified GST-CT333 (10 μg) or GST (10 μg) were incubated with pre-washed Glutathione Sepharose 4 Fast Flow (GE Healthcare) in 0.5 mL of Interaction Buffer (100 mM NaCl, 0.05% NP-40, 5 mM MgCl2, 10% Glycerol, 20 mM Tris-HCL, pH 8.0) for 1h at 4°C with end-over-end mixing. 200 μL of Xenopus egg extract (“extract”) or ELB buffer (“−”) was diluted with 200 μL Interaction Buffer, and then was added to the beads. After 2-hr incubation with rotation at 4°C, beads were washed with Interaction Buffer two times. The bead-bound proteins were subjected to SDS-PAGE followed by silver staining or immunoblotting analysis. For experiments in Fig. 3C and 3D, similar GST pulldown assays were performed with minor changes: (1) purified GST, GST-Importin β, or GST-ΔCT100 was coupled to beads; (2) recombinant WT Myc-TopBP1 was added to egg extracts (“Extract”, Fig. 3C) or ELB buffer (“Buffer”, Fig. 3D).
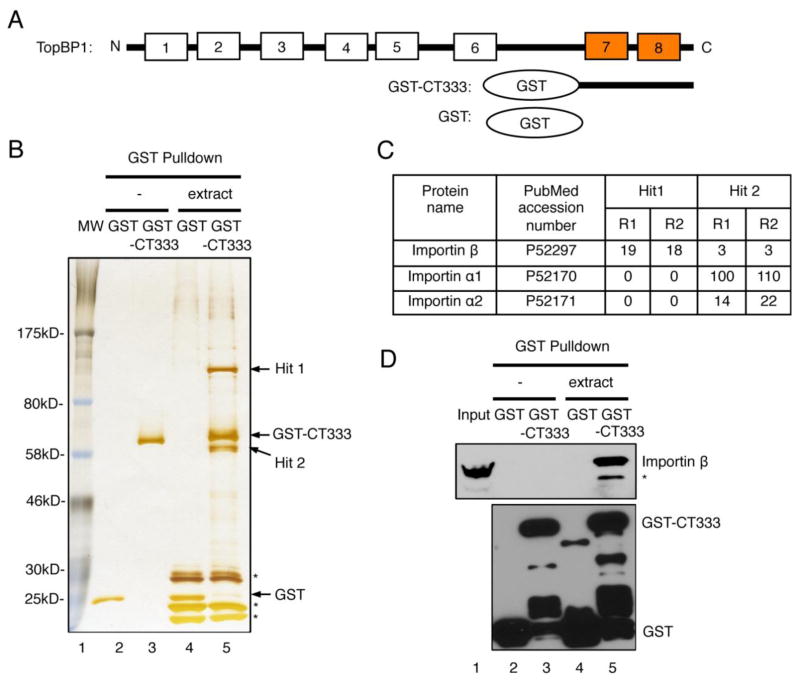
Fig. 1.
Identification and verification of Importin β as a new interacting protein of TopBP1 C-terminus. (A) Schematic diagram of TopBP1 and its C-terminus. (B) SDS-PAGE analysis followed by silver staining of GST pulldown samples with GST or GST-CT333 from either buffer (“−”) or egg extract (“extract”). * represent proteins bound to beads from extract nonspecifically. (C) The diagram shows the numbers of polypeptide fragments from two separate runs (R1 and R2) of mass spectrometry analysis of “Hit 1” and “Hit 2”. (D) Immunoblotting analysis of GST pulldown samples from (B) with antibodies against GST and Importin β, respectively. 1 μL of egg extract was used as “Input”. * represents a protein which can be recognized by anti-Importin β antibodies nonspecifically.
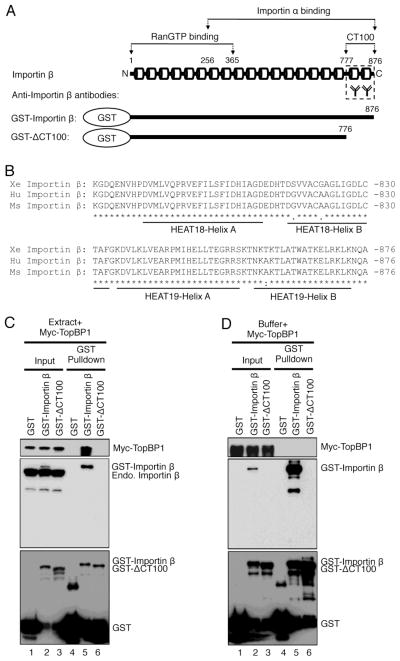
Fig. 3.
The CT100 fragment of Importin β is required for TopBP1 association. (A) Schematic presentation of Importin β protein, anti-Importin β antibodies, recombinant GST-Importin β and GST-ΔCT100 (GST-Importin β without the 100 amino acids at its extreme C-terminus). The numbers indicate the positions of respective amino acid residues. The cylinders represent the HEAT motifs in Importin β protein. (B) Sequence alignment of the extreme C-terminus 100 amino acids of Importin β protein from frog (Xe Importin β, ABY76052.1), human (Hu Importin β, NP_002256.2) and mouse (Ms Importin β, NP_032405.3) using the Clustal Omega software. *, identical residues; :, highly conserved residues;., moderately conserved residues. Conserved Helix motifs of HEAT18 and HEAT19 domains are underlined. (C) GST-Importin β, but neither GST nor GST-ΔCT100, associates with Myc-TopBP1 in egg extracts. GST pulldown assays were performed using egg extracts as described in Materials and Methods. The “GST Pulldown” samples and “Input” (1 μL of egg extract) were examined via immunoblotting using antibodies against Myc (top panel), Importin β (middle panel), and GST (bottom panel), respectively. (D) Similar GST Pulldown assays and immunoblotting were performed as (C) except that control egg lysis buffer was used as described in Materials and Methods.
2.4. Coimmunoprecipitation (Co-IP) assays
For Co-IP assay in Fig. 2A, antibodies against TopBP1 (1μg), Importin β (1μg), or control IgG antibodies (1μg) was added into 100 μL Interaction Buffer containing 15 μL of protein A agarose beads (GE Healthcare). After one-hour incubation, the beads were supplemented with 100 μL of Xenopus extract respectively. The beads were isolated by centrifugation and washed three times by Interaction Buffer. Bead-bound proteins were examined via immunoblotting analysis. For Co-IP assay in Fig. 2D, anti-Myc antibodies (1μg) were coupled to protein A agarose beads, followed by incubation with 100 μL of TopBP1-depleted egg extracts supplemented with either recombinant WT TopBP1 or ΔCTM TopBP1. The beads were washed three times by Interaction Buffer and bead-bound proteins were examined via immunoblotting.
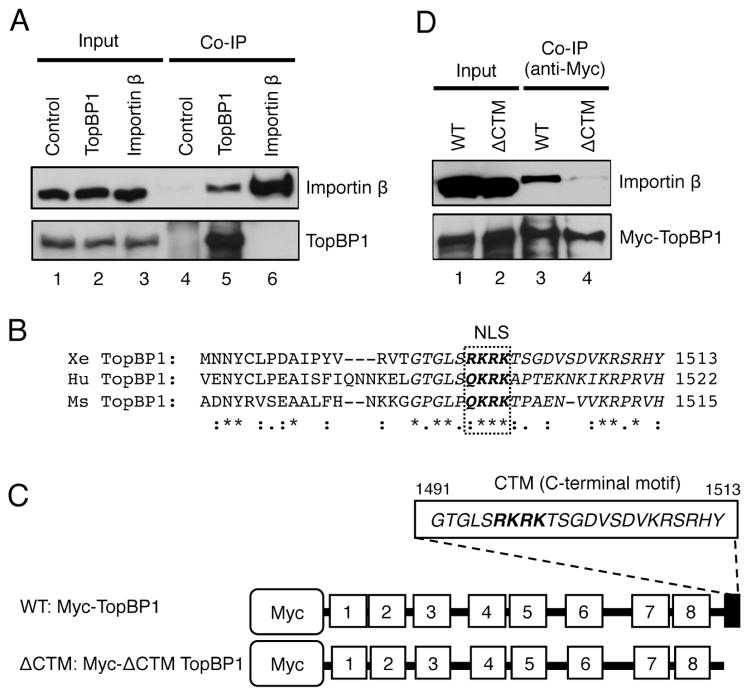
Fig. 2.
The CTM motif of TopBP1 is important for Importin β interaction. (A) Co-IP assay using control IgG or antibodies against TopBP1 or Importin β. 1 μL of egg extract before the addition of antibodies in the assays was collected and used as “Input”. The individual bead-bound fractions were examined as “Co-IP” samples. Immunoblotting was used to examine TopBP1 and Importin β as indicated. (B) Amino acid sequence alignment of the C-terminus of TopBP1 proteins in frog, human, and mouse using the Clustal Omega software. Abbreviations and GenBank accession no.: (1) Xe TopBP1, AAP03894.1; (2) Hu TopBP1, NP_008958.2; (3) Ms TopBP1, NP_795953.2. -, gaps in the alignment; *, identical residues; :, highly conserved residues;., moderately conserved residues; the dashed rectangle was used to illustrate the putative NLS. (C) Schematic diagram of the WT TopBP1 and ΔCTM TopBP1. The ΔCTM TopBP1 lacks the CTM motif (i.e., the last 23 amino acids in the extreme C-terminus of TopBP1). (D) Co-IP assays demonstrated that the CTM motif of TopBP1 was required for the interaction with Importin β. Anti-Myc antibodies were added to TopBP1-depleted egg extract supplemented with either WT TopBP1 or ΔCTM TopBP1 as indicated. The bead-bound proteins from Co-IP assays (“Co-IP”) were examined for Importin β and Myc-tagged TopBP1 via immunoblotting.
2.5. Immunofluorescence microscopy
10μL of extract with nuclei was added to 200 μL of 1% formaldehyde, which was made freshly in XBE (50 mM KCl, 0.05 mM CaCl2, 1 mM MgCl2, 2.5 mM EGTA, 25 mM sucrose, 5 mM HEPES, pH7.6). The mixture was resuspended gently and incubated for 12 minutes at room temperature. The mixture was spin through 4 mL cushion of 30% glycerol (v/v) in XBE for 10 minutes at 5,000 rpm in a spin-down tube containing a Poly-L-Lysine-coated coverslip. The coverslip was recovered from the tube and was washed by PBS and blocked with blocking solution (3% BSA/PBS) for at least one hour at room temperature. The coverslip was incubated with anti-TopBP1 antibodies (1:200 (v/v) with blocking solution) overnight at 4°C. After washing by PBS, the coverslip was then incubated with donkey anti-rabbit IgG linked with rhodamine (1:200 (v/v) with blocking solution) for one hour at room temperature. Next, the coverslip was stained by Hoechst 33258 (1 μg/mL) for 15 minutes at room temperature. After further washing by PBS, the coverslip was mounted to 4 μL of 2% n-propyl gallate (w/v) on a microscopic slide followed by fluorescence microscope examination. The isolated nuclei/chromatin fractions were typically stained with Hoechst 33258 for fluorescence microscopy analysis.
3. Results
3.1. Importin β interacts with TopBP1
To test the hypothesis that TopBP1’s multiple roles in DNA metabolism may be carried out through different domains and their respective interacting protein (s), we sought to identify new interacting protein (s) of the TopBP1 C-terminus via GST pulldown followed by mass spectrometry. We previously expressed and purified recombinant GST-CT333 from E. coli, which includes the C-terminal 333 amino acid domain of Xenopus TopBP1 with a GST tag on the N-terminus (Fig. 1A) [26]. Recombinant GST or GST-CT333 was used to perform pulldown experiments from either control egg lysis buffer (“−”) or Xenopus egg extract (“extract”). We further examined the eluates from the pulldown via silver staining. As expected, we detected only GST or GST-CT333 in pulldown samples from buffer (Fig. 1B, lanes 2 and 3). Interestingly, we found two unique bands, termed “Hit 1” and “Hit 2”, which strongly associated with GST-CT333, but not GST, in pulldown samples from the extract but not from the control buffer (Fig. 1B). Two runs of mass spectrometry analysis (R1 and R2) showed a number of peptide fragments of Importin β (19 and 18) for Hit1 and Importin α1 (100 and 110) and Importin α2 (14 and 22) for Hit 2 (Fig. 1C). The molecular weights of previously characterized Xenopus Importin β (97 kDa), Importin α1 (54 kDa) and Importin α2 (56 kDa) are consistent with the positions of Hit 1 and Hit 2 on the gel (Fig. 1C) [47]. We confirmed that Hit 1 was Importin β by immunoblotting with anti-Importin β antibodies and that endogenous Importin β from extract associated with GST-CT333, but not GST (Fig. 1D). In summary, we identified Importin β as a previously uncharacterized interacting protein of the TopBP1 C-terminus.
3.2. The CTM motif of TopBP1 mediates its interaction with Importin β
To further characterize the interaction between TopBP1 and Importin β under more physiologically relevant conditions, we tested whether TopBP1 associates with Importin β via coimmunoprecipitation assays. TopBP1 and Importin β were found in the immunoprecipitate complex using anti-TopBP1 antibodies, but not with control antibodies (Fig. 2A). However, anti-Importin β antibodies immunoprecipitated Importin β but not TopBP1 in a reciprocal coimmunoprecipitation experiment. This observation may indirectly indicate that our anti-Importin β antibodies bind to the TopBP1-interaction domain of Importin β, thereby preventing an association between TopBP1 and Importin β. We next sought to identify the possible domain of TopBP1 that mediates its interaction with Importin β. NucPred, a NLS predicting software (http://www.sbc.su.se/~maccallr/nucpred/cgi-bin/single.cgi), predicts that a four-amino acid motif (R1496K1497R1498K1499) in the extreme C-terminus of Xenopus TopBP1 is a potential NLS. This putative NLS of TopBP1 is conserved in Xenopus, human, and mouse (Fig. 2B). We previously generated a recombinant full-length wild type TopBP1 with a Myc-tag on the N-terminus (Fig. 2C, WT TopBP1) [26]. To test whether the NLS of TopBP1 is important for its interaction with Importin β, we generated a CTM deletion mutant of TopBP1 (Fig. 2C, ΔCTM TopBP1), which lacks the extreme C-terminal 23 amino acids. We added recombinant WT or ΔCTM TopBP1 to TopBP1-depleted egg extracts, and performed immunoprecipitation assays with anti-Myc antibodies. As expected, anti-Myc antibodies immunoprecipitated WT TopBP1 and ΔCTM TopBP1 (Fig. 2D). However, Importin β was found in the immunoprecipitate complex with the presence of WT TopBP1, but not ΔCTM TopBP1, suggesting the extreme C-terminal 23 amino acid motif is required for TopBP1 to associate with Importin β. Hereafter, we refer to this fragment as the CTM motif of TopBP1 in this study.
3.3. The CT100 fragment of Importin β is required for TopBP1 association
Importin β contains nineteen tandemly repeated HEAT motifs, which mediate interactions with RanGTP (HEAT 1–8, amino acid 1–365) and Impoitin α (HEAT 7–19, amino acid 256–876) (Fig. 3A) [48]. Our anti-Importin β antibodies was raised against a synthetic peptide from the extreme C-terminal 100 amino acids of human Importin β. Ninety seven out of one hundred amino acids (97%) are identical in Importin β protein from frog, human and mouse, suggesting the eighteenth and nineteenth HEAT motifs of Importin β are very conserved during evolution (Fig. 3B). From the observation in Fig. 2A, we speculate that our anti-Importin β antibodies may bind TopBP1-binding site on Importin β, thereby preventing the association between endogenous Importin β and TopBP1. To directly test whether the CT100 (i.e., the C-terminal 100 amino acids) of Xenopus Importin β is required for TopBP1 association, we performed GST pulldown assays with GST-Importin β and GST-ΔCT100 (CT100 deletion mutant of GST-Importin β) when recombinant WT Myc-TopBP1 was added to egg extracts (with a final concentration similar to that of endogenous TopBP1). As expected, anti-Importin β antibodies recognized endogenous Importin β (endo. Importin β) and GST-Importin β, but not GST-ΔCT100 (Fig. 3C, middle panel). Both GST-Importin β and GST-ΔCT100 were recognized by anti-GST antibodies (Fig. 3C, bottom panel). Notably, WT Myc-TopBP1 was observed in the pulldown sample using GST-Importin β, but not GST-ΔCT100 or GST, suggesting the CT100 fragment of Importin β is required for the association with TopBP1 in egg extracts (Fig. 3C, top panel).
To test the idea whether or not TopBP1 associates with Importin β directly, we performed GST pulldown assays with WT Myc-TopBP1, GST-Importin β and GST-ΔCT100 in control egg lysis buffer (“Buffer”). Under our experimental conditions, we did not observe any noticeable WT Myc-TopBP1 from the pulldown sample using GST-Importin β although GST-Importin β is indeed efficiently bound to beads (Fig. 3D). This observation suggests that TopBP1 doesn’t associate with Importin β directly or, alternatively, their association is significantly decreased when their adaptor proteins (such as Importin α1/α2) are absent in buffer in comparison to the situation of egg extracts.
3.4. The CTM motif of TopBP1 is required for nuclear import from outside of assembled nuclei
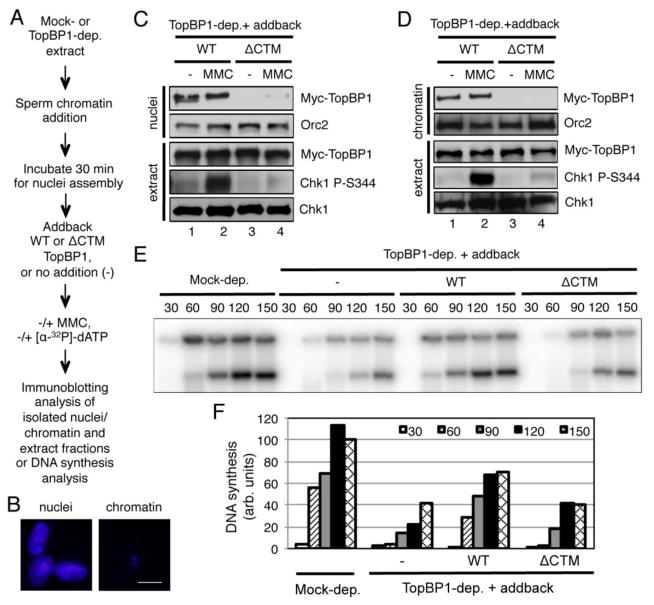
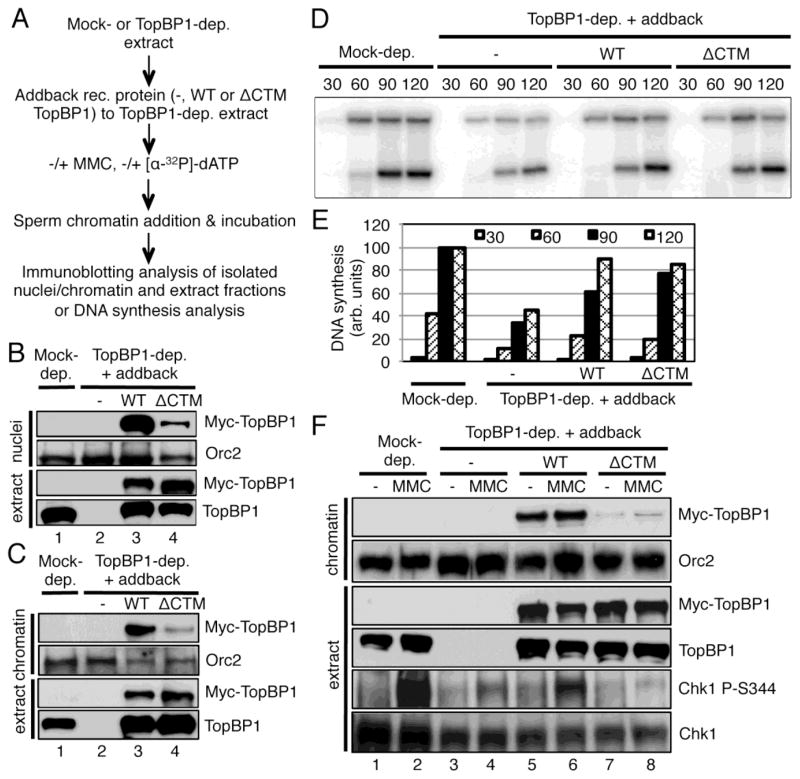
To reveal the biological significance of the TopBP1-Importin β interaction, we sought to test whether the CTM motif of TopBP1 is important for its function in DNA replication and ATR-Chk1 checkpoint signaling. We added sperm chromatin into TopBP1-depleted egg extract and incubated the reaction for 30 minutes to allow for nuclear membrane formation (Fig. 4A). We then added either WT or ΔCTM TopBP1 and interstrand crosslink reagent MMC. Subsequent immunoblotting analyses demonstrated that WT TopBP1, but not ΔCTM TopBP1, was shuttled into the nuclei from outside of assembled nuclei and subsequent onto chromatin, and was sufficient in triggering Chk1 phosphorylation in response to MMC treatment (Fig. 4C and D). To distinguish isolated nuclei fraction from isolated chromatin fraction, we typically examined them via fluorescence microscope, respectively. Our representative results showed that the nuclear envelope was intact in the isolated nuclei fraction (Fig. 4B, nuclei), but the nuclear envelope was broken in the isolated chromatin fraction (Fig. 4B, chromatin). Consistent with others, we have demonstrated previously that TopBP1 is required for DNA replication initiation via recruiting Cdc45 onto chromatin [49,50]. Thus, we tested the possible role of the CTM-dependent nuclear import of TopBP1 in DNA replication. TopBP1 depletion significantly decreased DNA synthesis, as expected (Fig. 4E and F). WT TopBP1, but not ΔCTM TopBP1, partially rescued the defects in DNA synthesis due to TopBP1 depletion (Fig. 4E and F). These data suggest that the CTM-dependent TopBP1 nuclear import plays important roles in both DNA replication and ATR-Chk1 checkpoint signaling.
Fig. 4.
The CTM motif of TopBP1 is important for its shuttling into nuclei. (A) Experimental design of the assays. (B) Nuclear membrane from the isolated nuclei fraction, but not chromatin fraction, was intact. Sperm chromatin was added to egg extracts. After 60 minutes incubation, nuclei fraction or chromatin fraction was isolated respectively and was further examined via fluorescence microscope after Hoechst 33258 staining. The scale bar represents 10 μm. (C) Nuclei fraction (“nuclei”) and total extract (“extract”) were examined at 90 minutes for the indicated proteins via immunoblotting as indicated. (D) Chromatin fraction (“chromatin”) and total extract (“extract”) were examined as (C). (E) DNA synthesis was examined as indicated. After WT or ΔCTM TopBP1 was added to appropriate egg extracts containing formed nuclei, [α-32P]-dATP was added for incorporation into the freshly replicated DNA. Mock-depleted egg extract was used as positive control. At different time points (minutes), samples were collected and examined on agarose gel followed by Phosphorimager analysis. (F) Quantification analysis of the DNA synthesis of the samples from (E).
3.5. Importin β-specific inhibitor importazole compromised the nuclear import of TopBP1
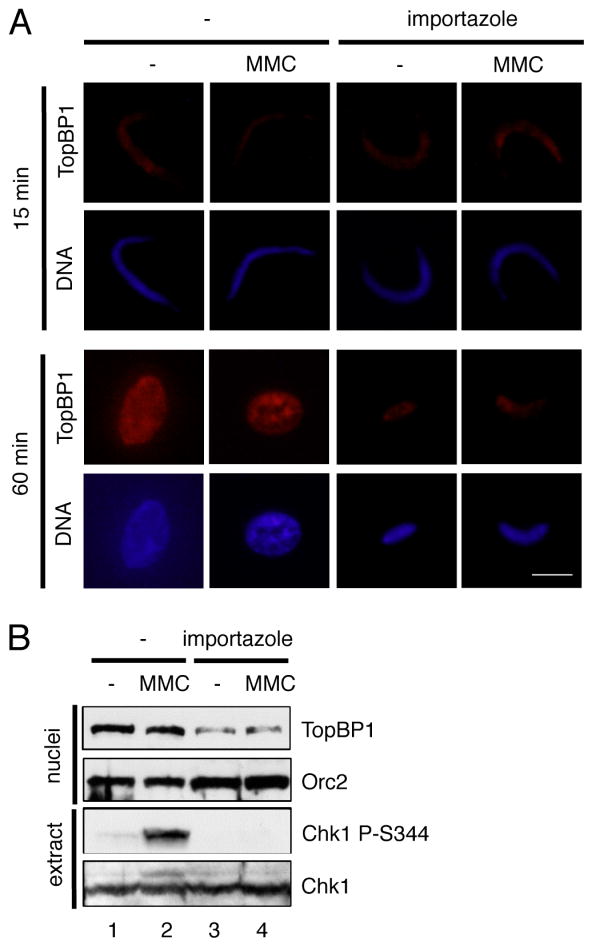
Our data demonstrate that TopBP1 interacts with Importin β and that the CTM motif of TopBP1 is important for the nuclear import of TopBP1. To further elucidate the importance of Importin β for TopBP1’s nuclear import, we reason that an Importin β-specific inhibitor may also affect the nuclear import of TopBP1. Importazole (N-(1-Phenylethyl)-2-(pyrrolidin-1-yl) quinazoline-4-amine) was recently shown to specifically block Importin β-mediated, but transportin and CRM1-independent, nuclear import of cargo proteins both in Xenopus egg extracts and cultured cells [51]. We added importazole to Xenopus egg extracts supplemented with sperm chromatin and MMC, and examined the nuclear import of TopBP1 via immunofluorescence microscopy and nuclei fraction for TopBP1 import and total extract for Chk1 phosphorylation via immunoblotting analysis (Fig. 5). Consistent with a previous study [49], TopBP1 started to be recruited to chromatin after 15 minutes of incubation (Fig. 5A, 15 min panel). At 15 minutes incubation, there was no noticeable difference with regard to the sperm chromatin morphology (“DNA”) and the nuclear import of TopBP1 (“TopBP1”) regardless of MMC treatment and importazole addition (Fig. 5A). At 60 minutes incubation with the absence of importazole, condensed sperm chromatin was assembled into a round nucleus and grew bigger in comparison to that at 15 min incubation (Fig. 5A). TopBP1 seems localized to the whole nucleus homogenously under normal condition and might re-localize to some specific “foci” in nucleus after MMC treatment at 60 minutes, although it was efficiently imported into the assembled nucleus under both conditions (Fig. 5A). The importazole addition compromised the round nucleus formation and size growth at 60 minutes, suggesting that importazole may prevent DNA replication as presumably importazole blocks all Importin β-mediated nuclear import of cargo proteins (Fig. 5A, importazole panel). The localization of TopBP1 to nucleus with the presence of importazole at 60 minutes was compromised significantly but not completely regardless of MMC treatment (Fig. 5A). These observations suggest Importin β is important for TopBP1’s nuclear transport. However, we can’t distinguish the deficient nuclear import of either TopBP1 or other Importin β-mediated cargo proteins leads to the DNA replication deficiency because importazole presumably blocks all Importin β-mediated nuclear import. Nevertheless, Importin β-specific inhibitor importazole compromised the nuclear import of TopBP1.
Fig. 5.
Importin β-specific inhibitor importazole inhibits the nuclear import of TopBP1. (A) Immunofluorescence microscopy analysis of TopBP1 in nucleus in Xenopus egg extracts with the presence or absence of MMC and importazole at 15 minutes and 60 minutes incubation. The “TopBP1” panel indicates its localization in chromatin/nucleus while the “DNA” panel represents chromatin/nucleus. The scale bar represents 10 μm. (B) Sperm chromatin was added to egg extracts supplemented with MMC and importazole. After 60 minutes incubation, nuclei fraction and total egg extract were isolated and examined via immunoblotting analysis for the indicated proteins.
Consistent with the findings from immunofluorescence microscopy, our immunoblotting blotting analysis showed that there was no noticeable difference for TopBP1’s import to nuclei fraction at 60 minutes under normal and MMC conditions and that importazole addition decreased TopBP1’s import into the nuclei fraction (Fig. 5B, nuclei panel). The loading control Orc2 showed almost equal loading, suggesting at least Orc2 is imported into nucleus normally after importazole addition (Fig. 5B, nuclei panel). Notably, the MMC-triggered Chk1 phosphorylation was inhibited by importazole addition (Fig. 5B, extract panel). From this observation, we speculate the nuclear import of TopBP1 may be important for the MMC-induced Chk1 phosphorylation, but this approach does not rule out the possibility that the compromised MMC-induced Chk1 phosphorylation after importazole addition is indirectly resulted from the defective DNA replication or Importin β-mediated nuclear import of other DDR proteins. Because of the limited technical advantage of importazole, we will address this question directly through an alternative approach in the next section. Collectively, Importin β-specific inhibitor importazole inhibits TopBP1’s nuclear import, DNA replication, and the MMC-induced ATR-Chk1 checkpoint.
3.6. The CTM-dependent nuclear import of TopBP1 is required for ATR-Chk1 checkpoint signaling, not due to its requirement for DNA replication
Our data indicate that MMC-induced Chk1 phosphorylation is compromised when the CTM of TopBP1 is lacking or when importazole is added (Fig. 4C and 5B). In both cases, DNA replication was either deficient or inhibited to some extent. We then asked whether the requirement of the CTM-dependent nuclear import of TopBP1 for checkpoint signaling is simply due to its requirement for DNA replication or whether TopBP1’s CTM-dependent nuclear import is necessary for checkpoint signaling with ongoing DNA replication. To directly address this question, we added WT TopBP1 or ΔCTM TopBP1 to TopBP1-depleted egg extracts before the addition of sperm chromatin (Fig. 6A). We observed that WT TopBP1 was shuttled into nuclei and bound to chromatin (Fig. 6B and C). A small amount of ΔCTM TopBP1 was also found in the nuclei fraction and chromatin fraction although it was not as efficiently as the WT TopBP1 (Fig. 6B and C). These observations suggest that TopBP1 binds to chromatin in a CTM-independent manner before nuclear membrane formation and that the CTM motif of WT TopBP1 may mediate efficient nuclear import after nuclear envelope assembly. Notably, ΔCTM TopBP1 rescued DNA synthesis as efficient as WT TopBP1 in TopBP1-depleted egg extracts using this approach (Fig. 6A, D and E). This observation is reminiscent of the S-CDK-independent recruitment of TopBP1, which is also a small amount of TopBP1 recruitment to chromatin in the very beginning but is sufficient for its role in DNA replication initiation [49]. With ongoing DNA replication machinery (Fig. 6D and E), WT TopBP1, but not ΔCTM TopBP1, rescued MMC-induced Chk1 phosphorylation in TopBP1-depleted egg extracts (Fig. 6F). These data suggest that the CTM-dependent nuclear import of TopBP1 is required for the ATR-Chk1 checkpoint signaling and that this requirement is not due to its requirement for DNA replication.
Fig. 6.
The CTM-dependent shuttling TopBP1 into nuclei and onto chromatin is required for checkpoint activation, not due to its requirement in DNA replication. (A) Experimental design of the experiments in this figure. (B) Nuclei fraction and total extract were examined at 90 minutes via immunoblotting as indicated. (C) Chromatin fraction and total extract were examined at 90 minutes via immunoblotting as indicated. (D) DNA synthesis was examined at different time points (minutes) using the similar procedure as Fig. 4E. (E) Quantification analysis of the DNA synthesis of the samples from (D). (F) MMC was used to trigger DNA damage response in different types of egg extracts. Then chromatin fraction and total extract were analyzed via immunoblotting at 90 minutes for the indicated proteins.
3.7. Nuclear import of TopBP1 plays a distinct regulatory role in the DNA damage response
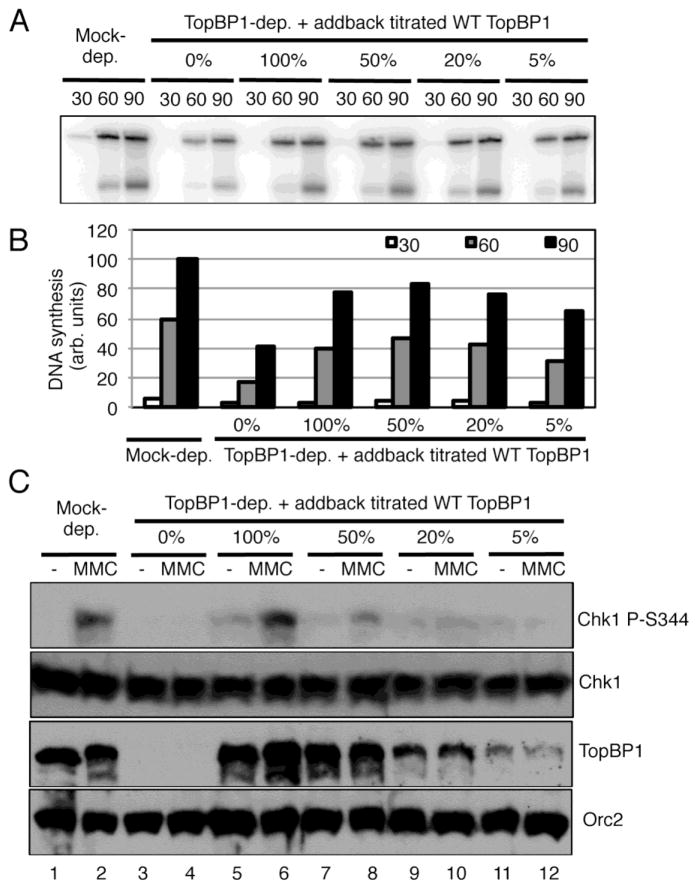
Our above observations also suggest that checkpoint signaling may require more TopBP1 on chromatin than DNA replication needs and that the efficient CTM-dependent nuclear import of TopBP1 plays an early regulation of the ATR-Chk1 checkpoint. To test this idea, we titrated recombinant WT TopBP1 to TopBP1-depleted egg extract and asked whether there is a concentration of TopBP1 that supports DNA replication but not checkpoint signaling. We observed that 20% TopBP1, in comparison of endogenous TopBP1 concentration (designated as 100%), is sufficient for rescuing DNA replication as efficient as 100% TopBP1 (Fig. 7A and B). However, 100% TopBP1, but neither 20% nor 5% TopBP1, rescued MMC-induced Chk1 phosphorylation in TopBP1-depleted egg extracts (Fig. 7C). In addition, 50% TopBP1 may partially, but not completely, rescued MMC-induced Chk1 phosphorylation in TopBP1-depleted egg extracts (Fig. 7C). These observations suggest that approximately 20% TopBP1 is sufficient for DNA replication but deficient in ATR-Chk1 checkpoint signaling. Therefore, efficient CTM-dependent nuclear import of TopBP1 via Importin β may provide a distinct regulation of the DNA damage response via supplying enough TopBP1 for necessary use upon stress.
Fig. 7.
Nuclear import of TopBP1 plays a distinct regulatory role in the ATR-Chk1 checkpoint signaling. (A) Different concentrations of WT TopBP1 (i.e., 0%, 100%, 50%, 20%, and 5% in comparison to endogenous TopBP1) were added to TopBP1-depleted egg extracts followed with the addition of sperm chromatin. [α-32P]-dATP was added and DNA synthesis were examined as Fig. 4E. (B) Quantification analysis of the DNA synthesis of the samples from (A). (C) Different concentrations of WT TopBP1 were added to TopBP1-depleted egg extracts supplemented with sperm chromatin as (A). MMC was used to damage chromatin, and checkpoint proteins were examined via immunoblotting as indicated.
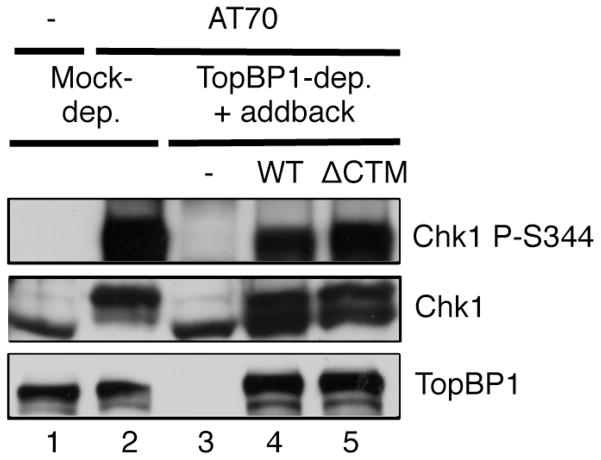
Moreover, if the sole role of TopBP1’s CTM motif lies in shuttling into the nucleus, we anticipate that the CTM motif is dispensable for a DNA damage response in a nucleus-free checkpoint activating system. Previous studies showed that checkpoint-activating structure AT70 triggers a robust ATR-Chk1 signaling independent of DNA replication machinery including polymerase alpha and MCMs [26,27]. We then asked if the CTM motif of TopBP1 is required for TopBP1’s role in AT70-induced Chk1 phosphorylation. We noted that both WT TopBP1 and ΔCTM TopBP1 rescued AT70-induced Chk1 phosphorylation in TopBP1-depleted egg extracts (Fig. 8). This observation suggests that CTM-dependent nuclear import of TopBP1 via Importin β may play a distinct role in the DDR in eukaryotic cells containing nucleus.
Fig. 8.
The CTM motif of TopBP1 is dispensable for checkpoint activation in a nucleus-free AT70 system. ATR checkpoint activation structure AT70 was added to Mock- or TopBP1-depleted egg extracts supplemented with WT TopBP1 (WT), ΔCTM TopBP1 (ΔCTM), or no addition (−). The total egg extracts were examined via immunoblotting as indicated.
4. Discussion
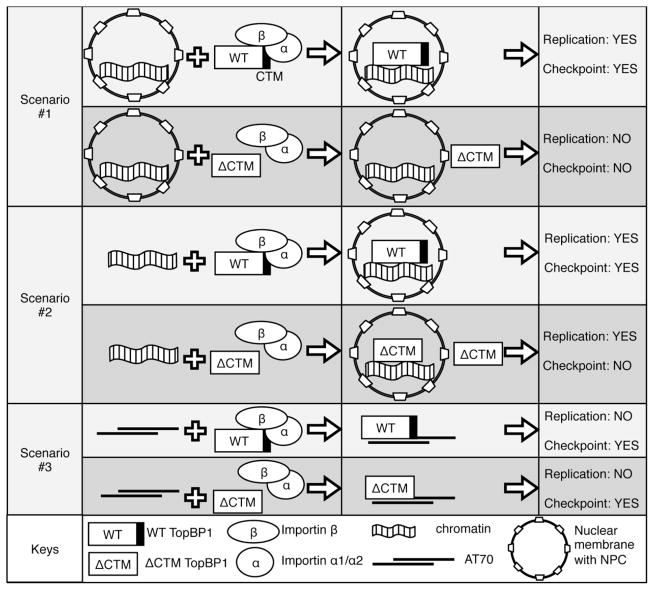
A variety of studies have revealed molecular mechanisms of TopBP1’s expression, stability, degradation, and recruitment onto chromatin. Together with MCPH1 (Microcephalin), E2F1 transcription factor regulates the expression of TopBP1 in DNA damage response [52,53]. In response to gamma irradiation, PML (Promyelocytic leukemia protein) associates and stabilizes TopBP1 [54]. E3 ubiquitin ligase hHYD and HectH9 interact with TopBP1 and mediate its proteasome-mediated degradation [55,56]. We and others have characterized three modes of TopBP1 recruitment to chromatin: Mode I is the earliest binding and independent on S-CDK kinase activity, which is sufficient for its role in DNA replication initiation; Mode II is S-CDK-dependent as it is after Cdc45 recruitment and CDK activation; and Mode III is CDK-independent but stress-dependent [49,50,57]. These different modes of TopBP1 recruitment to chromatin indicate that regulatory mechanisms may control and regulate these dynamic processes. Two recent studies suggested that the MRN complex (Mre11-Rad50-Nbs1) might play an essential role in TopBP1 recruitment to a defined ATR-activating structure or stalled DNA replication forks [58,59]. Here, our findings define Importin β-dependent importing TopBP1 into nucleus as an upstream regulation of TopBP1 function in ATR-Chk1 checkpoint signaling (Fig. 9): After nuclear membrane is formed and equipped with NPC, the CTM motif of TopBP1 is required for shuttling TopBP1 into nucleus, which in turn plays essential roles for DNA replication and checkpoint signaling (Fig. 4; Fig. 9, Scenario #1). When recombinant TopBP1 was added back to TopBP1-depleted extracts before nuclear envelope assembly, a small amount of TopBP1 may participate nuclear envelope assembly and is sufficient for its role in DNA replication, consistent with previous findings [26,60,61]. A portion of TopBP1 in the vicinity of demembranated chromatin may bind to chromatin and participate nuclear envelope assembly in a similar way as other NLS-containing proteins such as nucleoplasmin and histones [62]. However, compromised nuclear import of TopBP1 leads to deficiency in checkpoint activation (Fig. 6; Fig. 9, Scenario #2). The CTM motif of TopBP1, which contains a putative NLS motif, is dispensable for checkpoint activation in a nucleus-free AT70 system, suggesting eukaryotic cells may regulate ATR-Chk1 checkpoint signaling via shuttling checkpoint protein such as TopBP1 through their nuclear membrane (Fig. 8; Fig. 9, Scenario #3). Therefore, the TopBP1-Importin β interaction plays an early and important role in the DDR by mediating nuclear import of TopBP1 in eukaryotic cells.
Fig. 9.
Schematic summary of the nuclear import of TopBP1 for DNA replication (Replication) and DNA damage response (Checkpoint) in three different scenarios: (1) Scenario #1, recombinant WT or ΔCTM TopBP1 was added to egg extracts after nuclear membrane is formed; (2) Scenario #2, recombinant WT or ΔCTM TopBP1 was added to egg extracts before nuclear membrane is formed; and (3) Scenario #3, recombinant WT or ΔCTM TopBP1 was added to TopBP1-depleted extracts supplemented with ATR activation structure AT70, which did not form any nuclei. “YES” represents sufficiency, and “NO” represents deficiency.
We and other investigators previously demonstrated that the recruitment of TopBP1 to chromatin under stressful conditions (i.e., stalled DNA replication forks) is almost, if not completely, as efficient as that under normal conditions [43,57,61,63]. In current study, our data suggest that the CTM of TopBP1, which contains a putative NLS, is required for TopBP1’s nuclear import and subsequent chromatin recruitment. In addition, we demonstrated that the nuclear import of TopBP1 after MMC treatment was as efficient as that under normal conditions (Fig. 4C and 5B). In other words, the nuclear import of TopBP1 is dependent on its CTM motif and Importin β, but not DNA damage, which is a unique characteristic of TopBP1. Then a question may be what changes DNA damage can cause to TopBP1. It has been suggested that TopBP1 is phosphorylated possibly by ATM in response to double-strand breaks and replication stress and will re-localize to damage sites with BRCA1 [64,65]. Our data also suggest that TopBP1 may re-localize to some specific foci after MMC treatment although there is no noticeable increase of the overall nuclear import of TopBP1 by MMC treatment (Fig. 5A and B).
Consistent with our observations in Xenopus, a human TopBP1 mutant with a ~150 amino acid BRCT8 domain depletion was localized in cytoplasm, as was shown via immunofluorescence microscopy [66]. In our study of the nuclear import of Xenopus TopBP1, we showed several advancements with more molecular details: We identified Importin β as a new TopBP1 interacting protein de novo via mass spectrometry. We narrowed down the requirement of TopBP1 for nuclear import to the CTM motif (i.e., 23-amino-acid fragment at its C-terminus). Taking advantage of Xenopus egg extracts system, we demonstrated the biological significance of TopBP1-Importin β interaction for ATR-Chk1 checkpoint, which is not due to TopBP1’s role in DNA replication. Our compelling data suggest more TopBP1 is needed for ATR-Chk1 checkpoint than DNA replication requires. In addition, Importin β-specific inhibitor importazole prevents TopBP1’s nuclear import and compromises the MMC-induced Chk1 phosphorylation. Overall, we believe our investigation on the TopBP1-Importin β association for ATR-Chk1 checkpoint provides a better understanding of the nuclear import of TopBP1 in the DDR.
Our experiments also suggest that Importin α1 and Importin α2 are previously uncharacterized TopBP1-interacting proteins (Fig. 1B and C). In Xenopus, Importin α1/α2 and Importin β were identified and characterized as important mediators for nuclear protein import and docking of import substrate to distinct nucleoporins including Nup214, Nup153, and Nup98 [47]. Our data from Fig. 3D suggest that TopBP1 may associate with Importin β indirectly. Previous studies have demonstrated that the C-terminal 106 amino acids of Importin β are required for the binding to Importin α [67,68]. Therefore, we reason that TopBP1 associates with Importin β indirectly via Importin α1/α2. Because of the lack of suitable antibodies against Xenopus Importin α1/α2, we only followed up the TopBP1-Importin β interaction. Recently, we reported that WD40-repeat protein WDR18 associates with TopBP1 C-terminus using the similar GST-pulldown assay and that WDR18 collaborates with TopBP1 to promote DNA damage response [31]. Therefore, the finding of TopBP1-Importin β interaction and its role in ATR-Chk1 checkpoint signaling will add extra feature to the biological significance of TopBP1 C-terminus.
Accumulating data suggest a trend that DDR proteins are shuttled into nuclei via NLS-dependent interaction with Importin β and/or Importin α. A NLS motif in the C-terminal region of human Rad9 is also required for the nuclear transport of the Rad9-Rad1-Hus complex, thereby contributing to G2 checkpoint control [69]. Interestingly, a C-terminal NLS motif of Drosophila Rad9 regulates its localization to nuclear membrane, which in turn affects meiotic checkpoint [70]. Importin KPNA2 (Importin α) is identified as a Chk2-interacting protein via yeast two-hybrid screening assay and is required for protein nuclear localization and multiple functions of NBS1 [71,72]. The nuclear target and cell cycle regulatory function of BARD1, a BRAC1-interacting checkpoint protein, is also mediated through its NLS motif [73]. Nuclear import of 53BP1 is mediated by nucleoporin NUP153 via Importin β [74]. We demonstrate that the role of TopBP1’s nuclear import in ATR-Chk1 checkpoint is not dependent on its role in DNA replication (Fig. 6). Additionally, our observation of importazole’s inhibitory effect on TopBP1’s nuclear import may also be applied to studies of other DDR proteins if their nuclear import is Importin β-dependent.
A variety of DDR genes have been identified and characterized in breast cancer susceptibility, such as BRCA1, BRCA2, p53, and ATM [75]. As TopBP1 has sequence homology and functional similarities with BRCA1, Karppinne and co-workers proposed that TopBP1 is a plausible susceptibility gene for hereditary breast and ovarian cancer [76]. It was demonstrated that TopBP1 was expressed almost exclusively in the nuclei of normal breast epithelium. However, TopBP1 was aberrantly expressed in breast carcinomas and a significant percentage of TopBP1 was found in cytoplasm of breast cancer tissues [38,39]. Unfortunately, it is not fully understood how exactly the cytoplasmic mislocalization of TopBP1 is associated with breast cancer development. Our current study of the TopBP1-Importin β interaction in DNA damage response in Xenopus egg extracts provides a clue to a better understanding of the link between TopBP1 and cancer development. In other words, cytoplasmic localization of TopBP1 may lead to deficiency in ATR-Chk1 checkpoint but not DNA replication. Further studies are needed to further reveal whether the Importin β-dependent nuclear import of TopBP1 is a conserved mechanism in human cell lines and whether breast cancer patients have mutations in TopBP1’s CTM motif. If this is the case, the cytoplasmic localization of TopBP1 could be a biomarker for breast cancer and anti-cancer drugs may be developed via manipulating TopBP1’s nuclear import.
5. Conclusion
We identified and verified Importin β as a new interacting protein of TopBP1 C-terminus and mapped the nuclear localization requirement of TopBP1 to its extreme C-terminal 23 amino acids (CTM). The CTM-dependent nuclear import of TopBP1 is required for its role in DNA replication and ATR-Chk1 checkpoint signaling. In addition, Importin β-specific inhibitor importazole inhibits TopBP1’s nuclear import, DNA replication and ATR-Chk1 checkpoint signaling. Our data also suggest that checkpoint activation requires more TopBP1 protein than DNA replication does. All together, we conclude that the Importin β-mediated nuclear import of TopBP1 plays an important role in the ATR-Chk1 checkpoint signaling in Xenopus egg extracts.
Highlights.
Importin β has been identified as a new interacting protein of TopBP1 C-terminus
A C-terminal motif in TopBP1 is important for its nuclear import
Importazole inhibits TopBP1’s nuclear import and ATR-Chk1 checkpoint signaling
With ongoing DNA replication, nuclear import of TopBP1 is required for checkpoint
Checkpoint activation requires more TopBP1 than DNA replication does
Acknowledgments
We thank Dr. Tamara Slenn for critical reading of the manuscript and Drs. Norman Lefebvre, Chandra. D. Williams, and Yvette M. Huet and the University of North Carolina at Charlotte Vivarium staff for caring our frogs. We also thank Dr. Sunil Hwang and the CHS Mass Spectrometry and Proteomics Core Facility for the mass spectrometry analysis, and Dr. Douglass Forbes for invaluable reagents. The Yan lab is supported in part by funds provided by University of North Carolina at Charlotte and a grant from the NIH/NIGMS (R15 GM101571). The Michael lab is supported in part by a grant from the NIH/NIGMS (R01 GM067735).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jackson SP, Bartek J. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciccia A, Elledge SJ. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison JC, Haber JE. Annu Rev Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 4.Branzei D, Foiani M. Nat Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 5.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 6.Cuadrado M, Martinez-Pastor B, Murga M, Toledo LI, Gutierrez-Martinez P, Lopez E, Fernandez-Capetillo O. J Exp Med. 2006;203:297–303. doi: 10.1084/jem.20051923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shiotani B, Zou L. Mo Cell. 2009;33:547–558. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, O’Driscoll M, Jeggo PA. EMBO J. 2006;25:5775–5782. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cimprich KA, Cortez D. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 12.Guo Z, Kumagai A, Wang SX, Dunphy WG. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Sanchez Y. DNA Repair (Amst) 2004;3:1025–1032. doi: 10.1016/j.dnarep.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Toledo LI, Murga M, Fernandez-Capetillo O. Mol Oncol. 2011;5:368–373. doi: 10.1016/j.molonc.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith J, Tho LM, Xu N, Gillespie DA. Adv Cancer Res. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- 16.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.You Z, Shi LZ, Zhu Q, Wu P, Zhang YW, Basilio A, Tonnu N, Verma IM, Berns MW, Hunter T. Mol Cell. 2009;36:954–969. doi: 10.1016/j.molcel.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giannattasio M, Follonier C, Tourriere H, Puddu F, Lazzaro F, Pasero P, Lopes M, Plevani P, Muzi-Falconi M. Mol Cell. 2010;40:50–62. doi: 10.1016/j.molcel.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Willis J, Patel Y, Lentz BL, Yan S. Proc Natl Acad Sci USA. 2013;110:10592–10597. doi: 10.1073/pnas.1301445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cortez D, Guntuku S, Qin J, Elledge SJ. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 21.Zou L, Elledge SJ. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 22.Lieberman HB. J Cell Biochem. 2006;97:690–697. doi: 10.1002/jcb.20759. [DOI] [PubMed] [Google Scholar]
- 23.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumagai A, Lee J, Yoo HY, Dunphy WG. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 25.Mordes DA, Glick GG, Zhao R, Cortez D. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan S, Lindsay HD, Michael WM. J Cell Biol. 2006;173:181–186. doi: 10.1083/jcb.200601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumagai A, Dunphy WG, Claspin Mol Cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- 28.Sokka M, Parkkinen S, Pospiech H, Syvaoja JE. Subcell Biochem. 2010;50:119–141. doi: 10.1007/978-90-481-3471-7_7. [DOI] [PubMed] [Google Scholar]
- 29.Garcia V, Furuya K, Carr AM. DNA Repair (Amst) 2005;4:1227–1239. doi: 10.1016/j.dnarep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Yamane K, Kawabata M, Tusuruo T. Eur J Biochem. 1997;250:794–799. doi: 10.1111/j.1432-1033.1997.00794.x. [DOI] [PubMed] [Google Scholar]
- 31.Yan S, Willis J. Biochem Biophys Res Commun. 2013;431:466–471. doi: 10.1016/j.bbrc.2012.12.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong Z, Kim JE, Leung CC, Glover JN, Chen J. Mol Cell. 2010;37:438–446. doi: 10.1016/j.molcel.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu C, Paik JC, wang B, Lin FT, Lin WC. EMBO J. 2006;25:4795–4807. doi: 10.1038/sj.emboj.7601355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu K, Bellam N, Lin HY, Wang B, Stockard CR, Grizzle WE, Lin WC. Mol Cell Biol. 2009;29:2673–2693. doi: 10.1128/MCB.01140-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi JH, Lindsey-Boltz LA, Kemp M, Mason AC, Wold MS, Sancar A. Proc Natl Acad Sci USA. 2010;107:13660–13665. doi: 10.1073/pnas.1007856107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanahan D, Weinberg RA. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Bartkova J, Horejsí Z, Koed K, Krämer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland J, Lukas C, Orntoft T, Lukas J, Bartek J. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 38.Forma E, Krzeslak A, Bernaciak M, Romanowicz-Makowska H, Brys M. Mol Biol Rep. 2012;39:7795–7804. doi: 10.1007/s11033-012-1622-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Going JJ, Nixon C, Dornan ES, Boner W, Donaldson MM, Morgan IM. Histopathology. 2007;50:418–424. doi: 10.1111/j.1365-2559.2007.02622.x. [DOI] [PubMed] [Google Scholar]
- 40.Mosammaparast N, Pemberton LF. Trends Cell Biol. 2004;14:547–556. doi: 10.1016/j.tcb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 41.Harel A, Forbes DJ. Mol Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 42.Chook YM, Suel KE. Biochim Biophys Acta. 2011;1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan S, Michael WM. J Cell Biol. 2009;184:793–804. doi: 10.1083/jcb.200810185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van C, Yan S, Michael WM, Waga S, Cimprich KA. J Cell Biol. 2010;189:233–246. doi: 10.1083/jcb.200909105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willis J, Destephanis D, Patel Y, Gowda V, Yan S. J Vis Exp. 2012:e4449. doi: 10.3791/4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delmar VA, Chan RC, Forbes DJ. BMC Cell Biol. 2008;9:14. doi: 10.1186/1471-2121-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Radu A, Blobel G, Moore MS. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cingolani G, Petosa C, Weis K, Muller CW. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 49.Hashimoto Y, Takisawa H. EMBO J. 2003;22:2526–2535. doi: 10.1093/emboj/cdg238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Hatten RA, Tutter AV, Holway AH, Khederian AM, Walter JC, Michael WM. J Cell Biol. 2002;159:541–547. doi: 10.1083/jcb.200207090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soderholm JF, Bird SL, Kalab P, Sampathkumar Y, Hasegawa K, Uehara-Bingen M, Weis K, Heald R. ACS Chem Biol. 2011;6:700–708. doi: 10.1021/cb2000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoshida K, Inoue I. Oncogene. 2004;23:6250–6260. doi: 10.1038/sj.onc.1207829. [DOI] [PubMed] [Google Scholar]
- 53.Yang SZ, Lin FT, Lin WC. EMBO Rep. 2008;9:907–915. doi: 10.1038/embor.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu ZX, Timanova-Atanasova A, Zhao RX, Chang KS. Mol Cell Biol. 2003;23:4247–4256. doi: 10.1128/MCB.23.12.4247-4256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Honda Y, Tojo M, Matsuzaki K, Anan T, Matsumoto M, Ando M, Saya H, Nakao M. J Biol Chem. 2002;277:3599–3605. doi: 10.1074/jbc.M104347200. [DOI] [PubMed] [Google Scholar]
- 56.Herold S, Hock A, Herkert B, Berns K, Mullenders J, Beijersbergen R, Bernards R, Eilers M. EMBO J. 2008;27:2851–2861. doi: 10.1038/emboj.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yan S, Michael WM. Cell Cycle. 2009;8:2877–2884. doi: 10.4161/cc.8.18.9485. [DOI] [PubMed] [Google Scholar]
- 58.Duursma AM, Driscoll R, Elias JE, Cimprich KA. Mol Cell. 2013;50:116–122. doi: 10.1016/j.molcel.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee J, Dunphy WG. Mol Biol Cell. 2013;24:1343–1353. doi: 10.1091/mbc.E13-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumagai A, Shevchenko A, Dunphy WG. Cell. 2010;140:349–359. doi: 10.1016/j.cell.2009.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hashimoto Y, Tsujimura T, Sugino A, Takisawa H. Genes Cells. 2006;11:993–1007. doi: 10.1111/j.1365-2443.2006.00998.x. [DOI] [PubMed] [Google Scholar]
- 62.Lu Q, Lu Z, Liu Q, Guo L, Ren H, Fu J, Jiang Q, Clarke PR, Zhang C. Cell Res. 2012;22:1562–1575. doi: 10.1038/cr.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee J, Kumagai A, Dunphy WG. J Biol Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- 64.Yamane K, Wu X, Chen J. Mol Cell Biol. 2002;22:555–566. doi: 10.1128/MCB.22.2.555-566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoo HY, Kumagai A, Shevchenko A, Dunphy WG. J Biol Chem. 2007;282:17501–17506. doi: 10.1074/jbc.M701770200. [DOI] [PubMed] [Google Scholar]
- 66.Liu K, Lin FT, Ruppert JM, Lin WC. Mol Cell Biol. 2003;23:3287–3304. doi: 10.1128/MCB.23.9.3287-3304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kutay U, Izaurralde E, Bischoff FR, Mattaj IW, Gorlich D. EMBO J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rollenhagen C, Muhlhausser P, Kutay U, Pante N. Mol Biol Cell. 2003;14:2104–2115. doi: 10.1091/mbc.E02-06-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hirai I, Wang HG. J Biol Chem. 2002;277:25722–25727. doi: 10.1074/jbc.M203079200. [DOI] [PubMed] [Google Scholar]
- 70.Kadir R, Bakhrat A, Tokarsky R, Abdu U. PLoS One. 2012;7:e38010. doi: 10.1371/journal.pone.0038010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zannini L, Lecis D, Lisanti S, Benetti R, Buscemi G, Schneider C, Delia D. J Biol Chem. 2003;278:42346–42351. doi: 10.1074/jbc.M303304200. [DOI] [PubMed] [Google Scholar]
- 72.Tseng SF, Chang CY, Wu KJ, Teng SC. J Biol Chem. 2005;280:39594–39600. doi: 10.1074/jbc.M508425200. [DOI] [PubMed] [Google Scholar]
- 73.Schuchner S, Tembe V, Rodriguez JA, Henderson BR. J Biol Chem. 2005;280:8855–8861. doi: 10.1074/jbc.M413741200. [DOI] [PubMed] [Google Scholar]
- 74.Moudry P, Lukas C, Macurek L, Neumann B, Heriche JK, Pepperkok R, Ellenberg J, Hodny Z, Lukas J, Bartek J. Cell Death Differ. 2012;19:798–807. doi: 10.1038/cdd.2011.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oldenburg RA, Meijers-Heijboer H, Cornelisse CJ, Devilee P. Crit Rev Oncol Hematol. 2007;63:125–149. doi: 10.1016/j.critrevonc.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 76.Karppinen SM, Erkko H, Reini K, Pospiech H, Heikkinen K, Rapakko K, Syvaoja JE, Winqvist R. Eur J Cancer. 2006;42:2647–2652. doi: 10.1016/j.ejca.2006.05.030. [DOI] [PubMed] [Google Scholar]