Abstract
Fusion of the viral and host cell membranes is a necessary first step for infection by enveloped viruses and is mediated by the envelope glycoprotein. The transmembrane subunits from the structurally defined “class I” glycoproteins adopt an α-helical “trimer-of-hairpins” conformation during the fusion pathway. Here, we present our studies on the envelope glycoprotein transmembrane subunit, GP2, of the CAS virus (CASV). CASV was recently identified from annulated tree boas (Corallus annulatus) with inclusion body disease and is implicated in the disease etiology. We have generated and characterized two protein constructs consisting of the predicted CASV GP2 core domain. The crystal structure of the CASV GP2 post-fusion conformation indicates a trimeric α-helical bundle that is highly similar to those of Ebola virus and Marburg virus GP2 despite CASV genome homology to arenaviruses. Denaturation studies demonstrate that the stability of CASV GP2 is pH dependent with higher stability at lower pH; we propose that this behavior is due to a network of interactions among acidic residues that would destabilize the α-helical bundle under conditions where the side chains are deprotonated. The pH-dependent stability of the post-fusion structure has been observed in Ebola virus and Marburg virus GP2, as well as other viruses that enter via the endosome. Infection experiments with CASV and the related Golden Gate virus support a mechanism of entry that requires endosomal acidification. Our results suggest that, despite being primarily arenavirus like, the transmembrane subunit of CASV is extremely similar to the filoviruses.
Abbreviations: CASV, CAS virus; GGV, Golden Gate virus; LCMV, lymphocytic choriomeningitis virus; LASV, Lassa virus; EBOV, Ebola virus; SUDV, Sudan virus; MARV, Marburg virus; NHR, N-terminal heptad repeat; CHR, C-terminal heptad repeat; PBS, phosphate-buffered saline; GdnHCl, guanidine hydrochloride; IBD, inclusion body disease; MoMLV, Moloney murine leukemia virus; ASLV, avian sarcoma leukosis virus; NIH, National Institutes of Health
Keywords: viral membrane fusion, inclusion body disease, filovirus, arenavirus
Graphical abstract
Highlights
-
•
CASV is a novel arenavirus with a filovirus-like glycoprotein.
-
•
Arenaviruses and filoviruses are significant human pathogens.
-
•
The stability of the CASV GP2 post-fusion structure is dependent on pH.
-
•
CASV infection requires endosomal acidification.
-
•
The structure and function of CASV GP2 is similar to filovirus GP2.
Introduction
Infection by enveloped viruses, whose infectious particles are surrounded by a lipid bilayer, requires fusion of the host cell and viral membranes; this process is facilitated by one or more viral envelope glycoproteins [1]. Although details of this mechanism vary among viruses, viral glycoproteins typically consist of a surface subunit, which binds to a host cell receptor, and a transmembrane subunit responsible for drawing host and viral membranes together via the formation of a stable post-fusion conformation [2]. The “class I” viral fusion proteins, which include those of the human immunodeficiency viruses, influenza, Ebola viruses [exemplified by Ebola virus (EBOV) and Sudan virus (SUDV)] and Marburg virus (MARV), are defined by the formation of a core, trimeric α-helical bundle by the ectodomain of the transmembrane subunit during membrane fusion [1], [3]. The post-fusion conformations consist of a “trimer-of-hairpins” motif in which the ectodomain N- and C-terminal segments are folded back onto one another; this configuration draws the two membranes into proximity and the energy gained from refolding provides the driving force for membrane fusion. Here, we report the crystal structure of the transmembrane subunit, GP2, from the CAS virus (CASV). CASV was recently isolated from captive boid snakes with inclusion body disease (IBD) and is a putative causative agent [4]. Although CASV is primarily arenavirus like, with a similar genomic organization and a homologous nucleoprotein and RNA polymerase, its envelope glycoprotein is more similar to those of EBOV, SUDV and MARV.
EBOV, SUDV and MARV belong to the family Filoviridae (filoviruses) of enveloped, negative-strand RNA viruses [5], [6]. These are important human pathogens that cause severe hemorrhagic fever associated with high mortality rates [7]. Viral entry has been extensively characterized for EBOV and is mediated by its envelope glycoprotein, GP, which is composed of trimers of GP1–GP2 heterodimers [8]. Entry is initiated by GP1 binding to cell surface receptors that trigger viral uptake into the endocytic pathway [9], [10]. Within endosomes, GP is cleaved by host cysteine proteases, including cathepsin B and cathepsin L, which cleave GP1 to remove all but an ~17- kDa N-terminal fragment [11]. Interaction of this fragment with the viral receptor Niemann-Pick C1 cholesterol transporter [12], [13], [14] initiates the fusion reaction, whereby GP2 undergoes a large-scale conformational change resulting in the insertion of a hydrophobic fusion peptide into the host cell membrane. This conformation, where GP2 spans the host and viral membranes, is referred to as the “extended” or “pre-hairpin” intermediate. Next, the N-terminal heptad repeat (NHR) and the C-terminal heptad repeat (CHR) regions of GP2 fold into a thermostable six-helix bundle, or “trimer-of-hairpins”, promoting membrane fusion by bringing the two membranes into proximity and inducing lipid mixing of the outer leaflets [3], [15].
Members of the family Arenaviridae of enveloped viruses with ambisense single-strand RNA genomes (arenaviruses) are also known to cause significant human disease. There are currently 25 arenavirus species recognized by the International Committee for Taxonomy of Viruses, over 30% of which contain members that are known to infect humans [16], [17]. These species can be delineated into two groups based on their antigenic properties and geographic location: the Old World viruses, most notably the lymphocytic choriomeningitis virus (LCMV) and Lassa virus (LASV), and the New World viruses [16], [17]. Multiple Old and New World arenaviruses, including LASV, and a number of the new world viruses cause severe hemorrhagic fevers in humans associated with significant morbidity and mortality [17]. LCMV is endemic in Mus musculus (the common house mouse) worldwide; although LCMV generally causes a mild viral syndrome in immunocompetent persons, infection has been associated with meningitis, encephalitis and, sometimes, death in immunocompromised patients and with severe birth defects and mental retardation in congenital infection [18], [19], [20], [21], [22], [23].
Until recently, all arenaviruses have been tightly linked to rodent hosts as their primary reservoir [24]. Documented human infection has occurred through exposure to infected animals, either by direct contact of compromised skin with infectious materials or through mucosal exposure to aerosolized viral particles. Recently, however, Stenglein et al. identified CASV and two other novel arenavirus-like agents in boid snakes with IBD [4]. This condition is characterized symptomatically by neurological abnormalities, regurgitation and secondary infections and histologically by cellular eosinophilic inclusions [4], [25]. These novel viruses, CASV, the Golden Gate virus (GGV) and Collierville virus, represent the first arenavirus-like viruses to be identified in a reptilian species and suggest that the species tropism of the Arenaviridae family may be much broader than previously thought. Indeed, additional variants of these arenavirus-like viruses have subsequently been identified in boid snakes with IBD in the Netherlands [26] and in Finland [27], suggesting that such viruses may be present in snakes worldwide.
In the CASV GP2 post-fusion structure reported here, the overall backbone conformation of CASV GP2 is highly similar to those of EBOV and MARV GP2. Previous studies have shown that both EBOV and MARV GP2 post-fusion bundles exhibit a marked pH-dependent stability, with higher thermal and chemical stability observed at lower pH [28], [29], [30]. It is hypothesized that this feature of the post-fusion α-helical bundle is reflective of the endosomal mechanism of entry for these viruses; stabilization of conformations required for membrane fusion (e.g., the post-fusion six-helix bundle) is promoted under conditions of appropriate cellular compartments. Similar pH-dependent stability was observed for CASV GP2. Infectivity experiments using CASV and GGV in the presence of chemical inhibitors of endosomal acidification support the hypothesis that endosomal acidification is required for entry. This work is the first structural characterization of a membrane glycoprotein from this newly identified group of viruses.
Results
Protein design
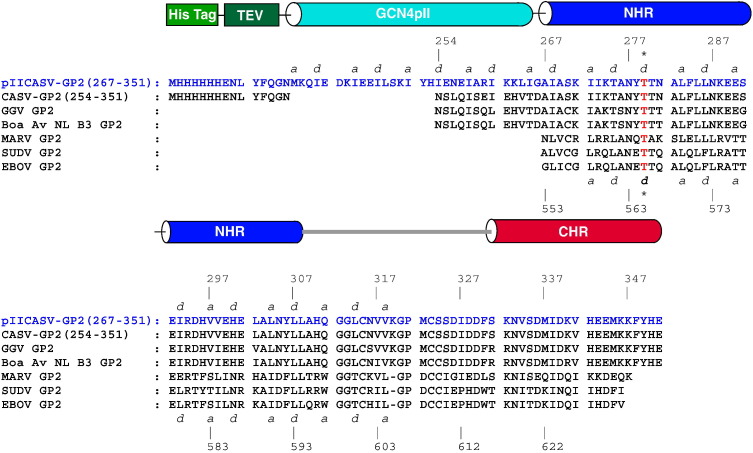
Alignment of GP2 sequences from CASV and related boid snake viruses with those of EBOV, SUDV and MARV suggests that the core domain consists of residues ~ 254–351 (CASV numbering; Fig. 1 ). Based on this sequence comparison, we prepared two protein constructs: one consisting of CASV GP2 residues 254–351 [“CASV-GP2(254–351)”] and another one consisting of residues 267–351 containing an N-terminal fusion to a trimeric variant of GCN4 coiled coil (“pII”) [“pIICASV-GP2(267–351)”]. The shortened CASV segment in pIICASV-GP2(267–351) was predicted based on the boundary of the α-helical NHR segment in EBOV and MARV GP2 [3], [15], [30]. The pII GCN4 coiled coil contains isoleucine side chains at the core heptad repeat positions (a and d of the canonical abcdefg repeat) that designate preferential homotrimer formation [31], [32]. Chimeras of pII and other trimeric coiled coils with fragments of class I fusion protein enhance solubility and have facilitated structural studies in the absence of the transmembrane segment in some cases [15], [30]. In both CASV GP2 constructs, the cysteines at positions 270 and 324 were mutated to serine. The corresponding residues in filovirus GP2 form disulfide bonds within the fusion loop (C270) and to the GP1 subunit (C324), both of which were omitted from the core domain construct. Cysteines 315 and 323 were left as native identity, as the corresponding residues form an intramolecular disulfide bond that stabilizes the loop regions of EBOV and MARV GP2 core domains [3], [15], [30].
Fig. 1.
CASV GP2 protein design. The sequences of the CASV GP2 constructs [pIICASV-GP2(267–351) and CASV-GP2(254–351)] are shown in alignment to the sequences of EBOV, SUDV, MARV, GGV and Boa Av NL B3 GP2. Segments corresponding to α-helical segments for the previously reported EBOV GP2 and MARV GP2 structures, as well as for the pIICASV-GP2(267–351) structure, are represented as colored cylinders, with the colors of the cylinders matching the structural elements depicted in Fig. 5a for the pIICASV-GP2(267–351) structure. The numbering for the EBOV GP2 residues is shown below the sequences; the numbering for the CASV GP2 residues is shown above the sequences. The a and d positions for the heptad repeat are indicated for the NHR region, with the heptad repeat stutter residue (T565 in EBOV; T279 in CASV) indicated with asterisks. The GCN4 pII segment is shown for pIICASV-GP2(267–351) (cyan-colored cylinder).
A heptad repeat stutter is predicted in the CASV GP2 NHR region at T279, by analogy to stutters found at corresponding positions of EBOV and MARV GP2 [15], [30]. Heptad repeat stutters result from a disruption in the heptad repeat pattern caused by insertion of an additional defg segment (abcdefg defgabc). The stutters in the EBOV and MARV GP2 NHR segment cause a distortion of the NHR α-helix at the stutter position and a slight local unwinding of the triple-stranded superhelix. For pIICASV-GP2(267–351), the pII segment was engineered to maintain registry of the heptad repeat pattern with the N-terminal segment of the NHR α-helix, thereby accounting for effects of the stutter on hydrophobic periodicity (Fig. 1). Notably, heptad stutters are found in a number of viral glycoproteins from both class I (α-helical) and class III (mixed α/β structure) categories and have been hypothesized to play a role in viral entry because of this conservation across phylogenetically distant species [33]. However, the precise mechanistic role of the heptad repeat stutter in viral fusion remains unknown.
Both CASV constructs were designed to contain an N-terminal hexahistidine tag, for purification purposes, separated from the rest of the sequence by the tobacco etch virus protease cleavage sequence (~ ENLYFQG ~). CASV-GP2(254–351) and pIICASV-GP2(267–351) were expressed in Escherichia coli BL21(DE3) cells. The proteins were purified by nickel-affinity chromatography and reverse-phase high-performance liquid chromatography; refolding was achieved by stepwise dialysis.
Solution characterization of CASV-GP2(254–351) and pIICASV-GP2(267–351)
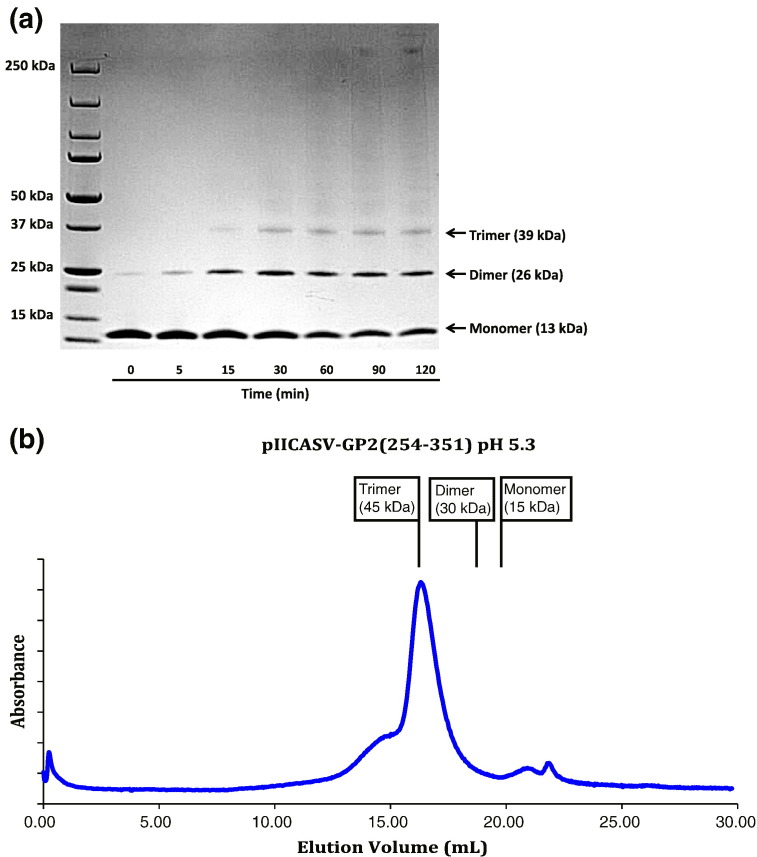
All class I viral fusion proteins that have been biochemically characterized exist as trimers in their fusion-active state [1], [2]. Glutaraldehyde cross-linking experiments with CASV-GP2(254–351) revealed bands with molecular masses corresponding to monomer (13 kDa), dimer (26 kDa) and trimer (39 kDa) (Fig. 2a). The band intensity shifted from the monomer to the trimer with increasing reaction time. Additionally, gel-filtration analysis by FPLC (fast protein liquid chromatography) with pIICASV-GP2(267–351) (with an expected molecular mass for a trimer of 45.4 kDa) revealed an observed molecular mass of 45.1 kDa (Fig. 2b). These data indicate that CASV-GP2(254–351) and pIICASV-GP2(254–351) exist as trimers in solution.
Fig. 2.
Glutaraldehyde cross-linking and FPLC analysis of CASV GP2 constructs. (a) Glutaraldehyde cross-linking gel with CASV-GP2(254–351). Bands were observed at 13 kDa, 26 kDa and 39 kDa, corresponding to monomer, dimer and trimer. The band intensity shifts from monomer to trimer with increasing reaction time. (b) FPLC size-exclusion chromatography trace for pIICASV-GP2(267–351). The observed molecular mass estimate is 45.1 kDa. Expected elution volumes for the monomer, dimer and trimer are indicated.
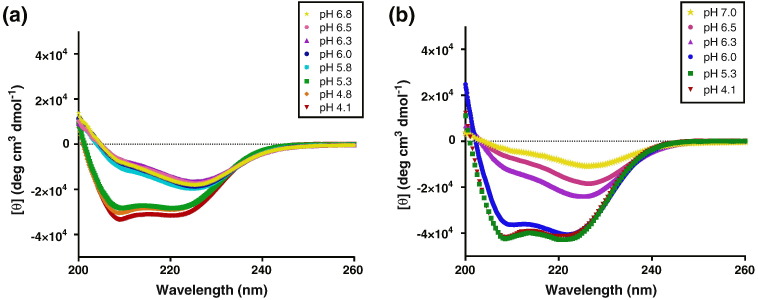
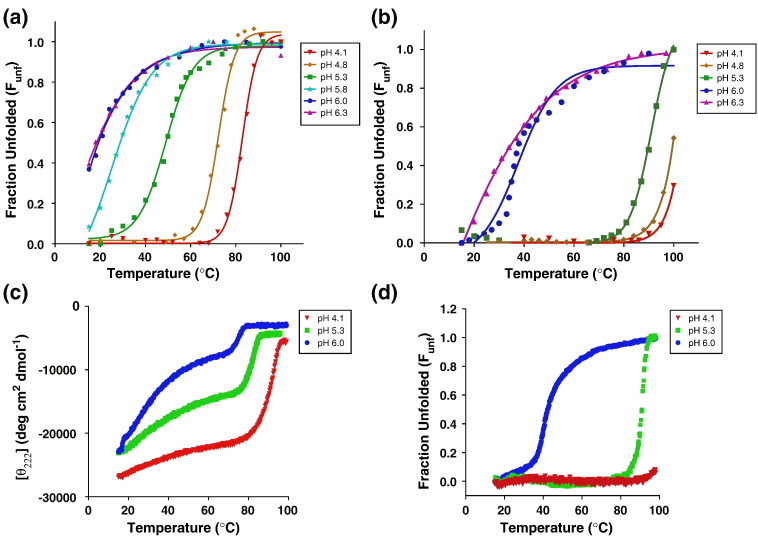
Circular dichroism (CD) indicates that both CASV-GP2(254–351) and pIICASV-GP2(267–351) are α-helical, with double minima observed at 208 and 222 nm in 10 mM sodium acetate (pH 4.1–6.5). However, the quaternary packing of the α-helices varies with pH for both CASV GP2 constructs, as indicated by the shifted signal intensity and the ratio of 208- to 222-nm peaks under different pH conditions (Fig. 3 ). Thermal denaturation revealed a significant pH dependence of thermal stability for both constructs. The thermal midpoint (T m) of CASV-GP2(254–351) was 82.9 ± 0.7 °C at pH 4.1 but was significantly lower (~ 30 °C) at pH 6.3 (Fig. 4 and Table 1 ). The overall thermal stability of pIICASV-GP2(267–351) was higher, as expected due to the contribution from the pII segment, but the general pH-dependent trend in thermal stability was still apparent. At pH 4.1, pIICASV-GP2(267–351) could not be fully unfolded even at ~ 100 °C, but at pH 6.3, the melting temperature was ~ 35 °C (Fig. 4 and Table 1). Above pH 6.5, both proteins were observed to precipitate out of solution. We have previously observed such pH-dependent behavior for both EBOV and MARV GP2 [28], [29], [30]. The pH-dependent stability of CASV GP2 follows the same pattern as observed with filovirus GP but is significantly more pronounced with CASV GP2 showing marked instability at pH values above pH ~ 5.3 and protein precipitation above pH 6.5. Addition of 500 mM salt (NaCl) resulted in an unusual pattern of thermal denaturation, with a biphasic melting profile (Fig. 4c); 500 mM NaCl appears to stabilize the bundle at pH 4.1, 5.3 and 6.0, but pH-dependent stability is still observed.
Fig. 3.
Full-wavelength CD scans. CD spectra of CASV-GP2(254–351) (a) and pIICASV-GP2(267–351) (b) in 10 mM sodium acetate (pH 4.1–7.0).
Fig. 4.
Thermal stability of CASV-GP2(254–351) and pIICASV-GP2(267–351) is pH dependent as monitored by molar ellipticity at 222 nm. Thermal denaturation curves of CASV-GP2(254–351) (a) and pIICASV-GP2(267–351) (b) in 10 mM sodium acetate at varying pH values. (c) Thermal denaturation curves of CASV-GP2(254–351) in 10 mM sodium acetate (pH 4.1, 5.3 and 6.0 in the presence of 500 mM NaCl). (d) Thermal denaturation curves of pIICASV-GP2(267–351; C323S) in 10 mM sodium acetate (pH 4.1, 5.3 and 6.0). Thermal midpoints (Tm values) are provided in Table 1.
Table 1.
Melting temperatures for CASV GP2 constructs
| Buffering condition | Tm (°C)a |
|---|---|
| CASV-GP2(254–351) | |
| 10 mM sodium acetate, pH 4.1 | 82.9 ± 0.7 |
| 10 mM sodium acetate, pH 4.8 | 72.3 ± 0.7 |
| 10 mM sodium acetate, pH 5.3 | 49.0 ± 0.1 |
| 10 mM sodium acetate, pH 5.8 | 25.1 ± 3.7 |
| 10 mM sodium acetate, pH 6.0 | ~ 35 (ambiguous fit) |
| 10 mM sodium acetate, pH 6.3 | ~ 30 (ambiguous fit) |
| pIICASV-GP2(267–351) | |
| 10 mM sodium acetate, pH 4.1 | ~ 99 (ambiguous fit) |
| 10 mM sodium acetate, pH 4.8 | ~ 97 (ambiguous fit) |
| 10 mM sodium acetate, pH 5.3 | 91.0 ± 1.3 |
| 10 mM sodium acetate, pH 6.0 | ~ 40 (ambiguous fit) |
| 10 mM sodium acetate, pH 6.3 | ~ 35 (ambiguous fit) |
| pIICASV-GP2(267–351; C323S) | |
| 10 mM sodium acetate, pH 4.1 | ~ 100 (ambiguous fit) |
| 10 mM sodium acetate, pH 5.3 | 90.8 ± 0.1 |
| 10 mM sodium acetate, pH 6.0 | 40.9 ± 0.1 |
Errors listed here represent 95% confidence intervals from the data fitting.
Dependence of CASV and GGV entry on endosomal pH
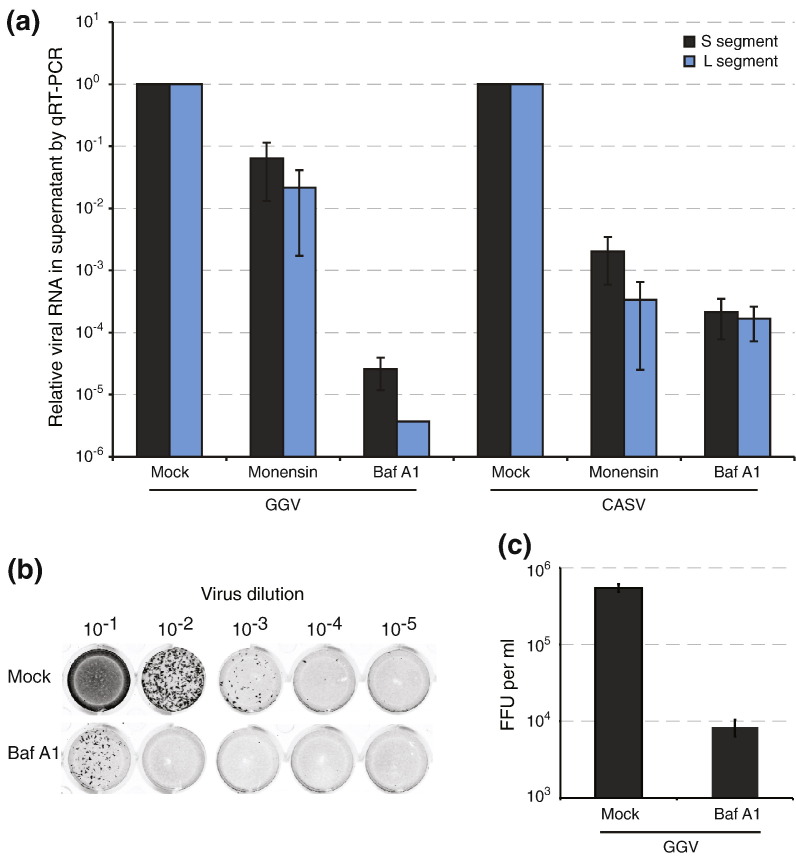
We hypothesized that CASV enters host cells through the endocytic pathway because of the observed pH-dependent stability of the CASV GP2 post-fusion conformation. We performed two types of experiments to test this hypothesis. First, we quantified virus particle production from infected cells by measuring supernatant viral RNA levels by quantitative real-time PCR (Fig. 5 ). We used CASV and the distantly related GGV that was identified along with CASV in annulated tree boas with IBD to perform these experiments. CASV and GGV share ~ 50% overall pairwise amino acid identity [4]. Pretreatment of JK cells (derived from boa constrictor) with monensin, a lysosomotropic ionophore, decreased GGV production by ~ 20- to 40-fold and CASV production by ~ 1000-fold. Pretreatment with bafilomycin A1 (Baf A1), a specific inhibitor of the vacuolar proton pump, had an even more dramatic effect, reducing production of both viruses by approximately 4 logs (Fig. 5a). We also measured the infectivity of individual GGV particles in a fluorescent-focus assay, which reflects the virus titer and the probability that an individual particle will initiate a productive infection under the experimental conditions (Fig. 5b). Note that we were unable to perform the fluorescent-focus assay using CASV due to a lack of suitable antibody reagents. Treatment of JK cells with Baf A1 reduced the number of GGV focus-forming units by ~ 65-fold (Fig. 5c). The results of these experiments suggest that an acidic endosomal environment is necessary for efficient infectivity by CASV and GGV.
Fig. 5.
CASV and GGV infectivity requires an acidic endosomal environment. (a) Chemical inhibitors of endosomal acidification decrease CASV and GGV production. Quantification of supernatant viral RNA levels following treatment with indicated drugs. The mean and standard deviation of duplicate experiments is shown. (b) Fluorescent-focus assay. Serial dilutions of GGV were used to inoculate monolayers of JK cells, which were covered with agarose overlays to prevent diffusion. After 7 days of incubation, foci of infection were stained with anti-GGV-NP antibody and counted. (c) Quantification of fluorescent-focus assays. The mean and standard deviation of duplicate experiments is shown.
Crystal structure of pIICASV-GP2(267–351)
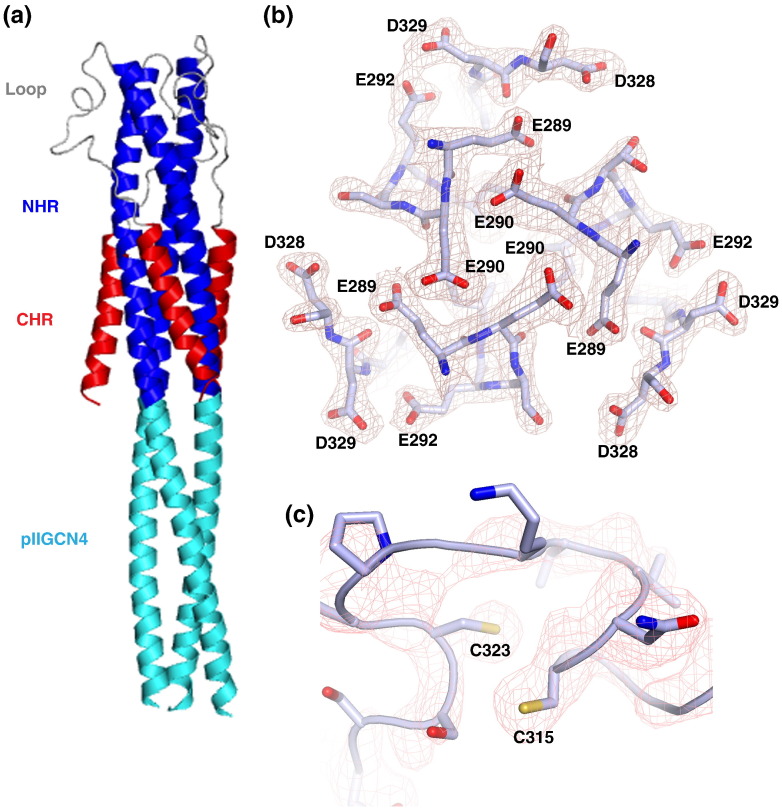
pIICASV-GP2(267–351) crystallized readily, with the best crystals diffracting to 2.0 Å. The MARV GP2 post-fusion structure (PDB ID 4G2K) was used as a molecular replacement model [30]. Initially, diffraction data were processed in rhombohedral (R3) space group, with one chain per asymmetric unit, but it soon became apparent that crystals demonstrate strong pseudo-symmetry. The structure was then solved and refined in monoclinic (C2) space group (Table 2 ), revealing a trimer of pIICASV-GP2(267–351) chains in the asymmetric unit. However, electron density in the loop areas was not interpretable, even though the rest of the structure was well defined. Only refinement in the lowest symmetry triclinic (P1) space group, with two trimers per asymmetric unit, produced traceable electron density within the loop areas (Table 2 and Fig. 6c). This conformational variability might be due partial disulfide bond formation between C315 and C323.
Table 2.
Data collection and refinement statistics for the pIICASV-GP2(254–351) crystal structure
| PDB | 4N23 | 4N21 |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.9791 | 0.9791 |
| Space group | C2 | P1 |
| Unit cell dimensions | ||
| a (Å) | 80.87 | 46.68 |
| b (Å) | 46.72 | 46.71 |
| c (Å) | 109.47 | 109.45 |
| α, β (°) | 90, 104.28 | 102.32, 89.99 |
| γ (°) | 90 | 119.98 |
| Resolution range (Å) | 20.0-2.0 | 20.0-2.0 |
| Observed reflections | 100,037 | 102,320 |
| Unique reflections | 27,014 | 51,572 |
| Completeness (%)a | 99.8 (99.9) | 97.7 (96.9) |
| Rmerge (I)b | 0.072 (0.491) | 0.059 (0.447) |
| I/σI | 11.1 (3.0) | 11.0 (2.9) |
| Structure refinement | ||
| Rcryst (%)c | 0.176 | 0.168 |
| Rfree (%)c | 0.228 | 0.212 |
| Protein nonhydrogen atoms | 3017 | 6057 |
| Water molecules | 131 | 232 |
| Average B-factor (Å2) | 52.4 | 53.9 |
| RMSD values from ideal value | ||
| Bonds (Å) | 0.009 | 0.009 |
| Angles (°) | 1.12 | 1.14 |
| Torsion angles (°) | 17.5 | 17.2 |
Values in parentheses indicate statistics for the high-resolution bin (2.05–1.99 Å).
Rmerge = ∑∑j|Ij(hkl) − 〈I(hkl)〉|/SSj|〈I(hkl)〉|, where Ij is the intensity measurement for reflection j and 〈I〉 is the mean intensity over j reflections.
Rcryst/(Rfree) = ∑||Fo(hkl)| − |Fc(hkl)||/S|Fo(hkl)|, where Fo and Fc are observed and calculated structure factors, respectively. No s-cutoff was applied. We excluded 5% of the reflections from refinement and used them to calculate Rfree.
Fig. 6.
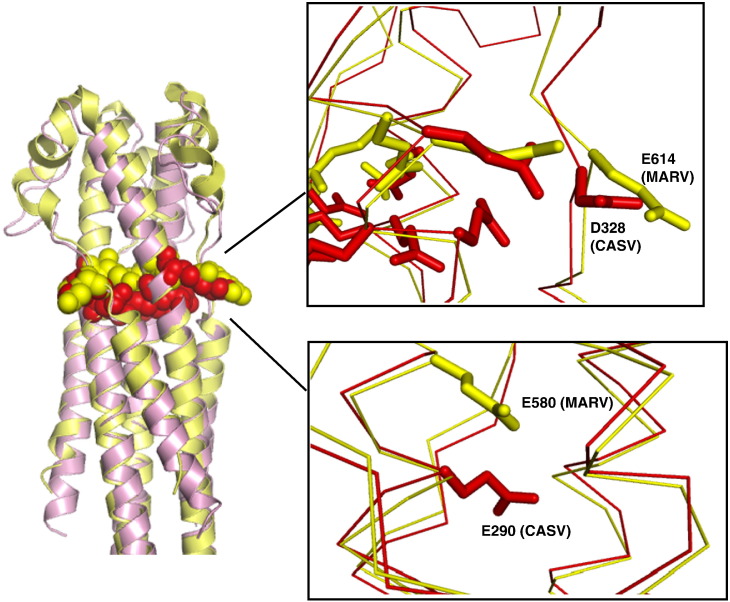
CASV GP2 post-fusion bundle structure. (a) Overall depiction of the CASV GP2 post-fusion bundle fold, with the secondary structural elements colorcoded according to the cylinders in Fig. 1. (b) Sample electron density, showing residues that comprise the “anion stripe”: E289, E290, E292 and D328. (c) Electron density in the loop region, focused around residues C315 and C323.
The overall architecture of pIICASV-GP2(267–351) is a trimeric coiled coil, spanning approximately 122 Å, ~ 55 Å of which corresponds to the pII coiled-coil segment (Fig. 6). T279, the predicted heptad repeat stutter position, points inward toward the core of the helical bundle. There are no breaks in the α-helical periodicity from the pII segment to the CASV GP2 bundle, indicating that a stutter is present in CASV GP2 as predicted. The NHR segments of CASV form the center of the bundle, while the CHR segments pack along the outside of the bundle, spanning approximately 50% of the total CASV GP2 trimer length. The NHR and CHR segments are separated by a loop region: the polypeptide chain undergoes a sharp turn at residue H310 to reverse direction at the membrane-distal side and lead into the helix–turn–helix motif that precedes the CHR α-helix. However, significant portions of this intervening segment had poorly resolved electron density.
In MARV GP2, the pH-dependent stability is primarily the result of specific anion–anion (AniAni) interactions that destabilize the post-fusion conformation. A “stripe” of acidic residues is present at the center of the post-fusion structure, consisting of both core (E580) and surface-exposed (E579 and E614) positions. Presumably, these residues are deprotonated at neutral pH, resulting in unfavorable interactions between anionic side chains and significant density of local negative charge that destabilizes the post-fusion structure. Under low pH conditions, however, these residue side chains would be protonated forming the conjugate acid, thereby alleviating such repulsive interactions. AniAni interactions were implicated in pH-dependent behavior of designed proteins containing shorter segments from the EBOV GP2 core domain, though these residues were found mostly at the surface of the six-helix bundle [28].
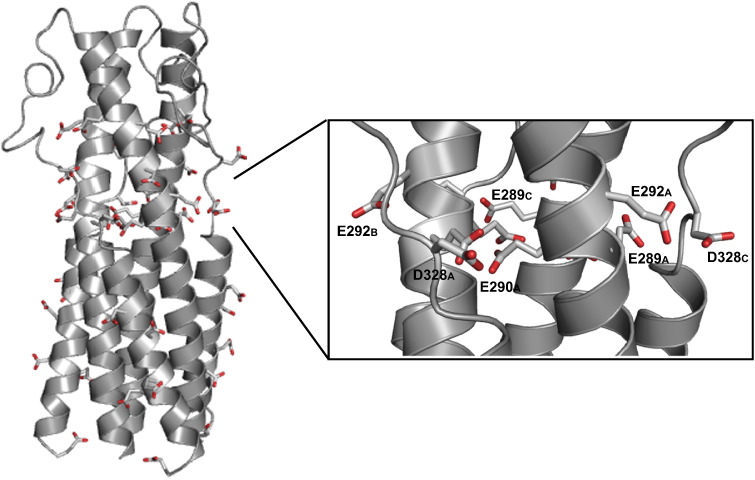
CASV GP2 has a high anionic character, containing seven glutamic acid residues and six aspartic acid residues per monomeric chain (21 glutamic acids and 18 aspartic acids per trimer) (Fig. 7 ). A large number of these anionic residues are concentrated in membrane-distal half of the α-helical bundle, separate from the NHR–CHR helical packing region. Several of the anionic residues in CASV form a stripe at the midsection of the CASV GP2 bundle, analogous to the anion stripe from MARV GP2 (Figs. 6b and 8 ). The anion stripe in CASV consists of residues E289, E290, E292 and D328. The electron density for these residues is well defined (Fig. 6b) and strongly supports the modeled side-chain orientation of these residues directly toward one another in close proximity. The high density of acidic residues results in pronounced negative electrostatic surface potential in this region of the protein (Fig. 9 ). The distances between these residues are as follows: E289 Oδ from Chain A to E290 Oδ from Chain C, 3.62 Å; E290 Oδ from Chain A to E289 Oδ from Chain B, 3.24 Å; E292 Oδ from Chain A to D328 Oγ from Chain C, 2.53 Å; D328 Oγ from Chain A to E292 Oδ from Chain B, 2.70 Å.
Fig. 7.
Anionic residues in CASV GP2 that may influence ph-dependent stability. Anionic residues (Asp and Glu) in the CASV GP2 bundle are shown as stick representations. The inset shows a close-up view of the anion stripe, consisting of residues E289, E290, E292, and D328.
Fig. 8.
Comparison of anion stripes between MARV and CASV. Alignment of CASV GP2 (red) with MARV GP2 (yellow) with the residues involved in the anion stripes in both proteins represented in spacefill. Inset shows blow-up of anionic stripes, shown in sticks. The residues in the stripe that are located on the surface of the bundles overlay quite well (top inset). However, the internally facing residues differ by one α-helical turn (lower inset).
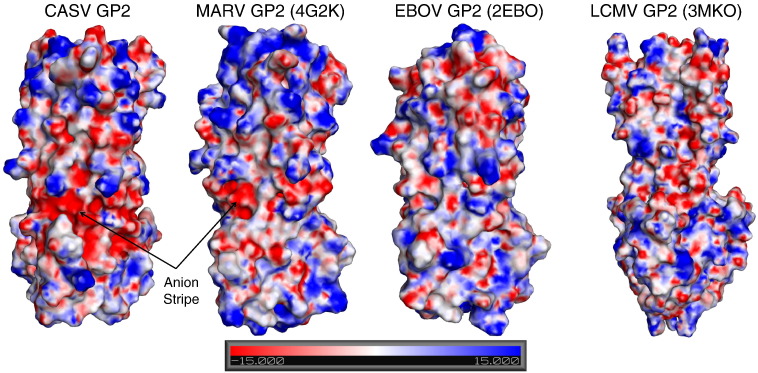
Fig. 9.
Calculated electrostatic surface potential for CASV GP2, MARV GP2 (PDB ID 4G2K), EBOV GP2 (PDB ID 2EBO) and LCMV (PDB ID 3MKO). The PBEQ server was used to create the surface representations with the generic presets and the solvent-accessible surface representation is shown with an electrostatic potential map (+ 15 kcal/mol of e in blue and − 15 kcal/mol of e in red).
It is likely that deprotonation causes unfavorable repulsions among the anionic residues and contributes to the dramatic pH-dependent stability of CASV GP2. Of note, the anion stripes in the CASV and MARV GP2 bundles are located in similar regions, at about the mid-section of the NHR core trimer (Fig. 8). Interestingly, the MARV GP2 anion stripe includes a core d residue, MARV E580, whereas the CASV GP2 anion stripe contains a core a residue, CASV E290. Therefore, the core residues of the CASV GP2 anion stripe are located ~ 1 α-helical turn closer to the N-terminus of the protein (Fig. 8). As a result, the E580 in MARV GP2 points almost directly into the core NHR trimer, whereas E290 in CASV GP2 is oriented toward the adjacent NHR α-helical axis. This contrast in side-chain orientation reflects the non-equivalent projection of the Cα–Cβ bond vector relative to the α-helical axis for a and d residues in trimeric coiled coils [32]. While this difference in a and d positions has a strong influence on coiled-coil topology for hydrophobic residues [31], [32], it is interesting to note that, for electrostatic effects, they have similar functional consequences. While the core acidic residues differ between CASV and MARV GP2, the surface residues of the two anion stripes are located at the same level within the coiled coils and are nearly superimposable (Fig. 8). These features highlight the conservation of this structural mechanism for pH-dependent stability between CASV and MARV GP2.
A cluster of residues exists surrounding the anion stripe in CASV, consisting of E299, H296 and H300 from one chain and E301 from the neighboring chain. The electron density for these residues is well defined. H300 is modeled in a single conformation, with the N1 atom directed toward E301conformation, with the with the N1 atom directed toward E301 Oδ from the neighboring chain. H296 is modeled with two side-chain orientations, one of which is directed toward E299 and E301. When the histidine residues are protonated, presumably below pH 6.0, they may act as counterions to balance the negative charge from the two glutamic acid residues. However, once the histidines become deprotonated at higher pH, this counterion effect would be removed, destabilizing the protein.
Anion binding pockets have been observed in a number of other viral GP2 proteins, but no anion binding pocket was observed in the CASV GP2 helical bundle. Chloride ions are bound to asparagine residues at the NHR trimer core in the EBOV GP2, MARV GP2, Moloney murine leukemia virus (MoMLV) GP2 and LCMV GP2 bundles at N586, N587, N71 and N325, respectively [3], [15], [30], [33], [34]. There are no asparagine residues pointed toward the internal core of CASV GP2, which is likely why no anion is bound. At CASV position 300, which corresponds to N586 in EBOV and N587 in MARV, there is a histidine residue. This H300 is conserved in GGV and Boa Av NL B2 GP2 as well (Fig. 1). The anion binding pocket to stabilize the other α-helical bundles [35] and the absence of the bound anion in CASV GP2 may contribute to this bundle's relative instability at higher pH.
Disulfide bonds within the loop region of CASV GP2
In EBOV and MARV GP2, the unstructured loop region is stabilized by an intrachain disulfide bond between C601 and C608 in EBOV and C602 and C609 in MARV [3], [15], [30]. By analogy, a stabilizing disulfide bond was predicted to form between CASV C315 and C323; however, no such disulfide bond was observed. Although the side chains of C315 and C323 appear to point toward one another in the crystal structure, the sulfur atoms are separated by 4.96 Å, twice the distance of a disulfide bond (Fig. 6c). Residue 324, a cysteine in the native GP2 sequence but a serine in our construct, is modeled near C315. It is therefore possible that a disulfide bond forms in the native protein between C324 and C315. However, this is conjecture only, as the electron density throughout the loop region is poorly defined, and the side-chain orientation cannot be definitively assigned. To explore this hypothesis, we generated an additional CASV GP2 construct, pIICASV-GP2(267–351; C323S), that contained the identity of wild-type cysteine side chain at positions 315 and 324 but a cysteine-to-serine mutation at position 323. We hypothesized that, if the native disulfide bond was between C324 and C315, then pIICASV-GP2(267–351; C323S) would display higher thermal stability than pIICASV-GP2(267–351). pIICASV-GP2(267–351; C323S) was expressed and purified, and thermal denaturation was performed as described for pIICASV-GP2(267–351). We observed that the T m values for pIICASV-GP2(267–351; C323S) were within error of the T m values for pIICASV-GP2(267–351) (Fig. 4d and Table 1). These data suggest either that a disulfide bond is not formed in the C323S construct or that a disulfide bond is formed but does not contribute to an increase in stability of this protein construct.
Salt bridges that may stabilize CASV GP2
A number of interchain and intrachain salt bridges appear to contribute to the stabilization of the CASV GP2 post-fusion structure (Table 3 ). These salt bridges are found in two clusters within the structure: five salt bridges are found stabilizing the NHR–CHR bundle portion of the protein, while two salt bridges are found in the membrane-proximal half of the bundle. Within the first cluster, salt bridges are observed between E344 Oδ and K347 Nε, E343 Oδ and K346 Nε and D336 Oγ and K340 Nε within a monomer (Table 3). Additional salt bridges are seen between neighboring monomers, consisting of residues E344 and K274, as well as between E351 and K271. The electron density for the lysine residues in these interactions is well defined to the Nε atom, and the majority of the residues involved in these salt bridges are modeled in single conformations. K274 in chain B was modeled in two conformations, one of which is not optimal for salt bridge formation because of the orientation of its Nε atom away from E344 Oδ. However, K274 in Chains A and C are modeled in a single conformation with the Nε atom directed toward E344 Oδ. D336 is also modeled in a double conformation, only one of which is consistent with salt bridge formation. The second cluster of salt bridges consists of D295 and R294 within the same chain and D328 and K288 of neighboring chains. The electron density for these residues is all well defined and modeled in a single conformation. Several possible salt bridges were identified in the EBOV and MARV GP2 structures, although salt bridge formation does not appear to be a major mechanism of stabilization for filovirus GP2 [3], [15], [30]. Interestingly, four stabilizing salt bridges per chain were observed in the LCMV GP2 structure, and mutational studies have shown that these salt bridges seem to play an important role in the stabilization of most arenavirus GP2s [33].
Table 3.
Salt bridges that may stabilize the CASV GP2 post-fusion structure
| Salt bridges observed in CASV GP2 post-fusion structure | |||
|---|---|---|---|
| Residue pairing | Interatomic distances (Å) |
||
| Chain A | Chain B | Chain C | |
| Glu344Oδ ↔ Lys274Nε | 2.7 | 4.2 | 3.5 |
| Glu344Oδ ↔ Lys347Nε | 3.3 | 3.6 | 2.7 |
| Glu351Oδ ↔ Lys271Nε | 3.0 | 2.7 | 3.5 |
| Glu343Oδ ↔ Lys346Nε | 3.7 | 4.6 | 3.3 |
| Asp336Oγ ↔ Lys340Nε | 5.1 | 3.2 | 4.4 |
| Asp328Oγ ↔ Lys288Nε | 4.1 | 4.3 | 3.9 |
| Asp295Oγ ↔ Arg294 guanidinium N | 4.1 | 4.2 | 3.8 |
Discussion
CASV is a member of a newly discovered class of arenavirus-like viruses that may be responsible for IBD in boas and pythons. Along with GGV, Collierville virus and similar viruses recently identified in the Netherlands, including Boa3 Av NL [26], and in Finland [27], these viruses represent the first arenavirus- or filovirus-like agents to be identified outside of a mammalian species, suggesting a previously unrecognized reservoir for potential human and/or mammalian pathogens. Our results are consistent with phylogenetic predictions that CASV GP2 resembles GP2 from EBOV and MARV, with a homologous overall backbone fold and an analogous pattern of pH-dependent thermostability. We confirmed the presence of a heptad repeat stutter in the coiled-coil region of the CASV GP2 sequence, at position T279. Such stutters have been observed in numerous class I and class III viral fusion proteins, notably in viruses that infect host cells through the endocytic pathway [33]. Alignment of CASV, GGV and Boa3 AV NL indicates that CASV T279 is conserved across these three viruses (Fig. 1).
We demonstrated that an acidic endosomal environment is necessary for efficient infectivity by CASV and GGV. These findings are consistent with our observations that CASV GP2 is structurally similar to filovirus GP2. Additional experiments will be required to more finely delineate the endocytic pathways and cellular factors involved in CASV entry. The JK tissue culture virus replication systems described here provide a platform for such investigations. The recent sequencing of the boa constrictor genome will facilitate the identification of host factors involved in entry [36].
We have identified an anion stripe, consisting of residues E289, E290, E292 and D328, at the mid-section of the CASV GP2 helical bundle. This stripe is analogous to the anion stripe observed in MARV GP2; interaction between residues in the stripe may be responsible for the pH-dependent stability of the CASV GP2 post-fusion bundle. These residues are surface exposed and result in significant negative electrostatic surface potential in this portion of the bundle with a solvation energy of − 7175 kcal/mol, indicating that the surface of this protein is highly charged (Fig. 9). Comparison of the solvation energy of CASV GP2 to EBOV GP2 (− 3178 kcal/mol), MARV GP2 (− 4163 kcal/mol) and LCMV GP2 (− 6162 kcal/mol) suggests that CASV GP2 contains the highest surface charge of these four proteins (Fig. 9). Several studies have demonstrated the importance of electrostatics in stabilizing viral glycoprotein post-fusion bundles, particularly for those that are stabilized by low pH [28], [29], [35], [37], [38], [39], [40]. The anion stripe found in CASV GP2 has significantly more anionic character than the analogous MARV GP2 anion stripe. This analysis provides a potential rationale for the pH-dependent stabilization of CASV GP2, which is stabilized at lower pH than either MARV GP2 or EBOV GP2 [28], [29]. We observed that 500 mM salt significantly stabilized the bundle from pH 4.1 to pH 6.0 (Fig. 4c), presumably by shielding the charges on the surface of CASV GP2, supporting the hypothesis that the anion stripe is at least partially responsible for the destabilization of the bundle at higher pH.
As an interesting comparison, the post-fusion structure of the transmembrane subunit from the avian sarcoma leukosis virus (ASLV) envelope glycoprotein (TM) was recently reported and shares high structural similarity to EBOV and MARV GP2 (backbone RMSD values of 1.6 and 1.5 Å, respectively) [41]. ASLV enters cells using an unusual two-step mechanism that has both pH-independent and pH-dependent features. Receptor binding and exposure of the fusion loop occur at neutral pH; however, viral reverse transcripts are not detected in cells treated with bafilomycin, suggesting that endosomal pH is required for infection [42]. Aydin et al. found that the ASLV TM bundle was equally stable at low and neutral pH, with only small differences in T m, over a range of pH 5.0–8.5 [41]. This behavior contrasts with the marked pH-dependent stability of the EBOV, MARV and CASV GP2 post-fusion α-helical bundles. Mutational studies on ASLV TM suggest that two histidine residues (H490 and H492) in the chain reversal region confer stability at low pH either by direct electrostatic interaction with a neighboring acidic residue (H492 and D496) or by overall stabilization of a putative helical dipole at the membrane-distal end of the core NHR trimer. No AniAni interactions were identified in ASLV TM, consistent with the finding that the post-fusion conformation is not sensitive to pH [41].
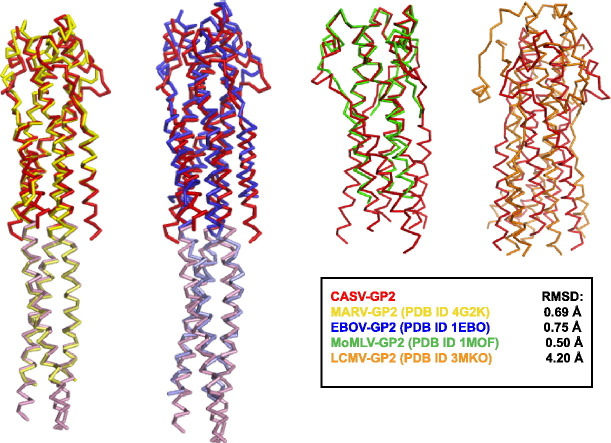
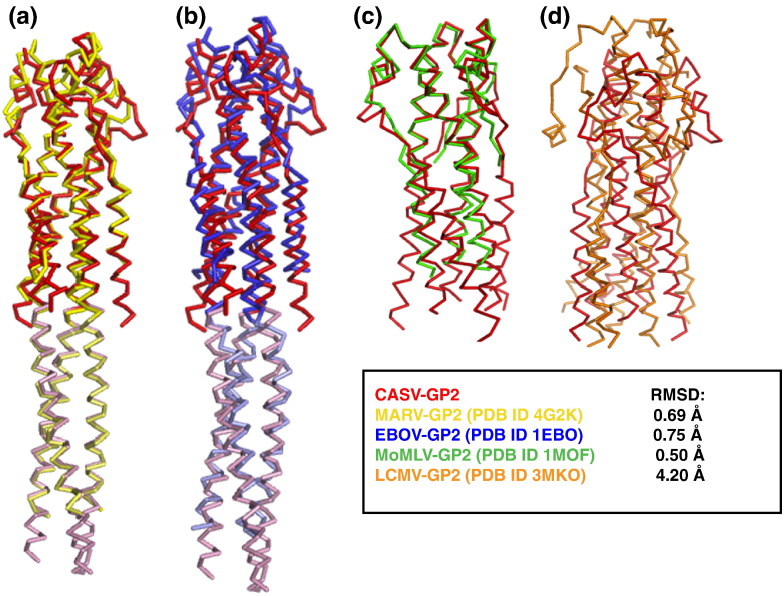
To compare the overall backbone folds of CASV GP2 to other viral glycoprotein fusion subunits, we examined the global backbone RMSD values of the ectodomain segments using the SuperPose server [43]. The pII (GCN4) segments for pII-GP fusions were omitted from the RMSD calculation. The overall backbone conformation of the CASV GP2 trimer is extremely similar to the backbone from both MARV and EBOV GP2 (RMSD values of 0.69 Å with MARV and 0.75 Å with EBOV) (PDB ID 2EBO) (Fig. 10 ). This striking similarity is somewhat surprising, given that the sequences share < 26% identity (Table 4 ). The backbone conformation of CASV GP2 also displays marked similarity to the fusion protein from MoMLV, a retrovirus that causes cancer in some murine hosts. The structure that is available for the MoMLV fusion protein (TM) is truncated before the CHR α-helical segments [34]; however, the trimeric NHR coiled-coil core of MoMLV aligns well with the CASV GP2 helical core (RMSD = 0.50 Å) with particular similarity observed in the membrane-proximal half of the bundle, including the loop region (Fig. 10). The CASV, MARV and EBOV GP2 structures all contained the pII (GCN4) trimerization motif. Although this segment was not included in the alignment, it should nonetheless be considered whether it is in part responsible for the similarity among these three GP2s. None of the pII fusion structures contain long-range interactions between GP2 segments and pII. Furthermore, two structures of EBOV GP2 exist, one containing the pII fusion and one without, and the EBOV GP2 segments are in strong agreement with one another and both the MARV and CASV segments [3], [15], [30]. Therefore, these structures likely represent the native GP2 post-fusion conformations.
Fig. 10.
CASV GP2 Post-Fusion Bundle Structural Alignment. Global alignments of CASV GP2 with MARV GP2 (a, PDB ID 4G2K), EBOV GP2 (b, PDB ID 1EBO), MoMLV TM (c, PDB ID 1MOF), and LCMV GP2 (d, PDB ID 3MKO). The pII GCN4 segments have been shown in lighter colors (a and b) or omitted from the CASV GP2 structure (c and d). RMSD calculations were performed excluding the pII GCN4 segments for CASV, EBOV, and MARV GP2.
Table 4.
Sequence alignment of viral glycoprotein transmembrane subunits with CASV GP2
| Sequence alignment of viral glycoprotein transmembrane subunits to CASV GP2 | ||
|---|---|---|
| Virus | % Identity | % Similarity |
| GGV | 80 | 92 |
| Boa Av NL B3 | 79 | 92 |
| EBOV | 26 | 45 |
| SUDV | 25 | 42 |
| MARV | 28 | 46 |
| LCMV | 13 | 24 |
| LASV | 14 | 27 |
Interestingly, however, global alignment of the CASV and LCMV GP2 post-fusion structures results in a high RMSD (4.20 Å; Fig. 10). The packing of the CHR segments relative to the NHR core trimers differs dramatically, and the loop region connecting the NHR and CHR segments is significantly different. Nonetheless, despite the low sequence homology (13%; Table 4) and poor global structural alignment, the architecture of CASV GP2 does share some similar features with LCMV GP2. Both structures contain a long core NHR trimer including a heptad repeat stutter at similar locations (Fig. S1 of the Supplementary Data), an overall the trimer-of-hairpins motif containing shorter CHR segments and the presence of many potential salt bridges. Additionally, the CASV GP2 and EBOV/MARV GP2 bundles display features that are remarkably similar to those in the fusion proteins of other unrelated viruses, such as the human T-lymphotropic virus type 1 GP21 and SARS (severe acute respiratory syndrome) coronavirus S2 [44], [45]. The conservation of the general architecture of the fusion proteins across these various viral families suggests that the structure plays a crucial role in viral entry.
It has been previously hypothesized that arenavirus and filovirus glycoproteins arose from a common RNA virus ancestor because of similarities in the overall structure and amino-terminal residues [46]. The evolutionary origin of the CASV family of viruses remains unknown, but the similarity of CASV GP2 with EBOV and MARV GP2 raises the interesting possibility that these viruses, or at least these glycoprotein genes, do indeed share a common ancestor. It is striking that the mechanism for pH dependence is similar for both CASV and MARV GP2, involving an anion stripe, but that the locations of participating residues are not identical. Thus, the specific interactions giving rise to pH-dependent behavior in these two cases may have resulted from independent co-evolution or may be the result of a single, common ancestor. Either way, the results presented here demonstrate that the GP2 structural architecture and pH-dependent stability of the post-fusion state are critical to function across multiple viruses.
Materials and Methods
Expression, purification and refolding of the CASV GP2 constructs
Synthetic DNA fragments encoding “CASV-GP2(254–351)” and “pIICASV-GP2(267–351)” (described in Results) were obtained from a commercial supplier (Genewiz, South Plainfield, NJ). The genes were cloned into pET22b vectors (Novagen, Madison, WI) using NdeI and XhoI restriction sites, producing the expression plasmids pJFK3 and pJFK4, respectively. pJFK3 and pJFK4 were transformed into E. coli BL21(DE3) cells (Invitrogen, Carlsbad, CA) at 42 °C. These cells were grown in 1 l cultures of 2 × YT or LB broth at 37 °C to an OD600 of ~ 0.6. Protein expression was then induced by the addition of 1 mM isopropyl-β-d-1-thiogalactopyranoside. The culture was incubated at 37 °C while shaking at 220 rpm for an additional 14–18 h. The cells were harvested by centrifugation and lysed in 6 M guanidine hydrochloride (GdnHCl) for 1–2 h at room temperature. The cell debris was pelleted by ultracentrifugation, and the supernatant was incubated with Ni-NTA resin (Qiagen, Valencia, CA) for 1–2 h at room temperature. The resin was washed with 10 column volumes of 6 M GdnHCl in phosphate-buffered saline (PBS) containing 50 mM imidazole. The proteins CASV-GP2(254–351) and pIICASV-GP2(254–351) were eluted with 6 M GdnHCl in PBS containing 250 mM imidazole. The second purification step was performed on the eluted protein by reverse-phase high-performance liquid chromatography on a Vydac (Hesperia, CA) C18 column (10 μM, 250 mm × 21.2 mm) with water/trifluoroacetic acid and acetonitrile mobile phases. The collected pure protein fractions were combined and lyophilized, then redissolved in 10 ml of 6 M GdnHCl in PBS. The proteins were refolded by stepwise dialysis into 100 mM glycine (pH 3.5) with 1 mM tris(2-carboxyethyl)phosphine (~ 4 h), followed by dialysis into 10 mM sodium acetate [pH 4.1 and pH 5.3 for CASV-GP2(254–351) and pIICASV-GP2(254–351), respectively] (~ 18 h). The protein yield from 10 l of cultures was ~ 0.3 mg and ~ 4.0 mg for CASV-GP2(254–351) and pIICASV-GP2(254–351), respectively.
CD spectroscopy
CD measurements were performed on a Jasco J-815 spectrometer with a 1-cm quartz cuvette for the thermal denaturation experiments and a 0.1-cm quartz cuvette for the chemical denaturation experiments. Protein concentrations for CD ranged from 1.2 to 3.6 μM, as determined by the absorbance at 280 nm. Full-wavelength scans were obtained with a 1-nm step size and a 2-s averaging time and represent the average of three scans. The signal was converted to mean molar ellipticity (θ) using the equation: θ (deg cm2 dmol− 1) = (millidegrees × mean residue weight)/(pathlength in millimeters × protein concentration in milligrams per milliliter), where the mean residue weight is the molecular weight divided by the number of backbone amides.
Thermal denaturation was monitored at 222 nm (θ222). Protein samples were added to the appropriate buffer (10 mM sodium acetate in PBS; pH 4.1, 4.8, 5.3, 5.8, 6.0 or 6.5) in a 1-cm-pathlength quartz cuvette and equilibrated for 5 min before beginning measurements at 15 °C. The sample was stirred constantly as the temperature was increased and data points were collected every 3–5 °C after equilibration for ~ 2 min at each temperature. θ222 was plotted as a function of temperature and converted to fraction unfolded (F UNF) using the following equation: F UNF = (θ222 − θUNF)/(θFOL − θUNF), where θUNF and θFOL are the CD signals for the unfolded and folded states, respectively.
Glutaraldehyde cross-linking
The cross-linking reaction was initiated by mixing purified CASV-GP2(254–315) and 25% glutaraldehyde (Sigma-Aldrich, St. Louis, MO) stock solutions to final concentrations of 0.15 mg/ml protein and 0.01% glutaraldehyde in 10 mM sodium acetate and 50 mM NaCl (pH 5.3). The reaction was quenched at 0, 5, 15, 30, 60, 90 and 120 min with reducing SDS sample buffer. The quenched samples were immediately boiled at 100 °C for ~ 5 min before being separated by SDS-PAGE.
Crystallization
CASV-GP2(254–351) and pIICASV-GP2(267–351) were concentrated to ~ 3.0 mg/ml in 10 mM sodium acetate (pH 4.1 and 5.3, respectively). The constructs were screened for crystallization using MCSG screening kits (Microlytic, Burlington, MA) in 96-well sitting-drop trays. Crystals for pIICASV-GP2(267–351) exhibiting the best diffraction were grown in MCSG4-D10 [0.2 M lithium sulfate, 0.1 M Mes (4-morpholineethanesulfonic acid)-NaOH (pH 6.0) and 35% methyl-2,4-pentanediol]. The crystals were mounted in cryo-loops directly from the crystallization droplet and flash-cooled in liquid nitrogen. Diffraction data were collected on a Rayonix 225 HE CCD detector (Rayonix LLC, Evanston, IL) with 0.979 Å wavelength radiation on the Lilly Research Laboratories Collaborative Access Team beamline (Advanced Photon Source, Illinois). Intensities were integrated using the HKL2000 program and reduced to amplitudes using the SCALEPACK2MTZ program [47], [48].
The structure of pIICASV-GP2(267–351) was determined by molecular replacement with PHASER [49]. The MARV GP2 post-fusion structure (PDB ID 4G2K) was used as a molecular replacement model [30]. Model building and refinement were performed with the programs Coot and REFMAC [47], [50]. Stereochemistry was checked with the program MolProbity [51]. LSQKAB and SSM algorithms were used for structural superpositions [47], [52]. Structural figures were prepared using PyMOL (Schrödinger, LLC)
Virus production assays
JK cells were cultured as described previously [4]. JK cells were plated in 12-well plates at a density of 100,000 per well. After 1 day of incubation, media in wells was replaced with media containing 100 nM bafilomycin A1 (Tocris Bioscience, Bristol, United Kingdom) or 10 μM monensin (Sigma-Aldrich). After 1 h of drug treatment, clarified virus stocks were added to wells at a multiplicity of infection of ~ 1/40. Six hours post-inoculation, media was replaced with drug-free media. Supernatant was replaced 2 days post-infection. Supernatant was collected 4 days post-infection, clarified and stored at − 80 °C until further analysis.
Fluorescent-focus assay
JK cells were seeded in 12-well plates at a density of 200,000 per well. After 1 day of incubation, media was replaced with media containing 100 nM bafilomycin A1 (Tocris Bioscience). After 1 h of drug treatment, wells were inoculated with 10-fold serial dilutions of GGV stocks. Dilutions were prepared in serum-free modified Eagle's medium with 25 mM Hepes (pH 7.4), from a culture supernatant stock that had been clarified by centrifugation at 10,000g for 1 min. After 3 h of incubation at 30 °C, culture medium was replaced with complete media supplemented with molten 0.75% low-melting point agarose (Life Technologies, Carlsbad, CA). After 7 days of incubation, overlays were removed and cells fixed in 4% formaldehyde in PBS. Wells were washed 3 times for 5 min in PBS, then in 0.1% Triton X-100 in PBS twice, then in PBST (PBS + 0.1% Tween-20). Cells were blocked by incubating in PBST + 1% bovine serum albumin for 30 min at 20 °C, then incubated in anti-GGV-NP polyclonal antibody [4] diluted 1:2000 into blocking buffer for 30 min. Cells were washed 4 times in PBST, then incubated in goat anti-rabbit secondary antibody (LI-COR Biosciences, Lincoln, NE) diluted 1:5000 in blocking buffer, then washed again as before. A final wash in PBS was performed before imaging wells on a LI-COR Odyssey scanner.
Quantitative real-time PCR
RNA was extracted from culture supernatant using the ZR Viral RNA kit (Zymo Research, Irvine, CA). Reverse transcription of RNA was performed in 10 μl reactions containing 1 × reaction buffer, 10 mM DTT, 1 mM each dNTP, 100 pmol random hexamer, 100 U of Superscript III RT (Life Technologies) and 5 μl RNA template. cDNA was diluted to 80 μM in 10 mM Tris (pH 7.5) and 0.1 mM ethylenediaminetetraacetic acid. Quantitative PCR reactions contained 5 μl diluted cDNA, 0.25 μM each primer, 10 mM Tris (pH 8.8), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM each dNTP, 5% glycerol, 0.08% NP-40, 0.05% Tween-20, 0.15 μl Taq DNA polymerase and 1 × Sybr Green (Life Technologies) in each 15 μl reaction. Primer sequences were as follows (5′–3′): GGV-S MDS-554 (CGG TGAATCCTAGTGAGGAG) and MDS-555 (CTACCTTGGACCCACTGGAA); GGV-L: MDS-532 (CGRCTCCACCGCCATT) and MDS-533 (GAGTGCTAGTGARGAAAGAGATCC); CASV-S MDS-556 (TTCGAAAGAAGCGCAGTGAT) and MDS-557 (GCCTCTTGGTGAAAATTCTGC); CASV-L MDS-548 (GGCACAAACAGCATTGGTATC) and MDS-549 (TTCATCAGTGAGAGAGACTCAAGTG). Thermocycling conditions were 95 °C for 30 min followed by 40 cycles of 95 °C for 10 min, 60 °C for 12 min and 72 °C for 12 min on a Lightcycler 480 instrument (Roche, Indianapolis, IN).
Accession codes
The X-ray coordinates and structure factors for pIICASV-GP2(267–351) have been deposited in Protein Data Bank IDs 4N21 and 4N23.
Acknowledgements
This work was funded by the Albert Einstein College of Medicine and the National Institutes of Health (NIH) (R01-AI090249 to J.R.L., R01-AI088027 to K.C. and U54-GM094662 and P30-CA013330 to S.C.A.). J.F.K. was supported in part by NIH Medical Scientist Training Grant T32-GM007288. J.L.D. is supported by the Howard Hughes Medical Institute. M.D.S. is supported by the Pacific Southwest Regional Center of Excellence (NIH Grant U54-AI06539). J.S.H. was supported by NIH postdoctoral training grant through the Lineberger Comprehensive Cancer Center at University of North Carolina (T32-CA009156).
Edited by T. J. Smith
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2013.12.009.
Appendix A. Supplementary data
Supplementary material.
References
- 1.Harrison S.C. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malashkevich V.N., Schneider B.J., McNally M.L., Milhollen M.A., Pang J.X., Kim P.S. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-Å resolution. Proc Natl Acad Sci USA. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stenglein M.D., Sanders C., Kistler A.L., Ruby J.G., Franco J.Y., Reavill D.R. Identification, characterization, and in vitro culture of highly divergent arenaviruses from boa constrictors and annulated tree boas: candidate etiological agents for snake inclusion body disease. MBio. 2012;3:e00180-12. doi: 10.1128/mBio.00180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhn J.H., Becker S., Ebihara H., Geisbert T.W., Johnson K.M., Kawaoka Y. Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations. Arch Virol. 2010;155:2083–2103. doi: 10.1007/s00705-010-0814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhn J.H., Bao Y., Bavari S., Becker S., Bradfute S., Brister J.R. Virus nomenclature below the species level: a standardized nomenclature for laboratory animal-adapted strains and variants of viruses assigned to the family Filoviridae. Arch Virol. 2013;158:1425–1432. doi: 10.1007/s00705-012-1594-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldmann H., Geisbert T.W. Ebola haemorrhagic fever. Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J.E., Saphire E.O. Ebolavirus glycoprotein structure and mechanism of entry. Future Virol. 2009;4:621–635. doi: 10.2217/fvl.09.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alvarez C.P., Lasala F., Carrillo J., Muniz O., Corbi A.L., Delgado R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J Virol. 2002;76:6841–6844. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kondratowicz A.S., Lennemann N.J., Sinn P.L., Davey R.A., Hunt C.L., Moller-Tank S. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc Natl Acad Sci USA. 2011;108:8426–8431. doi: 10.1073/pnas.1019030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carette J.E., Raaben M., Wong A.C., Herbert A.S., Obernosterer G., Mulherkar N. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller E.H., Obernosterer G., Raaben M., Herbert A.S., Deffieu M.S., Krishnan A. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J. 2012;31:1947–1960. doi: 10.1038/emboj.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cote M., Misasi J., Ren T., Bruchez A., Lee K., Filone C.M. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weissenhorn W., Carfi A., Lee K.H., Skehel J.J., Wiley D.C. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 16.Charrel R.N., de Lamballerie X., Emonet S. Phylogeny of the genus Arenavirus. Curr Opin Microbiol. 2008;11:362–368. doi: 10.1016/j.mib.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Emonet S.F., de la Torre J.C., Domingo E., Sevilla N. Arenavirus genetic diversity and its biological implications. Infect Genet Evol. 2009;9:417–429. doi: 10.1016/j.meegid.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Centers for Disease Control and Prevention Lymphocytic choriomeningitis virus infection in organ transplant recipients—Massachusetts, Rhode Island, 2005. MMWR Morb Mortal Wkly Rep. 2005;54:537–539. [PubMed] [Google Scholar]
- 19.Centers for Disease Control and Prevention Brief report: lymphocytic choriomeningitis virus transmitted through solid organ transplantation—Massachusetts, 2008. MMWR Morb Mortal Wkly Rep. 2008;57:799–801. [PubMed] [Google Scholar]
- 20.Amman B.R., Pavlin B.I., Albarino C.G., Comer J.A., Erickson B.R., Oliver J.B. Pet rodents and fatal lymphocytic choriomeningitis in transplant patients. Emerg Infect Dis. 2007;13:719–725. doi: 10.3201/eid1305.061269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonthius D.J. Lymphocytic choriomeningitis virus: an underrecognized cause of neurologic disease in the fetus, child, and adult. Semin Pediatr Neurol. 2012;19:89–95. doi: 10.1016/j.spen.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonthius D.J., Wright R., Tseng B., Barton L., Marco E., Karacay B. Congenital lymphocytic choriomeningitis virus infection: spectrum of disease. Ann Neurol. 2007;62:347–355. doi: 10.1002/ana.21161. [DOI] [PubMed] [Google Scholar]
- 23.Palacios G., Druce J., Du L., Tran T., Birch C., Briese T. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med. 2008;358:991–998. doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]
- 24.Charrel R.N., de Lamballerie X. Zoonotic aspects of arenavirus infections. Vet Microbiol. 2010;140:213–220. doi: 10.1016/j.vetmic.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 25.Li-Wen Chang E.R.J. Inclusion body disease, a worldwide infectious disease of boid snakes: a review. J Exotic Pet Med. 2010;19:216–225. [Google Scholar]
- 26.Bodewes R., Kik M., Stalin Raj V., Schapendonk C., Haagmans B., Smits S.L. Detection of novel divergent arenaviruses in boid snakes with inclusion body disease in the Netherlands. J Gen Virol. 2013;94:1206–1210. doi: 10.1099/vir.0.051995-0. [DOI] [PubMed] [Google Scholar]
- 27.Hetzel U., Sironen T., Laurinmaki P., Liljeroos L., Patjas A., Henttonen H. Isolation, identification and characterization of novel Arenaviruses, the etiological agent of boid inclusion body disease. J Virol. 2013;87:10918–10935. doi: 10.1128/JVI.01123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrison J.S., Higgins C.D., Chandran K., Lai J.R. Designed protein mimics of the Ebola virus glycoprotein GP2 alpha-helical bundle: stability and pH effects. Protein Sci. 2011;20:1587–1596. doi: 10.1002/pro.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrison J.S., Koellhoffer J.F., Chandran K., Lai J.R. Marburg virus glycoprotein GP2: pH-dependent stability of the ectodomain alpha-helical bundle. Biochemistry. 2012;51:2515–2525. doi: 10.1021/bi3000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koellhoffer J.F., Malashkevich V.N., Harrison J.S., Toro R., Bhosle R.C., Chandran K. Crystal structure of the Marburg virus GP2 core domain in its postfusion conformation. Biochemistry. 2012;51:7665–7675. doi: 10.1021/bi300976m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harbury P.B., Kim P.S., Alber T. Crystal structure of an isoleucine-zipper trimer. Nature. 1994;371:80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- 32.Harbury P.B., Zhang T., Kim P.S., Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 33.Igonet S., Vaney M.C., Vonhrein C., Bricogne G., Stura E.A., Hengartner H. X-ray structure of the arenavirus glycoprotein GP2 in its postfusion hairpin conformation. Proc Natl Acad Sci USA. 2011;108:19967–19972. doi: 10.1073/pnas.1108910108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fass D., Harrison S.C., Kim P.S. Retrovirus envelope domain at 1.7 angstrom resolution. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 35.Lamb D., Schuttelkopf A.W., van Aalten D.M., Brighty D.W. Charge-surrounded pockets and electrostatic interactions with small ions modulate the activity of retroviral fusion proteins. PLoS Pathog. 2011;7:e1001268. doi: 10.1371/journal.ppat.1001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bradnam K.R., Fass J.N., Alexandrov A., Baranay P., Bechner M., Birol I. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience. 2013;2:10. doi: 10.1186/2047-217X-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang A., Hackett B.A., Winter C.C., Buchholz U.J., Dutch R.E. Potential electrostatic interactions in multiple regions affect human metapneumovirus F-mediated membrane fusion. J Virol. 2012;86:9843–9853. doi: 10.1128/JVI.00639-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C.Y., Kielian M. E1 mutants identify a critical region in the trimer interface of the Semliki forest virus fusion protein. J Virol. 2009;83:11298–11306. doi: 10.1128/JVI.01147-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vanderlinden E., Goktas F., Cesur Z., Froeyen M., Reed M.L., Russell C.J. Novel inhibitors of influenza virus fusion: structure-activity relationship and interaction with the viral hemagglutinin. J Virol. 2010;84:4277–4288. doi: 10.1128/JVI.02325-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrison J.S., Higgins C., O'Meara M.J., Koellhoffer J.F., Kuhlman B.A., Lai J.R. Role of electrostatic repulsion in controlling pH-dependent conformational changes of viral fusion proteins. Structure. 2013;21:1085–1096. doi: 10.1016/j.str.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aydin H., Smrke B.M., Lee J.E. Structural characterization of a fusion glycoprotein from a retrovirus that undergoes a hybrid 2-step entry mechanism. FASEB J. 2013;27:5059–5071. doi: 10.1096/fj.13-232371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mothes W., Boerger A.L., Narayan S., Cunningham J.M., Young J.A. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell. 2000;103:679–689. doi: 10.1016/s0092-8674(00)00170-7. [DOI] [PubMed] [Google Scholar]
- 43.Maiti R., Van Domselaar G.H., Zhang H., Wishart D.S. SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res. 2004;32:W590–W594. doi: 10.1093/nar/gkh477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duquerroy S., Vigouroux A., Rottier P.J., Rey F.A., Bosch B.J. Central ions and lateral asparagine/glutamine zippers stabilize the post-fusion hairpin conformation of the SARS coronavirus spike glycoprotein. Virology. 2005;335:276–285. doi: 10.1016/j.virol.2005.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenberg A.R., Delamarre L., Pique C., Pham D., Dokhelar M.C. The ectodomain of the human T-cell leukemia virus type 1 TM glycoprotein is involved in postfusion events. J Virol. 1997;71:7180–7186. doi: 10.1128/jvi.71.10.7180-7186.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gallaher W.R., DiSimone C., Buchmeier M.J. The viral transmembrane superfamily: possible divergence of Arenavirus and Filovirus glycoproteins from a common RNA virus ancestor. BMC Microbiol. 2001;1:1. doi: 10.1186/1471-2180-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr Sect D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 48.Otwinowski Z.M., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 49.Storoni L.C., McCoy A.J., Read R.J. Likelihood-enhanced fast rotation functions. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krissinel E., Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.