Summary
Rifampicin resistance, a defining attribute of multidrug resistant tuberculosis, is conferred by mutations in the β subunit of RNA polymerase. Sequencing of rifampicin resistant (RIF-R) clinical isolates of Mycobacterium tuberculosis revealed, in addition to RIF-R mutations, enrichment of potential compensatory mutations around the double-psi β-barrel domain of the β’ subunit comprising the catalytic site and the exit tunnel for newly synthesized RNA. Sequential introduction of the resistance allele followed by the compensatory allele in isogenic M. smegmatis showed that these mutations respectively caused and compensated a starvation enhanced growth defect by altering RNA polymerase activity. While specific combinations of resistance and compensatory alleles converged in divergent lineages, other combinations recurred among related isolates suggesting transmission of compensated RIF-R strains. These findings suggest nutrient poor growth conditions impose larger selective pressure on RIF-R organisms that results in the selection of compensatory mutations in a domain involved in catalysis and starvation control of RNA polymerase transcription.
Keywords: Tuberculosis, Rifampicin, RNA polymerase, Compensatory mutation
Introduction
Tuberculosis (TB) continues to exact a vast toll of human lives and the specter of multidrug-resistant (MDR) and now extensively drug-resistant (XDR) TB threatens to undermine global control efforts (WHO, 2011). MDR TB is defined by the acquisition of resistance to isoniazid and rifampicin (RIF), XDR by additional resistance to any fluoroquinolone and an injectable agent other than streptomycin. Although surveillance is very poor throughout most of the world there are an estimated 630,000 cases of MDR TB currently (WHO, 2012). Mycobacterium tuberculosis (Mtb) acquires such resistance primarily by the accumulation of chromosomally encoded mutations in drug targets. The evolution of an XDR TB strain from a drug-sensitive progenitor therefore requires at least four different mutations be selected and fixed. The acquisition of drug resistance often comes with a cost to strain fitness and this is frequently invoked in estimating the magnitude of the threat (Borrell & Gagneux, 2009, Cohen & Murray, 2004). Because of limited access to accurate drug-susceptibility testing facilities, current guidelines for treating patients who fail first-line therapy with isoniazid, RIF, pyrazinamide and ethambutol, or for those who relapse after completing six months of this first line therapy, are to simply retreat such patients with these agents (WHO, 2009). Thus in most cases if there is an initial fitness defect accrued by the organism the selective pressure is maintained for a very long time, allowing time for compensatory mutations to arise that may alleviate this defect and fix the resistance allele in the population. Controlling the emergence and spread of MDR TB requires an understanding of the fitness and transmissibility of such strains and what evolutionary pressures lead to adaptation of strains and stable fixation of resistance alleles in the population.
RIF is the backbone of modern short-course chemotherapy for TB, its introduction effectively halved the duration of chemotherapy (Barry, 2011). RIF exerts its antibacterial effect by binding to the β subunit of RNA polymerase (RNAP) about 12Å from the active site of the enzyme where it affects an early block of elongation of short RNA transcripts, when they reach 2–3 nucleotides in length (Campbell et al., 2001). Acquisition of RIF resistance generally occurs through mutations in the rpoB gene encoding the β subunit and alters residues that form direct contacts with the drug. Many strains bearing these mutations have slightly reduced overall growth rates in vitro in rich growth media (often referred to as “fitness”) (Gagneux et al., 2006). RIF-resistant (RIF-R) strains isolated serially from treated patients show mixed degrees of fitness in this assay, some appearing equivalent in fitness to their drug sensitive progenitor while others show little effect, despite that the identical mutation in laboratory-selected resistant isolates shows a clear growth defect. These results suggested that the apparent fitness defect engendered by the initial mutation had been suppressed by a second site compensatory mutation.
While most of the fitness phenotypes have been determined in vitro, fitness in rich, defined laboratory growth media has not been shown to be correlated with fitness in the human host, and there are compelling reasons to expect this relationship to be complex. In Salmonella typhimurium strain LT2 resistant to either streptomycin or fusidic acid selection of compensatory alleles either in vitro (in rich growth medium) or in vivo (in mice) led to selection of different alleles and different frequencies of reversion to a wild type allele (favored in vivo) compared to acquisition of suppressor mutations (favored in vitro; (Bjorkman et al., 2000)). Indeed in some cases mutants with no apparent fitness defect in vitro in rich media showed profound fitness defects in mice, suggesting specific environmental conditions in the host could affect fitness costs.
The evolutionary dynamics of fitness-impairing resistance acquisition, compensation and reversion has been studied extensively in many bacterial species with an eye to reversing resistance by modulating the application of antibiotics (nicely reviewed in (Andersson & Hughes, 2010)). Unfortunately there seem to be few broadly applicable themes on the removal of drug selective pressure on resistant bacterial populations. One confounding factor in many organisms is acquisition of resistance by horizontal gene transfer that may include co-selection for other advantageous plasmid-encoded genes. This is not a factor in Mtb where horizontal gene transfer has not been observed, nonetheless there are a few examples of well understood resistance dynamics in Mtb and these may well be specific to each antibiotic mechanism. These questions are of fundamental importance to TB control strategies since fixation of resistance alleles may well occur only on a time-scale of decades (Luciani et al., 2009) and may not be easily observable in routine epidemiologic studies that span much shorter intervals.
In this study we examine clinical isolates from a highly drug-treated population of patients from South Korea with extensive drug-resistance collected over a five-year period to obtain a snapshot of currently circulating alleles with a focus on understanding the evolution of RIF resistance and the acquisition of compensatory mutations in the extant pathogen population, and present evidence that the magnitude of the fitness defect depends on nutrient limitation, a growth condition that is highly relevant to the host environment.
Results
Preferential accumulation of rpoC mutations in drug-resistant isolates from South Korea
To obtain deeper insight into the genetic changes in clinical isolates associated with RIF resistance we determined the genome sequences of 33 recent clinical isolates from subjects enrolled in a prospective longitudinal cohort study (ClinicalTrials.gov identifier: NCT00341601). Subjects in this study were enrolled over a five-year period from 2005–2010 from a tertiary care hospital in South Korea that specializes in treating drug-resistant TB. Amongst the sequenced strains were 15 drug-sensitive and 18 drug-resistant Mtb isolates. Amongst the eighteen drug-resistant isolates were 2 strains resistant to multiple agents that were not formally MDR, 8 MDR, 6 pre-XDR (resistant to isoniazid and RIF as well as either an aminoglycoside or a fluoroquinolone but not both) and 2 XDR (complete details of isolates, subject characteristics, treatment outcomes and drug-susceptibility are available as Tables S1 and S2).
Sequences with an average coverage of 61x across the genome, as determined using the Illumina Genome Analyzer platform, were subjected to a cleanup algorithm and gene alignments that contained potentially problematic sequences were excluded. To understand mutations associated with drug-resistance in this rapidly evolving setting we calculated the relative risk of a protein to be mutated in the drug-resistant compared to the drug-sensitive isolates (Fig. 1; Supplementary Methods). Among the proteins with a high relative risk for experiencing mutation in drug-resistant strains were many of the known targets of the antimicrobials used to treat Mtb including GyrA and GyrB (associated with fluoroquinolone resistance; (Zhang & Yew, 2009)), GidB (associated with streptomycin resistance; (Wong et al., 2011)), PncA (associated with pyrazinamide resistance; (Zhang & Yew, 2009)), KatG, InhA and IniB (associated with isoniazid resistance; (Laurenzo & Mousa, 2011)), EmbB and EmbC (associated with ethambutol resistance; (Zhang & Yew, 2009)) and RpoB (associated with rifampicin resistance; (Zhang & Yew, 2009)). In addition to these resistance-causing mutations there were also mutations in AhpC, an alkyl hydroperoxidase implicated in compensating for loss of catalase function in some isoniazid resistant isolates (Sherman et al., 1996). There was also an increased risk for mutation in RpoC, the β‘ subunit of RNAP, in the drug-resistant genomes. Given its close functional association with the β subunit of RNAP, this was a promising candidate for the accumulation of compensatory mutations for RIF resistance.
Figure 1.
Relative risk for mutation in proteins from drug-resistant as compared to drug-susceptible clinical isolates from South Korea. We calculated the relative risk of mutation from 15 drug-sensitive and 18 drug-resistant South Korean isolates in each protein in the drug-resistant strains. Known genes associated with drug-resistance alleles are red, while genes associated with potential compensatory suppressor alleles are shown in blue.
Association of rpoC mutations with the RIF-resistant rpoB alleles
To understand the relationship between RIF resistance and compensatory mutations in a more global context, we used 41 previously completed genome sequences for comparison, including laboratory strains (Cole et al., 1998, Fleischmann et al., 2002, Zheng et al., 2008), highly drug-resistant strains from South Africa ((Ioerger et al., 2010, , Broad, 2013) and recent clinical isolates from the United States (Filliol et al., 2006). (Complete details of previously published genome data are provided in Table S3). Together with the strains from South Korea this new set contained 41 drug-susceptible and 35 drug-resistant strains (including some duplicate isolates and sequential isolates from a single patient; Fig. 2). Single nucleotide polymorphism (SNP)-based alignment of the new genomic sequences with those previously determined showed that the major lineages of the Beijing clade previously described from South Africa were represented in this data (Fig. S1). Of 31 independent isolates that had RpoB mutations, 30 of these were located in the rifampicin resistance-determining region (RRDR); the single exception was the mutation V251F in an XDR strain. Eleven of the 31 also carried an additional mutation in RpoC (Fig. 2). In contrast, amongst 36 isolates lacking amino acid modifying mutations in RpoB only 5 showed changes in RpoC (two-sided Fisher’s exact test, p = 0.048, odds ratio = 3.3). Mutations in Ser531 of RpoB (according to convention we will use the numbering of the E. coli enzyme when referring to mutations within the RRDR and according to the M. tuberculosis numbering when referring to mutations outside of that region) seemed to be disproportionately represented in strains carrying a further mutation in RpoC; 8 out of 13 such strains had secondary RpoC mutations compared to only 3 among 18 RIF resistant (RIF-R) strains with other mutations in RpoB (p = 0.02, odds ratio = 7.4).
Figure 2.
RpoB and RpoC protein sequence variation amongst 41 drug-susceptible and 35 drug-resistant isolates based on combining new data from South Korean strains with available published data. Amino acid substitutions relative to the consensus are displayed according to the key in the figure. Strains in green represent sequential evolution of drug-resistance within a single South Korean subject, strains in gold represent sequential evolution of drug resistance within previously reported sequenced strains from three individuals in the Western Cape from South Africa. The strains in red represent the likely clonal expansion of an XDR lineage from KwaZulu Natal, South Africa. The two pairs with black brackets represent sequences from sequential isolates in the same individual. More details concerning the source of the isolates can be found in Tables S1 and S2, and source and description of the previously sequenced isolates is in Table S3. The K at the start of a sequence name indicates a new sequence from South Korea. Sequences were obtained from 2 different laboratories in this study, and a few isolates shown here were sequenced twice, once in each laboratory. Those sequences obtained at the NIH are labled with an asterisk, the others were sequenced by Novartis. The country of origin for the other strains are indicated at the beginning of the name using the International Standard 2 letter designations: United States, US; South Africa, ZA; and Canada, CA. The year of ioslation is indicated when reported, and the region of origin for South African set is also indicated (Western Cape, WeC; Durban, Dur; and Tugela Ferry TUF). The two letter code at the end of each isolate name indicates the reported drug profile as defined and detailed in Table S2. DS indicates drug sensitive; XR, XDR; MR, MDR; PR, pre-XDR; IR, INH resistant; and DR, drug resistant with a specfic profile that is not readily categorized. Fig. S1 is a companion to this figure, showing the same mutational patterns shown highlighted here, but presented in the context of a SNP-based phylogeny using all available sequence data to illustrate the genetic relatedness of the strains.
Within this group of strains were highly related isolates with identical patterns of secondary mutations in rpoB and rpoC genes. Sequential isolates from a single subject (K33) were obtained six months apart during which progression from pre-XDR to XDR disease was observed (green in Fig. 2 and Fig. S1), showing increasing RIF-R can build in vivo on an rpoB/rpoC framework. Among isolates from the Western Cape (Ioerger et al., 2010) were three highly related strains from three separate subjects with progressive levels of drug resistance, from MDR to pre-XDR (resistance to an injectable but not a fluoroquinolone), and finally to XDR (gold in Fig. 2 and Fig. S1). These isolates all carried the same distinctive RpoB S531L and RpoC V483G mutations, consistent with this particular RIF-R strain being transmitted and serving as a backbone for continuing acquisition of drug resistance. The distinctive triple RpoB mutant D516G, L533P and I1187T also likely represents the transmission an XDR strain, and clonal expansion of the 605 and R506 Beijing lineages in South Africa -- notably 8 additional cases of R506 were identified in the original study (Ioerger et al., 2009) (red in Fig. 2 and Fig. S1).
Putative compensatory mutations in rpoA, rpoB, and rpoC genes
The bacterial RNAP core enzyme is comprised of 5 subunits, α2ββ’ω, encoded by rpoA, rpoB, rpoC and rpoZ respectively, and suppressor alleles of the RIF-R mutations that impair the transcriptional activity of RNAP could, in theory, occur in any of those genes. Several rpoA alleles were noted in RIF-R Mtb isolates as possible candidates for compensatory mutation (Casali et al., 2012, Comas et al., 2012, de Vos et al., 2013). To investigate the nature of compensatory evolution in rifampicin resistance more thoroughly the entire sequences of rpoA, rpoB, rpoC and rpoZ were determined from an additional 170 RIF-R strains collected in the South Korean cohort study (Fig. 3A, Table S4).
Figure 3.
Compensatory mutation of the S531L allele is driven by both transmission and convergent evolution. A) Sequencing of 170 RIF-resistant strains from South Korea showing linkage between rpoB and rpoC alleles. Distinct amino acids substitutions are indicated by colored tickmarks as detailed in the inset. The mutations are summarized by strain in Table S4 and shown relative to the protein sequences in Fig. S2. (B) A Neighbor-joining tree of the strains based on 15-locus MIRU-VNTR patterns showing that specific combinations of alleles occurred in clusters, while others appeared to have evolved convergently. Strains carrying the rpoB S531L mutation are marked with symbols, while being colored for the alleles that occurred in multiple strains. Gray circles indicate strains with all other additional mutations in rpoB outside of the RRDR and rpoC.
In rpoC 42 unique, non-synonymous mutations (including one insertion and one deletion) in 34 loci were identified in 60 strains, while 20 unique, non-synonymous mutations in 20 loci outside of the RRDR of rpoB were observed in 27 strains. The G594E substitution in RpoC has been reported in rifampicin-sensitive strains (Comas et al., 2012) and was therefore excluded from further analysis. In addition, 7 unique non-synonymous mutations of rpoA were found in 7 strains. There was no mutational change found in rpoZ from these strains. Overall, in addition to the RIF-R mutations in RRDR of rpoB, at least one secondary mutation was found in 84 strains (49.4%) in either rpoC, rpoB, or rpoA.
The occurrence of those secondary mutations was strongly associated with specific RIF-R mutations, with RpoB S531L appearing highly correlated. Out of 92 strains harboring the RpoB S531L mutation 64 strains (67%) also had secondary mutations, compared to only 18 (23%) of the remaining 78 isolates (p < 0.0001). Mutations in RpoC showed the strongest association with the RpoB S531L allele, and were found in 50 (54%) out of the 92 strains harboring the RpoB S531L allele compared to only 8 (10%) among 78 isolates carrying other RIF-resistant mutations (p < 0.0001).
Convergent evolution and transmission of compensated strains
In this dataset there were several mutations that occurred in multiple strains (Fig. 3A, Table S4). The RpoC F452L mutation was observed in 13 strains, RpoC V483G in 4 strains, RpoB P45S in 4 strains, and RpoC N416S, RpoC L507V, RpoC E750G, RpoB T52P, and RpoB F503S each in two strains, respectively, suggesting convergent evolution or transmission of those strains. To explore the possibility of convergent compensatory changes or transmission events among these strains we looked at their genetic relationship using a standard method of mycobacterial interspersed repetitive unit-variable number of tandem repeat (MIRU-VNTR) typing that has been validated in this geographic context ((Shamputa et al., 2010); Fig. 3B; Table S5). All 13 of the isolates with RpoB S531L that had acquired the F452L mutation in rpoC were localized in one branch (blue circles in Fig. 3B). Inspection of the 24 MIRU-VNTR elements among the isolates revealed identical numbers of repeated elements at every locus in eleven strains. Only in a single locus were differences observed in the two remaining strains (Table S5). This level of clonality in MIRU-VNTR patterns appears to be most consistent with a linked transmission-chain that originated from a single index case. Another cluster of three cases, (RpoB S531L RpoB P45S, green circles) and multiple pairs of highly related strains (RpoB S531L RpoC L507V, cyan circles; RpoB S531L RpoC E750G, light green circles) were also found (shown arranged by DNA, protein and MIRU-VNTR phylogeny in Fig. S3).
In contrast, four strains bearing the RpoB S531L mutation independently appeared to acquire the distinct RpoC V483G mutation since they were greatly separated in the phylogenetic analysis implying possible convergent compensation of RpoB S531L (red stars in Fig. 3B, Table S5). Four other apparent unrelated pairs of strains with linked mutations were also found; RpoB S531L RpoC N416S (brown stars), RpoB S531L RpoB F503S (light pink stars), RpoB S531L RpoB T52P (pink stars) and RpoB S531L RpoC V1252L (yellow stars). Strains carrying RpoB S531L and RpoB P45S were unique in that one of those was separated from the other three that formed a cluster. These data suggest that both convergent suppression of the RIF-R RpoB S531L mutation and successful transmission of the compensated strains are both contributing to the high rate of mutation observed in RpoC.
Culture condition-dependent fitness costs of the mutations
To investigate the fitness costs of these genetic changes we used M. smegmatis as a surrogate host to assess the growth phenotype of changes in the Mtb rpoB and rpoC mutations. We constructed a recombinant strain of M. smegmatis that expressed the wild type alleles of the Mtb rpoB and rpoC under its native promoter at the attP integration site, after which the wild type M. smegmatis rpoB and rpoC genes were deleted by homologous recombination. Similarly we also constructed individual M. smegmatis strains bearing the rpoB C1349T (RpoB S531L) rpoC (wild type), the rpoB C1349T (RpoB S531L) rpoC T1354C (RpoC F452L) (a transmitted combination in the South Korean cohort) and the rpoB C1349T (RpoB S531L) rpoC T1448G (RpoC V483G) (a convergent combination in the South Korean cohort) alleles.
Upon plating of these transformants onto simple 7H10 growth media a growth defect of the strain carrying the RpoB S531L was immediately evident (Fig. 4A) as was a partial complementation of this growth defect by the F452L substitution and a full compensation of this growth defect by the V483G substitution in rpoC. When we attempted to quantify this difference by measuring the growth rate in liquid culture in rich broth media the effect became much less apparent. The growth rate in 7H9 media of the strain carrying the RpoB S531L mutation showed only a slight decrease of about 13% compared to wild type, which appeared to be restored by the putative compensatory mutations in RpoC, F452L and V483G (open bars in Fig. 4B). This discrepancy between the fitness impacts on organisms grown on solid media compared to liquid media led us to explore the effect of carbon limitation or starvation for both biotin and iron. The growth defect imposed by the RpoB S531L mutation became much larger under these more stringent growth conditions. Under the most stringent conditions, including a very low carbon source and starvation for biotin and iron (black and dark gray bars in Fig. 4B), the growth defect was almost two-fold and again only partially corrected by the F452L compensatory change in RpoC. The V483G compensatory change in RpoC, however, fully restored the growth defect even under these stringent growth conditions. The fitness defect of the mutations was also examined by pairwise competition assay. As with the difference in growth rate observed in single culture, the relative fitness of the RpoB S531L mutation varied depending upon the nutrient level. The relative fitness defect imparted by the RpoB S531L mutation was distinctively higher under stringent growth conditions, and this defect was corrected by both the compensatory changes in RpoC, F452L and V483G (Fig. 4C).
Figure 4.
Fitness defect of the rpoB S531L mutant and compensation by rpoC alleles. (A) The growth of M. smegmatis strains in which the wild type rpoB and rpoC have been replaced by the corresponding rpoB and rpoC genes from Mtb on 7H10 plates. The leftmost panel shows the growth phenotype of the wild type Mtb alleles, the second panel from the left shows the diminished growth by the rpoB S531L allele and the two right panels show compensation of this defect by the corresponding rpoC alterations at F452L and V483G. (B) The fitness impact is muted in 7H9 media supplemented with 0.2% glucose (open bars) and 7H9 with reduced iron and biotin levels (light gray bars) but this defect is amplified under conditions of carbon source limitation (0.002% glucose, black bars) and under conditions of restriction of carbon source, biotin and iron deprivation (dark gray bars). (C) Competitive fitness of the rpoB S531L mutant and two compensated strains by either rpoC F452L or rpoC V483G when co-cultured with the strain carrying wild type Mtb rpoB. Culture conditions indicated as bars are same as in (A).
Restoration of RNA polymerase activity by compensatory mutations
Although there have been several reports that described compensatory mutations in RIF resistance in Mtb (Casali et al., 2012, Comas et al., 2012, de Vos et al., 2013), none of these have examined the functional consequences of such mutations directly on RNAP activity. To show that the secondary mutations in RpoC compensate for compromised enzymatic activity of RNAP bearing the RIF-R RpoB S531L mutation we performed in vivo transcription assays on the same set of M. smegmatis strains heterologously expressing the Mtb rpoB and rpoC, either wild type or mutant. Expression of the inducible acetamidase gene, amiE, and a constitutive recA gene was measured by quantitative real-time PCR, and transcription efficiency was represented as the kinetics of production of amiE transcript relative to recA transcript (to adjust for differences in growth rates). In all strains production of amiE was increased until 2 hours post induction. While the transcriptional efficiency of the strains carrying the RpoB S531L mutation was half the wild type strain, that of the strains carrying the compensatory mutations, RpoC F452L and RpoC V483G were 65% and 83%, respectively, of the wild type (Fig 5).
Figure 5.
Restoration of RNA polymerase activity by rpoC F452L and rpoC V483G alleles. M. smegmatis strains carrying the respective Mtb rpoB and rpoC genes were induced for amiE gene expression, and the slope of the line plotting the proportional change in 2ȒΔΔC† per minute of the induced amiE transcript relative to a constitutive recA transcript was represented as transcription efficiency.
Discussion
This study of 170 isolates from South Korea allowed us to document both convergent selection of the same compensatory allele and transmission of compensated strains in the same medical setting. We identified 13 isolates with nearly identical MIRU-VNTR patterns that had acquired the RpoC F452L mutation in addition to the RpoB S531L mutation. This level of clonality in MIRU-VNTR patterns appears to be most consistent with nosocomial transmission from a single index case (none of the subjects lived in the same city and shared only the tertiary referral care center where they were in-patients as a common factor). In contrast, four strains bearing the RpoB S531L mutation appeared to acquire the distinct RpoC V483G mutation independently since they were widely separated in the phylogenetic analysis, a result most consistent with convergent compensation. The transmissibility of such compensated mutants was also recently described in isolates from the Western Cape (de Vos et al., 2013) and these authors earlier identified examples of convergently selected compensatory alleles (Comas et al., 2012). In our reanalysis of early data from the Western Cape (Ioerger et al., 2010) we identified three individuals carrying highly related isolates, based on SNP phylogenies, each of which carried the same RpoB S531L RpoC V483G mutations (Fig. 2, Fig. S1). These isolates had distinct drug resistance levels: one was MDR, one pre-XDR, and the other XDR. This is consistent with the hypothesis that transmission of fitness compensated RIF resistant strains can in some cases provide a foundation for the eventual acquisition of XDR mutations at the population level.
The pace of fixation of these compensatory changes implies a relatively strong selective pressure, at odds with the observed modest impact on bacterial growth rates in vitro that were maximally only 30% reduced. When we constructed isogenic pairs of M. smegmatis strains expressing only the Mtb rpoB and rpoC alleles we found the same subtle growth defects reported for the RIF-R RpoB S531L mutation in rich growth media. These results were unexpected given the large difference in colony size we observed for the same organisms leading us to explore the impact of differences in growth media on the magnitude of the growth defect. We found that under most stringent conditions, with limited availability of nutrients, a very strong growth defect was observed. Similarly, a decrease in the relative competitive fitness upon prolonged incubation into stationary phase was observed in rich media (data not shown). Growth condition-specific fitness costs have previously been reported for Salmonella typhimurium selected for streptomycin or fusidic acid resistance either in vitro or in mice (Bjorkman et al., 2000). These authors reported examples of mutants that appeared fully competent for growth in vitro that nonetheless were severely compromised in animals. In addition it has been previously reported that RIF resistant Mtb strains carrying rpoB mutations showed a stronger growth impairment in macrophages than in rich media in vitro (Mariam et al., 2004). It has been suggested that there is a significant correlation between fitness cost of the mutations and the frequencies; the smaller the fitness cost the higher the frequency and that the small fitness cost associated with certain alleles (including RpoB S531L) implied that there was little likelihood of spontaneous reversion to RIF susceptible (Billington et al., 1999). Our study suggests that the fitness cost of specific alleles would be better assessed in conditions relevant to the host environment. The strong growth defect under stringent conditions imposed by the RpoB S531L mutation was alleviated by the compensatory mutations in RpoC, F452L and V483G. Transcriptional efficiency mirrored the growth phenotypes of the M. smegmatis strains and was impaired by the RpoB S531L mutation and restored by the compensatory mutations in RpoC.
RNAP has a characteristic “crab claw” shape with the upper pincer formed from the β subunit and the lower from the β‘ subunit (Zhang et al., 1999). The RIF binding site lies entirely within the β subunit but this lies at the inner surface of the deep cleft of the claw that is near the site of RNA synthesis. The RIF binding pocket lies along the exit tunnel for newly synthesized RNA formed at the interface of β and β‘ subunits (Fig. 6A displays a model of the Mtb enzyme based upon the structure of the T. thermophilus enzyme, PDB: 2O5J) and Ser531 forms a critical hydrogen bond to the napthol core of RIF. Newly synthesized RNA is extruded through an exit tunnel (Fig. 6B) which is formed at an interface of β and β‘ subunits. Three α helices at the end of this exit tunnel in the β‘ subunit are the focus of most of the compensatory mutations coupled to changes in Ser531 of the β subunit (cyan in Fig. 6). These a helices contain both the compensatory mutations, RpoC F452L and V483G (the transmitted and convergent compensatory mutations highlighted in Fig. 3B). Substitution of Ser531 with leucine, a slightly larger residue, may impede the movement of RNA through the exit tunnel, reducing the overall efficiency of transcription (Fig. 6C and D); consistent with the hypothesis that the restoration of fitness may be a consequence of the slight expansion of the elongation tunnel on the opposite side due to the mutations in β’ subunit. Notably a strain bearing a combination of 3 RpoB mutations, D516G, L533C and I1187T, with no RpoC mutations (red in Fig. 2 and Fig. S1) was reported from an outbreak of XDR-TB in KwaZulu-Natal (Ioerger et al., 2009). Although the RpoB I1187T mutation is in the β subunit, it is structurally immediately proximal to the cluster of proposed compensatory mutations in the β‘ subunit (Fig. 6D), raising the possibility that transmission of fitness-compensated RIF resistant forms may have contributed to the expansion of the Tugela Ferry XDR outbreak.
Figure 6.
Compensatory mutations in RpoC might reduce impedance of RNA exit from RIF-resistant mutant RNA polymerase. (A) A model of the RNAP from Mtb (based on the structure from T. thermophiles PDB: 2O5J) is shown with the DNA in blue and RNA in orange. The β subunit is shown in space filling representation, while the β’ subunit is shown in magenta ribbon for contrast (except the three helix loop in cyan which is the focus of many of the potential compensatory mutations). RIF resistance inducing mutations are shown in red and noted, and RIF is shown in green. (B) The exit tunnel for RNA is highlighted, while in (C) the critical contacts of this three-α-helix loop at the exit of the tunnel is shown compared to Ser531 of β subunit. (D) The position of the F452L compensatory mutation in the β’ subunit is shown, as well as the proximity of the I1187 residue in the β subunit.
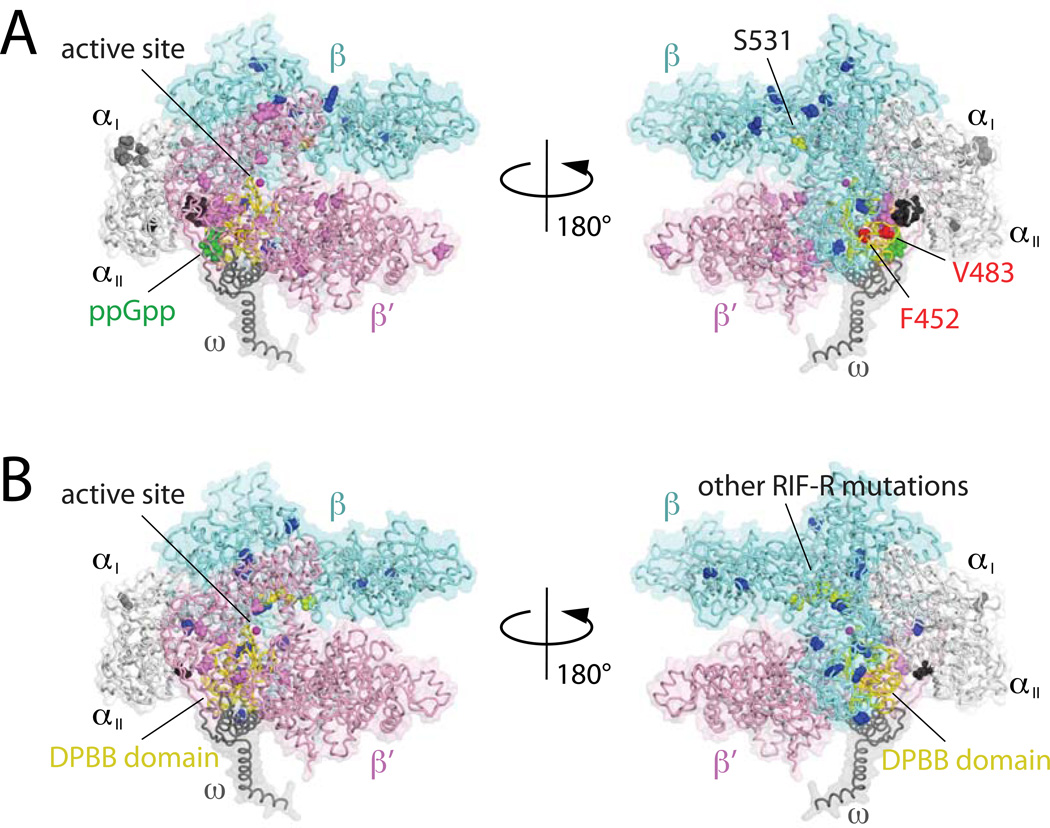
The condition-specific fitness defect and changes in RNAP transcription activity suggest additional nuance in the molecular mechanism of compensation. Most of the compensatory mutation sites are located on a double-psi β-barrel (DPBB) domain of the β’ subunit (amino acid residues 423–565 and 348–490 in Mtb and E. coli β’ subunits, respectively; Fig. 7 shows the location of these mutations on the E. coli enzyme structure from PDB: 4JKR), which forms the active site of RNAP coordinating the catalytic Mg2+ by the -DFDGD- motif, found in all cellular RNAPs (Iyer et al., 2003). In E. coli, the DPBB domain is also involved in binding ppGpp which signals the stringent response of the E. coli RNAP transcription regulation, and the ppGpp binding may provide an allosteric signal that modifies RNAP activity (Mechold et al., 2013, Ross et al., 2013, Zuo et al., 2013). There is no report indicating direct binding of ppGpp to the Mtb RNAP to initiate the stringent response, but the proximity of the compensatory mutations to this region on the DPBB domain, coupled with the linkage between nutrient starvation and the magnitude of the fitness defect, make it tempting to speculate that they restore RNAP activity by modulating some starvation control mechanism that operates in this region of the polymerase. Many other compensatory mutations found in the α (all mutants except S82R) and β subunits (H835R, I1035V, T1078I and K1102T) are also located in this region and these mutants may also similarly modify the starvation response of RNAP. Notably a relMtb deficient strain of Mtb that is unable to synthesize ppGpp is severely compromised for in vivo survival in mice supporting the possible relevance of stringent conditions for assessing fitness consequences of drug-resistance associated alleles (Dahl et al., 2003). Interestingly compensatory mutations selected for the RpoB S531L mutation are enriched in the DPBB domain of the β’ subunit, while suppressors to other RIF-R mutations are scattered throughout other subunits (Fig. 7). Therefore, there may be subtle differences in the mechanism of impairment of RNAP activity by other RIF-R mutations and the compensation of these impairments.
Figure 7.
Three-dimensional representation of the RIF-resistance conferring and compensatory mutations on the crystal structure of the E. coli RNAP-RIF complex (PDB: 4JK1, (Mechold et al., 2013)). (A) The RIF-resistance conferring rpoB S531L mutation and its compensatory mutations. The RNAP core enzyme is depicted as an α-carbon backbone trace (α subunit, white; β subunit, cyan; β‘ subunit, pink; ω subunit, gray). The Ser531 residue of the β subunit is shown as yellow sphere. Compensatory mutations found on αI, αIIβ and β‘ subunits are highlighted as gray, black, blue and dark pink, respectively. Two compensatory mutations, rpoC F452L and rpoC V483G, characterized in the in vitro transcription assay were shown as red. The ppGpp binding site is indicated as green sphere and the DPBB domain of the β‘ subunit containing the RNAP active site (magenta sphere) is highlighted in yellow. (B) RIF-resistance associated mutations other than rpoB S531L and their compensatory mutations. Rifampicin resistant mutations are shown as yellow spheres, and compensatory mutations found on αI, αIIβ and β‘ subunits are highlighted as gray, black, blue and dark pink, respectively.
In a highly complex, multi-subunit macromolecule such as RNAP, both intragenic and intergenic suppression could be associated with changes in structural, functional or regulatory interactions between (or within) the subunits (Maisnier-Patin & Andersson, 2004). Prior analyses of genome sequences from Mtb clinical isolates have also identified rpoC as a gene experiencing mutation preferentially in drug-resistant isolates, consistent with our findings (Comas et al., 2012, de Vos et al., 2013). As in these reports, we also find that potential compensatory mutations also occurred in the β subunit, outside of the canonical RIF-binding region, as well as in the α subunit. Many of these substitutions are bioinformatically predicted (Choi et al., 2012, Kumar et al., 2009) to have functional consequences but biochemical confirmation remains lacking. Similarly in Samonella enterica, Brandis at al. (2012) selected compensatory mutations by serial passage (60 generations) of a strain carrying rpoB R529C with or without continued RIF selection. Interestingly, they never observed reversion to the wild type rpoB sequence but instead found acquisition of secondary mutations in rpoA, rpoB and rpoC. When they repeated this starting from a strain carrying rpoB S531L they obtained similar results but with little overlap in compensatory alleles (Brandis & Hughes, 2013). Given the complexity of the structural and regulatory interactions that modulate RNAP activity (Korzheva et al., 2000, Mechold et al., 2013, Vassylyev et al., 2007) it is likely that there are many potential mechanisms to compensate fitness defects and these may well have allele specificity.
Overall our study supports and extends recent findings of the importance of mutations in rpoC in compensating fitness defects incurred by RIF-R mutations in rpoB. We show that these fitness defects are directly linked to impaired RNAP transcriptional activity, particularly during nutrient-limited growth, and are rescued by mutations in rpoC. The strong fitness defect observed under stringent growth conditions fits with the rapid fixation of these compensatory alleles in the bacterial population, supporting that Mtb in humans experiences a stringent environment during pathogenesis. We have shown that even large fitness defects can be compensated by alterations in other subunits of the polymerase, setting the stage for acquisition of further drug resistance resulting in strains able to both cause fulminant disease in humans and transmit to new hosts.
Experimental Procedures
DNA sequencing of rpo genes from clinical isolates
Genomic regions of clinical isolates were amplified by PCR using Solg™ Pfu-X DNA polymerase (Solgent, South Korea) to cover the entire sequences of the rpo genes of the Mtb H37Rv genome (GenBank Accession Number, AL123456): rpoA (Rv3457c), 3,877,347-3,878,558; rpoB (Rv0667), 759,573–763,416; rpoC (Rv0668), 763,173–767,522; rpoZ (Rv1390), 1,564,863-1,565,690. Sequence chromatogram files obtained by commercial sequencing of the purified PCR products (Solgent, South Korea) were analyzed for sequence polymorphisms in comparison with corresponding gene sequences of the H37Rv genome using Sequencher 4.5 (Gene Codes Corp., USA).
Phylogenetic analysis of the isolates
The standardized MIRU-VNTR typing was done as previously described (Shamputa et al., 2010) and the Neighbor-joining tree was constructed using MIRU-VNTRplus (http://www.miru-vntrplus.org).
Construction of M. smegmatis strains
Plasmids and strains used in the construction of M. smegmatis strains are listed in Table S6. DNA fragments carrying Mtb rpoB and rpoC genes and its flanking sequences, corresponding to nt 758,663–767,673 of the Mtb H37Rv genome (GenBank Accession Number, AL123456), were amplified by PCR (Solg™ Pfu-X DNA polymerase; Solgent, South Korea) using genomic DNA prepared from a RIF-sensitive clinical isolate and RIF-R isolates bearing rpoB C1349T (S531L) rpoC (wild type), rpoB C1349T (S531L) rpoC T1354C (F452L) and rpoB C1349T (S531L) rpoC T1448G (V483G) alleles, respectively. The purified PCR products were separately cloned in pTS421, a derivative of pMV306, between XbaI and HpaI sites, resulting in pTS422, pTS423, pTS424 and pTS429. Codons for six histidine residues were incorporated at the 3' end of rpoC during PCR. Following transformation with the plasmid constructs and selection for hygromycin-resistance (50 µg ml−1) M. smegmatis mc2 155 strains, TS102, TS103, TS105 and TS112 were verified for the integration of M. tuberculosis rpoB and rpoC carrying the respective alleles by DNA sequencing of PCR products. The temperature-sensitive sacB delivery system was used to delete M. smegmatis rpoB and rpoC from the strains as described previously (Pelicic et al., 1997, Raman et al., 2004). Approximately 1-kb DNA fragments, either upstream 923-bp plus 45-bp at 5' end of rpoB and 21-bp at 3' end of rpoC plus downstream 1,003-bp, were joined with zeocin-resistance cassette on pRH1351 to obtain pTS428. TS102, TS103, TS105 and TS112 were transformed separately with pTS428, selected sequentially for zeocin-resistance (30 µg ml−1) at 32°C, sucrose-resistance (10% sucrose), and absence of XylE activity (white colony in the presence of 1% catechol). The resulting strains, TS106, TS108, TS110 and TS113 were verified for the deletion of M. smegmatis rpoB and rpoC by PCR, Southern blot and DNA sequencing of the genomic regions.
Growth phenotype
Growth of the resulting M. smegmatis strains (TS106, TS108, TS110 and TS113) was observed on 7H10 agar plates supplemented with Middlebrook OADC enrichment (BD, USA) 4 days at 37°C after plating serially diluted cultures in regular 7H9 media (BD, USA). For comparison of the growth in different nutrient conditions 7H9 broth base was prepared from the constituents either complete or devoid of biotin and iron, and supplemented with either 0.2% glucose or 0.002% glucose. One mililiter of 4-day culture of the strains in regular 7H9 media, after washing three times with the 7H9 broth base devoid of biotin and iron, was transferred to 50 ml of fresh media in different nutrient conditions, and the growth was monitored at 37°C by measuring optical density at 600 nm. Specific growth rate was calculated in the linear range of logarithmic growth in triplicates.
Pairwise competition assay
M. smegmatis strains were cultured in 7H9 media supplemented with 0.2% glucose for 4 days at 37°C, and 0.5 ml culture each of TS108, TS110 and TS113 was mixed with an equal volume of the culture of TS106 separately. After washing three times with the 7H9 broth base devoid of biotin and iron, 150 µl portions of mixture was taken to inoculate 30 ml of fresh media in different nutrient conditions. The competition cultures were incubated with shaking for 4 day at 37°C, after which 150 µl portions of the cultures were transferred to 30 ml of fresh media of the same conditions for further incubation for 4 days. The baseline mixtures, and the end point cultures were counted for CFUs on 7H10 agar plates with or without rifampicin at 25 µg ml−1, and the numbers of RIF-susceptible strain, TS106 were calculated by subtracting the CFUs of the RIF-resistant strains, TS108, TS110 and TS113 respectively. The relative competitive fitness of TS108, TS110 and TS113 to TS106 was calculated following the formula of Gagneux et al. (2006).
Transcription efficiency assay
M. smegmatis strains were grown in 7H9 broth to OD600 1.0 and washed with saline supplemented with 0.05% Tween 80. Two milligram of wet cells were retained for baseline, and the rest of the washed cells were incubated in pre-warmed minimal medium without glucose containing 2 g of acetamide ml−1 at 37°C with shaking for the induction of the acetamidase gene, amiE (Narayanan et al., 2000). Cell pellets harvested 20, 40, 60, 90 and 120 min postinduction were resuspended in 1 ml TRIzol (Invitrogen, USA) with FastPrep Lysing Matrix and disrupted on a FastPrep-24 instrument (MP Biomedicals, USA). Following purification by extraction with chloroform and precipitation by isopropanol, the total RNA preparation was treated with RNase-free DNase (Promega, USA). Synthesis of cDNA and real-time PCR were performed using DiaStar™ RT kit and Solg™ Real-Time PCR kit (Solgent, South Korea) according to the manufacture’s instruction. Quantification of cDNA was performed on LightCycler 480 (Roche, Switzerland) using EvaGreen™ dye in reaction mixtures containing the pairs of primers specific to amiE and recA, respectively: amiE, GGTTATTCAGGCATCTTCGCG and ACGTTCGCGCTCGTTCCAT; recA, AAGTTCTACGCCTCGG-TCCG and TGAACCAGGACCCGGACTT. PCR conditions were as follows: 95°C for 15 min followed by 45 cycles of 95°C for 20 s, 54°C for 40 s, 72°C for 20 s followed by elevation from 40°C to 90°C for 1 min to observe the melting curve‥ Expression of amiE relative to the constitutive expression of recA was calculated by the 2- ΔΔCT method (Livak & Schmittgen, 2001). The slope of the line obtained from plotting the proportional change in 2-ΔΔCT over time was represented as transcription efficiency.
Molecular modeling
The crystal structure of the T. thermophilus RNA polymerase elongation complex (PDB: 2O5J) was used to model the TB RNAP structure. Sequence identities were 48% and 41% respectively for the β and β’ subunits, respectively, allowing a straightforward homology modeling using an in-house developed approach particularly suitable for large nucleoprotein complexes (Tung & Sanbonmatsu, 2004). The modeled structure of the RpoB/RpoC complex was subjected to a minimization procedure using AMBER (Case et al., 2005). The structure of the mRNA in 2O5J was transferred to the TB elongation complex directly and the structure of the longer DNA molecule (compared to 2O5J) from the E. coli 3IYD complex was also transferred.
Supplementary Material
Acknowledgements
This work was supported (in part) by the Intramural Research Program of the NIAID, NIH, (in part) by continuous support from the Korean Centers for Disease Control of the Korean Ministry of Health and Welfare to the International Tuberculosis Research Center, and (in part) by NIH Grant GM087350- A1 (K.S.M.). We would like to thank the subjects who enrolled in this research study for their active participation and donation of specimens (ClinicalTrials.gov identifier: NCT00341601) and the clinical staff who supported that trial.
Footnotes
The authors of this study declare that they have no conflicts of interest with respect to any aspect of this research.
References
- Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- Barry CE. Lessons from seven decades of antituberculosis drug discovery. Curr Top Med Chem. 2011;11:1216–1225. doi: 10.2174/156802611795429158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billington OJ, McHugh TD, Gillespie SH. Physiological cost of rifampin resistance induced in vitro in Mycobacterium tuberculosis . Antimicrob Agents Chemother. 1999;43:1866–1869. doi: 10.1128/aac.43.8.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkman J, Nagaev I, Berg OG, Hughes D, Andersson DI. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science. 2000;287:1479–1482. doi: 10.1126/science.287.5457.1479. [DOI] [PubMed] [Google Scholar]
- Borrell S, Gagneux S. Infectiousness, reproductive fitness and evolution of drug-resistant Mycobacterium tuberculosis . Int J Tuberc Lung Dis. 2009;13:1456–1466. [PubMed] [Google Scholar]
- Brandis G, Hughes D. Genetic characterization of compensatory evolution in strains carrying rpoB Ser531Leu, the rifampicin resistance mutation most frequently found in clinical isolates. J Antimicrob Chemother. 2013 doi: 10.1093/jac/dkt224. [DOI] [PubMed] [Google Scholar]
- Broad Institute of Harvard and MIT. Mycobacterium tuberculosis. 2013 Diversity Sequencing Project [WWW Document]. URL http://www.broadinstitute.org/annotation/genome/mycobacterium_tuberculosis_diversity/MultiHome.html.
- Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- Casali N, Nikolayevskyy V, Balabanova Y, Ignatyeva O, Kontsevaya I, Harris SR, Bentley SD, Parkhill J, Nejentsev S, Hoffner SE, Horstmann RD, Brown T, Drobniewski F. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res. 2012;22:735–745. doi: 10.1101/gr.128678.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr., Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. Journal of computational chemistry. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen T, Murray M. Modeling epidemics of multidrug-resistant M. tuberculosis of heterogeneous fitness. Nat Med. 2004;10:1117–1121. doi: 10.1038/nm1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, Galagan J, Niemann S, Gagneux S. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet. 2012;44:106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JL, Kraus CN, Boshoff HI, Doan B, Foley K, Avarbock D, Kaplan G, Mizrahi V, Rubin H, Barry. CE., 3rd The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci U S A. 2003;100:10026–10031. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos M, Muller B, Borrell S, Black PA, van Helden PD, Warren RM, Gagneux S, Victor TC. Putative compensatory mutations in the rpoC gene of rifampin-resistant Mycobacterium tuberculosis are associated with ongoing transmission. Antimicrob Agents Chemother. 2013;57:827–832. doi: 10.1128/AAC.01541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filliol I, Motiwala AS, Cavatore M, Qi W, Hazbon MH, Bobadilla del Valle M, Fyfe J, Garcia-Garcia L, Rastogi N, Sola C, Zozio T, Guerrero MI, Leon CI, Crabtree J, Angiuoli S, Eisenach KD, Durmaz R, Joloba ML, Rendon A, Sifuentes-Osornio J, Ponce de Leon A, Cave MD, Fleischmann R, Whittam TS, Alland D. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J Bacteriol. 2006;188:759–772. doi: 10.1128/JB.188.2.759-772.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, Peterson J, DeBoy R, Dodson R, Gwinn M, Haft D, Hickey E, Kolonay JF, Nelson WC, Umayam LA, Ermolaeva M, Salzberg SL, Delcher A, Utterback T, Weidman J, Khouri H, Gill J, Mikula A, Bishai W, Jacobs WR, Jr., Venter JC, Fraser CM. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol. 2002;184:5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux S, Long CD, Small PM, Van T, Schoolnik GK, Bohannan BJ. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis . Science. 2006;312:1944–1946. doi: 10.1126/science.1124410. [DOI] [PubMed] [Google Scholar]
- Ioerger TR, Feng Y, Chen X, Dobos KM, Victor TC, Streicher EM, Warren RM, Gey van Pittius NC, Van Helden PD, Sacchettini JC. The non-clonality of drug resistance in Beijing-genotype isolates of Mycobacterium tuberculosis from the Western Cape of South Africa. BMC Genomics. 2010;11:670. doi: 10.1186/1471-2164-11-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioerger TR, Koo S, No EG, Chen X, Larsen MH, Jacobs WR, Jr., Pillay M, Sturm AW, Sacchettini JC. Genome analysis of multi- and extensively-drug-resistant tuberculosis from KwaZulu-Natal, South Africa. PLoS One. 2009;4:e7778. doi: 10.1371/journal.pone.0007778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Aravind L. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct Biol. 2003;3:1. doi: 10.1186/1472-6807-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, Darst SA. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Laurenzo D, Mousa SA. Mechanisms of drug resistance in Mycobacterium tuberculosis and current status of rapid molecular diagnostic testing. Acta Trop. 2011;119:5–10. doi: 10.1016/j.actatropica.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luciani F, Sisson SA, Jiang H, Francis AR, Tanaka MM. The epidemiological fitness cost of drug resistance in Mycobacterium tuberculosis . Proc Natl Acad Sci U S A. 2009;106:14711–14715. doi: 10.1073/pnas.0902437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisnier-Patin S, Andersson DI. Adaptation to the deleterious effects of antimicrobial drug resistance mutations by compensatory evolution. Res Microbiol. 2004;155:360–369. doi: 10.1016/j.resmic.2004.01.019. [DOI] [PubMed] [Google Scholar]
- Mariam DH, Mengistu Y, Hoffner SE, Andersson DI. Effect of rpoB mutations conferring rifampin resistance on fitness of Mycobacterium tuberculosis . Antimicrob Agents Chemother. 2004;48:1289–1294. doi: 10.1128/AAC.48.4.1289-1294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechold U, Potrykus K, Murphy H, Murakami KS, Cashel M. Differential regulation by ppGpp versus pppGpp in Escherichia coli . Nucleic Acids Res. 2013;41:6175–6189. doi: 10.1093/nar/gkt302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan S, Selvakumar S, Aarati R, Vasan SK, Narayanan PR. Transcriptional analysis of inducible acetamidase gene of Mycobacterium smegmatis . FEMS microbiology letters. 2000;192:263–268. doi: 10.1111/j.1574-6968.2000.tb09392.x. [DOI] [PubMed] [Google Scholar]
- Pelicic V, Jackson M, Reyrat JM, Jacobs WR, Jr., Gicquel B, Guilhot C. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis . Proc Natl Acad Sci U S A. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S, Hazra R, Dascher CC, Husson RN. Transcription regulation by the Mycobacterium tuberculosis alternative sigma factor SigD and its role in virulence. J Bacteriol. 2004;186:6605–6616. doi: 10.1128/JB.186.19.6605-6616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell. 2013;50:420–429. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamputa IC, Lee J, Allix-Beguec C, Cho EJ, Lee JI, Rajan V, Lee EG, Min JH, Carroll MW, Goldfeder LC, Kim JH, Kang HS, Hwang S, Eum SY, Park SK, Lee H, Supply P, Cho SN, Via LE, Barry. CE., 3rd Genetic diversity of Mycobacterium tuberculosis isolates from a tertiary care tuberculosis hospital in South Korea. J Clin Microbiol. 2010;48:387–394. doi: 10.1128/JCM.02167-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman DR, Mdluli K, Hickey MJ, Arain TM, Morris SL, Barry CE, 3rd, Stover CK. Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis . Science. 1996;272:1641–1643. doi: 10.1126/science.272.5268.1641. [DOI] [PubMed] [Google Scholar]
- Tung CS, Sanbonmatsu KY. Atomic model of the Thermus thermophilus 70S ribosome developed in silico. Biophysical journal. 2004;87:2714–2722. doi: 10.1529/biophysj.104.040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- WHO. Treatment of tuberculosis: guidelines. 4th ed. Geneva: WHO Press; 2009. [Google Scholar]
- WHO. Towards universal access to diagnosis and treatment of multidrug-resistant and extensively drug-resistant tuberculosis by 2015: WHO progress report 2011. Geneva: WHO Press; 2011. [Google Scholar]
- WHO. Global tuberculosis report 2012. Geneva: WHO Press; 2012. [Google Scholar]
- Wong SY, Lee JS, Kwak HK, Via LE, Boshoff HI, Barry CE., 3rd Mutations in gidB confer low-level streptomycin resistance in Mycobacterium tuberculosis . Antimicrob Agents Chemother. 2011;55:2515–2522. doi: 10.1128/AAC.01814-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yew WW. Mechanisms of drug resistance in Mycobacterium tuberculosis . Int J Tuberc Lung Dis. 2009;13:1320–1330. [PubMed] [Google Scholar]
- Zheng H, Lu L, Wang B, Pu S, Zhang X, Zhu G, Shi W, Zhang L, Wang H, Wang S, Zhao G, Zhang Y. Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLoS One. 2008;3:e2375. doi: 10.1371/journal.pone.0002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Wang Y, Steitz TA. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol Cell. 2013;50:430–436. doi: 10.1016/j.molcel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.